Introduction
Cancer is one of the leading causes of death
worldwide (1). The outcomes of the
current therapeutic modalities, including surgery, radiotherapy,
chemotherapy, biotherapy and palliative treatments, are far from
satisfactory. Therefore, it is essential to identify novel markers
with high sensitivity and specificity in order to improve the early
diagnosis and prognosis of patients with cancer. Long non-coding
RNAs (lncRNAs) have gained considerable attention as reliable
cancer biomarkers. LncRNAs are transcripts longer than 200
nucleotides that lack an open reading frame and protein-coding
capacity (2,3). They constitute >80% of the entire
transcriptome and have long been considered as transcriptional
‘noise’ (4,5). However, more recent studies have
indicated that lncRNAs have significant roles in cancer progression
(6,7). The gene encoding the lncRNA cancer
susceptibility candidate 9 (CASC9) is located on the human
chromosome 8q21.11 and has four isoforms (CASC9-001 to -004), of
which CASC9-004 is commonly referred to as CASC9 due to its
relatively high prevalence (8,9). CASC9
was initially identified in esophageal squamous cell carcinoma
(ESCC) (10) and has since then been
detected at aberrantly high levels in nasopharyngeal carcinoma
(11), oral squamous cell carcinoma
(OSCC) (12), lung squamous cell
carcinoma (13), breast cancer (BC)
(14), gastric cancer (GC) (15), hepatocellular carcinoma (9), pancreatic ductal adenocarcinoma
(16), colorectal cancer (CRC)
(17), ovarian cancer (OC) (18) and cervical cancer (19). Furthermore, high expression of CASC9
is associated with poor prognosis and unfavorable
clinicopathological parameters in cancer patients (12,15,17,18,20-23).
Since individual studies are limited by relatively small sample
sizes and discrete outcomes, a systematic analysis is necessary in
order to obtain more insight into the role of CASC9 in human
cancers. To this end, a meta-analysis was performed in the present
study to evaluate the prognostic and clinicopathological
significance of CASC9 in cancer.
Materials and methods
Literature search strategy
Relevant articles published in the English language
till November 30, 2019 were retrieved from the PubMed, Embase,
Cochrane Library and Web of Science databases by two independent
researchers (HYD and XD). According to the Cochrane systematic
evaluation manual 5.1, the following MeSH terms and free-text words
were combined for the literature search: ‘long noncoding RNA CASC9’
OR ‘lncRNA CASC9’ OR ‘long non-coding RNA CASC9’ OR ‘cancer
susceptibility 9 lncRNA’ OR ‘cancer susceptibility 9 long noncoding
RNA’ OR ‘CASC9 non-coding long RNA’ and ‘Neoplasia’ OR ‘Neoplasias’
OR ‘Neoplasm’ OR ‘Neoplasms’ OR ‘Tumors’ OR ‘Tumor’ OR ‘Cancer’ OR
‘Cancers’ OR ‘Malignancy’ OR ‘Malignancies’. The bibliography of
the selected studies was manually examined to identify any further
relevant studies. In the case of insufficient data, the authors
were contacted to request the relevant information via e-mail. The
articles were preliminarily screened on the basis of their titles
and abstracts, and their eligibility for the meta-analysis was
determined by two researchers after full-text analysis. The
meta-analysis was performed according to the Preferred Reporting
Items for Systematic Reviews and Meta-Analyses guidelines (24). Any disagreements were resolved by a
third researcher (HSL).
Literature selection
The inclusion criteria for the studies were as
follows: i) Published clinical research, ii) published in the
English language, iii) including patients with histo-pathologically
confirmed cancer, iv) reporting on the association between CASC9
expression and clinicopathological features or prognosis of cancer
patients. Studies that did not fulfill the inclusion criteria,
literature reviews, editorials, letters and case reports, as well
as studies lacking full text or relevant data, were excluded.
Data extraction
Basic information on the studies, including the
first author's name, year of publication, country, sample size,
tumor type, method of detection, cut-off value, survival analysis
type and Newcastle-Ottawa Scale (NOS) scores, as well as
clinicopathological features of the patients, including sex, age,
tumor size, depth of invasion, tumor differentiation, lymph node
metastasis and clinical stage, and outcome indicators such as
overall survival (OS), disease-free survival (DFS) and
progression-free survival (PFS), were extracted. Certain outcome
indicators were directly acquired from the studies and others were
extracted from Kaplan-Meier survival curves using hazard ratio (HR)
digitizer software Engauge Digitizer 4.1(25).
Quality assessment
The NOS (26) was
used for evaluating the quality of included studies. This quality
assessment method evaluated studies by eight items, which were
categorized into three aspects: Patient selection (4 items), study
comparability (1 item) and study outcome (3 items) (27). An answer of ‘yes’ to each question of
the items was counted as the score of 1; otherwise, a score of 0
was awarded (26). The NOS scores
ranged from 0 to 9 and studies with scores of ≥6 were considered to
be of high quality. Any disagreement concerning the quality
assessment of studies was resolved by discussion until consensus
was reached among the researchers.
Statistical analyses
Review Manager 5.2 (The Cochrane Institute) and
Stata 12.0 software (StataCorp.) were used for statistical
analysis. Since the meta-analysis was performed on previously
published studies, no ethical approval was required. The
association between CASC9 expression and OS rates was evaluated by
determining the HR and 95% CI, and that between CASC9 expression
and clinicopathological features by determining odds ratios (ORs)
and 95% CIs. A χ2-based Q-test and I2
statistics were used to estimate the heterogeneity of the included
studies. A fixed-effects model was used in case of insignificant
heterogeneity (I2<50%), and otherwise, the
random-effects model was used for meta-analysis. Sensitivity
analysis was performed using Stata 12.0 software to determine the
robustness of the meta-analysis results. The potential publication
bias was assessed using Begg's funnel plot and Egger's test
(28). All P-values were two-tailed
and P<0.05 was considered to indicate statistical
significance.
