|
1
|
Shimura T, Shibata M, Gonda K, Kofunato Y,
Okada R, Ishigame T, Kimura T, Kenjo A, Kono K and Marubashi S:
Significance of circulating galectin-3 in patients with
pancreatobiliary cancer. Anticancer Res. 37:4979–4986.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ohya A, Yamanoi K, Shimojo H, Fujii C and
Nakayama J: Gastric gland mucin-specific o-glycan expression
decreases with tumor progression from precursor lesions to
pancreatic cancer. Cancer Sci. 108:1897–1902. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Park J, Choi Y, Namkung J, Yi SG, Kim H,
Yu J, Kim Y, Kwon MS, Kwon W, Oh DY, et al: Diagnostic performance
enhancement of pancreatic cancer using proteomic multimarker panel.
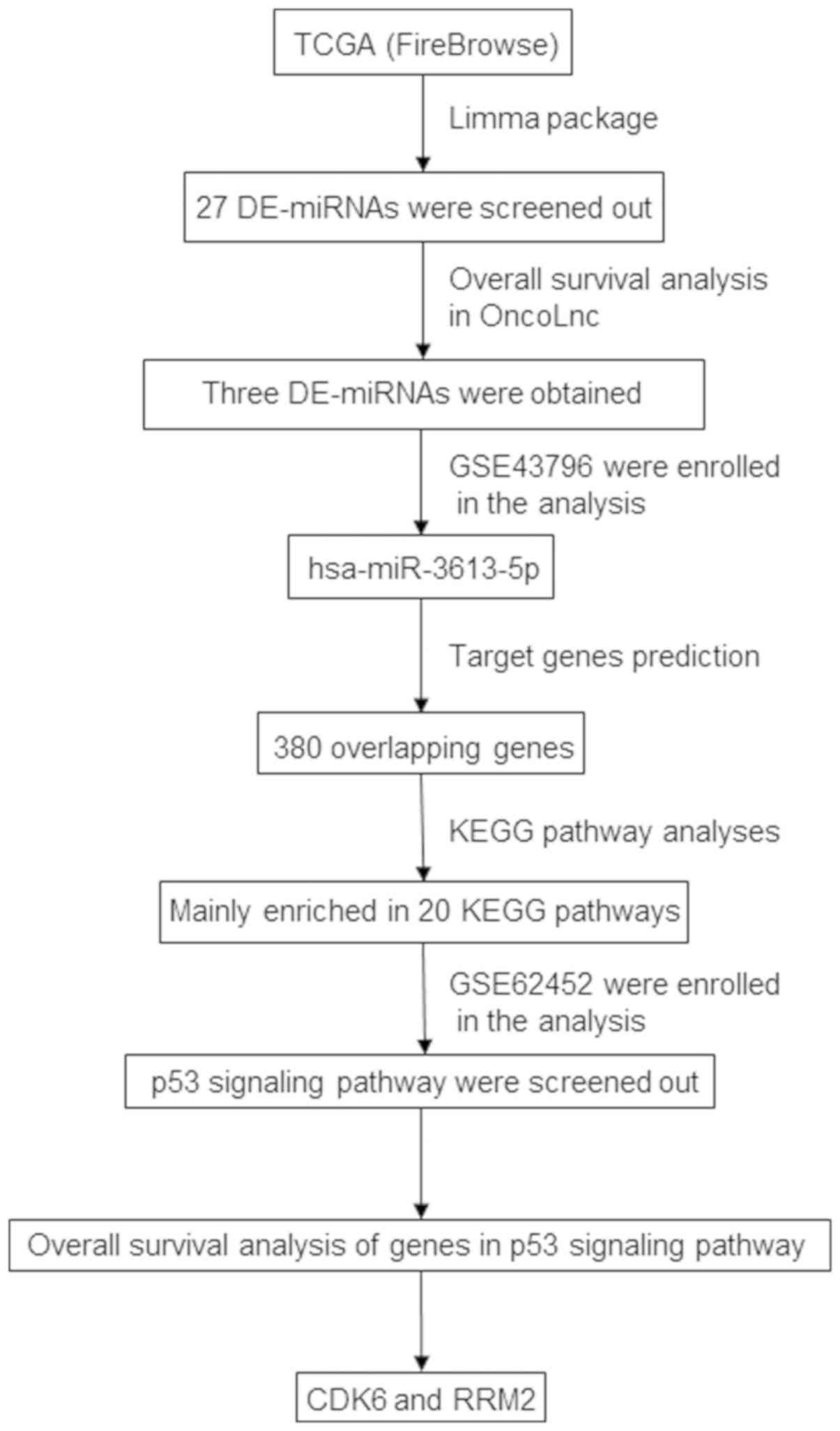
Oncotarget. 8:93117–93130. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kwon HM, Kang EJ, Kang K, Kim SD, Yang K
and Yi JM: Combinatorial effects of an epigenetic inhibitor and
ionizing radiation contribute to targeted elimination of pancreatic
cancer stem cell. Oncotarget. 8:89005–89020. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Han T, Zhuo M, Hu H, Jiao F and Wang LW:
Synergistic effects of the combination of 5-Aza-CdR and
suberoylanilide hydroxamic acid on the anticancer property of
pancreatic cancer. Oncol Rep. 39:264–270. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Brooks J, Fleischmann-Mundt B, Woller N,
Niemann J, Ribback S, Peters K, Demir IE, Armbrecht N, Ceyhan GO,
Manns MP, et al: Perioperative, spatiotemporally coordinated
activation of T and NK cells prevents recurrence of pancreatic
cancer. Cancer Res. 78:475–488. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Mirkin KA, Hollenbeak CS and Wong J:
Greater lymph node retrieval and lymph node ratio impacts survival
