Introduction
Peroxidases are widely distributed in nature
including plants and animals (1).
Peroxidase is a heme-containing oxidoreductase, which oxidatively
degrades peroxide structure to two hydroxyl groups (2). Peroxidase reacts to hydrogen peroxide
as a substrate, and also reacts to organic peroxides to protect the
body from oxidative stress (3).
Moreover, salivary peroxidase and myeloperoxidase are known for
antibacterial activity against oral bacteria (4,5).
Salivary peroxidase catalyzes to produce hypothiocyanite ion
(OSCN-) with antibacterial activity from thiocyanate ion
(SCN-) derived from diets and hydrogen peroxide
(H2O2) produced by oral cells and oral
commensal bacteria (6,7).
Previous indications have pointed to the possibility
that horseradish peroxidase (HRP) adsorbed to the membrane of
certain bacterial cells, such as oral streptococci, including
Streptococcus mutans (S. mutans) and Streptococcus
sanguinis (S. sanguinis) (8). However, the mechanism of interaction
between HRP and bacterial cell walls is unknown, although HRP is
popularly used as tools in biochemical and pathological studies,
diagnostic examinations, and in tests to treat various industrial
effluents (9).
Dental plaques as biofilm containing oral bacteria
and bacterial products are developed on the dental pellicle with
salivary glycoproteins sheathing the tooth surface (10-13).
Dental plaque causes oral diseases including periodontitis
(14-16).
Thus, it is essential for the prevention and treatment of oral
diseases that dental plaques be controlled (17,18).
‘Dental plaque disclosing agents’ are applied to visualize dental
plaques and to control dental plaques on dental therapy (19). However, current ‘dental plaque
disclosing agents’ stain not only dental plaques, but also dental
pellicles. Thus, it is difficult to evaluate just dental plaque by
‘dental plaque disclosing agents’ (20,21).
In this present study, we have demonstrated that HRP
interacted with the cell wall peptidoglycan (PGN) from the
gram-positive bacterium, but not the cell wall lipopolysaccharide
(LPS) from the gram-negative bacterium, and clearly disclosed the
dental plaques as the biofilms developed by the major gram-positive
oral streptococcal species S. sanguinis, Streptococcus
salivarius (S. salivarius), and the major gram-positive
rod Lactobacillus casei (L. casei), and slightly
disclosed the biofilm by the major gram-negative bacterium,
Escherichia coli (E. coli). The results obtained in
this study suggest possibilities that the adsorption activity of
HRP not only contributes to the evaluation of dental plaque, but
that the enzymatic activity of HRP may also improve oral flora and
environment, and dental health.
Materials and methods
Bacterial culture
Streptococcus salivarius ATCC 13419,
Streptococcus sanguinis ATCC 10556, Lactobacillus
casei ATCC 334 and Escherichia coli strain K12 were
cultivated with Brain Heart Infusion broth (BD Bioscience) at
37˚C on shaking platforms.
Adsorptions of HRP to bacterium
Bacteria from 100 µl of bacterial cultures at
optical density (OD)=0.2 at 560 nm were washed twice with
physiological phosphate buffered saline without calcium ion
[PBS(-)] followed by centrifugations at 12,000 x g at room
temperature (RT). The 1.5 ml test tubes (Eppendorf, Germany) were
blocked with 4% bovine serum albumin fraction V (BSA)
(Sigma-Aldrich) for 3 days at 4˚C. Washed bacteria were
incubated with 100 µl of 100 µg/ml HRP (Wako, Japan) in PBS(-)
containing 4% BSA for 30 min at RT, and washed 4 times with PBS(-).
HRP adsorbed to bacterium was developed with 100 µl of the
chromogenic substrate, tetramethylbenzidine (TMB) solution (BD
Bioscience) for 30 min at RT, fixed with 50 µl of 2 N sulfuric
acid, and then ODs of supernatants by centrifugation at 12,000 x g
were measured at 450 nm. The BSA-coated tubes were used in all the
steps.
Interactions between HRP and bacterial
cell wall components
Eight mg/ml (100 µl/well) of LPS purified by
gel-filtration from E. coli (O26:B6 strain) (L8274;
Sigma-Aldrich) in PBS(-) supplemented with 25% ethanol was
immobilized on 96 well cell culture plate (3599; Corning Costar) at
4˚C overnight, and then the plates were blocked with 4%
BSA at 4˚C overnight. 10 mg PGN from Staphylococcus
aureus (Sigma-Aldrich) were washed twice with PBS(-) by
centrifugations at 12,000 x g in the 1.5 ml test tube blocked with
BSA. The LPS-immobilized well and PGN in the test tube were
incubated with 100 µl of 100 µg/ml HRP in PBS(-) containing 4% BSA
for 30 min at RT, and washed four times with PBS(-). HRP interacted
to LPS and PGN were developed with 100 µl of the chromogenic TMB
solution for 30 min at RT, fixed with 50 µl of 2 N sulfuric acid,
and then ODs of supernatants by centrifugation at 12,000 x g were
measured at 450 nm.