Results
Characteristics and quality of
included studies
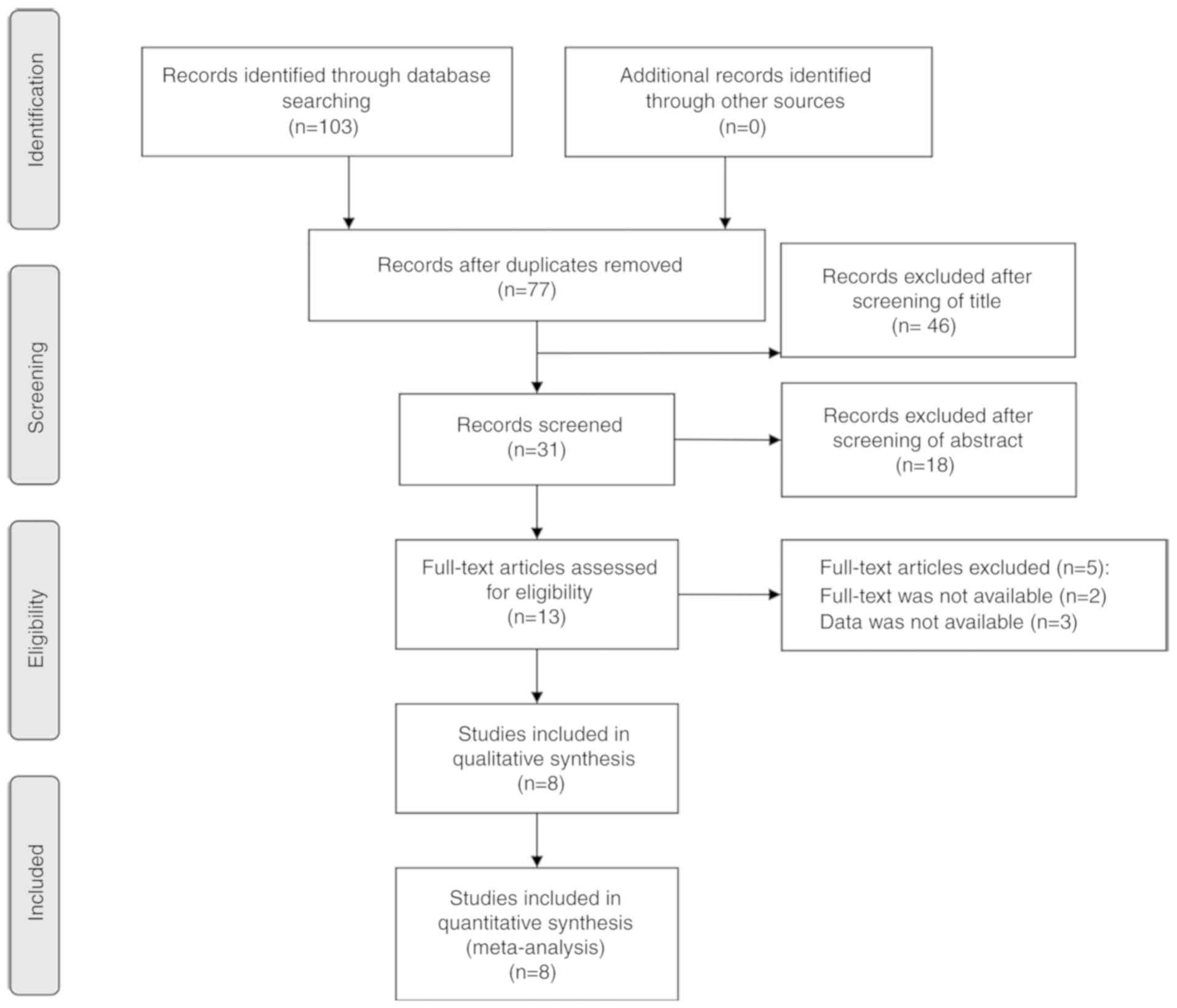
Based on the search strategy, 103 papers were
initially retrieved, of which 13 full-text articles were evaluated
for their eligibility after preliminary screening of titles and
abstracts. Finally, eight studies in including 565 patients were
selected (12,15,17,18,20-23).
The screening procedure is outlined in Fig. 1. The included studies were published
between 2016 and 2019, and the most recent publication was from
July 2019. The mean sample size of these studies was 121 (range,
20-128). A total of 3 studies were on ESCC (21-23),
and one study each was on OSCC (12), GC (15), CRC (17), OC (18) and esophageal cancer (EC) (20). The expression levels of CASC9 had
been detected by reverse transcription-quantitative PCR (15,17,18,20-23)
and in situ hybridization (12). The internal controls differed across
the studies and included 18S ribosomal RNA (20), β-actin (17), U6(18)
and GAPDH (12,22,23).
