Introduction
Conventional drug therapy cannot activate neurons in
the brain (1). To treat neurological
diseases such as encephalopathy, the only effective form of
treatment is neural stem cell transplantation (1-3).
Neural stem cells are undifferentiated pluripotent or committed
cells (4), the transplantation of
which has been documented as an effective method to repair and
replace damaged brain tissue by reconstructing partial loop and
function (5). Kochanek et al
(6) transplanted neural stem cells
into the damaged brain tissue of a Parkinson's disease rat model
and demonstrated that tremor symptoms were significantly mitigated,
in a manner that may be associated with the production of dopamine
in midbrain neural stem cells. In another study, Ogawa et al
(7) cultured E14.5 stem cells
derived from embryonic spinal cords in vitro, after which
histopathological analysis was performed 5 days after
transplantation. The results revealed that treated rats had
significantly recovered motor function. However, immune reactions
induced by neural stem cell application and the resulting rejection
continue to be the main challenges obstructing their current
clinical application (8,9). Therefore, it is important to understand
the mechanism underlying the suppression of immune-related cell
activity during neural stem cell transplantation (10,11).
Exosomes serve as important tools for the exchange
of biologically active substances between cells (12). Most, if not all, mammalian stem cells
release exosomes containing certain characteristics of their source
parent cells (13). Exosomes contain
cell-specific proteins, lipids and nucleic acids, which act as
carriers for signal transduction, information exchange between
cells and participate in the development of normal physiological
processes and diseases (14). Li
et al (15) previously
reported that human umbilical cord mesenchymal stem cell exosomes
significantly inhibit the ratio of peripheral blood
CD3+CD4+ T cells and
CD3+CD8+ T cells in normal humans. Exosomes
with effective immunosuppressive functions have been demonstrated
to provide a novel target for immunotherapy in treating tumors and
autoimmune diseases (16,17).
Previous studies have suggested that autophagy
participates in the regulation of inflammation to prevent the
development of autoimmune and inflammatory diseases (18). Autophagy not only eliminates
macromolecules in autophagic cells, but also clears damaged
organelles to maintain intracellular homeostasis (19). Microglia are an important type of
neuroimmune cell, which in their activated state, induce tissue
repair and neuroprotection by releasing neurotrophic factors and
phagocytizing damaged nerve cells (20). In cases of acute trauma to the
central nervous system, including traumatic brain/spinal injury,
hypoxia or ischemic brain damage, microglia rapidly initiate an
immune response (21). Appropriate
activation of microglia is beneficial for wound repair and
microenvironmental reconstruction, which serves an important role
in a number of nerve cell repair processes (22). The occurrence of autophagy in
microglia also serves an important role in the differentiation,
survival and homeostasis maintenance of transplanted stem cells
(23). A study by Wang et al
indicated that bone marrow-derived neural progenitor cells can
differentiate into neurons, the transplantation of which in
vivo can effectively promote motor function in rats following
brain injury (24).
In previous studies, bone marrow-derived neural
progenitor cells have been characterized, revealing that these
cells have the potential to differentiate into neurons (25-27).
However, progress has been slow regarding investigation into the
treatment of brain injury using neural stem cell transplantation,
which may be due to changes in the intracranial microenvironment
following brain injury (26). A
series of studies have reported that the autophagy of microglia
serves an important role in brain injury, involving cranial nerve
inflammation, cerebral ischemia and cerebral hypoxia (28-30).
Stem cells that are transplanted into the body frequently fail and
do not result in tissue repair (31). This may be due to the fact that stem
cell transplantation is an exogenous procedure. Whether this
process activates microglia autophagy, or whether microglia
autophagy is associated with this process is yet to be fully
elucidated. Observation and study on this series of problems are
therefore urgently required for future clinical work on cell
transplantation. To expand on previous studies assessing bone
marrow-derived neural progenitor cell-mediated tissue repair
(28-30,32),
the present study systematically characterized the structure and
size of bone marrow-derived neural progenitor exosomes using
optical technology, analyzed its content using second-generation
sequencing technology and investigated the molecular mechanism
underlying microglia autophagy induced by the exosomes from bone
marrow-derived neural progenitor cells using molecular and cell
biology techniques. The present study provided theoretical
information on neural progenitor cell survival and differentiation
following the transplantation of bone marrow-derived neural
progenitor cells, in addition to offering mechanistic and
experimental support for the future clinical application of cell
transplantation.
Materials and methods
Materials
All reagents and chemicals were purchased and used
directly without further purification. The bone marrow stromal cell
line was collected from the rat model of our team (28-30),
whilst the BV-2 microglial cell line was provided by CHI Scientific
Inc. (cat. no. 7-1502). All aqueous solutions were prepared in
deionized water and triple distilled water was used for all in
vitro procedures. MTT, pancreatin and trypsin were purchased
from Sigma-Aldrich; Merck KGaA. DMEM/F12 and FBS were purchased
from Thermo Fisher Scientific, Inc. and Zhejiang Tianhang
Biological Technology Co., Ltd., respectively. ExoQuick™ reagent
(cat. no. EXOQ5A-1; Guangzhou Ruijing Information Technology Co.,
Ltd.), bicinchoninic acid (BCA) protein assay kit (cat. no. P0012S;
Beyotime Institute of Biotechnology) and Ultrafiltration centrifuge
tubes (cat. no. UFC901096) were purchased from Guangzhou Ruijing
Information Technology Co., Ltd. Rabbit antibodies for disabled
homolog 2-interacting protein (DAB2IP; cat. no. ab87811), Beclin1
(cat. no. ab62557), microtubule-associated protein 1A/1B-l light
chain 3 (LC3; cat. no. ab48394), p62 (cat. no. ab109012), CD81
(cat. no. ab124717), CD9 (cat. no. ab223052) and β-actin (cat. no.
ab179467) were purchased from Abcam. The present study was approved
by the Ethics Committee of Guangdong Provincial People's Hospital
(approval no. GDREC2018042A).
Isolation and culture of bone
marrow-derived neural progenitor cells
The methods and procedures for the isolation,
culture and identification of bone marrow-derived neural progenitor
cells were performed based on previous studies (25-27).
Three Sprague-Dawley rats (sex, male; age, 5 weeks; weight, 115-150
g) were housed in a temperature of 18-25˚C and 30-60% humidity.
Temperature 18-25˚C; humidity, 30-60%. Animals were purchased from
the Guangdong Medical Laboratory Animal Center. The rats were
sacrificed by cervical dislocation and placed in a beaker
containing 75% alcohol for 10 min. The bilateral femurs and tibias
were cut, and the soft tissues were removed. The medullary cavities
of the bones were washed with α-MEM supplemented with 10% fetal
bovine serum. Centrifugation occurred at 200 x g for 20 min. The
cloudy cell layers were plated on attachment plastic tissue culture
dishes, and the α-MEM containing 10% FBS was changed 4 days later
and renewed every 3 days. The cells were plated to obtain adherent
rat bone marrow stromal cells (MSCs), which were harvested when
cells reached 60-80% confluence.
Extraction of exosomes from bone
marrow-derived neural progenitor cells
Neural progenitor cells were cultured in a 5%
CO2 incubator at 37˚C. When the confluence reached
70-80%, cells were washed with physiological saline and DMEM/F12
medium was replaced. The medium was changed after 48 h to collect
the cell supernatant, followed by centrifugation (5,000 x g for 20
min, 4˚C) to remove cells or cell debris. The exosomal supernatant
(12 ml) was then added to the ultrafiltration centrifuge tube, and
another 12 ml of the supernatant was added into another
ultrafiltration centrifuge tube and centrifuged at 5,000 x g for 40
min (4˚C). The supernatant was subsequently transferred to a 1.5 ml
sterile, enzyme-free centrifuge tube, which was covered well.
ExoQuick Exosome reagent was subsequently added. The tube was
turned upside down to ensure adequate mixing and incubated at 4˚C
for 12 h. Subsequently, the mixture was centrifuged at 1,500 x g
for 30 min (4˚C), before the supernatant was carefully removed and
the remaining mixture was centrifuged (5,000 x g, 25˚C) for another
5 min. All the liquid was carefully discarded, leaving only the
white portion deposited at the bottom. Finally, 10% original volume
of DEPC water was used to resuspend the pellet and mixed prior to
storage at 4˚C for further use.
Identification of exosomes from bone
marrow-derived neural progenitor cells and microRNA (miRNA)
sequencing
The morphology of exosomes was observed by
transmission electron microscopy. A total of 10 µl of the 50 µl
freshly extracted exosomal suspension was obtained for a hanging
drop preparation on a copper mesh for 10 min and dried using filter
paper. Subsequently, 5 µl of 2% phosphotungstic acid negative dye
solution was added for 5 min and washed twice with ultrapure water.
Excess water was absorbed with filter paper. The copper mesh was
then incubated at room temperature until the sample dried
naturally. Samples were then placed into sample boxes for
observation under a transmission electron microscope at 20˚C and
30% humidity (Hitachi H-7650, Hitachi, Ltd.). Exosomes were fixed
with 90% ethanol at 20˚C for 10 min. Data were analyzed using
DigitalMicrograph 3.9 (Gatan, Inc.).