Adsorptions of fluorescence-labeled
HRP to bacterium
HRP were labelled with Dylight Dye 488 by a protein
labeling kit (Thermo Fisher Scientific, Inc.). S.
salivarius, S. sanguinis and E. coil were washed
twice with PBS(-) via centrifugation at 14,000 rpm. The bacteria
were incubated with 1 µl of 2 mg/ml fluorescence-labeled HRP in 30
µl PBS(-) containing 4% BSA for 25 min at RT, washed twice with
PBS(-), and bacterium adsorbed with fluorescence-labeled HRP were
embedded and dried on a slide glass. The fluorescence-labeled HRP
adsorbed to bacterium was observed by Axio Observer (Carl Zeiss)
and imaged by a charge-coupled device camera (Nippon Roper).
Mimics of the artificial tooth surface
and the artificial dental pellicle
6% (wt/vol) carbonate apatite (CA) was prepared by
mixing 8 l of 2 M calcium nitrate solution and 2 l of 1.2 M
disodium hydrogen phosphate solution containing 1.2 M disodium
carbonate for 3 days at 100˚C and pH 9.0±0.1(22). The pH was maintained constant by
automatic addition of dilute sodium hydroxide. The precipitate was
washed 10 times with de-ionized distilled water, freeze-dried, and
then sieved with mesh (0.125 mm). Sieved samples were placed in a
metal mold (10x10x50 mm), remolded at 15 MPa and further compacted
isostatically at 200 MPa. The sintered CA specimens, which
contained about 3% wt carbonate, were produced by heating compacted
samples at 1,100˚C for 2 h with a temperature increase and
subsequent decrease of 5˚C/min. Approximately 2 mm thick plates
(2x9x9 mm) were cut from the sintered specimens (9x9x45 mm) by a
diamond saw, and the artificial tooth surfaces were made up with
no. 2000 water-proof sandpaper. The dental pellicle was mimicked on
the autoclaved artificial tooth surface by soaking it with 2.8
mg/ml 0.45 µm filter-sterilized mucin type I from bovine
submaxillary glands (Sigma-Aldrich) in PBS containing 1.6 mM
calcium chloride (CaCl2) [PBS(+)] overnight.
Developments and disclosing of the
dental plaque
The dental plaques were developed by static culture
on the artificial tooth surface with the dental pellicle in BHI
broth overnight at RT. The dental plaque were rinsed with 1 ml of
100 µg/ml HRP in PBS(-) for 30 min at RT, and then gently washed
twice with PBS(-). Next, the dental plaque was disclosed with 10
mg/ml diaminobenzidine (DAB) (Dako), the chromogenic substrate, in
50 mM Tris-HCl (pH 7.6) supplemented with 7.5 µl/ml of 3% hydrogen
peroxide for 5 min at RT. Meanwhile, the dental pellicle and the
dental plaque were disclosed with a major ‘dental plaque disclosing
agent’ (1.5% (w/w) D&C Red no. 28; Sunstar), as control
experiments.
Statistical analysis
All analyses were performed by using statistical
software (SPSS 11.0 software package; SPSS Inc., Chicago, IL, USA).
Data are expressed as means ± standard deviation (SD). The
Student's t-test was applied to determine the significance
of differences between two groups. One-way analysis of variance was
applied to determine the significance of differences among overall
groups, followed by the Tukey-Kramer test for multiple
comparisons.