Based on the cut-off threshold, all patients with cancer were
divided into the CASC9 high and CASC9 low groups in each included
study. The characteristics of the studies are summarized in
Table I and the functional roles or
molecular mechanisms of CASC9 in Table
II. The survival outcomes of cancer patients were assessed in
terms of OS (12,18,21-23),
DFS (22) and PFS (18) in 5 (62.5%), 1 (12.5%) and 1 (12.5%)
studies, respectively. The quality of all included studies was
good, each with a NOS score ≥6 (Table
III).
 | Table ICharacteristics of studies included
in the meta-analysis. |
Table I
Characteristics of studies included
in the meta-analysis.
| First author
(year) | Country | Sample size | Tumor type | Cut-off value | Detection
method | Outcomes | Survival
analysis | NOS scores | (Refs.) |
|---|
| Yang (2019) | China | 84 | OSCC | NA | ISH | CP, OS | Kaplan-Meier | 7 | (12) |
| Pan (2016) | China | 44 | EC | NA | RT-qPCR | CP | Not reported | 6 | (20) |
| Wu (2017) | China | 91 | ESCC | Median | RT-qPCR | CP, OS | Kaplan-Meier | 7 | (21) |
| Liang (2018) | China | 115 | ESCC | Median | RT-qPCR | CP, OS | Kaplan-Meier | 6 | (22) |
| Gao (2018) | China | 128 | ESCC | Median | RT-qPCR | CP, OS, DFS | Kaplan-Meier | 8 | (23) |
| Fang (2019) | China | 20 | GC | NA | RT-qPCR | CP | Not reported | 7 | (15) |
| Luo (2019) | China | 40 | CRC | NA | RT-qPCR | CP | Not reported | 6 | (17) |
| Hu (2019) | China | 43 | OC | Median | RT-qPCR | CP, OS, PFS | Kaplan-Meier | 8 | (18) |
 | Table IIFunction and molecular mechanisms of
CASC9 in included studies. |
Table II
Function and molecular mechanisms of
CASC9 in included studies.
| Tumor type | Direction of
expression | Functional
roles | Interacting
targets | Signaling
pathway | (Refs.) |
|---|
| OSCC | Upregulation | Proliferation,
apoptosis | p-AKT, p-mTOR, P62,
BCL-2, BAX | AKT/mTOR
pathway | (12) |
| EC | Upregulation | Invasion,
migration | Not reported | Not reported | (20) |
| ESCC | Upregulation | Proliferation, cell
cycle | PDCD4, EZH2,
H3K27me3 | Not reported | (21) |
| ESCC | Upregulation | Invasion,
migration | LAMC2, CBP | PI3K/AKT
signaling | (22) |
| ESCC | Upregulation | Proliferation,
invasion, migration | Not reported | EMT pathway | (23) |
| GC | Upregulation | Apoptosis | BMI1 | Not reported | (15) |
| CRC | Upregulation | Proliferation,
apoptosis | CPSF3, TGF-β2,
TERT, Smad3 | TGF-β
signaling | (17) |
| OC | Upregulation | Proliferation,
invasion, migration | miR-758-3p,
LIN7A |
CASC9/miR-758-3p/LIN7A axis | (18) |
 | Table IIIQuality assessment of included
studies using the Newcastle-Ottawa Scale. |
Table III
Quality assessment of included
studies using the Newcastle-Ottawa Scale.
| First author
(year) | Selection | Comparability | Outcome | Total scores | (Refs.) |
|---|
| Yang (2019) | 4 | 1 | 2 | 7 | (12) |
| Pan (2016) | 4 | 1 | 1 | 6 | (20) |
| Wu (2017) | 3 | 1 | 3 | 7 | (21) |
| Liang (2018) | 2 | 1 | 3 | 6 | (22) |
| Gao (2018) | 4 | 1 | 3 | 8 | (23) |
| Fang (2019) | 4 | 2 | 1 | 7 | (15) |
| Luo (2019) | 4 | 1 | 1 | 6 | (17) |
| Hu (2019) | 3 | 2 | 3 | 8 | (18) |
Association between CASC9 expression
and OS
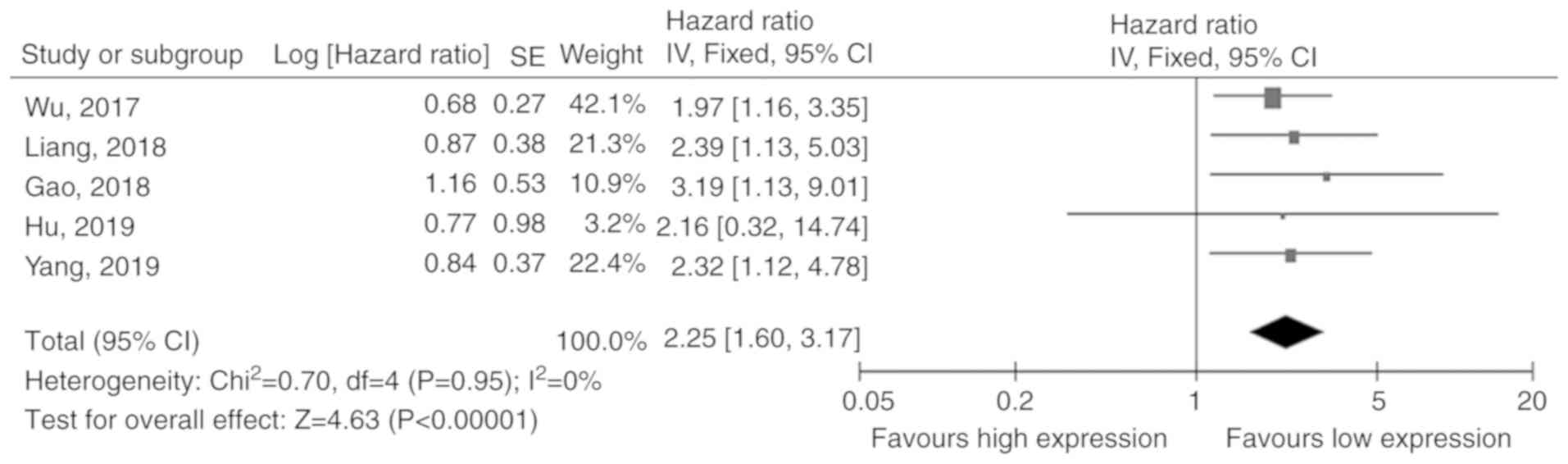
A total of 5 studies (12,18,21-23)
including 461 patients reported on the association between CASC9
expression and OS rates. Since there was no obvious heterogeneity
in these studies (I2=0%), the fixed-effects model was
applied. The pooled results revealed that cancer patients with high
CASC9 expression had a shorter OS compared to those with low levels
(HR=2.25, 95% CI: 1.60-3.17, P<0.001; Fig. 2).