Exosome morphology was observed using atomic force
microscopy (AFM). Freshly extracted exosomal suspension was first
diluted with 0.9% physiological saline. A freshly cleaved mica
plate was then fixed onto the surface of a glass slide, where 50 µl
sample solution was dropped slowly onto to the mica plate and
allowed to dry. Finally, the sample was examined at room
temperature using a silicon probe in tapping mode (Dimension Edge;
Bruker Corporation).
The size and distribution of the freshly prepared
exosomes (300 µl) were measured using a particle size analyzer
(Zetasizer Nano ZS90; Malvern Instruments, Ltd.). The ID50 was
obtained and calculated once the cumulative size distribution
percentage reaches 50% of the grain size.
For on-column DNase digestion, exosome sample
solution (400 µl) was added to 400 µl 2X RNAgents®
Denaturing Solution (cat. no. Z5651; Promega Corporation), mixed by
vortexing and incubated on ice. An equal volume of
phenol/chloroform was added, mixed by vortexing and centrifuged
(5,000 x g for 20 min) at 4˚C. The supernatant was then pipetted
into absolute ethanol (1 ml), mixed and transferred to a column for
centrifugation (12,000 x g for 10 min) at 4˚C. A total of 350 µl
miRNA wash solution 1 was added, followed by further centrifugation
(12,000 x g for 5 min at 4˚C) and the supernatant was discarded.
DNase I (10 µl) was mixed with 70 µl RDD buffer (Qiagen GmbH)
before being transferred to the spin column and placed at room
temperature for 15 min. A total of 350 µl miRNA wash solution 1 was
added before further centrifugation (12,000 x g for 5 min at 25˚C)
to discard the supernatant. Wash solution 2/3 (500 µl) was then
added to rinse the column twice. The spin column was then placed
into a new collection tube, where 100 µl pre-warmed elution
solution was added to the center of the column, followed by
centrifugation (12,000 x g for 5 min at 25˚C). The resulting liquid
in the collection tube was the extracted total RNA and the sample
was stored at -80˚C.
Total RNA was initially used in a chip experiment,
which was subjected to quality inspection using NanoDrop ND-2000
spectrophotometer (Thermo Fisher Scientific, Inc.) and Agilent
Bioanalyzer 2100. Qualified RNA was then subjected to subsequent
chip experiments. High-throughput analysis of the miRNA expression
profiles was performed using Agilent miRNA chip technology (Rat
miRNA 8x15k v21.0 microarray; Agilent Technologies GmbH). Sinotech
Genomics performed miRNA sequencing. The chip results were scanned
using the Agilent Microarray Scanner, where the data were read by
the Feature Extraction software 10.7.1.1 (Agilent Technologies
GmbH), followed by normalization using the R language package.
Bone marrow-derived neural progenitor
exosome-induced microglia autophagy
For autophagy detection in BV2 cells, different
amounts of exosomes were added to BV2 cells (5 µl, 360 ng/µl; 15
µl, 360 ng/µl) at 37˚C. After 48 h, an inverted fluorescence
microscope (TE2000-E; Nikon Corporation) was used to capture
images. The culture medium (DMEM/F12) was then removed, cells were
washed once with PBS and fixed with 2.5% glutaraldehyde solution at
4˚C overnight. The fixative was removed, the sample was rinsed
three times with PBS and fixed with 1% osmic acid solution for 2 h
at 37˚C. The sample was subsequently dehydrated using an ascending
ethanol gradient (30, 50, 70, 80, 90 and 95%) and treated with pure
acetone for 20 min at 25˚C. The sample was treated with a mixture
of epikote resin and acetone (v/v=1/1) for 1 h, followed by the
same mixture but at a higher concentration of the embedding agent
(v/v=3/1) for 3 h before incubation with 100% embedding agent
overnight at 25˚C. The treated sample was embedded and heated at
70˚C overnight to obtain an embedded sample. The samples were
sectioned using a Leica EM UC7 ultramicrotome (Leica Microsystems
GmbH) where 70-90 nm sections were obtained. The sections were
stained with 10% lead citrate solution for 10 min at 25˚C and 50%
uranyl acetate for 10 min at 25˚C before observation under a
transmission electron microscope (Hitachi H-7650, Hitachi,
Ltd.).
For colony formation assays, BV cells were seeded
into six-well plates at 500 cells/well. Following 24 h incubation
at 37˚C with 5% CO2, DMEM was then replaced and exosomes
were added for 14 days (5 µl, 360 ng/µl). The control group was not
treated with exosomes. The cells were then fixed with 90% ethanol
for 30 min at 25˚C and washed with PBS three times. After further
incubation for 30 min at 37˚C in the dark and staining with 1X
Giemsa solution at 25˚C for 20 min, images was obtained on an
inverted microscope (TE2000-E; Nikon Corporation) at x100
magnification.
MTT assay
Inhibition of cell growth by exosomes was measured
using an MTT assay. The cells were exposed to exosomes at different
times (12, 24 and 48 h). Briefly, BV cells were seeded in 96-well
tissue culture plates (5x103 cells/per well) for 12 h at
37˚C with 5% CO2. After exosome (5 µl, 360 ng/µl)
treatment, BV2 cells were incubated for 24 h at 37˚C with 5%
CO2. After incubation, 20 µl/well MTT solution (5.0
mg/ml in PBS) was added and incubated for 4 h at 37˚C with 5%
CO2. The medium was aspirated and replaced with 100
µll/well DMSO to dissolve the formazan salt. The absorbance
intensity was measured by using a microplate spectrophotometer at a
wavelength of 570 nm (Thermo Fisher Scientific, Inc.).
Molecular analysis of autophagy
induction
The number of exosomes used for molecular mechanism
analysis was determined by measuring the total protein
concentration in the exosomes, which was obtained using the BCA kit
(360 ng/µl).
Western blotting
The exosomes secreted by the bone marrow-derived
neural progenitor cells and the bone marrow-derived neural
progenitor cells of the normal control group were first collected
by centrifugation at 12,000 x g for 10 min at 4˚C. A total of 70 µl
RIPA buffer (cat. no. P0013C; Beyotime Institute of Biotechnology)
was then added and incubated on ice for 30 min. The protein was
quantified using the BCA method, following which the protein mass
concentration was adjusted to 5 g/l using RIPA buffer and stored at
-80˚C for further use. For electrophoresis, 20 µg protein was
separated by 10% SDS-PAGE, before being transferred onto
nitrocellulose membranes at 100V for 1 h and blocked in blocking
solution (cat. no. P0216; Beyotime Institute of Biotechnology) for
1 h at 37˚C. The membranes were then incubated with an appropriate
amount of primary antibodies against DAB2IP, Beclin1, p62, CD81,
CD9 and LC3 (all, 1:1,000) overnight, following which the membranes
were incubated with corresponding secondary antibody (alkaline
phosphatase-labeled goat anti-rabbit immunoglobulin G; 1:1,000;
cat. no. A0239; Beyotime Institute of Biotechnology) for 1 h at
room temperature. The protein bands were visualized by ECL
luminescence, following which densitometric analysis was performed.
Protein bands were visualized using GelDoc XR (BioRad Laboratories,
Inc.).
Calcium ion detection
Different amounts of exosomes were added to BV2
cells (5 µl, 360 ng/µl; 15 µl, 360 ng/µl), which were then
incubated for 24 h at 37˚C. Fluo-3 AM working solution (1 µM; cat.
no. S1056; Beyotime Institute of Biotechnology) was added to
sufficiently cover the cells. After incubation for 30 min at 37˚C,
the fluorescence of Fluo-3 was measured (excitation wavelength, 488
nm; emission wavelength, 530 nm) using a fluorescence microplate
reader (Varioskan Flash; ThermoFisher Scientific, Inc.) to assess
any changes in intracellular calcium ion concentrations.
Luciferase assay
The miRNA and 3'-UTR binding site (accession no.
NM_032552.3; Homo sapiens DAB2 interacting protein, DAB2IP,
transcript variant 1, mRNA) was predicted by BLAST (https://docs.microsoft.com/en-us/previous-versions/hh397746(v=msdn.10)?redirectedfrom=MSDN).
Upon reaching a confluence of ~70%, 293T cells (American Type
Culture Collection; incubated at 37˚C with 5% CO2) were
seeded into six-well plates at a density of 2x106
cells/ml in DMEM. Following incubation at 37˚C in 5% CO2
for 16 h, cell confluency typically reached ~80%. The culture
medium was then pipetted into each well, where 2 ml Opti-MEM was
added to each well, and culture continued in a constant temperature
incubator at 37˚C. Plasmids (Guangzhou RiboBio Co., Ltd.; 2 µg; WT
or mut-psi-CHECK2 and scrambled RNA, mimic-miR-32-3P or
inhibitor-miR-32-3P; 1:1 ratio) was mixed with 0.5 ml Opti-MEM and
5 µl Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) diluted in 0.5 ml of opti-MEM.
miR-32 mimic was transfected into 293T cells using
Lipofectamine (Invitrogen; Thermo Fisher Scientific, Inc.). The
sequences of the miRNAs (synthesized by Guangzhou RiboBio Co.,
Ltd.) were as follows: mimic-miR-32-3p,
5'-CAATTTAGTGTGTGTGATATTT-3'; inhibitor-miR-32-3p,
5'-AAATATCACACACACTAAATTG-3' and scramble RNA (reference),
5'-GATAGACGCGCAGGAAGTAGA-3'. The plasmid mixture and the
transfection reagent mixture were then mixed and incubated for 20
min prior to addition to the cells. Following 6 h culture at 37˚C,
cells were added with 2 ml complete medium and incubated at 37˚C
for a further 24 h. The medium of the six-well plate was then
removed, washed twice with PBS and lysed with 1 X PLB. After
incubation for 15 min at room temperature, 20 µl lysates was
transferred to a 96-well plate, where 100 µl Luciferase Assay
Reagent II (LAR II) was added to each well. The fluorescence value
was detected using a chemiluminescence apparatus. A total of 100 µl
Stop & Glo® buffer was added to each well and the
fluorescence was measured for a second time. Renilla
luciferase activity was used as an internal control.
