Introduction
Tuberculosis (TB), caused by Mycobacterium
tuberculosis, is one of the most common infectious diseases
worldwide and causes more mortalities than any other pathogen
(1). The causative bacteria has
developed complex mechanisms to affect the innate and the acquired
immune systems of the host (2). It
is estimated that more than one third of the world's population is
infected with Mycobacterium tuberculosis, but 90% of
infected people never develop clinical symptoms, suggesting a
natural immunity to TB, due to genetic factors in the host
(3). However, the molecular
identity and function of these genetic factors remain largely
unknown.
Naive immunological responses to Mycobacterium
tuberculosis are particularly important in the lung, as
inhalation of this harmful bacteria into the alveolar macrophage is
a key event in the pathogenesis of the disease (4). Pulmonary surfactant, as a
surface-active lipoprotein complex, is composed of 90% lipids and
10% surfactant proteins (SFTPs). Surfactant protein C (SFTPC), a
hydrophobic membrane protein, is required for biophysical function,
and disruption affects the health of the normal lung (5,6).
Therefore, we hypothesized that SFTPC contributes toward the
progression of TB. It is well-known that mycobacteria are
characterized by a complex cell wall, and various polysaccharides
and glycolipids in the cell wall of the mycobacterial species serve
significant roles in immune recognition (7-9),
including lipoarabinomannan (LAM), which is considered to be a
critical virulence factor. LAM from unique strains of mycobacteria
have been reported to account for triggering different immune
responses (10,11). Furthermore, Sidobre et al
(7) reported that the mannosylated
LAM was a putative ligand for the attachment of the human pulmonary
surfactant protein to the pathogenic Mycobacterium
tuberculosis. Mutations in the SFTPC gene have been
identified in pulmonary surfactant metabolism dysfunction and were
associated with interstitial lung diseases (5,12-17),
and multiple genetic variants in the SFTP genes were identified in
TB (18,19). However, until now, the association
between the SFPTC gene and TB has not been investigated in
well-defined populations.
In the present study, in order to provide genetic
evidence regarding the effect of SFTPC polymorphisms on TB
in the Chinese Han population, a set of single nucleotide
polymorphisms (SNPs) within SFTPC were genotyped in 900
patients with TB and 1,534 healthy control subjects, and the
association between the risk of TB and the clinical characteristics
of active tuberculosis, and selected SFTPC polymorphisms was
investigated.
Materials and methods
Study subjects
A total of 900 patients with TB (mean age 42.51
years, range 20-92 years, 542 males) and 1,534 healthy control
subjects (mean age 37.96 years, range 17-80 years, 821 males) were
included in the present case-controlled study. All samples were
obtained from the ‘Tuberculosis Researches’ Bio-Bank located within
the West China Hospital between January 2014 and February 2016 in
the Department of Laboratory Medicine, West China Hospital, Sichuan
University, (China). All the patients with TB, enrolled in the
present study, were newly diagnosed by two independent experienced
respiratory physicians. The patients with active TB were diagnosed
according to their clinical symptoms, sputum smear tests, sputum
culture, and radiological and histological pathological
examination. Patients with immunodeficiency disease, hepatitis
virus infection, human immunodeficiency virus-infection, or other
lung diseases were excluded from the study. All the healthy control
subjects were recruited from a population of healthy controls, who
had not previously suffered from TB and had negative chest
radiographs. All participants were of Han ethnicity and were not
related to each other.
Demographic data of the enrolled population were
reviewed from the medical information system of the West China
Hospital of Sichuan University. A total of 2 ml EDTA-anticoagulated
blood was collected from each of the subjects for genotyping.
Written informed consent was provided by each participant and the
study was approved by the Clinical Trials and Biomedical Ethics
Committee of West China Hospital, Sichuan University (China) and
conducted according to the Declaration of Helsinki.
SNP selection
The human SFTPC gene, which is located on
chromosome 8p21.3 is ~7.4 kb nucleotides long, with six exons. The
genetic polymorphism data of the whole sequence of SFTPC was
obtained from the dbSNP database (http://asia.ensembl.org/Homo_sapiens/Variation/). All
SNPs were filtered according to the minor allele frequency (MAF;
>0.05) in the Han Chinese population in Beijing. Subsequently,
the SNPs were preferentially selected if they were located in
potentially functional regions, including an exon, the promoter, an
intron and potential regulatory regions (within 2 kb of the genes).