Association between CASC9 expression
and clinicopathological features
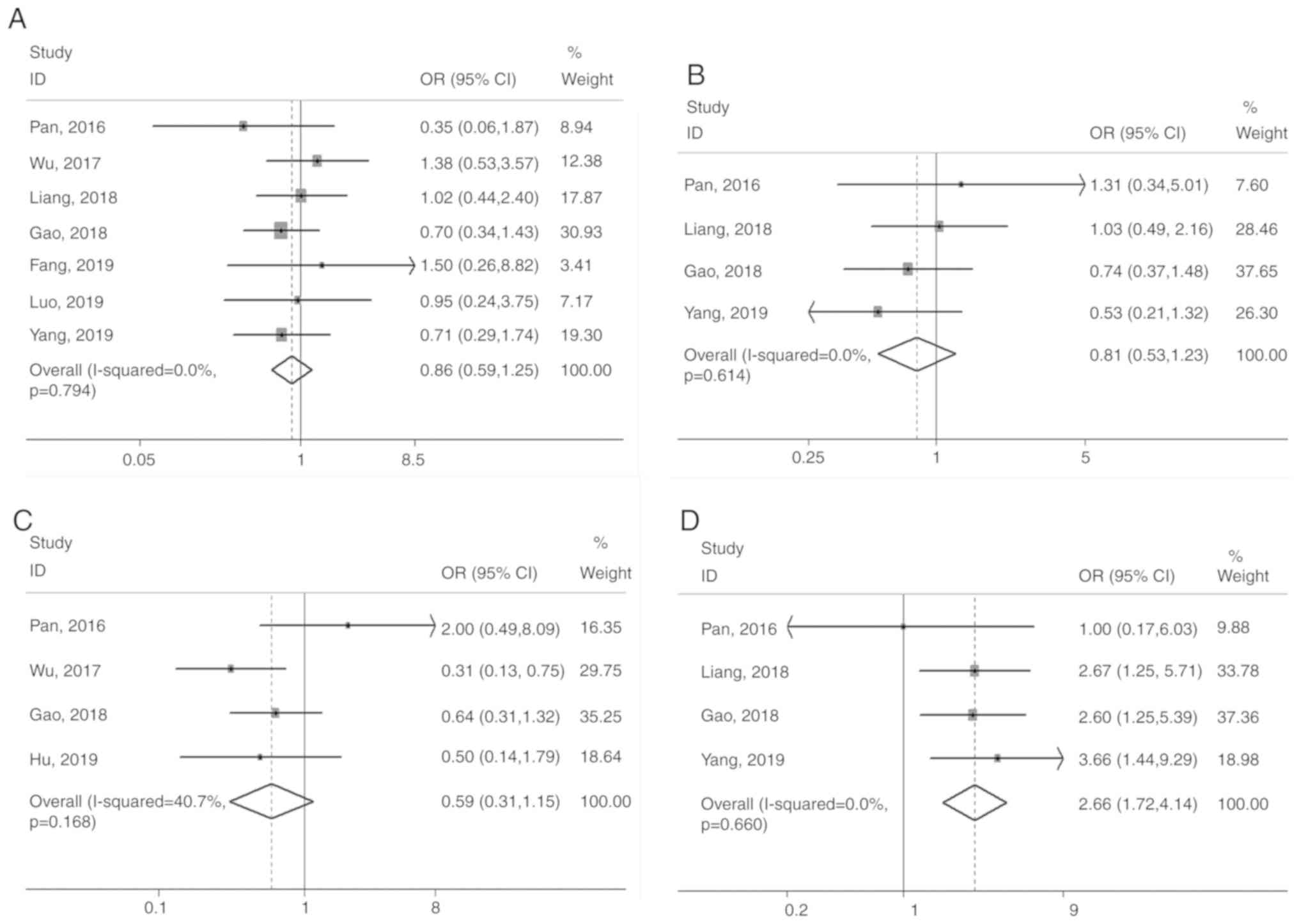
The association of CASC9 expression with sex (male
vs. female), age (<60 vs. ≥60 years), tumor size (≤4 vs. >4
cm), depth of invasion (T3+T4 vs. T1+T2), degree of tumor
differentiation (poor vs. well/moderate), lymph node metastasis
(presence vs. absence) and clinical stage (III+IV vs. I+II) were
systematically evaluated. The depth of invasion (OR=2.66, 95% CI:
1.72-4.14, P<0.001), tumor differentiation (OR=2.44, 95% CI:
1.24-4.78, P=0.009), lymph node metastasis (OR=3.42, 95% CI:
1.98-5.92, P<0.001) and clinical stage (OR=3.21, 95% CI:
2.21-4.66, P<0.001) were significantly associated with high
levels of CASC9, whereas sex, age and tumor size did not exhibit
any significant association (P>0.05; Fig. 3).