Reverse transcription-quantitative PCR
(RT-qPCR). BV2 cells were seeded in six-well plates
(1x106 cells/well) for 12 h at 37˚C, then different
amounts of exosomes were added to BV2 cells (5/15 µl, 360 ng/µl)
for 24 h at 37˚C. The BV2 cells were collected, total RNA was then
extracted using RNAprep Pure Cell/Bacteria kit (cat. no. DP430;
Tiangen Biotech Co., Ltd.) in accordance with the manufacturer's
protocol, following which RNA purity was measured using a NanoDrop
2000 spectrophotometer (Thermo Fisher Scientific, Inc.). The
sequences used were as follows: U6 forward, 5'-CTCGCTTCGGCAGCACA-3'
and reverse, 5'-AACGCTTCACGAATTTGCGT-3'; miR-32-3p,
3'-CAATTTAGTGTGTGTGATAT-5'; miR-211-3p,
3'-GCAAGGACAGCAAAGGGGGGC-5'; miR-188-5p,
3'-CATCCCTTGCATGGTGGAGGG-5'; miR-466b-5p,
3'-TGATGTGTGTGTACATGTACAT-5'.
RNA was added with polyA tail and reverse
transcribed into cDNA (RevertAid First Strand cDNA synthesis kit;
cat. no. K1622; Thermo Fisher Scientific, Inc.), following which
the expression of four potential target genes were detected via
qPCR (GoTaq qPCR and RT-qPCR Systems; cat. no. A6001; Promega
Corporation). Reactions were performed on a fluorescence qPCR
instrument (CFX Connect; cat. no. 1855200; BioRad Laboratories,
Inc.). The following thermocycling conditions were used for the
qPCR: Initial denaturation at 95˚C for 10 min, followed by 50
cycles of 95˚C for 15 sec and 60˚C for 1 min. The relative mRNA
levels of target samples to control samples were calculated
according to 2-ΔΔCq method (33).
Statistical analysis
Data are expressed as the mean ± standard error of
the mean based on at least three independent experiments.
Statistical analysis was performed using one-way ANOVA, followed by
a Bonferroni post hoc test for multiple group comparisons or a
Student's unpaired t-test for pairwise comparisons. P<0.05 was
considered to indicate a statistically significant difference
unless otherwise indicated. All analyses were performed using
GraphPad Prism Software version 6.0 (GraphPad Software, Inc.).
Results
Isolation of bone marrow-derived
neural progenitor cells and the effect in BV2 cells
The methods and procedures for the isolation,
culture and identification of bone marrow-derived neural progenitor
cells were performed based on previous studies (25-27).
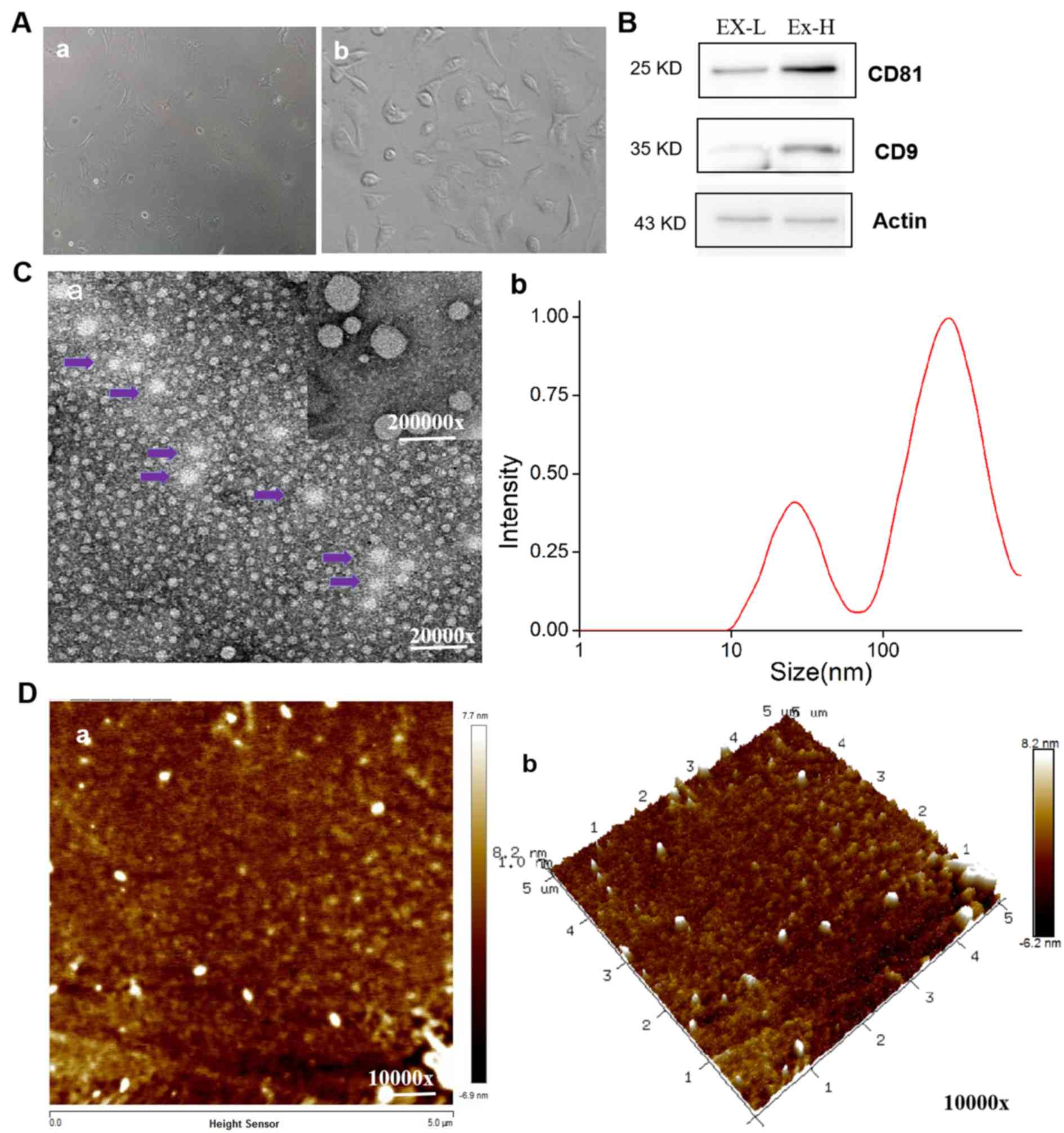
As shown in Fig. 1A-a, the
morphology of the isolated and identified bone marrow-derived
neural progenitor cells appeared to be slightly. The mouse
microglial cell line BV-2 was purchased and sub-cultured, which
were in fusiform states (Fig. 1A-b).
Exosome-associated target proteins, such as CD9 and CD81, are types
of biomarkers for exosomes (34). As
shown in Fig. 1B, the exosomes of
bone marrow-derived neural progenitor cells were first
characterized via western blotting, by examining the expression of
exosome-associated target proteins CD9 and CD81 (13-15).
Morphology and particle size
distribution of exosomes from bone marrow-derived neural progenitor
cells
The exosomes of bone marrow-derived neural
progenitor cells were collected via ultrafiltration concentration
centrifugation and observed by transmission electron microscopy
(Fig. 1Ca). They were found to be
either circular or elliptical in shape, of homogeneous
distribution, with double-layered lipid vesicle (arrows). The
vesicles secreted by bone marrow-derived neural progenitor cells
have clear vesicle membrane structures with higher internal
dioptres compared with the background, consistent with the exosome
features observed (34). Based on
this, the size of exosomes from bone marrow-derived neural
progenitor cells was then measured using a laser particle size
analyzer. The abscissa represents the diameter range, whilst the
ordinate represents the concentration/intensity of the exosomes
from the bone marrow-derived neural progenitor cells. The exosome
ID50 value of the bone marrow-derived neural progenitor
cells was 100 nm (Fig. 1Cb).
The surface morphology and three-dimensional
structures of these exosomes were analyzed further using AFM
(Fig. 1D). The average diameter of
the exosomes was measured to be 28.63±11.59 nm (n=3), where a small
number of vesicles >1,000 nm in diameter were observed. The
results indicated that the exosomes of bone marrow-derived neural
progenitor cells were successfully isolated, where the diameter of
most exosomes was <100 nm.
Sequencing of miRNA found in exosomes
from bone marrow-derived neural progenitor cells
The exosomes of bone marrow-derived neural
progenitor cells were collected and total exosomal RNA was
extracted. High-throughput analysis of miRNA expression profiles
was performed using Agilent miRNA chip technology. An increasing
number of studies have previously demonstrated that miRNAs serve
important roles in various diseases and biological functions
(34,35). miRNAs can regulate a number of
physiological processes, including proliferation, cell cycle
progression, apoptosis, invasion and angiogenesis by inhibiting the
expression of target genes (36).