Considering the experimental conditions required for genotyping,
seven SNPs in SFTPC (rs1124, rs4715, rs8192309, rs4995702,
rs8192313, rs13248346 and rs2070686) were eventually included in
the present study.
DNA isolation and genotyping
Genomic DNA was extracted from the peripheral blood
samples using a QIAamp DNA blood mini kit (Qiagen GmbH), according
to the manufacturer's protocol. Candidate SNP genotyping was
performed using the improved multiplex ligation detection reaction
method (Genesky Biotechnologies, Inc.) as previously described
(20). Detailed information
regarding the primers is available upon request. For the quality
assurance of the genotyping, double distilled water was used as the
negative control in each reaction. Furthermore, blinded repeat
genotyping of ~10% of all the samples was included in the quality
control measures and the concordance rate was 100%.
Statistical analysis
The required sample size was calculated using PASS
statistical software version 11 (NCSS LLC), prior to data
collection, as previously reported (21). Goodness-of-fit χ2 test
was used to evaluate Hardy-Weinberg equilibrium (HWE) for each SNP
in the healthy control subjects using PLINK v1.0732 (http://zzz.bwh.harvard.edu/plink/).
Sex and age were compared between patients with TB
and healthy control subjects using a χ2 test and an
independent t-test, respectively. Dominant and recessive genetic
models were used to assess the allele and genotype distribution
difference between TB cases and healthy control subjects using
multivariate logistic regression, with sex and age as covariates.
The Bonferroni method was then used to correct for multiple
testing. The associations between the genetic variants and the
clinical features of TB were evaluated. The aforementioned
statistical methods were performed using SPSS version 19.0 software
(IBM Corp.). In addition, the Haploview software package version
4.2 (Broad Institute) was used for analyzing linkage disequilibrium
patterns. The haplotype was analyzed using the SHEsis online
program (http://analysis.bio-x.cn/) (22). A two-tailed P<0.05 was considered
to indicate a statistically significant difference.
Results
Demographic characteristics
As described in Table
I, there were significant differences in sex and age between TB
cases and healthy control subjects (P<0.001).
 | Table IDemographic characteristics of study
participants. |
Table I
Demographic characteristics of study
participants.
| Characteristic | TB (n=900) | HC (n=1534) |
P-valuea |
|---|
| Male, n (%) | 542 (60.22) | 821 (53.52) | <0.001 |
| Age years, mean ±
SD | 42.51±18.11 | 37.96±11.07 | <0.001 |
Genotyping results and single SNP
association analysis
The genotype call rates of all the selected SNPs
were 99.55%. All healthy control subjects were of Han ethnicity,
and genotype distributions of the selected SNPs within the
SFTPC gene in the controls did not deviate from HWE
(P>0.05), except for rs4995702 (P<0.001). The SNP functional
consequence, P-value for the HWE test, the MAF in the healthy
controls and the Chinese Han population in Beijing based on the
1,000 Genomes Project database are summarized in Table SI. The majority of MAFs in the
healthy controls were similar to that in the Chinese Han Beijing
population, but a few were different. However, the MAFs in the
healthy controls were consistent with those in the Southern Han
Chinese population based on the 1,000 Genomes Project database
(data not shown), which indicated that the approach of using the
healthy controls was valid in the present study.
There were no data available in the database and
reported literature regarding rs4995702 and in association with
disease. In addition, rs4995702 was not in HWE in the healthy
controls. Numerous factors may contribute toward the inconsistency
of the MAF results, including migration, selection and genotyping
errors. To produce more reliable conclusions and reduce the chance
of error, rs4995702 was removed in the subsequent association
analysis. Table II shows the
allele frequencies and genotype distributions of the remaining six
SNPs in the SFTPC gene between all the TB cases and the
healthy control subjects. The minor allele (A) frequency of rs1124
was 35.70 and 32.48% in the TB cases and healthy control subjects,
respectively, and was significant after adjusting for sex and age
[P=0.024; odds ratio (OR), 1.15; 95% confidence interval (CI),
1.02-1.31)], but was not significant after using Bonferroni
correction test (P=0.144). The frequency of the AA genotype for
rs1124 was 13.07 and 10.02% in TB cases and in the healthy
controls, respectively, with no statistical significance (P=0.097).