Results
HRP interacts with PGNs on
gram-positive bacteria
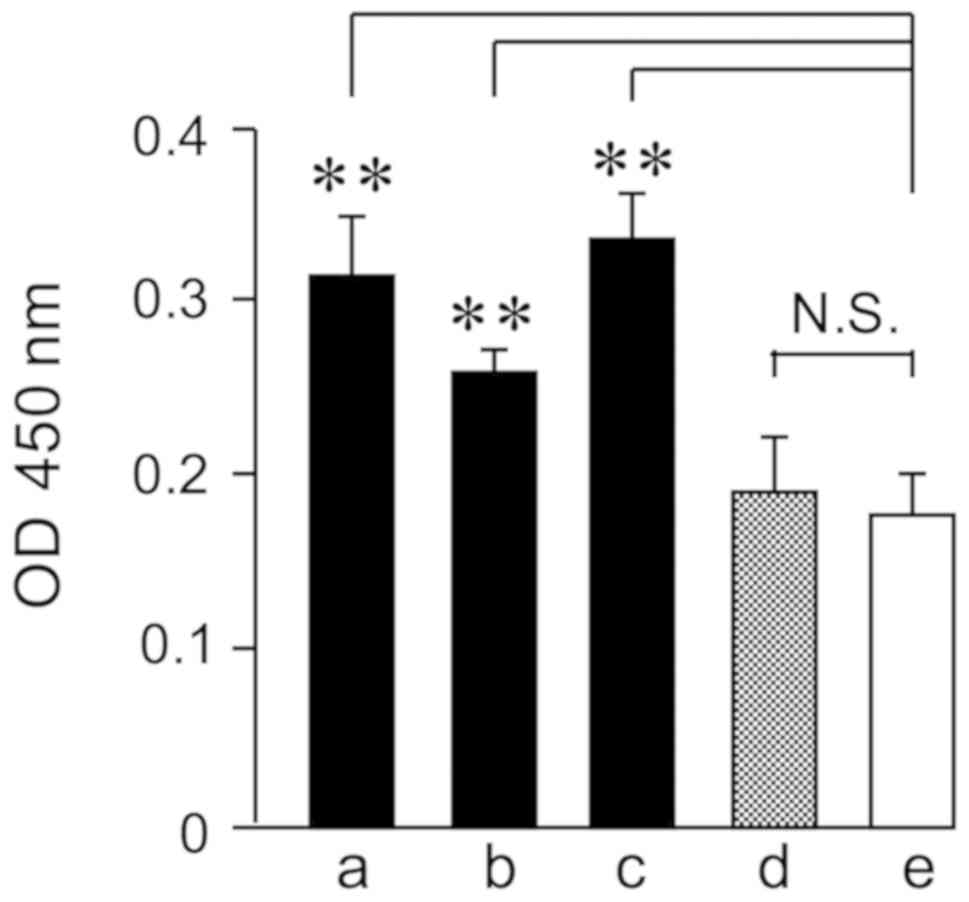
Adsorptions of HRP to gram-positive bacterium were
quantified by the chromogenic substrate (Fig. 1). S. sanguinis and S.
salivarius are major oral gram-positive oral streptococci. HRP
adsorbed significantly to S. sanguinis and S.
salivarius. Moreover, HRP adsorbed significantly to the major
gram-positive bacterium L. casei, but not to the major
gram-negative bacterium, E. coli. The HRP interaction with