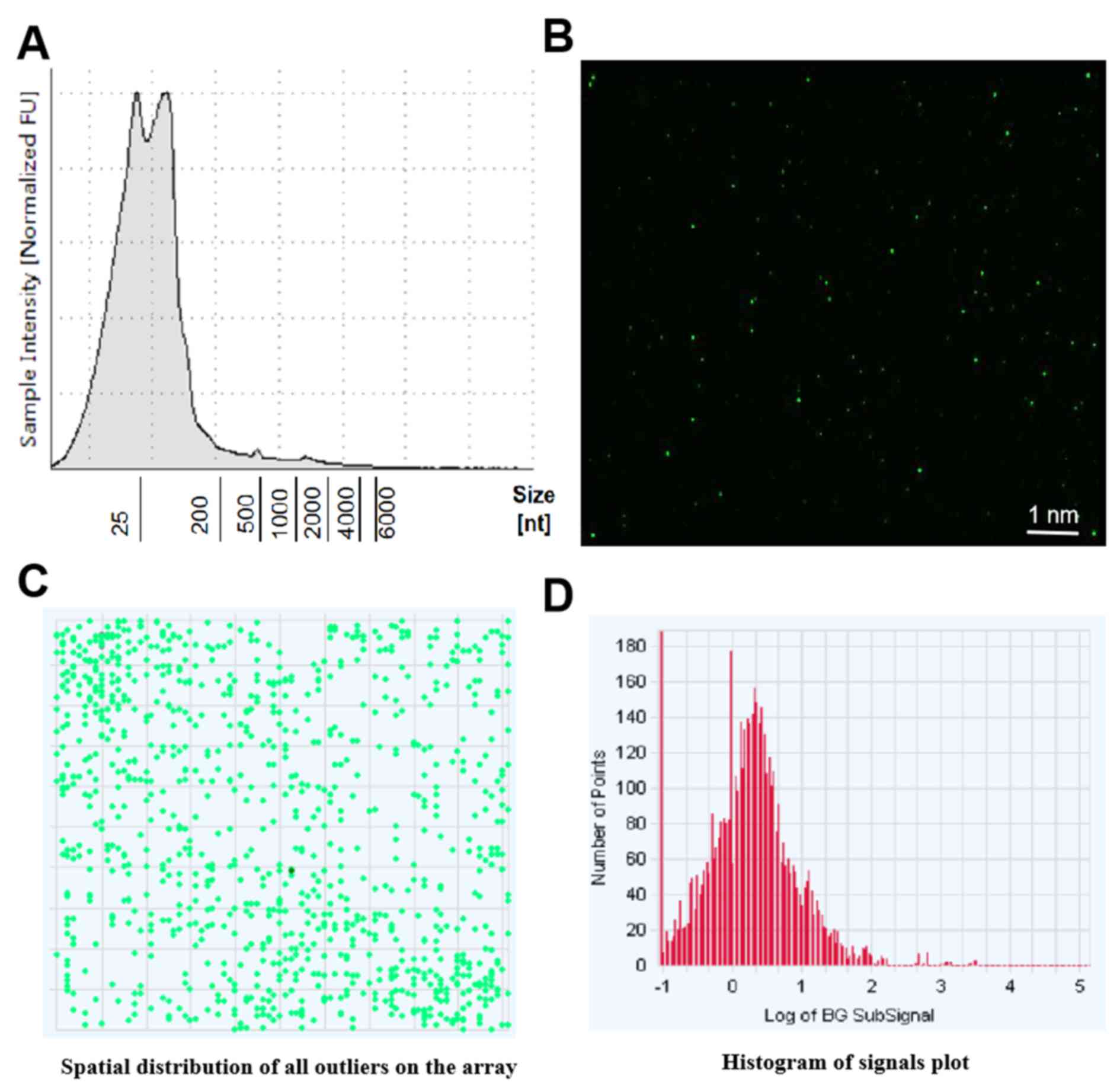
The results of sequencing of miRNA shown that 758 miRNAs found in
the exosomes met the quality control requirements of bone
marrow-derived neural progenitor cells. The original data of
chip-based detection is presented in Fig. 2B. The obtained data is demonstrated
in Fig. 2C. The signal value of the
original data miRNA probe is presented in Fig. 2D.
From the 758 miRNAs detected in the exosomes of bone
marrow-derived neural progenitor cells, the top 20 miRNAs according
to abundance were selected. Information, including type name, the
degree of correspondence between the probe and the original data,
in addition to their active sequence structure information, are
provided in Table I. After combining
literature reports (37-41),
four potential miRNAs which have been documented to serve a role in
exosome-induced microglia autophagy were identified: rno-miR-32-3p,
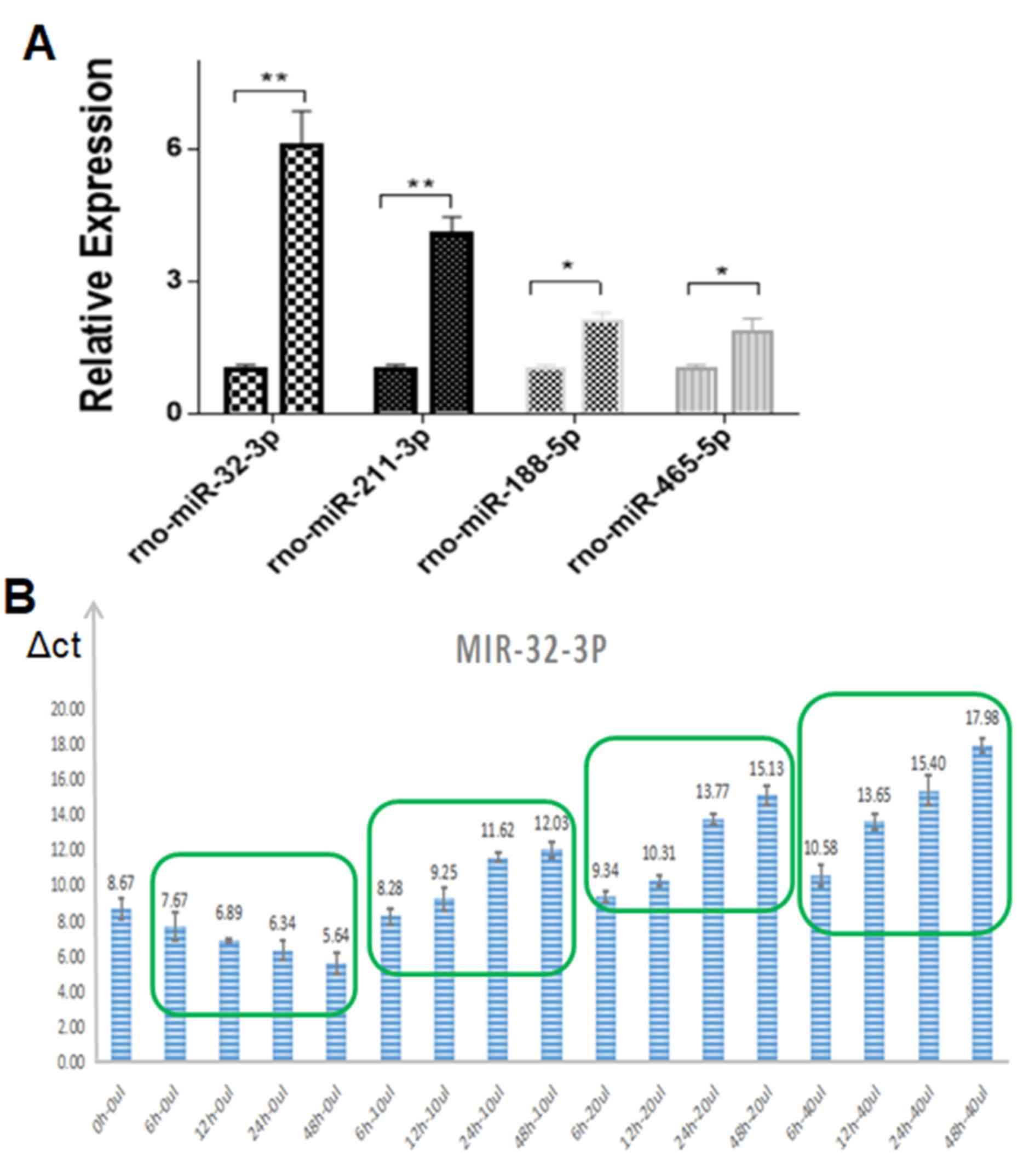
rno-miR-211-3p, rno-miR-188-5p and rno-miR-465-5p. RT-qPCR was
performed to investigate the differences in the relative expression
of these four potential target miRNAs in the microglia cells in the
presence or absence of bone marrow-derived neural progenitor
exosomes (Fig. 3).
 | Table IResults and information of miRNA chip
detection in exosomes (top 20). |
Table I
Results and information of miRNA chip
detection in exosomes (top 20).