Sensitivity analysis
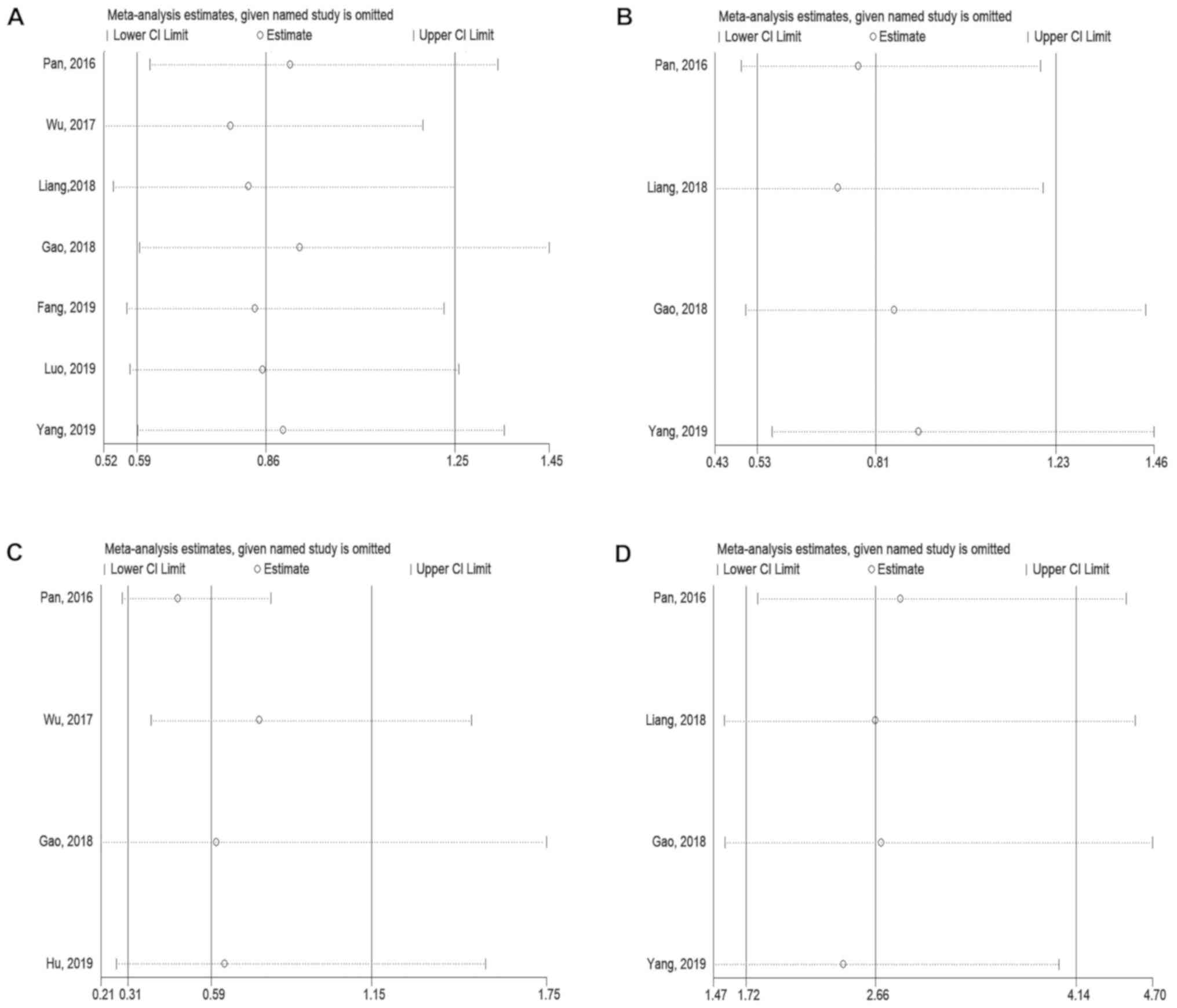
The sensitivity analysis indicated an insignificant
impact of omitting each study one by one on the overall results,
which validated the credibility of the meta-analysis (Fig. 4). Due to the small number of studies
reporting on DFS or PFS, sensitivity analysis was not performed for
these variables.
Publication bias
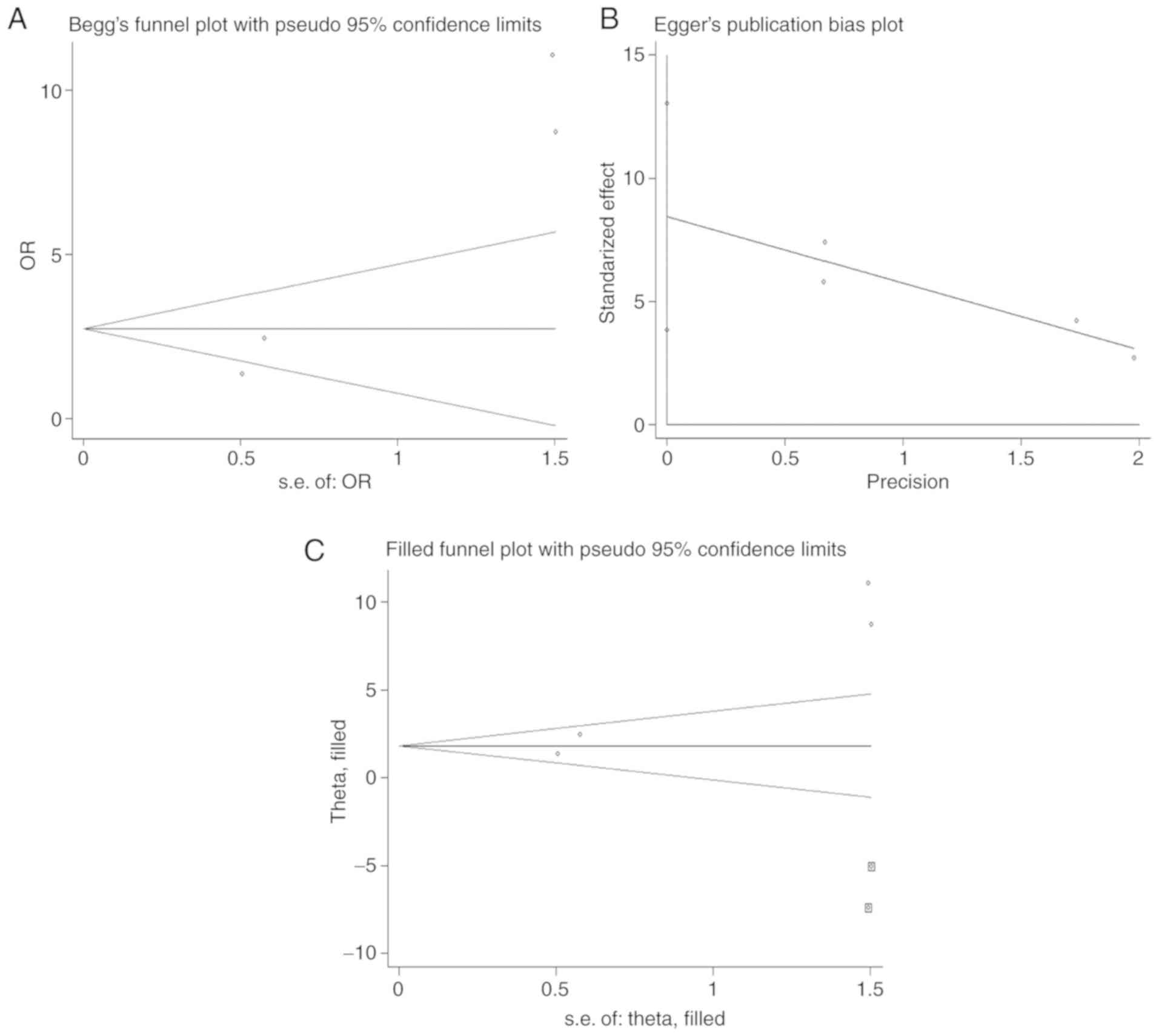
Begg's funnel plot and Egger's tests revealed no
publication bias for OS, sex, age, tumor size, depth of invasion,
lymph node metastasis and clinical stage (Table IV). However, the asymmetrical funnel
plot (Fig. 5A) and Egger's test
(P=0.016; Fig. 5B) suggested a
publication bias in terms of tumor differentiation. Therefore, the
‘trim and fill’ method (29) was
used to trace the possible influencing factors. As indicated in
Fig. 5C, two studies were identified
at the bottom of the adjusted funnel plot. The adjusted overall
effect size (OR=6.208, 95% CI: 3.196-12.056, P<0.001) for tumor
differentiation was not significantly different compared with that
before adjustment (OR, 8.449; 95% CI, 3.861-13.037; P=0.016), and
the funnel plot did not exhibit any evident asymmetry following
correction, indicating robust results.
 | Table IVPublication bias of all outcomes
assessed by Begg's test and Egger's test. |
Table IV
Publication bias of all outcomes
assessed by Begg's test and Egger's test.
| Outcome | Number of
studies/patients | Estimates | Begg's test
(P-value) | Egger's test
(P-value) | Publication
bias |
|---|
| Sex | 7/522 | OR+95% CI | 0.548 | 0.698 | Not
significant |
| Age | 4/371 | OR+95% CI | 0.734 | 0.620 | Not
significant |
| Tumor size | 4/306 | OR+95% CI | 0.734 | 0.402 | Not
significant |
| Depth of
invasion | 4/371 | OR+95% CI | 1.000 | 0.522 | Not
significant |
| Tumor
differentiation | 4/283 | OR+95% CI | 0.308 | 0.016 | Significant |
| Lymph node
metastasis | 7/474 | OR+95% CI | 0.548 | 0.152 | Not
significant |
| Clinical stage | 8/565 | OR+95% CI | 0.266 | 0.298 | Not
significant |
| OS | 5/461 | HR+95% CI | 0.462 | 0.350 | Not
significant |
Discussion
Studies increasingly indicate the involvement of