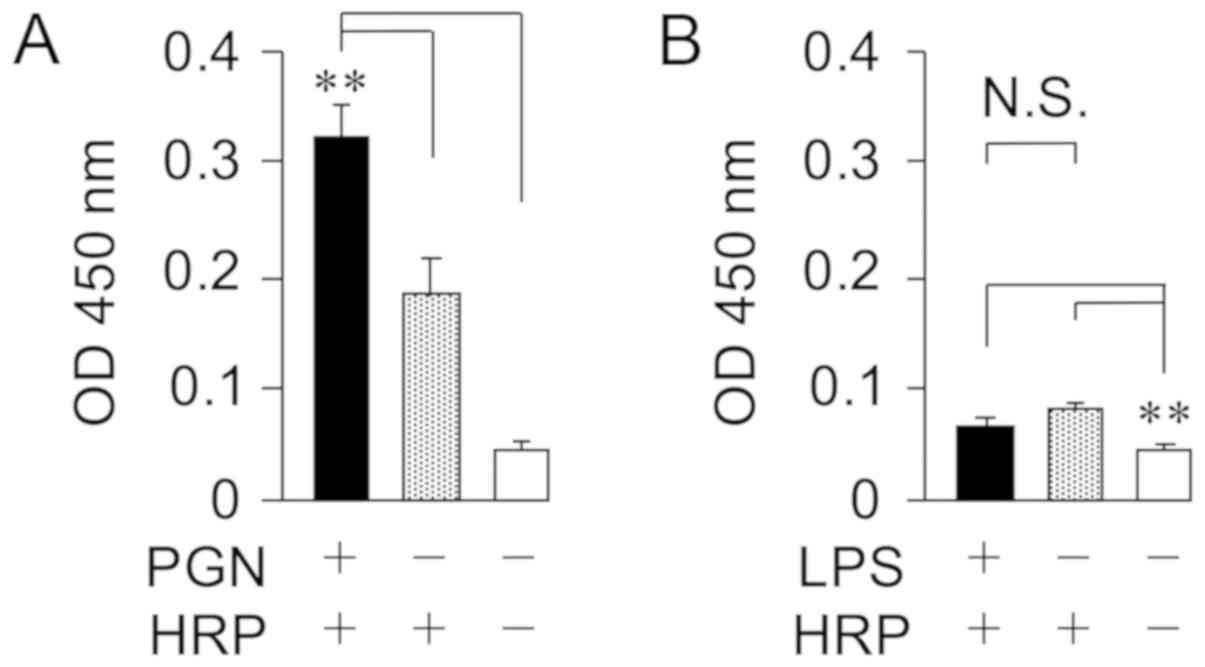
the membrane components from bacteria were quantified by the
chromogenic substrate (Fig. 2). HRP
interacted with the cell wall PGN from the major gram-positive
bacterium S. aureus, but not the cell wall LPS from the
major gram-negative bacterium, E. coli. In addition, it was
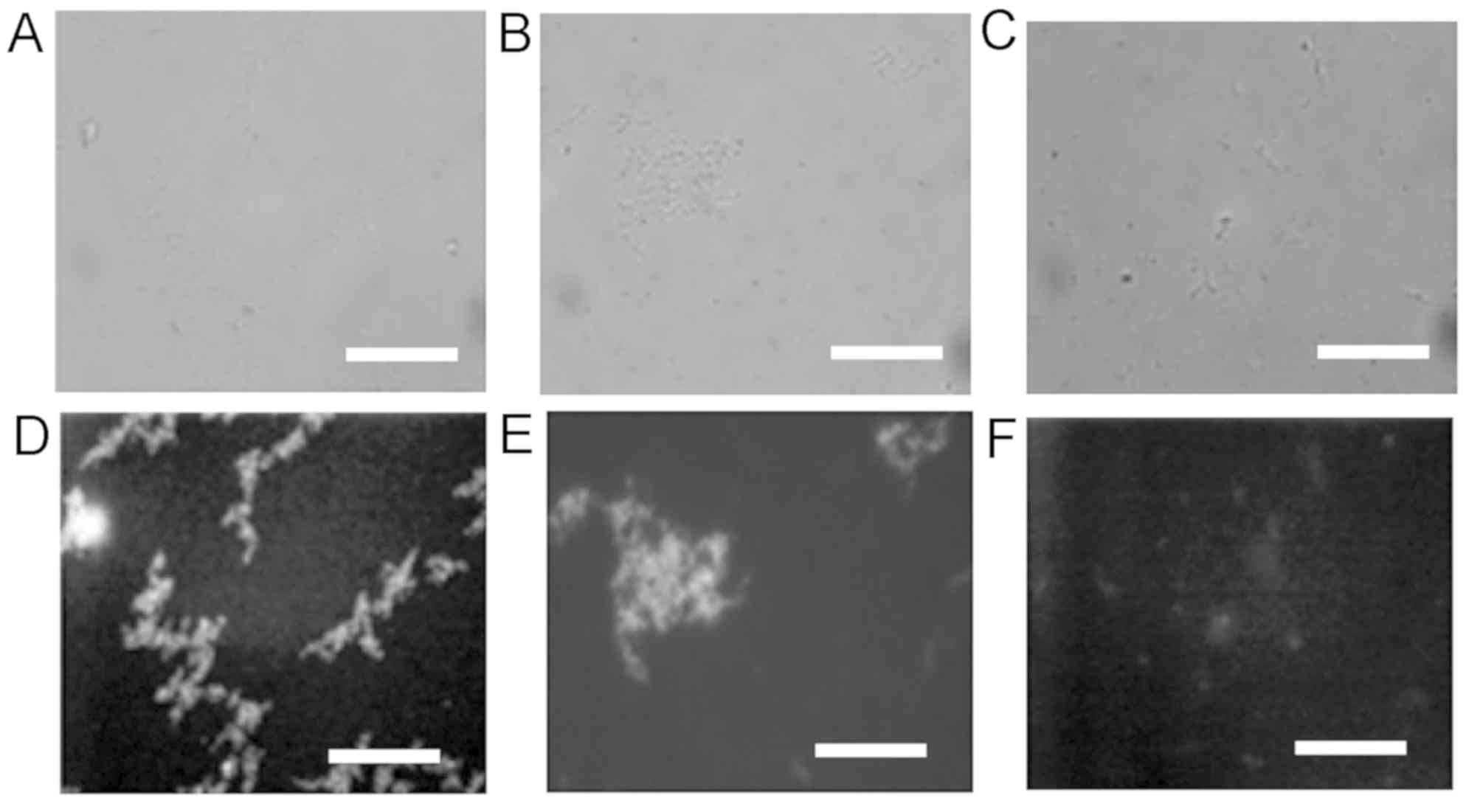
confirmed that the adsorbed HRPs labeled with the fluorescence
DyLight Dye 488 were detected on oral streptococci S.
sanguinis and S. salivarius, but not on E. coli
(Fig. 3).