| Systematic
name | Total probe signal
(raw) data | Total probe signal
(normalized) | Sequence |
|---|
| miR-1224 | 1511.147 | 10.561428 |
5'-GTGAGGACTCGGGAGGTGG-3' |
| miR-466b-5p | 737.30896 | 9.526125 |
5'-TATGTGTGTGTGTATGTCCATG-3' |
| miR-32-3p | 129.2438 | 7.0139513 |
5'-CAATTTAGTGTGTGTGATATTT-3' |
| miR-1896 | 82.5686 | 6.3675213 |
5'-TGGTGGGTGAGGAGGAGG-3' |
| miR-672-5p | 73.4067 | 6.1978397 |
5'-TGAGGTTGGTGTACTGTGTGTGA-3' |
| miR-1306-3p | 64.6163 | 6.013826 |
5'-ACGTTGGCTCTGGTGGTG-3' |
| miR-211-3p | 34.9854 | 5.128681 |
5'-GCAGGGACAGCAAAGGGGTGC-3' |
| miR-466c-5p | 33.4107 | 5.0622387 |
5'-TGTGATGTGTGTATGTACATG-3' |
| miR-466d | 31.84107 | 4.992817 |
5'-ATGTGTGTGTATGTCCTTTTGT-3' |
| miR-465-5p | 31.7184 | 4.987248 |
5'-TATTTAGAACGGTGCTGGTGTG-3' |
| miR-3473 | 31.4372 | 4.9744005 |
5'-TCTAGGGCTGGAGAGATGGCTA-3' |
| miR-3593-3p | 27.8128 | 4.797677 |
5'-CCATCACCACACGACTGGATTTTGG
CCTCCGCAGGGTTGAAGCTGCTAGAGA AGCTTCAACCTTAAGGGGGCCTCAGGG
AGAGAGGAGTGG-3' |
| miR-1249 | 26.360802 | 4.720322 |
5'-GGGAGGAGGGAGGAGATGGGCCAAGT |
| | | |
TCCCTCTGGCTGGAACGCCCTTCCCCCC
CTTCTTCACCTG-3' |
| miR-6216 | 25.43394 | 4.668683 |
5'-GATACACAGAGGCAGGAGGAGAA-3' |
| miR-3588 | 25.288 | 4.660381 |
5'-TCACAAGTTAGGGTCTCAGGGA-3' |
| miR-483-5p | 24.348429 | 4.6057568 |
5'-AAGACGGGAGGAAAGAAGGGAG-3' |
| miR-2985 | 23.6542 | 4.5640244 |
5'-ATCGCCACACCTAAAGGATCCTCATTA
AGGTGGGTGGAATAGTATAACAATGTGCT CAATGTTGTTATAGTATCCCACCTACCCTG
ATGTGTCTTTAAGACTCTAACGGT-3' |
| miR-296-5p | 20.11955 | 4.3305264 |
5'-AGGGCCCCCCCTCAATCCTGT-3' |
| miR-188-5p | 19.535389 | 4.2880177 |
5'-CATCCCTTGCATGGTGGAGGG-3' |
| miR-3562 | 18.44011 | 4.2047753 |
5'-TTGGGGCAGTGGCTGGATGGGA-3' |
It was revealed that that miR-32-3p exhibited the
highest expression, which meant it could be used as a potential
target molecule (Fig. 3A).
Therefore, the expression of miR-32-3p in microglia was examined
further using different volumes of exosomes and different treatment
times, the results of which are presented in Fig. 3B.
Bone marrow-derived neural progenitor
exosomes induce microglia autophagy
Using molecular biology and cell biology techniques,
the mechanism underlying microglia autophagy induced by exosomes
from bone marrow-derived neural progenitor cells was investigated
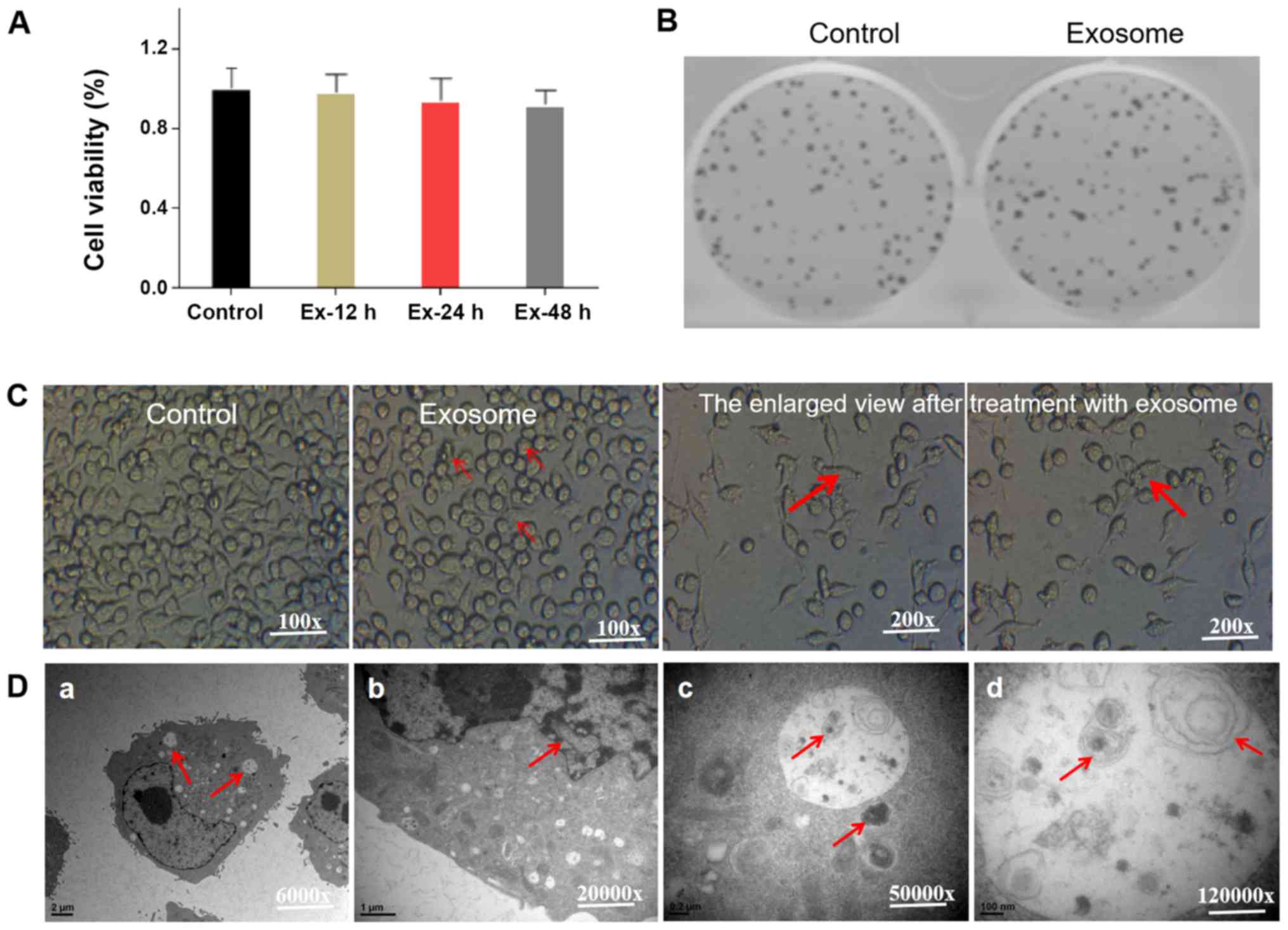
further. MTT and colony formation assays were performed to
investigate the effects of exosomes from bone marrow-derived neural
progenitor cells on the viability and proliferation of microglia.
Exosomes of bone marrow-derived neural progenitor cells were
collected during microglia growth (Fig.
4A). High-dose exosomes (Ex-H) had no effect on the growth of
microglia. No statistically significant differences were observed
in the survival rates between cells cultured for 12 h with exosomes
and cells without the addition of exosomes, nor between cells
cultured in the absence of exosome treatment and those treated with
exosomes for 24 and 48 h.
This suggested that exosomes of bone marrow-derived
neural progenitor cells had no significant effect on microglia cell
growth. This can be clinically relevant if performed without
affecting the growth of normal cells. By measuring the formation
rate of colonies, the effect of exosomes from bone marrow-derived
neural progenitor cells on the proliferation of microglia was
investigated (Fig. 4B). The number
of microglial colonies formed following the addition of exosomes
from bone marrow-derived neural progenitor cells was not found to
be markedly different from the control group, suggesting that the
addition of exosomes from bone marrow-derived neural progenitor
cells did not affect microglia proliferation.
Notably, the shape of microglial cells change from
roundness to fusiform after the addition of exosomes from bone
marrow-derived neural progenitors (Fig.
4C). Damaged organelles such as swollen and degenerated
mitochondria were observed under a transmission electron microscope
(Fig. 4D). Additionally, residue
that could not be fully degraded in the autolysosome was observed.
These results suggested that exosomes from bone marrow-derived
neural progenitor cells induced microglia autophagy without
affecting growth and proliferation.