in resected pancreatic cancer. J Surg Res. 220:12–24.
2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Khan MA, Azim S, Zubair H, Bhardwaj A,
Patel GK, Khushman M, Singh S and Singh AP: Molecular drivers of
pancreatic cancer pathogenesis: Looking inward to move forward. Int
J Mol Sci. 18(E779)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Du YX, Liu ZW, You L, Wu WM and Zhao YP:
Advances in understanding the molecular mechanism of pancreatic
cancer metastasis. Hepatobiliary Pancreat Dis Int. 15:361–370.
2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen L and Kang C: miRNA interventions
serve as ‘magic bullets’ in the reversal of glioblastoma hallmarks.
Oncotarget. 6:38628–38642. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Liu W, Ma R and Yuan Y:
Post-transcriptional regulation of genes related to biological
behaviors of gastric cancer by long noncoding RNAs and microRNAs. J
Cancer. 8:4141–4154. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Shirafkan N, Mansoori B, Mohammadi A,
Shomali N, Ghasbi M and Baradaran B: MicroRNAs as novel biomarkers
for colorectal cancer: New outlooks. Biomed Pharmacother.
97:1319–1330. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Huang L, Cai JL, Huang PZ, Kang L, Huang
MJ, Wang L and Wang JP: miR19b-3p promotes the growth and
metastasis of colorectal cancer via directly targeting ITGB8. Am J
Cancer Res. 7:1996–2008. 2017.PubMed/NCBI
|
|
15
|
Long M, Zhan M, Xu S, Yang R, Chen W,
Zhang S, Shi Y, He Q, Mohan M, Liu Q and Wang J: miR-92b-3p acts as
a tumor suppressor by targeting Gabra3 in pancreatic cancer. Mol
Cancer. 16(167)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bayraktar R, Van Roosbroeck K and Calin
GA: Cell-to-cell communication: Micrornas as hormones. Mol Oncol.