In addition, the minor allele (C) frequency of rs8192313 was 37.86
and 35.02% in the TB group and in the healthy control subjects,
respectively, and was significant after adjusting for sex and age
(P=0.045; OR, 1.13; 95% CI, 1.00-1.28); however, it was not
significant after Bonferroni correction (P=0.278). The frequency of
the CC genotype for rs8192313 was 15.37 and 11.87% in TB cases and
in healthy control subjects, respectively. The presence of CC was
more common in TB cases, but the difference was borderline
significant (P=0.05). The other four loci (rs4715, rs8192309,
rs13248346 and rs2070686) were not significantly different between
the TB cases and controls in either genotype or allele frequencies
(all P>0.05).
 | Table IIAssociation between genetic
polymorphisms in SFTPC and tuberculosis risk. |
Table II
Association between genetic
polymorphisms in SFTPC and tuberculosis risk.
| SNP | Variant | Case, n (%) | Control, n (%) | OR (95% CI) |
P-valuea |
P-valueb | Variant | Case, n (%) | Control, n (%) |
P-valuec |
|---|
| rs1124 | A | 639 (35.70) | 992 (32.48) | 1.15
(1.02-1.31) | 0.024 | 0.144 | AA | 117 (13.07) | 153 (10.02) | 0.097 |
| G>A | G | 1,151 (64.30) | 2,062 (67.52) | 1 | - | | AG | 405 (45.25) | 686 (44.92) | |
| | | | | | | | GG | 373 (41.68) | 688 (45.06) | |
| rs4715 | A | 548 (30.44) | 886 (29.03) | 1.07
(0.94-1.21) | 0.318 | | AA | 82 (9.11) | 118 (7.73) | 0.100 |
| C>A | C | 1,252 (69.56) | 2,166 (80.97) | 1 | - | | AC | 384 (42.67) | 650 (42.60) | |
| | | | | | | | CC | 434 (48.22) | 758 (49.67) | |
| rs8192309 | A | 268 (14.92) | 472 (15.48) | 0.95
(0.81-1.13) | 0.577 | | AA | 21 (2.34) | 40 (2.62) | 0.578 |
| G>A | G | 1,528 (85.08) | 2,578 (84.52) | 1 | - | | AG | 226 (25.17) | 392 (25.71) | |
| | | | | | | | GG | 651 (72.49) | 1,093 (71.67) | |
| rs8192313 | C | 680 (37.86) | 1,068 (35.02) | 1.13
(1.00-1.28) | 0.045 | 0.270 | CC | 138 (15.37) | 181 (11.87) | 0.050 |
| A>C | A | 1,116 (62.14) | 1,982 (64.98) | 1 | - | | CA | 404 (44.99) | 706 (46.30) | |
| | | | | | | | AA | 356 (39.64) | 638 (41.84) | |
| rs13248346 | A | 406 (22.61) | 639 (20.92) | 1.10
(0.95-1.26) | 0.211 | | AA | 50 (5.57) | 62 (4.06) | 0.265 |
| G>A | G | 1,390 (77.39) | 2,415 (79.08) | 1 | - | | AG | 306 (34.08) | 515 (33.73) | |
| | | | | | | | GG | 542 (60.36) | 950 (62.21) | |
| rs2070686 | A | 453 (25.25) | 799 (26.23) | 0.95
(0.83-1.08) | 0.427 | | AA | 54 (6.02) | 109 (7.16) | 0.452 |
| G>A | G | 1,341 (74.75) | 2,247 (73.77) | 1 | - | | AG | 345 (38.46) | 581 (38.15) | |
| | | | | | | | GG | 498 (55.52) | 833 (54.69) | |
The data regarding the dominant and recessive
genetic model analysis are shown in Table III. The results indicated that
rs1124 was significantly associated with an increased risk of TB in
the recessive model (AA vs. AG+GG), with an estimated OR of 1.33
(95% CI, 1.03-1.72; P=0.031 after adjusting for sex and age;
P=0.186 after Bonferroni correction). Furthermore, there were
similar results for rs8192313 using the recessive model (CC vs.