lncRNAs in biological processes, including the cell cycle,
proliferation, apoptosis and tumorigenesis (30-32).
There is also evidence confirming that lncRNAs are frequently
dysregulated in human cancers and that these abnormally expressed
lncRNAs are prognostically relevant (33-38).
Therefore, identifying novel cancer-associated lncRNAs may provide
predictors of the prognosis of cancer patients.
The lncRNA CASC9 is aberrantly expressed in various
malignancies and associated with cancer development and
progression, although its exact functional role depends on the
cancer type. For instance, CASC9 is an established oncogene in ESCC
cells, wherein it downregulates the programmed cell death 4,
upregulates recombinant laminin γ2 and promotes
epithelial-mesenchymal transition (21-23).
Furthermore, CASC9 knockdown inhibited the proliferation, cell
cycle, migration and invasion of ESCC cells. Silencing of CASC9 in
GC cells facilitated the degradation of B lymphoma Moloney murine
leukemia virus insertion region 1, leading to apoptosis (15). Furthermore, CASC9 promoted the
proliferation of CRC cells and inhibited apoptosis by interacting
with cleavage and polyadenylation specificity factor subunit 3 and
activating transforming growth factor-β2 signaling (17). Hu et al (18) indicated that CASC9 increased the
proliferation, invasion and migration of OC cells by upregulating
lin-7 homolog A via microRNA-758-3p. Jiang et al (39) revealed that CASC9 could promote the
growth, metastasis and chemoresistance of BC cells via binding to
enhancer of zeste homolog 2 and mediating the expression of
multidrug resistance protein 1. Yang et al (12) demonstrated
that high expression of CASC9 in OSCC cells enhanced proliferation
and inhibited apoptosis through the AKT/mTOR pathway. The results
of the present meta-analysis also suggested an oncogenic role of
CASC9, since its high expression levels in solid tumors were
associated with pro-oncogenic effects, including like proliferation
(12,17,18,21,23),
invasion (18,20,22,23),
migration (18,20,22,23) and
suppression of apoptosis (12,15,17), via
different targets or signaling pathways depending on the cancer
type.