11:1673–1686. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fujiwara T, Uotani K, Yoshida A, Morita T,
Nezu Y, Kobayashi E, Yoshida A, Uehara T, Omori T, Sugiu K, et al:
Clinical significance of circulating miR-25-3p as a novel
diagnostic and prognostic biomarker in osteosarcoma. Oncotarget.
8:33375–33392. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yu Y, Zuo J, Tan Q, Zar Thin K, Li P, Zhu
M, Yu M, Fu Z, Liang C and Tu J: Plasma miR-92a-2 as a biomarker
for small cell lung cancer. Cancer Biomark. 18:319–327.
2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Schwarzenbach H: Clinical relevance of
circulating, cell-free and exosomal microRNAs in plasma and serum
of breast cancer patients. Oncol Res Treat. 40:423–429.
2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Deng W, Wang Y, Liu Z, Cheng H and Xue Y:
HemI: A toolkit for illustrating heatmaps. PLoS One.
9(e111988)2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yang S, He P, Wang J, Schetter A, Tang W,
Funamizu N, Yanaga K, Uwagawa T, Satoskar AR, Gaedcke J, et al: A
novel MIF signaling pathway drives the malignant character of
pancreatic cancer by targeting NR3C2. Cancer Res. 76:3838–3850.
2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Park M, Kim M, Hwang D, Park M, Kim WK,
Kim SK, Shin J, Park ES, Kang CM, Paik YK and Kim H:
Characterization of gene expression and activated signaling
pathways in solid-pseudopapillary neoplasm of pancreas. Mod Pathol.
27:580–593. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Anaya J: OncoLnc: Linking TCGA survival
data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Science.
2(e67)2016.
|
|
24
|
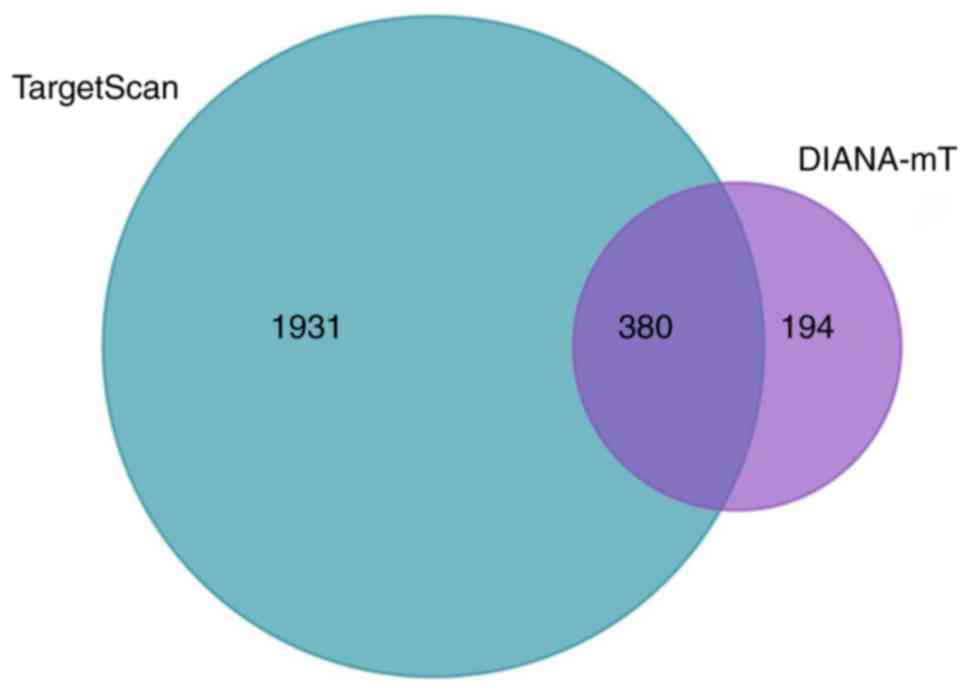
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4(7554)2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C,
Dalamagas T and Hatzigeorgiou AG: DIANA-microT web server v5.0:
Service integration into miRNA functional analysis workflows.