CA+AA), with an estimated OR of 1.36 (95% CI, 1.06-1.73; P=0.014
after adjusting for sex and age; P=0.084 after Bonferroni
correction).
 | Table IIIAssociation between 6 SNPs of
SFTPC and tuberculosis risk in Chinese Han population. |
Table III
Association between 6 SNPs of
SFTPC and tuberculosis risk in Chinese Han population.
| | Dominant model | Recessive
model |
|---|
| SNP | OR (95% CI) |
P-valuea |
P-valueb | OR (95% CI) |
P-valuea |
P-valueb |
|---|
| rs1124 G>A | 1.15
(0.98-1.37) | 0.095 | | 1.33
(1.03-1.72) | 0.031 | 0.186 |
| rs4715 C>A | 1.06
(0.90-1.26) | 0.471 | | 1.17
(0.87-1.58) | 0.302 | |
| rs8192309
G>A | 0.96
(0.19-1.15) | 0.639 | | 0.88
(0.51-1.51) | 0.640 | |
| rs8192313
A>C | 1.10
(0.93-1.30) | 0.286 | | 1.36
(1.06-1.73) | 0.014 | 0.084 |
| rs13248346
G>A | 1.07
(0.90-1.27) | 0.447 | | 1.40
(0.95-2.06) | 0.090 | |
| rs2070686
G>A | 0.97
(0.82-1.14) | 0.684 | | 0.82
(0.58-1.15) | 0.248 | |
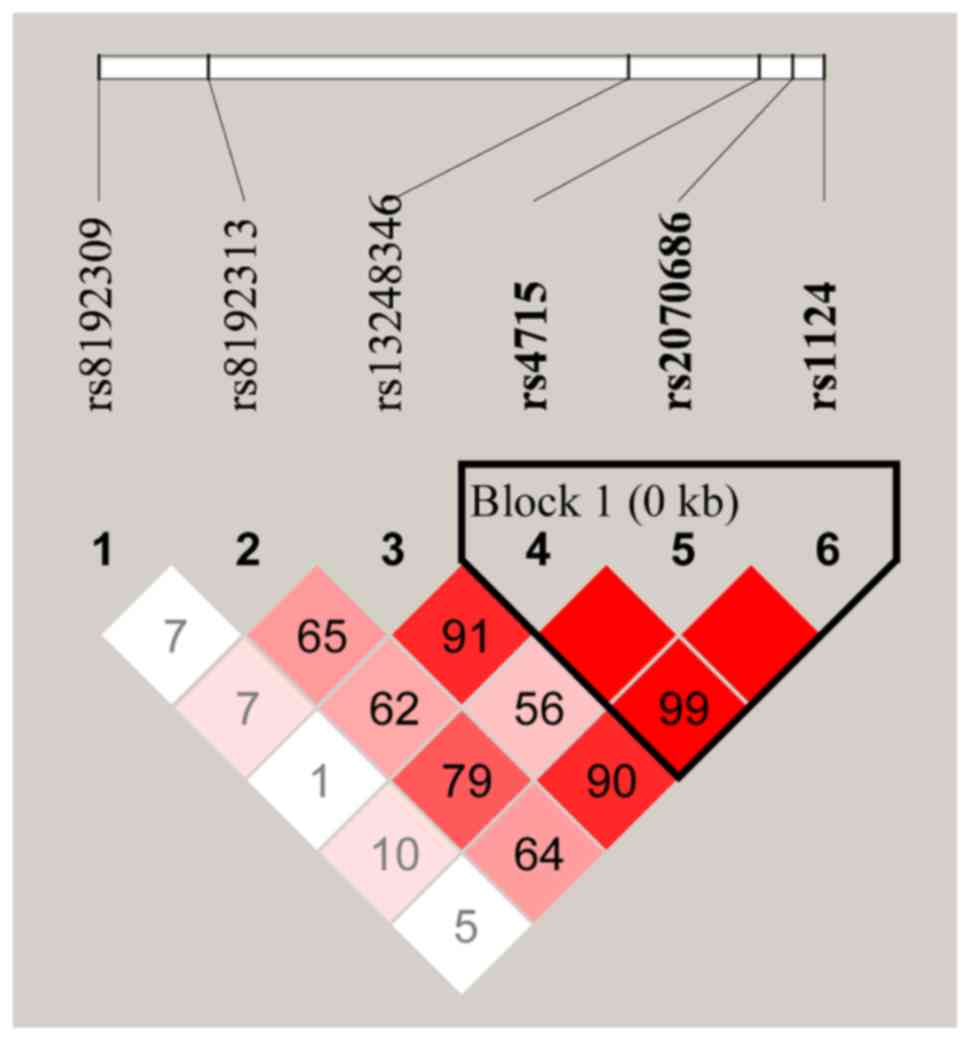
Linage analysis and haplotype
construction
The linkage disequilibrium (LD) plots of the
genotyped SNPs are displayed in Fig.