Western blotting and luciferase reporter gene assays
were subsequently performed to validate the potential targets of
miR-32-3p and associated regulatory mechanism. Following the
addition of exosomes from bone marrow-derived neural progenitor
cells, the expression of the DAB2IP protein in microglial cells was
found to be upregulated whilst that of p62 was significantly
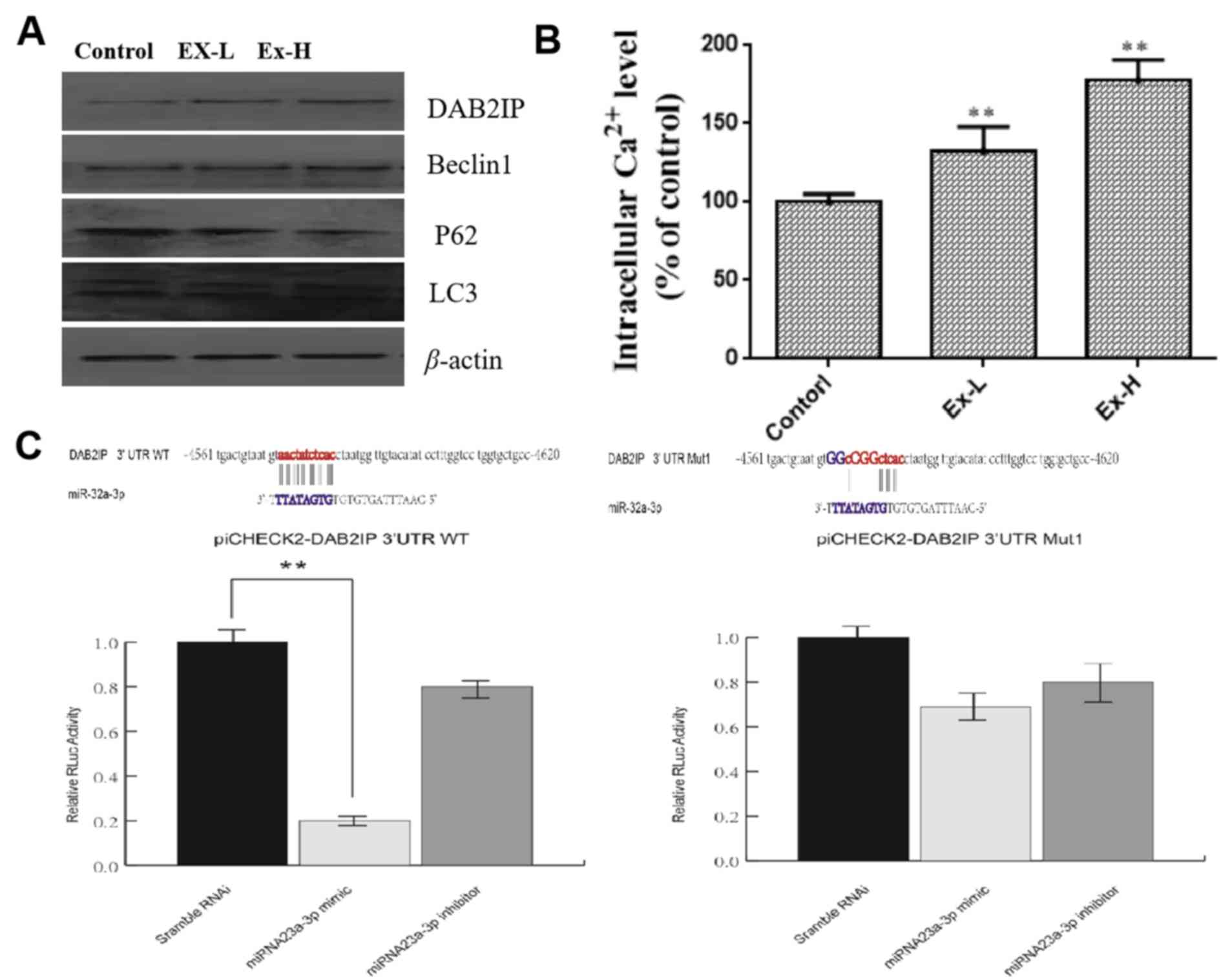
downregulated (Fig. 5A). The shear
band (the upper or lower band LC3 II/LC3 I) of the LC3 protein was
also found to be significantly increased. In addition, Beclin 1 was
observed to be significantly upregulated following the addition of
exosomes from bone marrow-derived neural progenitor cells (Fig. 5A). Different amounts of exosomes were
added to BV2 cells [Low dose (Ex-L), 5 µl, 360 ng/µl; high-dose
(Ex-H), 15 µl, 360 ng/µl], and the expression of proteins CD9 and
CD81 of Ex-H was markedly higher compared with Ex-L (Fig. 1B). These results suggested that
microglia autophagy may be associated with the target binding of
miR-32-3p and the DAR2IP (39),
alongside the subsequent activation of Beclin1(42).
The potential process of endoplasmic reticulum
stress was examined further. As presented in Fig. 5B, after treatment with exosomes, the
intracellular Ca2+ level in BV2 cells significantly
increased compared with controls. The potential interaction between
miR-32-3p and the DAB2IP protein was examined further using
luciferase reporter gene assays. As shown in Fig. 5C, miR-32 could bind DAB2IP by
directly targeting 3'-UTR, to regulate the expression of DAB2IP
mRNA.
Discussion
A number of studies have demonstrated the important
role of microglia autophagy in brain injury, including cranial
nerve inflammation, cerebral ischemia and cerebral hypoxia
(28-30).
Clinical stem cells transplantation frequently fails and does not
result in tissue repair (31). It is
therefore important to investigate the effect of microglia
autophagy in this process. Observation and study on this series of
problems will be helpful in exploring future clinical applications
regarding cell transplantation. Microglia autophagy serve an
important role in the differentiation, survival and homeostasis
maintenance of transplanted stem cells (43-46).
Previous studies have demonstrated that bone marrow-derived neural
progenitor cells can differentiate into neurons and their
transplantation in vivo can effectively promote the motor
function of rats following brain injury (25-27).
Microglial cell autophagy has also been found in the
process of bone marrow-derived neural progenitor cell
transplantation during the treatment of brain nerve injury in SD
mice in another previous study (47). Investigating the relationship and
mechanism between microglia autophagy and the survival and
differentiation of neural stem cells are of significance for
targeting microglia-associated inflammation, promoting the survival
of transplanted neural stem cells in damaged tissues and improving
their ability to repair tissues.
The number of studies into the role of exosomes on
biological processes is increasing where their importance is
gradually being revealed (48,49).
However, since the extraction yield of exosomes reported in
existing studies has generally been low, the specific molecular
mechanism of exosome involvement in intercellular communication
remains unclear (50).
Therefore, using information obtained from previous
studies, the present study extracted exosomes of high-yield
(51,52) and high purity from bone
marrow-derived neural progenitor cells. The molecular mechanism of
the effects of autophagy on the relationship between bone
marrow-derived neural progenitor cells and microglia was
subsequently analyzed by sequencing and western blotting. The
pathway of microglia autophagy that was affected by exosome miRNA
was then verified, where a potential molecular mechanism of action
was proposed. Results of present study provided a basic theoretical
basis for an in-depth study on the clinical application of RNA
molecules in exosomes secreted by bone marrow-derived neural
progenitor cells.
Among the 758 miRNAs detected in the exosomes of
bone marrow-derived neural progenitor cells, the top 20 miRNAs were
selected according to detection abundance. Their type name, the
extent of probe correspondence between original and normalized
data, and the active sequence structure information were obtained.
Combining bioinformatics analysis and literature reports, four
potential miRNAs (rno-miR-32-3p, rno-miR-211-3p, rno-miR-188-5p and
rno-miR-465-5p), which have been reported to serve a role in
exosome-induced microglia autophagy, were identified (53,54).
Using RT-qPCR, differences in the relative expression of these four
potential target molecules in microglia in the presence and absence
of exosomes of bone marrow-derived neural progenitor cells was
investigated further. Following the addition of exosomes from bone
marrow-derived neural progenitor cells, all four potential target
miRNAs exhibited significantly increased expressions in microglial
cells, where the expression of miR-32-3p, which is located in the
14th intron of the gene C9orf5, was found to be the highest,
suggesting that it is a potential target molecule. Previous studies
on the biological effects of miR-32 have mainly concentrated on
tumors, viral replication and androgen regulation, which
demonstrated that upon miR-32 overexpression, growth of the
androgen-dependent prostate cancer cell line LNCaP was promoted.
miR-32 has also been found to reduce apoptosis by directly
regulating the expression of the BTG2 gene (55). Upregulation of miR-32 has been
reported to inhibit the expression of apoptosis-inducing protein
phosphoinositide-3-kinase interacting protein 1 and regulate the
Ser/Thr protein kinase mitogen-activated protein kinase kinase,
thus serving an important role in a variety of different signal
transduction pathways (56). miR-32
serves a regulatory role in a number of physiological processes,
including cell growth, differentiation, inflammation, apoptosis,
vascular endothelial cell proliferation and neovascularization
(57,58). However, the role of miR-32 in
microglial growth, proliferation and apoptosis remains poorly
understood.
In the present study, molecular and cell biology
techniques revealed that exosomes from bone marrow-derived neural
progenitor cells exerted no significant effects on the growth and
proliferation of microglia. Notably, following the addition of
exosomes from bone marrow-derived neural progenitor cells,
microglia began to develop autophagy, with typical autophagy
features (59) observed under light
and transmission electron microscopy. The results suggested that
exosomes from bone marrow-derived neural progenitor cells can
induce microglia autophagy without affecting the growth and
proliferation of microglia.