Nucleic Acids Res. 41:W169–W173. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Reczko M, Maragkakis M, Alexiou P, Grosse
I and Hatzigeorgiou AG: Functional microRNA targets in protein
coding sequences. Bioinformatics. 28:771–776. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
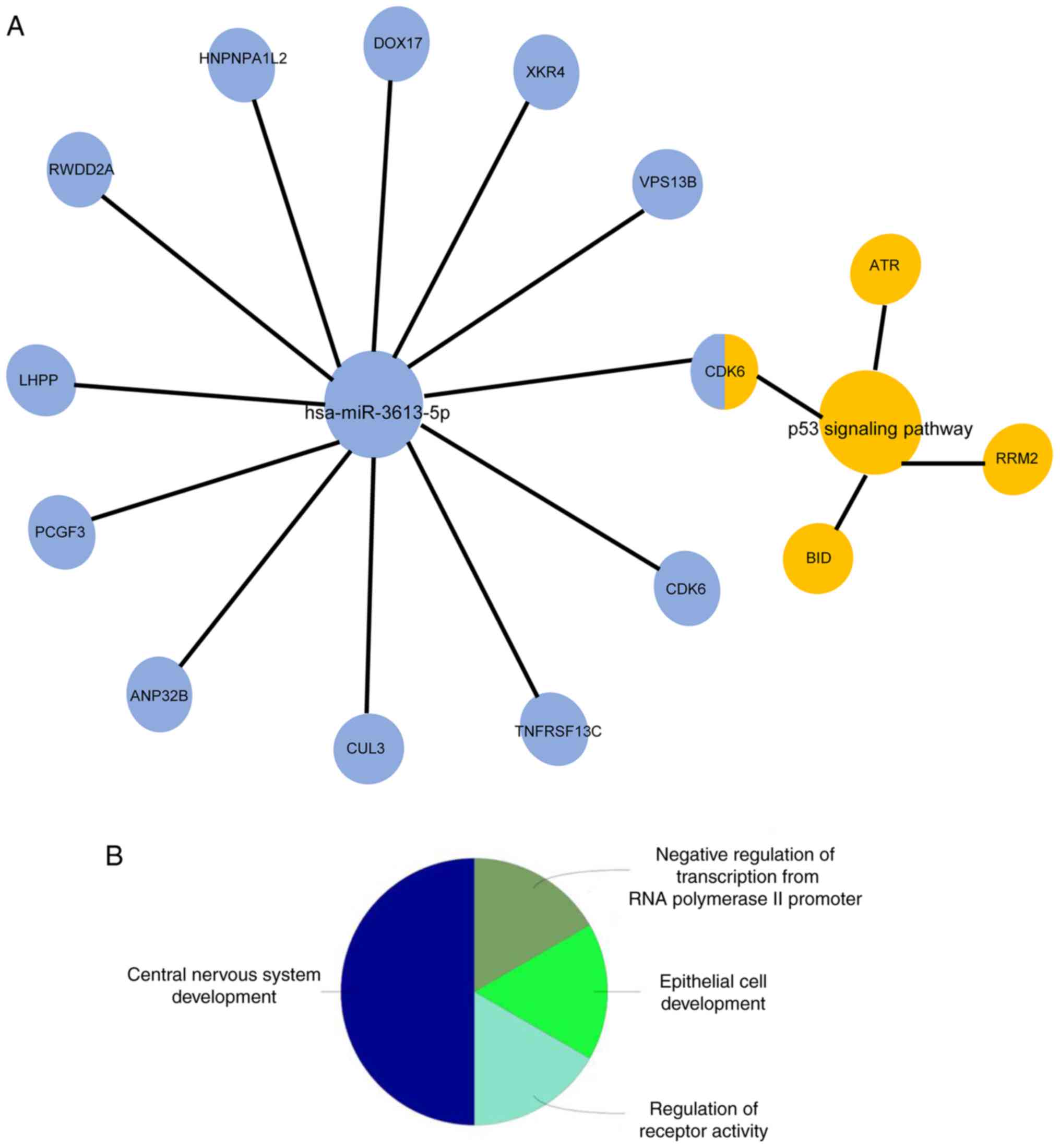
Shannon P, Ozier O, Baliga NS, Wang JT,
Ramage D, Amin N, Schwikowski B, Ideker T and Markiel A: Cytoscape:
A software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Pontén F, Jirström K and Uhlen M: The
human protein atlas-a tool for pathology. J Pathol. 216:387–393.