1. A total of 3 polymorphisms in SFTPC (rs4715,
rs2070686 and rs1124) were revealed to be in high LD with each
other. The haplotype frequencies and their associations with TB
predisposition are summarized in Table
IV. The results indicated that the CGA haplotype was
significantly associated with increased susceptibility to TB
[P=0.001 with an OR of 1.59 (95% CI, 1.20-2.12)] and remained
statistically significant after Bonferroni correction
(P=0.005).
 | Table IVHaplotype constructions of the
SFTPC variants associated with the risk of TB. |
Table IV
Haplotype constructions of the
SFTPC variants associated with the risk of TB.
| | Frequency | |
|---|
|
Haplotypea | All | TB cases | Healthy
controls | OR (95%CI) |
P-valueb |
P-valuec |
|---|
| AGA | 0.295 | 0.303 | 0.290 | 1.06
(0.94-1.21) | 0.340 | |
| CAG | 0.259 | 0.253 | 0.263 | 0.95
(0.93-1.09) | 0.441 | |
| CGA | 0.044 | 0.054 | 0.034 | 1.59
(1.20-2.12) | 0.001 | 0.005 |
| CGG | 0.404 | 0.39 | 0.412 | 0.91
(0.81-1.03) | 0.128 | |
Association between clinical
phenotypes and polymorphisms
The clinical symptoms and progression of active TB
may be affected by a particular genetic polymorphism as previously
reported (23). Therefore, the six
candidate SNPs were further analyzed with respect to the clinical
symptoms in the patients with TB. The erythrocyte sedimentation
rate (ESR) and C-reactive protein (CRP), which are considered to be
regular inflammatory markers for the host defense (24), were selected and evaluated. As
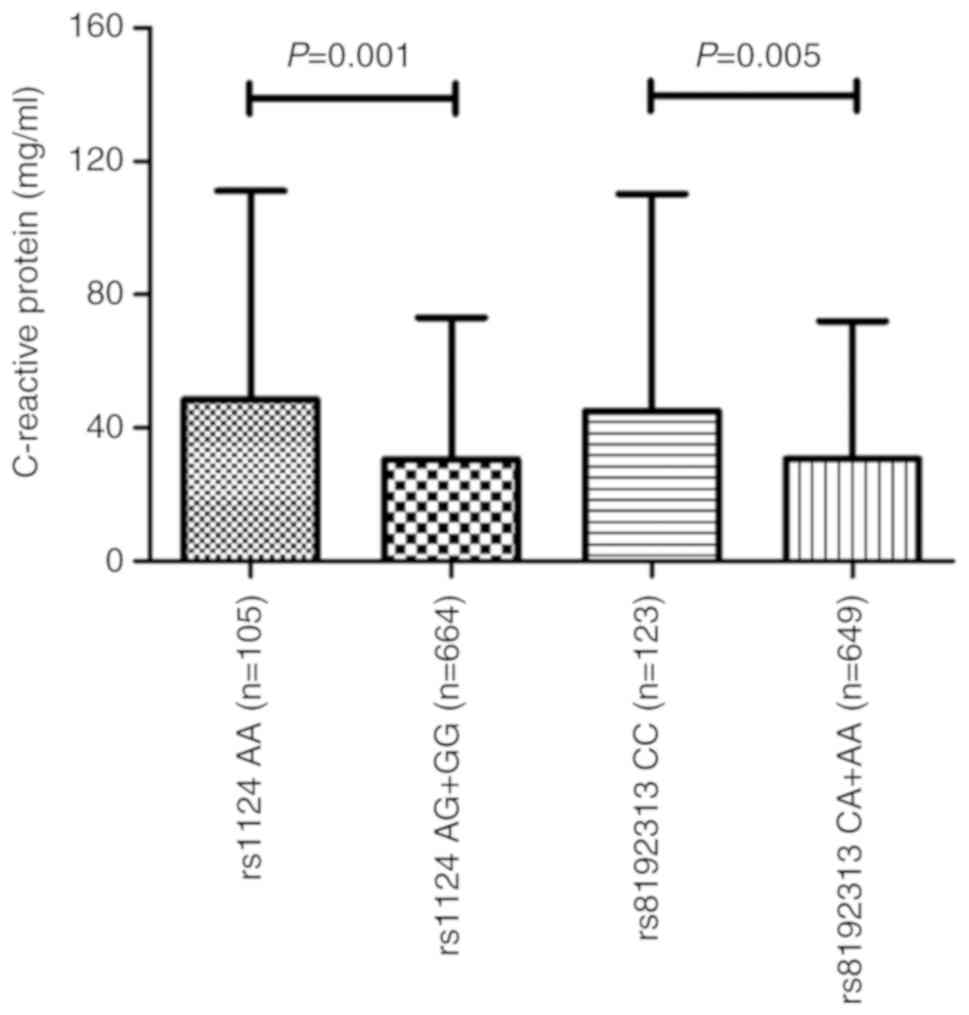
presented in Fig. 2, CRP levels
were significantly higher in those who are homozygous for the
rs1124 SNP (AA genotype) than in those who had the AG and GG
genotypes (P=0.001). A significant association in patients with TB
between the CC genotype for rs8192313 and CRP levels was also
identified (P=0.005). ESR levels were not associated with anyone
with the rs1124 or rs8192313 genotypes; therefore, with respect to
the other four SNPs, none of the clinical phenotypes were
statistically associated with the genotypes (data not shown).
Discussion
The associations between susceptibility and disease
traits in TB were preliminarily investigated in six candidate SNPs