HRP discloses dental plaques, but not
the dental pellicle
The artificial tooth surfaces with the pellicles
were mimicked by carbonate apatite (22), and a major salivary glycoprotein
mucin, as the dental pellicle formed by the mucin required calcium
ion. The combination of HRP and chromogenic substrate clearly
disclosed dental plaques and biofilm by oral streptococci S.
sanguinis and S. salivarius, and the major gram-positive
rod L. casei, and slightly disclosed the biofilm by the
major gram-negative bacterium E. coli, developed on the
artificial tooth surface (Fig. 4,
left column). Meanwhile, the combination of HRP and chromogenic
substrate neither detected the dental pellicle with the salivary
mucin nor stained the naked artificial tooth surface. On the other
hand, 1.5 % (w/w) D&C Red no. 28, a major ‘dental disclosing
agents’, disclosed also the dental pellicle not only the dental
plaques and biofilm developed on the artificial tooth surface by
bacteria, such as gram-positive bacteria (S. sanguinis,
S. salivarius, L. casei) and gram-negative bacterium
(E. coli) (Fig. 4, right
column).
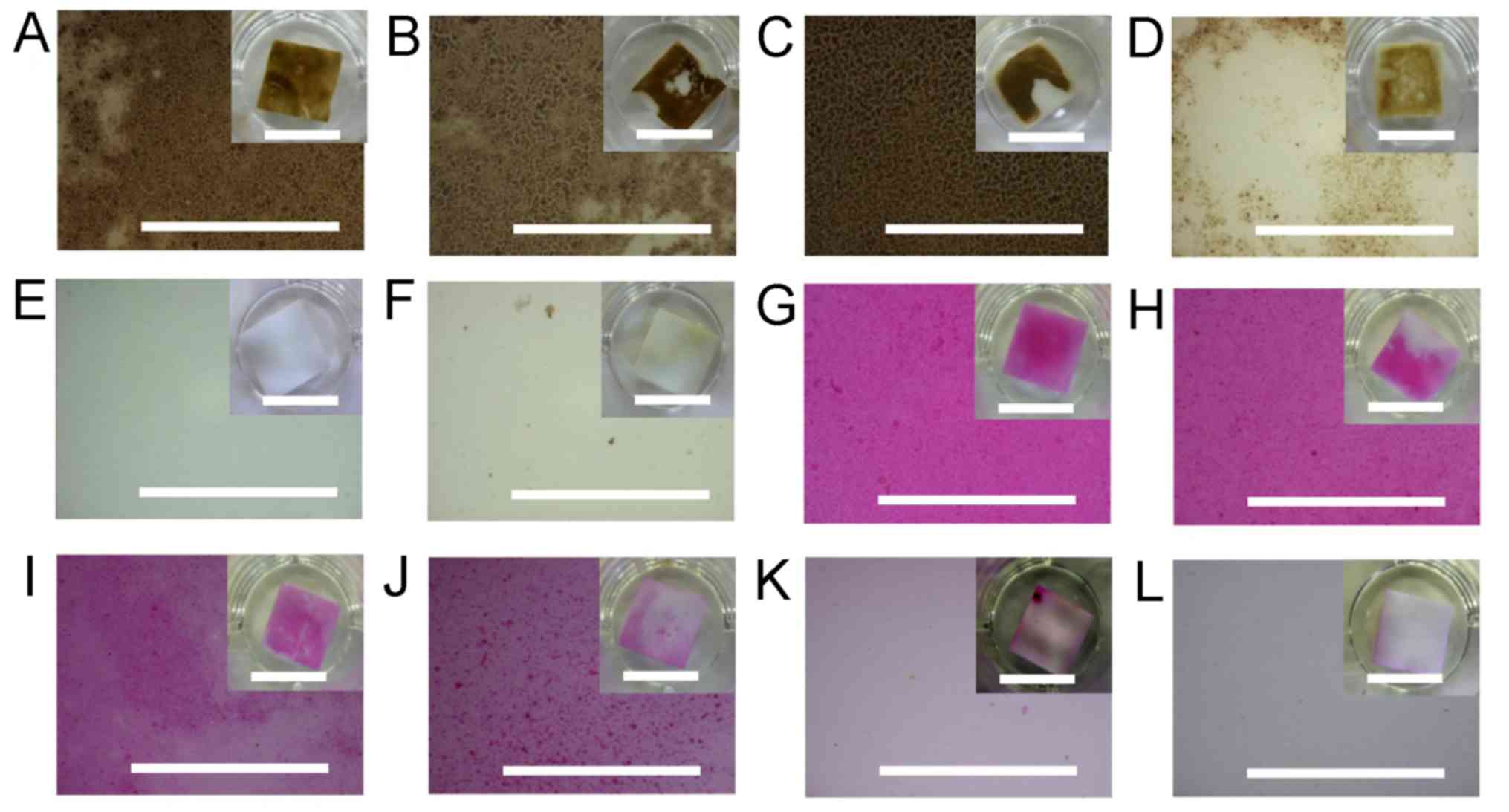 | Figure 4Disclosing of the dental plaque by
HRP. Dental plaques and biofilms by oral gram-positive bacteria (A)
S. sanguinis and (B) S. salivarius, (C) the major
gram-positive bacterium L. casei, and (D) the major
gram-negative bacterium E. coli were developed on the
artificial tooth surface. (E) The dental pellicle and (F) the naked
artificial tooth surface (f) were additionally applied. They were
subsequently stained by HRP with the chromogenic DAB substrate. (G)
S. sanguinis and (H) S. salivarius, (I) L. casei, (J)
E. coli and the (K) dental pellicle developed on surfaces of
the artificial tooth, and (L) the naked artificial tooth surface
was additionally stained by the commercially available ‘dental
plaque disclosing agent’ D&C Red No 28. The partial magnified
images of the artificial tooth surfaces are shown in large windows,
with the whole images shown in small windows at upper right. The
long scales indicate 1 mm, whereas, the short scales in each of the
small windows on the upper right indicate 10 mm. HRP, horseradish