Western blot and luciferase reporter gene assays
were performed to validate the potential targets of the miR-32-3p
associated regulatory mechanism. Following the addition of exosomes
from bone marrow-derived neural progenitor cells, DAB2IP protein
expression in microglia was found to be upregulated. The expression
of Beclin1, which has been recently determined to be involved in
the regulation of autophagy (60),
was also found to be upregulated.
These results suggested that autophagy in microglia
may be associated with miR-32-3p. In this process, miR-32-3p may be
produced by the introduction of exosomes from bone marrow-derived
neural progenitor cells. During the induction of autophagy in
microglia, the marker protein p62 was degraded in the current
study, whilst that of the LC3 protein shear band was increased,
suggesting that the conditions for cellular stress to cause the
endoplasmic reticulum to initiate autophagy.
The present study also examined the underlying
process of endoplasmic reticulum stress. Following the addition of
exosomes from bone marrow-derived neural progenitor cells,
microglial intracellular calcium concentrations were increased to
slightly increased. This suggested that the entry of exogenous
signals resulted in the induction of endoplasmic reticulum stress
in microglial cells, consistent with the degradation of p62 found
in western blotting. Combining these experimental results, the
mechanism underlying microglia autophagy induced by the exosomes of
bone marrow-derived neural progenitor cells was hypothesized.
First, miR-32-3p present in the exosomes secreted by bone
marrow-derived neural progenitor cells enter into microglia, where
they specifically bind to the DAB2IP to affect its expression.
Then, the increase in DAB2IP protein expression in turn promotes
the expression of the autophagy-inducing protein Beclin1 and
enhances the stress response in microglia. At the same time, the
endoplasmic reticulum initiates the stress response mechanism,
increasing the intracellular calcium concentration (43) and causing p62 degradation, LC3
cleavage and the formation of cell autophagosomes. In this process,
miR-32-3p was found to be a potential target molecule, whilst
DAB2IP protein was a potential target protein. Briefly, bone marrow
stromal cells (MSCs) are potentially useful for the treatment of a
number of brain injury diseases due to their abundant supply. An
increasing number of studies have reported their prospective
functions and applications (9,12-17).
In the present study, 758 miRNAs were detected in exosomes of MSCs
using Agilent miRNA chip technology, where rno-miR-32-3p,
rno-miR-211-3p, rno-miR-188-5p and rno-miR-465-5p were first found
in the exosomes of MSCs.
Since microglia autophagy serves an important role
in conditions associated with brain injury, including cranial nerve
inflammation, cerebral ischemia and cerebral hypoxia, the results
of the present study provided a theoretical reference for studies
into brain injury treatment based on neural stem cell
transplantation. In addition, the present study provided certain
theoretical basis for cell survival and differentiation following
transplantation of bone marrow-derived neural progenitor cells, in
addition to theoretical and experimental support for future
clinical cell transplantation. However, there are some limitations
to the present study. For example, it only focused on the
biological effects of exosomes from bone marrow-derived neural
progenitor cells on microglia, instead of the effects of microglial
metabolism. In addition, among the plethora of miRNAs found in the
exosome, only 4 potential molecules were studied by bioinformatics
analysis and the role of the remaining miRNAs on microglial
autophagy cannot be ruled out. The present study only investigated
the binding of miR-32-3p to DAB2IP instead of investigating its
potential role further, which are the key issues that need to be
addressed in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural
Science Foundation of China (NNSFC; grant no. 81871842), the
Guangdong Medical Research Fund Project (grant no. A2018510) and
the Guangdong Science and Technology Program (grant no.
2016A020214014).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FYY, MXZ, MSZ and JYO designed and directed the
experiments. FYY, MXZ, YHS, WFB and MHL performed the experiments.
FYY, WFB and MSZ wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Guangdong Provincial People's Hospital (Certificate
no. GDREC2018042A).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Khan AR, Yang XY, Fu MF and Zhai GX:
Recent progress of drug nanoformulations targeting to brain. J
Controlled Release. 291:37–64. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hakes AE and Brand AH: Neural stem cell
dynamics: The development of brain tumours. Curr Opin Cell Biol.
60:131–138. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chrostek MR, Fellows EG, Crane AT, Grande
AW and Walter C: Low efficacy of stem cell-based therapies for
stroke. Brain Res. 1722(146362)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Grochowski C, Radzikowska E and
Maciejewski R: Neural stem cells therapy-Brief review. Clin Neurol
Neurosurg. 173:8–14. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Huang L and Zhang LB: Neural stem cell
therapies and hypoxic-ischemic brain injury. Prog Neurobiol.
173:1–17. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kochanek PM, Jackson TC, Ferguson NM,
Carlson SW, Simon DW, Brockman EC, Ji J, Bayır H, Poloyac SM,
Wagner AK, et al: Emerging therapies in traumatic brain injury.
Semin Neurol. 35:83–100. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ogawa Y, Sawamoto K, Miyata T, Miyao S,
Watanabe M, Nakamura M, Bregman BS, Koike M, Uchiyama Y, Toyama Y
and Okano H: Transplantation of in vitro-expanded fetal neural
progenitor cells results in neurogenesis and functional recovery
after spinal cord contusion injury in adult rats. J Neurosci Res.
69:925–933. 2002.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Volpe G, Bernstock JD, Peruzzotti-Jametti
L and Pluchino S: Modulation of host immune responses following
non-hematopoietic stem cell transplantation: Translational
implications in progressive multiple sclerosis. J Neuroimmunol.
331:11–27. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gupta N, Henry RG, Kang SM, Strober J, Lim
DA, Ryan T, Perry R, Farrell J, Ulman M, Rajalingam R, et al:
Long-term safety, immunologic response, and imaging outcomes
following neural stem cell transplantation for pelizaeus-merzbacher
disease. Stem Cell Rep. 13:254–261. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang YH, Chen JA, Zhou J, Nong F, Lv JH
and Liu J: Reduced inflammatory cell recruitment and tissue damage
in spinal cord injury by acellular spinal cord scaffold seeded with
mesenchymal stem cells. Exp Ther Med. 13:203–207. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Adams KV and Morshead CM: Neural stem cell
heterogeneity in the mammalian forebrain. Prog Neurobiol. 170:2–36.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ning Y, Wang X, Zhang P, Liu A, Qi X, Liu
M and Guo X: Dietary exosome-miR-23b may be a novel therapeutic
measure for preventing Kashin-Beck disease. Exp Ther Med.
15:3680–3686. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yang Y, Bucan V, Baehre H, Ohe JVD, Otte A
and Hass R: Acquisition of new tumor cell properties by MSC-derived
exosomes. Int J Oncol. 47:244–252. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lee KS, Choi JS and Cho YW: Reprogramming
of cancer stem cells into non-tumorigenic cells using stem cell
exosomes for cancer therapy. Biochem Biophys Res Commun.
512:511–516. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Li L, Liu S, Xu Y, Zhang A, Jiang J, Tan
W, Xing J, Feng G, Liu H, Huo F, et al: Human umbilical
cord-derived mesenchymal stem cells downregulate inflammatory
responses by shifting the Treg/Th17 profile in experimental
colitis. Pharmacology. 92:257–264. 2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Pan D, Chang X, Xu M, Zhang M, Zhang S,
Wang Y, Luo X, Xu J, Yang X and Sun X: UMSC-derived exosomes
promote retinal ganglion cells survival in a rat model of optic
nerve crush. J Chem Neuroanat. 96:134–139. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Cho JA, Park HP, Lim EH and Lee KW:
Exosomes from breast cancer cells can convert adipose
tissue-derived mesenchymal stem cells into myofibroblast-like
cells. Int J Oncol. 40:130–138. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Pankiv S, Alemu EA, Brech A, Brech A,
Bruun JA, Lamark T, Overvatn A, Bjørkøy G and Johansen T: FYCO1 is
a Rab7 effector that binds to LC3 and PI3P to mediate microtubule
plus end-directed vesicle transport. Cell Biol. 188:253–269.
2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Demarco RS, Uyemura BS and Jones DL: Egfr
signaling stimulates autophagy to regulate stem cell maintenance
and lipid homeostasis in the drosophila testis. Cell Rep.
30:1101–1116. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Mizushima N, Yoshimori T and Levine B:
Methods in mammalian autophagy research. Cell. 140:313–326.
2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Polazzi E and Monti B: Microglia and
neuroprotection: From in vitrostudies to therapeutic applications.
Prog Neurobiol. 92:293–315. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hu X, Leak RK, Shi Y, Suenaga J, Gao Y,
Zheng P and Chen J: Microglial and macrophage polarization-new
prospects for brain repair. Nat Rev Neurol. 11:56–64.
2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li D, Huang S, Yin Z, Zhu J, Ge X, Han Z,
Tan J, Zhang S, Zhao J, Chen F, et al: Increases in miR-124-3p in
microglial exosomes confer neuroprotective effects by targeting
fip200-mediated neuronal autophagy following traumatic brain
injury. Neurochem Res. 44:1903–1923. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang Y, Zhou K, Li T, Xu Y, Xie C, Sun Y,
Zhang Y, Rodriguez J, Blomgren K and Zhu C: Inhibition of autophagy
prevents irradiation-induced neural stem and progenitor cell death
in the juvenile mouse brain. Cell Death Dis.