in the SFTPC gene within the Western Chinese Han population.
The results indicated that the target SFTPC polymorphisms
(rs1124 and rs8192313) were not only associated with an increased
risk of TB but may also affect the immune response of the host to
Mycobacterium tuberculosis. The results of the present study
suggested that genetic variants in SFTPC may act as
promising novel biomarkers for the pathogenesis of TB.
An improved understanding of the molecular
mechanisms underlying host-pathogen interaction would provide a
basis for the study of TB pathogenesis. The innate immune response
is crucial for the host defense against Mycobacterium
tuberculosis (25). During
Mycobacterium tuberculosis infection, the pathogen utilizes
diverse strategies to circumvent or evade the host innate immunity.
By contrast, the host orchestrates multiple signaling cascades to
initiate a large variety of innate immune defense functions
(26). For example, mannosylated
LAM was identified in pathogenic Mycobacterium tuberculosis,
which serves as a modulator of the host immune system (27). A study by Zhang et al
(11) reported that the pathogens
had divergent effects on DC maturation and cytokine responses based
on the amount and scaffold of mannose in LAMs with diverse
mannosylated structures. By contrast, genetic polymorphisms in
host-derived toll-like receptors were associated with
susceptibility to TB (23,28). Changes in the alveolar lining fluid,
including decreased binding ability of SFTPA and SFTPD to
Mycobacterium tuberculosis, may increase TB susceptibility
(29). It was reported that SFTPC
is a hydrophobic membrane protein and serves a significant role in
surfactant function (5), and
mutations in SFTPC may modulate recruitment and activation of key
myeloid cell populations in interstitial lung disease (30). However, there is very little data
supporting the effects of SFTPC genetic polymorphisms on
predisposition to and the clinical phenotype for TB, which is
highly prevalent in China. Therefore, the present case-control
study in the Western Chinese Han population was performed to
preliminarily investigate potential TB-associated SNPs within
SFTPC and to determine if these loci were associated with
clinical manifestations of TB.