peroxidase; S. sanguinis, Streptococcus sanguinis; S.
salivarius, Streptococcus salivarius; L. casei, Lactobacillus
casei; E. coli, Escherichia coli; DAB, diaminobenzidine. |
Discussion
In this present study, we have clarified that HRP
interacted with the cell wall PGN from the major gram-positive
bacterium S. aureus, but not the cell wall LPS from the
major gram-negative bacterium, E. coli. And, we have also
shown that HRP adsorbed to the major oral streptococci S.
sanguinis and S. salivarius, and the gram-positive rod
L. casei, but not to the major gram-negative bacterium,
E. coli. Moreover, we have also confirmed that the adsorbed
HRPs were detected on S. sanguinis and S. salivarius,
but not on E. coli, although previous indications pointed to
the possibility that HRP adsorbed to the membrane of certain
bacterial cells, such as oral streptococci, S. mutans and
S. sanguinis (8). We have
demonstrated that the HRP-adsorbed dental plaques and the biofilm
developed by S. sanguinis, S. salivarius and L. casei
on our established artificial tooth surface were clearly visualized
with the chromogenic substrate, while the biofilm developed by
E. coli was slightly visualized.
It has been reported within dental studies that
bacteria were adsorbed on a hydroxyapatite plate via various
methods, such as a hydroxyapatite plate coated by human saliva and
the artificial tooth surface with carbonate apatite mimics human
tooth surfaces more than artificial tooth surfaces with just
hydroxyapatite (22,23). A dental pellicle is formed through
calcium ion as scaffolding by salivary glycoproteins such as mucin,
and a dental plaque as biofilm is developed on the pellicle
sheathing tooth surface (24,25). In
this study, we mimicked the autoclaved artificial tooth surface by
utilizing No. 2000 water-proof sandpaper on the carbonate apatite,
and the mimicked pellicle was sheathed by the sterilized bovine
mucin and calcium ion. Various bactera, including S.
sanguinis and S. salivarius, then developed on the
artificial tooth surface with the dental pellicle. The artificial
tooth surface with the dental pellicle established in this present
study should be applicable to various dental studies.