2008.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Karmakar S, Kaushik G, Nimmakayala R,
Rachagani S, Ponnusamy MP and Batra SK: MicroRNA regulation of
K-ras in pancreatic cancer and opportunities for therapeutic
intervention. Semin Cancer Biol. 54:63–71. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lu Z, Lai ZQ, Leung AWN, Leung PS, Li ZS
and Lin ZX: Exploring brusatol as a new anti-pancreatic cancer
adjuvant: Biological evaluation and mechanistic studies.
Oncotarget. 8:84974–84985. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Choi M, Bien H, Mofunanya A and Powers S:
Challenges in ras therapeutics in pancreatic cancer. Semin Cancer
Biol. 54:101–108. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Halbrook CJ and Lyssiotis CA: Employing
metabolism to improve the diagnosis and treatment of pancreatic
cancer. Cancer Cell. 31:5–19. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wang L and Xie K: Nitric oxide and
pancreatic cancer pathogenesis, prevention, and treatment. Curr
Pharm Des. 16:421–427. 2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen WJ, Tang RX, He RQ, Li DY, Liang L,
Zeng JH, Hu XH, Ma J, Li SK and Chen G: Clinical roles of the
aberrantly expressed lncRNAs in lung squamous cell carcinoma: A
study based on RNA-sequencing and microarray data mining.
Oncotarget. 8:61282–61304. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Chen Y, Teng L, Liu W, Cao Y, Ding D, Wang
W, Chen H, Li C and An R: Identification of biological targets of
therapeutic intervention for clear cell renal cell carcinoma based
on bioinformatics approach. Cancer Cell Int. 16(16)2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Liang B, Li Y and Wang T: A three miRNAs
signature predicts survival in cervical cancer using bioinformatics
analysis. Sci Rep. 7(5624)2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Rachagani S, Macha MA, Heimann N,
Seshacharyulu P, Haridas D, Chugh S and Batra SK: Clinical
implications of miRNAs in the pathogenesis, diagnosis and therapy
of pancreatic cancer. Adv Drug Delivy Rev. 81:16–33.
2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chitkara D, Mittal A and Mahato RI: miRNAs
in pancreatic cancer: Therapeutic potential, delivery challenges
and strategies. Adv Drug Deliv Rev. 81:34–52. 2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li L, Li Z, Kong X, Xie D, Jia Z, Jiang W,
Cui J, Du Y, Wei D, Huang S and Xie K: Down-regulation of
microRNA-494 via loss of SMAD4 increases FOXM1 and β-catenin
signaling in pancreatic ductal adenocarcinoma cells.
Gastroenterology. 147:485–497. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Schultz NA, Dehlendorff C, Jensen BV,
Bjerregaard JK, Nielsen KR, Bojesen SE, Calatayud D, Nielsen SE,
Yilmaz M, Holländer NH, et al: MicroRNA biomarkers in whole blood
for detection of pancreatic cancer. JAMA. 311:392–404.