However, the prognostic significance of CASC9 in
human cancers remains largely inconclusive. According to the
present meta-analysis, increased expression of CASC9 portends
shorter OS, deeper tumor invasion, poor tumor differentiation,
lymph node metastasis and advanced clinical stage. Since only one
study (23) analyzed the impact of
increased CASC9 expression on the DFS of ESCC patients and one
study (18) reported the influence
of high CASC9 expression on the PFS of patients with OC, it was not
possible to determine a definite association between CASC9
expression and DFS/PFS. In addition, a publication bias in terms of
tumor differentiation was observed. Nevertheless, the ‘trim and
fill’ method indicated robust results, and the sensitivity analysis
additionally demonstrated the consistency and reliability of the
results.
To the best of our knowledge, the present study was
the first meta-analysis to evaluate the prognostic and
clinicopathological significance of CASC9 in multiple cancer types.
The results are highly relevant from a clinical point of view and
suggest that CASC9 may be a promising therapeutic target for
multiple cancer types. However, several limitations of the present
study should be taken into consideration. First, since all
populations in the included studies were Chinese, the results
should be extrapolated with caution to populations in other
regions. In addition, only articles published in the English
language were included, and relevant studies in other languages may
have been omitted. Furthermore, the number of included studies and
cancer types in the present meta-analysis was relatively small,
which may have affected the estimated predictive ability of CASC9.
Likewise, the HRs and 95% CIs from three studies were indirectly
extracted by extrapolating the Kaplan-Meier survival curves, which
may have reduced the accuracy of the results. Finally, there was no
consensus on the cut-off for CASC9 overexpression in tumor tissues
across the different studies, which may have led to potential
heterogeneity among these studies.
To conclude, CASC9 overexpression is predictive of
poor OS in cancer patients and is significantly associated with
deeper tumor invasion, poor tumor differentiation, lymph node
metastasis and advanced clinical stage. It is a promising
prognostic biomarker for cancer and requires to be validated by
multi-center studies with large patient cohorts.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HYD and HSL conceived and designed the current
study. HYD and XD conducted data collection and extraction. HYD
analyzed the data. HYD and HSL drafted the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ulitsky I and Bartel DP: lincRNAs:
genomics, evolution, and mechanism. Cell. 154:26–46.
2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen LL: Linking long noncoding RNA
localization and function. Trends Biochem Sci. 41:761–772.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hung T and Chang HY: Long noncoding RNA in
genome regulation: Prospects and mechanisms. RNA Biol. 7:582–585.
2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yamashita A, Shichino Y and Yamamoto M:
The long non-coding RNA world in yeasts. Biochim Biophys Acta.
1859:147–154. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Loewen G, Jayawickramarajah J, Zhuo Y and
Shan B: Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol.
7(90)2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Iguchi T, Uchi R, Nambara S, Saito T,
Komatsu H, Hirata H, Ueda M, Sakimura S, Takano Y, Kurashige J, et
al: A long noncoding RNA, lncRNA-ATB, is involved in the
progression and prognosis of colorectal cancer. Anticancer Res.
35:1385–1388. 2015.PubMed/NCBI
|
|
8
|
Flicek P, Ahmed I, Amode MR, Barrell D,
Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S,
et al: Ensembl 2013. Nucleic Acids Res. 41 (D1):D48–D55.
2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Klingenberg M, Groß M, Goyal A,
Polycarpou-Schwarz M, Miersch T, Ernst AS, Leupold J, Patil N,
Warnken U, Allgayer H, et al: The long noncoding RNA cancer
susceptibility 9 and RNA binding protein heterogeneous nuclear
ribonucleoprotein L form a complex and coregulate genes linked to
AKT signaling. Hepatology. 68:1817–1832. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cao W, Wu W, Shi F, Chen X, Wu L, Yang K,
Tian F, Zhu M, Chen G, Wang W, et al: Integrated analysis of long
noncoding RNA and coding RNA expression in esophageal squamous cell
carcinoma. Int J Genomics. 2013:480534–480543. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Su X, Li G and Liu W: The long noncoding
RNA cancer susceptibility candidate 9 promotes nasopharyngeal
carcinogenesis via stabilizing HIF1a. DNA Cell Biol. 36:394–400.
2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yang Y, Chen D, Liu H and Yang K:
Increased expression of lncRNA CASC9 promotes tumor progression by
suppressing autophagy-mediated cell apoptosis via the AKT/mTOR
pathway in oral squamous cell carcinoma. Cell Death Dis. 10:41–56.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gao L, Guo YN, Zeng JH, Ma FC, Luo J, Zhu
HW, Xia S, Wei KL and Chen G: The expression, significance and
function of cancer susceptibility candidate 9 in lung squamous cell
carcinoma: A bioinformatics and in vitro investigation. Int J
Oncol. 54:1651–1664. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Shao G, Wang M, Fan X, Zhong L, Wang Z,
Zhang P and Ji S: lncRNA CASC9 positively regulates CHK1 to promote
breast cancer cell proliferation and survival through sponging the
miR 195/497 cluster. Int J Oncol. 54:1665–1675. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Fang J, Chen W and Meng XL: lncRNA CASC9
Suppressed the Apoptosis of gastric cancer cells through regulating
BMI1. Pathol Oncol Res. 10:22–29. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yu X, Lin Y, Sui W, Zou Y and Lv Z:
Analysis of distinct long noncoding RNA transcriptional
fingerprints in pancreatic ductal adenocarcinoma. Cancer Med.