HRP interacted with the specific component PGN on
gram-positive bacteria, but did not interact with the specific
component LPS on gram-negative bacteria. HRP adsorbed to
gram-positive species S. sanguinis, S. salivarius and
L. casei, but not to the gram-negative bacterium, E.
coli. PGN forms a net-like sacculus made of glycan strands
crosslinked by peptides as the basic structure of the bacterial
cell wall (26). PGN is thicker in a
gram-positive bacteria than in a gram-negative bacteria (27). Therefore, the combination of HRP and
chromogenic substrate probably made the dental plaques visible and
the biofilm developed on the artificial tooth surface by S.
sanguinis, S. salivarius and L. casei more
clearly than did the biofilm by E coli. The major oral
gram-positive streptococci S. salivalis and S.
sanguinis developed more frequently than other oral bacteria in
both early and mature dental plaques (28). HRP should be able to adsorb to
earlier dental plaque, and not only to mature dental plaque, and
the combination of HRP and chromogenic substrate should also be
able to make visible an earlier dental plaque with high
sensitivity.
In present study, it was suggested the possibility
that the adsorption activity of HRP would contribute to the
evaluation of dental plaque. Dental plaques are mixtures including
both variously gram-negative bacteria and gram-positive bacteria,
such as S. mutans, S. salivarius, and S. sanguinis.
And dental plaques cause oral diseases including periodontitis
(14-16).
Thus, the evaluation of dental plaque is important for the
prevention of periodontal disease. Periodontitis is developed by
inflammation induced by LPS in cell walls on mainly oral
gram-negative bacteria, such as Porphyromonas gingivalis,
Prevotella intermedia, Tannerella forsythia, and
Aggregatibacter actinomycetemcomitans. On the other hand,
caries are developed by acids produced from mainly oral
gram-positive bacteria, such as S. mutans, which can be
adsorbed with HRP (8). Therefore,
evaluation of dental plaque with adsorptive activity of HRP to
S. mutans suggests applicable also to prevention of dental
caries though further study with S. mutans is necessary.
Salivary peroxidase and myeloperoxidase are known to
possess antibacterial activity against oral bacteria (4), but HRP is not known for antibacterial
activity against oral bacteria. Salivary peroxidase catalyzes
reaction to produce OSCN- with antibacterial activity
from hydrogen peroxide and thiocyanate produced by oral cells and
oral bacteria. HRP reacts with peroxides from oral tissues
(3). Thus, it is expected that HRP
should express an antibacterial effect when HRP is adsorbed to
bacteria in dental plaques. Biofilm in the oral cavity includes
exopolysaccharide (29), which may
inhibit penetration of HRP. Further study is required to determine
whether HRP will be conducive to plaque control and dental health
for dental therapy. The chromogenic substrate diaminobenzidine
(DAB) is potentially a carcinogen (30), and the fluorescent agent Dylight Dye
488 is visualized with ultraviolet light under dark conditions. It
is necessary to develop a nontoxic chromogenic substrate or an
agent visible under normal light in the oral cavity to apply during
dental therapy. Our results in this study suggests the possibility
that the adsorption activity of HRP not only contributes to the
evaluation of dental plaque, but that the enzymatic activity of HRP
may also contribute to the improvement of oral flora and
environment, and dental health.
Acknowledgements
The authors would like to thank Ms Mayumi Oda of the
Department of Medical Bioengineering, Graduate School of Natural
Science and Technology, Okayama University (Okayama, Japan), for
contributions to the experiments involving DyLight Dye 488.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
HM, ET and AS made substantial contributions to the
conception and design of the study, and the acquisition, analysis
and interpretation of data, and were involved in drafting the
manuscript and revising it critically for important intellectual
content. TI, YM, MA, KY, MN, SS, YD, MMK and NK made substantial
contributions to the conception and design of the study, and the
acquisition, analysis and interpretation of data. DE, TT and MM
made substantial contributions to the acquisition, analysis and
interpretation of data, and were involved in drafting the
manuscript and revising it critically for important intellectual
content. All authors gave final approval of the version to be
published, and agreed to be accountable for all aspects of the work
in ensuring the questions related to the accuracy or integrity of
any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Azevedo AM, Martins VC, Prazeres DM,
Vojinović V, Cabral JM and Fonseca LP: Horseradish peroxidase: A
valuable tool in biotechnology. Biotechnol Annu Rev. 9:199–247.
2003.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Nunavath H, Banoth C, Talluri VR and
Bhukya B: An analysis of horseradish peroxidase enzyme for effluent
treatment. Bioinformation. 12:318–323. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Poon HF, Calabrese V, Scapagnini G and
Butterfield DA: Free radicals and brain aging. Clin Geriatr Med.
20:329–359. 2004.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ihalin R, Loimaranta V, Lenander-Lumikari
M and Tenovuo J: The effects of different (pseudo) halide
substrates on peroxidase-mediated killing of Actinobacillus
actinomycetemcomitans. J Periodontal Res. 33:421–427.