There were no significant associations in the risk
of TB with rs4715, rs8192309, rs13248346 and rs2070686 within
SFTPC between TB cases and healthy control subjects in the
Western Chinese Han population. Age did not affect the frequency of
genotypes (all P>0.05), and there was no difference in
the frequency of genotypes between males and females (all
P>0.05) in patients with TB (Table SII). Furthermore, there was no
statistical significance in the frequency of genotypes between
patients with a positive and negative TB sputum smear test, except
for rs8192309 (P=0.025; data not shown). However, the differences
in A allele frequencies (P=0.024) and genotype distributions under
the recessive model (P=0.031) for the rs1124 SNP were statistically
significant. In addition, similar results were observed with the
rs8192313 SNP (P=0.045 and P=0.014 for the differences in C allele
frequencies and genotype distributions under the recessive model,
respectively). The differences reported for the rs1124 and
rs8192313 SNPs did not remain statistically significant after
Bonferroni correction. However, it was hypothesized that the A
allele or the C allele of rs1124 and rs8192313 SNPs, respectively,
within the SFTPC gene had a low susceptibility risk for the
development of TB based on the results of the present study. The
results also indicated that the CGA haplotype, formed by these
three SFTPC polymorphisms and possibly due to the A allele
for the rs1124 SNP, was significantly associated with increased TB
susceptibility (P=0.001) with an OR of 1.59 (95% CI, 1.20-2.12).
This finding was consistent with the individual rs1124 SNP
analysis. However, the homozygotic genotype CC/GG/AA was not
analyzed for TB susceptibility, due to the small homozygous sample
size.
In the present study, the rs1124 and rs8192313 SNPs
were significantly associated with differences in TB clinical
symptoms. The results suggested that carrying the AA genotype for
the rs1124 SNP was significantly associated with higher levels of
CRP (P=0.001) among patients with active TB, which reflected the
host inflammatory response to Mycobacterium tuberculosis
infection. This was consistent with the result from the genetic
analysis of susceptibility association, and further supported the
effect of the A allele in rs1124 on the risk of TB. The effect of
the rs8192313 SNP on the clinical features of TB and its effect on
the susceptibility to TB were also observed using the same method.
The TB susceptibility SNP rs8192313 was associated with CRP levels
in patients with active TB within the current dataset. High CRP
levels were significantly associated with the CC genotype, compared
with the A allele, including heterozygous and homozygous genotypes
(P=0.005). The results regarding the susceptibility SNPs to the
development of TB and genetic loci associated with TB clinical
phenotype indicated that the development of TB, progression and
disease manifestation may be affected by diverse genetic loci. The
specific molecular mechanisms remain unclear, but the results
suggested that SFTPC genetic variants may affect the host
defense responses against Mycobacterium tuberculosis and
lead to diverse clinical manifestations in patients with active
TB.
The SNPs, rs1124 and rs4715, are exonic splicing
enhancers, while rs8192309, rs8192313, rs13248346, rs2070686 and
rs4995702 are all transcription factor binding sites, as predicted
using the SNPinfo method, as previously reported (31) (Table
SI). It has also been reported that the rs4715 SNP was
associated with respiratory distress syndrome (5,14,16);
however, this was not identified in the present study. The results
also suggested that the SNP rs1124 may serve an important role in
disease development, which was in accordance with the results of
previous studies (5,13,14,16).
The SNPs, rs4715 and rs1124, are both missense, in which the amino
acids 138 (Thr/Asn) and 186 (Ser/Asn) are changed, respectively;
however, the potential mechanisms in which they affect function
remain unknown. The single SNP statistical analyses of the other
four SFTPC variants did not reveal any significant associations,
but the investigation of these polymorphisms supplemented the
knowledge on the association of SFTPC polymorphisms with the
susceptibility to TB. Based on the results of the present study,
the association between the SFTPC gene and TB should be interpreted
cautiously, as it is hypothesized that this gene was not the
primary cause of TB but serves a lesser role in the complex genetic
pattern.