6:673–680. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Luo K, Geng J, Zhang Q, Xu Y, Zhou X,
Huang Z, Shi KQ, Pan C and Wu J: lncRNA CASC9 interacts with CPSF3
to regulate TGF-β signaling in colorectal cancer. J Exp Clin Cancer
Res. 38:249–264. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hu X, Li Y, Kong D, Hu L, Liu D and Wu J:
Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p
to facilitate the malignancy of ovarian cancer. J Cell Physiol.
234:10800–10808. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhang J, Wang Q and Quan Z: Long
non-coding RNA CASC9 enhances breast cancer progression by
promoting metastasis through the meditation of miR-215/TWIST2
signaling associated with TGF-β expression. Biochem Biophys Res
Commun. 515:644–650. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Pan Z, Mao W, Bao Y, Zhang M, Su X and Xu
X: The long noncoding RNA CASC9 regulates migration and invasion in
esophageal cancer. Cancer Med. 5:2442–2447. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Wu Y, Hu L, Liang Y, Li J, Wang K, Chen X,
Meng H, Guan X, Yang K and Bai Y: Up-regulation of lncRNA CASC9
promotes esophageal squamous cell carcinoma growth by negatively
regulating PDCD4 expression through EZH2. Mol Cancer. 16:150–163.
2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang
K, Guan X, Yang K and Bai Y: lncRNA CASC9 promotes esophageal
squamous cell carcinoma metastasis through upregulating LAMC2
expression by interacting with the CREB-binding protein. Cell Death
Differ. 25:1980–1995. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gao GD, Liu XY, Lin Y, Liu HF and Zhang
GJ: lncRNA CASC9 promotes tumorigenesis by affecting EMT and
predicts poor prognosis in esophageal squamous cell cancer. Eur Rev
Med Pharmacol Sci. 22:422–429. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Moher D, Liberati A, Tetzlaff J, Altman DG
and Group P: PRISMA Group. Preferred reporting items for systematic
reviews and meta-analyses: The PRISMA statement. PLoS Med.
6(e1000097)2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tierney JF, Stewart LA, Ghersi D, Burdett
S and Sydes MR: Practical methods for incorporating summary
time-to-event data into meta-analysis. Trials. 8(16)2007.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Stang A: Critical evaluation of the
Newcastle-Ottawa scale for the assessment of the quality of
nonrandomized studies in meta-analyses. Eur J Epidemiol.
25:603–605. 2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wong WC, Cheung CS and Hart GJ:
Development of a quality assessment tool for systematic reviews of
observational studies (QATSO) of HIV prevalence in men having sex
with men and associated risk behaviours. Emerg Themes Epidemiol.
5:23–26. 2008.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Begg CB and Mazumdar M: Operating
characteristics of a rank correlation test for publication bias.
Biometrics. 50:1088–1101. 1994.PubMed/NCBI
|
|
29
|
Duval S and Tweedie R: Trim and fill: A
simple funnel-plot-based method of testing and adjusting for
publication bias in meta-analysis. Biometrics. 56:455–463.
2000.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Mathieu EL, Belhocine M, Dao LT, Puthier D
and Spicuglia S: Functions of lncRNA in development and diseases.
Med Sci (Paris). 30:790–796. 2014.PubMed/NCBI View Article : Google Scholar : (In French).
|
|
31
|
Peng WX, Koirala P and Mo YY:
lncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Gao P and Wei GH: Genomic insight into the
role of lncRNA in cancer susceptibility. Int J Mol Sci.
18:1239–1249. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yarmishyn AA and Kurochkin IV: Long
noncoding RNAs: A potential novel class of cancer biomarkers. Front
Genet. 6:145–154. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Evans JR, Feng FY and Chinnaiyan AM: The
bright side of dark matter: lncRNAs in cancer. J Clin Invest.
126:2775–2782. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
35
|
Zhao QS, Li L, Zhang L, Meng XW, Li LL, Ge
XF and Li ZP: Over-expression of lncRNA SBF2-AS1 is associated with
advanced tumor progression and poor prognosis in patients with
non-small cell lung cancer. Eur Rev Med Pharmacol Sci.
20:3031–3034. 2016.PubMed/NCBI
|
|
36
|
Qi P and Du X: The long non-coding RNAs, a
new cancer diagnostic and therapeutic gold mine. Mod Pathol.
26:155–165. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang JJ, Huang YQ, Song W, Li YF, Wang H,
Wang WJ and Huang M: Comprehensive analysis of the lncRNA
associated competing endogenous RNA network in breast cancer. Oncol
Rep. 42:2572–2582. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang X, Zhou X, Liu J, Liu Z, Zhang L,
Gong Y, Huang J, Yu L, Wang Q, Yang C, et al: Genome wide
investigation of the clinical implications and molecular mechanism
of long noncoding RNA LINC00668 and protein coding genes in
hepatocellular carcinoma. Int J Oncol. 55:860–878. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Jiang B, Li Y, Qu X, Zhu H, Tan Y, Fan Q,
Jiang Y, Liao M and Wu X: Long noncoding RNA cancer susceptibility
candidate 9 promotes doxorubicin resistant breast cancer by binding
to enhancer of zeste homolog 2. Int J Mol Med. 42:2801–2810.
2018.PubMed/NCBI View Article : Google Scholar
|