2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Bibi F, Naseer MI, Alvi SA, Yasir M,
Jiman-Fatani AA, Sawan A, Abuzenadah AM, Al-Qahtani MH and Azhar
EI: MicroRNA analysis of gastric cancer patients from Saudi Arabian
population. BMC Genomics. 17(751)2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen X, Lu P, Wang DD, Yang SJ, Wu Y, Shen
HY, Zhong SL, Zhao JH and Tang JH: The role of miRNAs in drug
resistance and prognosis of breast cancer formalin-fixed
paraffin-embedded tissues. Gene. 595:221–226. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Chong GO, Jeon HS, Han HS, Son JW, Lee YH,
Hong DG, Lee YS and Cho YL: Differential microrna expression
profiles in primary and recurrent epithelial ovarian cancer.
Anticancer Res. 35:2611–2617. 2015.PubMed/NCBI
|
|
44
|
Castro-Magdonel BE, Orjuela M, Camacho J,
García-Chéquer AJ, Cabrera-Muñoz L, Sadowinski-Pine S,
Durán-Figueroa N, Orozco-Romero MJ, Velázquez-Wong AC,
Hernández-Ángeles A, et al: miRNome landscape analysis reveals a 30
miRNA core in retinoblastoma. BMC Cancer. 17(458)2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Grygalewicz B, Woroniecka R, Rygier J,
Borkowska K, Rzepecka I, Łukasik M, Budziłowska A, Rymkiewicz G,
Błachnio K, Nowakowska B, et al: Monoallelic and biallelic
deletions of 13q14 in a group of CLL/SLL patients investigated by
CGH haematological cancer and SNP array (8x60K). Mol Cytogenet.
9(1)2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Tsiloulis T, Pike J, Powell D, Rossello
FJ, Canny BJ, Meex RC and Watt MJ: Impact of endurance exercise
training on adipocyte microRNA expression in overweight men. FASEB
J. 31:161–171. 2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Liu K, Zhao X, Gu J, Wu J, Zhang H and Li
Y: Effects of 12C6+ heavy ion beam irradiation on the p53 signaling
pathway in HepG2 liver cancer cells. Acta Biochim Biophys Sin
(Shanghai). 49:989–998. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Song D, Zhao J, Su C, Jiang Y and Hou J:
Etoposide induced NMI promotes cell apoptosis by activating the
ARF-p53 signaling pathway in lung carcinoma. Biochem Biophys Res
Commun. 495:368–374. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wu Z, Zhu X, Xu W, Zhang Y, Chen L, Qiu F,
Zhang B, Wu L, Peng Z and Tang H: Up-regulation of CIT promotes the
growth of colon cancer cells. Oncotarget. 8:71954–71964.
2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Mello SS, Valente LJ, Raj N, Seoane JA,
Flowers BM, McClendon J, Bieging-Rolett KT, Lee J, Ivanochko D,
Kozak MM, et al: A p53 super-tumor suppressor reveals a tumor
suppressive p53-Ptpn14-Yap axis in pancreatic cancer. Cancer Cell.
32:460–473. 2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Morita K, Noura M, Tokushige C, Maeda S,
Kiyose H, Kashiwazaki G, Taniguchi J, Bando T, Yoshida K, Ozaki T,
et al: Autonomous feedback loop of RUNX1-p53-CBFB in acute myeloid
leukemia cells. Sci Rep. 7(16604)2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Chen H, Qian J, Werner S, Cuk K, Knebel P
and Brenner H: Development and validation of a panel of five
proteins as blood biomarkers for early detection of colorectal
cancer. Clin Epidemiol. 9:517–526. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Masetti M, Acquaviva G, Visani M, Tallini
G, Fornelli A, Ragazzi M, Vasuri F, Grifoni D, Di Giacomo S,
Fiorino S, et al: Long-term survivors of pancreatic adenocarcinoma
show low rates of genetic alterations in KRAS, TP53 and SMAD4.
Cancer Biomark. 6:323–334. 2017.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Makohon-Moore A and Iacobuzio-Donahue CA:
Pancreatic cancer biology and genetics from an evolutionary
perspective. Nat Rev Cancer. 16:553–565. 2016.PubMed/NCBI View Article : Google Scholar
|