1998.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Pruitt KM, Mansson-Rahemtulla B, Baldone
DC and Rahemtulla F: Steady-state kinetics of thiocyanate oxidation
catalyzed by human salivary peroxidase. Biochemistry. 27:240–245.
1988.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tenovuo J and Pruitt KM: Relationship of
the human salivary peroxidase system to oral health. J Oral Pathol.
13:573–584. 1984.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Schultz CP, Ahmed MK, Dawes C and Mantsch
HH: Thiocyanate levels in human saliva: Quantitation by Fourier
transform infrared spectroscopy. Anal Biochem. 240:7–12.
1996.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lai CH, Listgarten MA and Rosan B:
Immunoelectron microscopic identification and localization of
Streptococcus sanguinis with peroxidase-labeled antibody:
Localization of surface antigens in pure cultures. Infect Immun.
11:193–199. 1975.PubMed/NCBI
|
|
9
|
Karigar CS and Rao SS: Role of microbial
enzymes in the bioremediation of pollutants: A review. Enzyme Res.
2011(805187)2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Addy M, Slayne MA and Wade WG: The
formation and control of dental plaque-an overview. J Appl
Bacteriol. 73:269–278. 1992.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Claydon NC: Current concepts in
toothbrushing and interdental cleaning. Periodontol 2000. 48:10–22.
2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Löe H: Oral hygiene in the prevention of
caries and periodontal disease. Int Dent J. 50:129–139.
2000.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Socransky SS and Haffajee AD: Dental
biofilms: Difficult therapeutic targets. Periodontol 2000.
28:12–55. 2002.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Loe H, Theilade E and Jensen SB:
Experimental gingivitis in man. J Periodontol. 36:177–187.
1965.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kelner RM, Wohl BR, Deasy MJ and Formicola
AJ: Ginigival inflammation as related to frequency of plaque
removal. J Periodontol. 45:303–307. 1974.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lang NP, Cumming BR and Löe H:
Toothbrushing frequency as it relates to plaque development and
gingival health. J Periodontol. 44:396–405. 1973.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Löe H: The gingival index, the plaque
index and the retention index systems. J Periodontol. 38:610–616.
1967.PubMed/NCBI View Article : Google Scholar
|
|
18
|
O'Leary TJ, Drake RB and Naylor JE: The
plaque control record. J Periodontol. 43(38)1972.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sumter S and Arnim BA: The use of
disclosing agents for measuring tooth cleanliness. J Periodontol.
34:227–245. 1963.
|
|
20
|
Block PL, Lobene RR and Derdivanis JP: A
two-tone dye test for dental plaque. J Periodontol. 43:423–426.
1972.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gallagher IH, Fussell SJ and Cutress TW:
Mechanism of action of a two-tone plaque disclosing agent. J
Periodontol. 48:395–396. 1977.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Doi Y, Koda T, Wakamatsu N, Goto T,
Kamemizu H, Moriwaki Y, Adachi M and Suwa Y: Influence of carbonate
on sintering of apatites. J Dent Res. 72:1279–1284. 1993.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lee HS, Myers C, Zaidel L, Nalam PC,
Caporizzo MA, A Daep C and Eckmann DM: Masters JG and Composto RJ:
Competitive adsorption of polyelectrolytes onto and into
pellicle-coated hydroxyapatite investigated by QCM-D and force
spectroscopy. ACS Appl Mater Interfaces. 9:13079–13091.
2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Scannopieco FA, Bergey EJ, Reddy MS and
Levine MJ: Characterization of salivary alpha-amylase binding to
Streptococcus sanguinis. Infect Immun. 57:2853–2863.
1989.PubMed/NCBI
|
|
25
|
Cukkemane N, Bikker FJ, Nazmi K, Brand HS
and Veerman EC: Identification and characterization of a
salivary-pellicle-binding peptide by phage display. Arch Oral Biol.
59:448–454. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Vollmer W and Seligman SJ: Architecture of
peptidoglycan: More data and more models. Trends Microbiol.
18:59–66. 2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Amako K, Takade A, Taniai H and Yoshida S:
Electron microscopic examination of uncultured soil-dwelling
bacteria. Microbiol Immunol. 52:265–269. 2008.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ritz HL: Microbial population shifts in
developing human dental plaque. Arch Oral Biol. 12:1561–1568.
1967.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gibbons RJ and van Houte J: On the
formation of dental plaques. J Periodontol. 44:347–360.
1973.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hu J, Mao Y and White K: Renal cell
carcinoma and occupational exposure to chemicals in Canada. Occup
Med (Lond). 52:157–164. 2002.PubMed/NCBI View Article : Google Scholar
|