8(e2694)2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bai WF, Feng Y, Huang H and Zhang MS:
Fifty-hertz electromagnetic fields facilitate the induction of rat
bone mesenchymal stromal cells to differentiate into functional
neurons. Cytotherapy. 15:961–970. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bai WF, Xu WC, Zhu HX, Huang H, Wu B and
Zhang MS: Efficacy of 50 Hz electromagnetic fields on human
epidermal stem cell transplantation seeded in collagen sponge
scaffolds for wound healing in a murine model. Bioelectromagnetics.
38:204–212. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bai WF, Zhang MS, Huang H, Zhu HX and Xu
WC: Effects of 50 Hz electromagnetic fields on human epidermal stem
cells cultured on collagen sponge scaffolds. Int J Radiat Biol.
88:523–530. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
He HY, Ren L, Guo T and Deng YH: Neuronal
autophagy aggravates microglial inflammatory injury by
downregulating CX3CL1/fractalkine after ischemic stroke. Neural
Regen Res. 14:280–288. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bie M, Lv Y, Ren C, Xing F, Cui Q, Xiao J
and So KF: Lycium barbarum polysaccharide improves bipolar pulse
current-induced microglia cell injury through modulating autophagy.
Cell Transplant. 24:419–28. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang XT, Ma J, Fu Q, Zhu L, Zhang ZL,
Zhang F, Lu N and Chen AM: Role of hypoxia-inducible factor-1α in
autophagic cell death in microglial cells induced by hypoxia. Mol
Med Rep. 15:2097–2105. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Rajendran JC, Bose and Robert F
Mattrey: Accomplishments and challenges in stem cell imaging in
vivo. Drug Discov Today. 24:492–504. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Li XP, Wang X, Bai LM, Zhao P and Zhang
MS: Exposure to 50 Hz electromagnetic fields enhances hair follicle
regrowth in C57BL/6 mice. Exp Biol Med (Maywood). 244:389–394.
2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yong TY, Zhang XZ, Bie NB, Zang HZ, Zang
XH, Li FL, Hakeem AH, Hu JH, Gan LG, Santos HAS and Yang XY: Tumor
exosome-based nanoparticles are efficient drug carriers for
chemotherapy. Nature Comm. 10(3838)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kyei B, Li L, Yang L, Zhan SY and Zhang
HP: CDR1as/miRNAs-related regulatory mechanisms in muscle
development and diseases. Gene. 730(144315)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kashyap D and Kaur H: Cell-free miRNAs as
non-invasive biomarkers in breast cancer: Significance in early
diagnosis and metastasis prediction. Life Sci.
2461(7417)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ozeki N, Hase N, Hiyama T, Yamaguchi H,
Kawai-Asano R, Nakata K and Mogi M: MicroRNA-211 and
autophagy-related gene 14 signaling regulate osteoblast-like cell
differentiation of human induced pluripotent stem cells. Exp Cell
Res. 352:63–74. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Meng F, Zhang S, Song R, Liu Y, Wang J,
Liang Y, Wang J, Han J, Song X, Lu Z, et al: NCAPG2 overexpression
promotes hepatocellular carcinoma proliferation and metastasis
through activating the STAT3 and NF-κB/miR-188-3p pathways.
EBioMedicine. 44:237–249. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liao H, Xiao Y, Hu Y, Xiao Y, Yin Z and
Liu L: MicroRNA-32 induces radioresistance by targeting DAB2IP and
regulating autophagy in prostate cancer cells. Oncol Lett.
10:2055–2062. 2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang D, Zeng T, Lin Z, Yan L, Wang F, Tang
L, Wang L, Tang D, Chen P and Yang M: Long non-coding RNA SNHG5
regulates chemotherapy resistance through the miR-32/DNAJB9 axis in
acute myeloid leukemia. Biomed Pharmacother.
123(109802)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Liu KX, Chen GP, Lin PL, Huang JC, Lin X,
Qi JC and Lin QC: Detection and analysis of apoptosis- and
autophagy-related miRNAs of mouse vascular endothelial cells in
chronic intermittent hypoxia model. Life Sci. 193:194–199.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yu L, Tumati V, Tseng SF, Hsu FM, Kim DN,
Hong D, Hsieh JT, Jacobs C, Kapur P and Saha D: DAB2IP regulates
autophagy in prostate cancer in response to combined treatment of
radiation and a DNA-PKcs inhibitor. Neoplasia. 14:1203–1212.
2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Rong Y, Liu W, Wang J, Fan J, Luo Y, Li L,
Kong F, Chen J, Tang P and Cai W: Neural stem cell-derived small
extracellular vesicles attenuate apoptosis and neuroinflammation
after traumatic spinal cord injury by activating autophagy. Cell
Death Dis. 10(340)2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kang I, Lee BC, Lee JY, Kim JJ, Sung EA,
Lee SE, Shin N, Choi SW, Seo Y, Kim HS and Kang KS: Stem
cell-secreted 14,15- epoxyeicosatrienoic acid rescues cholesterol
homeostasis and autophagic flux in Niemann-pick-type C disease. Exp
Mol Med. 50:1–14. 2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Sung K and Jimenez-Sanchez M: Autophagy in
astrocytes and its implications in neurodegeneration. J Mol Biol.
432:2605–2621. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Oliveira MC, Elias JB, Moraes DA, Simões
BP, Rodrigues M, Ribeiro AAF, Piron-Ruiz L, Ruiz MA and Hamerschlak
N: A review of hematopoietic stem cell transplantation for
autoimmune diseases: Multiple sclerosis, systemic sclerosis and
Crohn's disease. Hematol Transfus Cell. Ther:30032–30038.
2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Bai WF: Bone marrow derived neural
progenitor cells differentiate into neurons and promote
neurogenesis of brain injury rats (unpublished PhD thesis). Jinan
University, 2017.
|
|
48
|
Rahmati S, Shojaei F, Shojaeian A,
Rezakhani L and Dehkordi MB: An overview of current knowledge in
biological functions and potential theragnostic applications of
exosomes. Chem Phys Lipids. 226(104836)2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Shi ZY, Yang XX, Malichewe CY, Li YS and
Guo XL: Exosomal microRNAs-mediated intercellular communication and
exosome-based cancer treatment. Int J Biol Macromol. 158:530–541.
2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Su WT, Li HJ, Chen WW and Qin JH:
Microfluidic strategies for label-free exosomes isolation and
analysis. TrAC Trend Anal Chem. 118:686–698. 2019.
|
|
51
|
Riazifar M, Pone EJ, Lötvall J and Zhao W:
Stem cell extracellular vesicles: Extended messages of
regeneration. Annu Rev Pharmacol Toxicol. 57:125–154.
2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Reyes-Ruiz JM, Osuna-Ramos JF,
Jesús-González LAD, Hurtado-Monzón AM, Farfan-Morales CN,
Cervantes-Salazar M, Bolaños J, Cigarroa-Mayorga OE,
Martín-Martínez ES, Medina F, et al: Isolation and characterization
of exosomes released from mosquito cells infected with dengue
virus. Virus Res. 266:1–14. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Liang PH, Mao L, Zhang SH, Guo X, Liu G,
Wang L, Hou J, Zheng Y and Luo X: Identification and molecular
characterization of exosome-like vesicles derived from the Taenia
asiatica adult worm. Acta Trop. 198(105036)2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Li D, Huang S, Zhu J, Hu T, Han Z, Zhang
S, Zhao J, Chen F and Lei P: Exosomes from miR-21-5p-increased
neurons play a role in neuroprotection by suppressing
Rab11a-mediated neuronal autophagy in vitro after traumatic brain
injury. Med Sci Monit. 25:1871–1885. 2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Xia W, Shi R, Zheng WL and Ma WL: Lack of
association between cytotoxic T-lymphocyte antigen-4-318C/T
polymorphism and cancer risk: A meta-analysis of case-control
studies. Technol Cancer Res Treat. 12:565–574. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ng R, Hussain NA, Zhang Q, Chang C, Li H,
Fu Y, Cao L, Han W, Stunkel W and Xu F: miRNA-32 drives brown fat
thermogenesis and trans-activates subcutaneous white fat browning
in mice. Cell Rep. 19:1229–1246. 2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Jiang M, Wang H, Jin M, Yang X, Ji H,
Jiang Y, Zhang H, Wu F, Wu G, Lai X, et al: Exosomes from
MiR-30d-5p-ADSCs reverse acute ischemic stroke-induced,
autophagy-mediated brain injury by promoting M2
microglial/macrophage polarization. Cell Physiol Biochem.
47:864–878. 2018.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Chappell WH, Abrams SL, Lertpiriyapong K,
Fitzgerald TL, Martelli AM, Cocco L, Rakus D, Gizak A, Terrian D,
Steelman LS and McCubrey JA: Novel roles of androgen receptor,
epidermal growth factor receptor, TP53, regulatory RNAs,
NF-kappa-B, chromosomal translocations, neutrophil associated
gelatinase, and matrix metalloproteinase-9 in prostate cancer and
prostate cancer stem cells. Adv Biol Regul. 60:64–87.
2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Li X, Wang Y, Xiong Y, Wu J, Ding H, Chen
X, Lan L and Zhang H: Galangin induces autophagy via deacetylation
of LC3 by SIRT1 in HepG2 cells. Sci Rep. 6(30496)2016.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Politano G, Logrand F, Brancaccio M and Di
Carlo S: In-silico cardiac aging regulatory model including
microRNA post-transcriptional regulation. Methods. 124:57–68.
2017.PubMed/NCBI View Article : Google Scholar
|