To the best of our knowledge, the present study was
the first to investigate the association between SFTPC
genetic variations and TB in a Western Chinese Han population.
Furthermore, to minimize the bias of Mycobacterium
tuberculosis complex exposure, all subjects from the same
geographical area were recruited over the same study period.
However, there are several limitations to the present study. To
begin with, only a small number of SNPs within SFTPC and a
small sample size were investigated. Additionally, the association
between SFTPC SNPs and the risk of TB requires further
investigation using subgroup analysis. Furthermore, there were no
other clinical characteristics that were matched between the groups
for the risk of TB, including smoking and alcohol. Finally, the
specific population with latent TB infection among the disease-free
controls was not included in the present study. Therefore, further
comprehensive, in-depth and multi-center research is required, to
reveal the mechanisms of the genetic polymorphisms in SFTPC
in TB development and clinical manifestation.
The present study investigated the pathogenesis of
TB with respect to surfactant protein-based polymorphisms. A total
of 2 SNPs within the SFTPC (rs1124 and rs8192313) were
associated with TB susceptibility. Furthermore, the rs1124 AA and
rs8192313 CC genotypes were associated with higher CRP levels, in a
recessive model. The results indicated that SFTPC polymorphisms may
affect TB susceptibility and host immune response to
Mycobacterium tuberculosis and may be used as novel
biomarkers for the pathogenesis of TB.
Supplementary Material
Characteristics of the candidate
SNPs.
The genotype distributions between
male and female in patients with tuberculosis.
Acknowledgements
The authors would like to thank Dr Yu Wang from
Department of Respiratory and Critical Care Medicine, the First
Affiliated Hospital of Zhengzhou University for providing writing
assistance.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81672095 and
81501715) and the Projects of the Health and Family Planning
Commission in Sichuan Province (grant no. 16ZD004).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LM and BWY conceived and designed the present study.
JWZ and LJ performed the experiments, analyzed the data and wrote
the manuscript. MMG, LZ, XBW and SHG analyzed the data and
conceived the study. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Clinical
Trials and Biomedical Ethics Committee of West China Hospital,
Sichuan University. Written informed consent was obtained from each
participant and all experiments were performed according to the
Declaration of Helsinki.
Patient consent for publication
All patients provided written informed consent for
publication.
Competing interests
The patients declare that they have no competing
interests.
References
|
1
|
McShane H: Insights and challenges in
tuberculosis vaccine development. Lancet Respir Med. 7:810–819.
2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Orme IM, Robinson RT and Cooper AM: The
balance between protective and pathogenic immune responses in the
TB-infected lung. Nat Immunol. 16:57–63. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Arend SM, Engelhard AC, Groot G, de Boer
K, Andersen P, Ottenhoff TH and van Dissel JT: Tuberculin skin
testing compared with T-cell responses to Mycobacterium
tuberculosis-specific and nonspecific antigens for detection of
latent infection in persons with recent tuberculosis contact. Clin
Diagn Lab Immunol. 8:1089–1096. 2001.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Guilliams M, Lambrecht BN and Hammad H:
Division of labor between lung dendritic cells and macrophages in
the defense against pulmonary infections. Mucosal Immunol.
6:464–473. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lin Z, Thorenoor N, Wu R, DiAngelo SL, Ye
M, Thomas NJ, Liao X, Lin TR, Warren S and Floros J: Genetic
association of pulmonary surfactant protein Genes, SFTPA1, SFTPA2,
SFTPB, SFTPC, and SFTPD with cystic fibrosis. Front Immunol.
9(2256)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li B, Meng YQ, Li Z, Yin C, Lin JP, Zhu DJ
and Zhang SB: MiR-629-3p-induced downregulation of SFTPC promotes
cell proliferation and predicts poor survival in lung
adenocarcinoma. Artif Cells Nanomed Biotechnol. 47:3286–3296.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sidobre S, Nigou J, Puzo G and Rivière M:
Lipoglycans are putative ligands for the human pulmonary surfactant
protein A attachment to mycobacteria. Critical role of the lipids
for Lectin-carbohydrate recognition. J Bio Chem. 275:2415–2422.
2000.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Guenin-Macé L, Siméone R and Demangel C:
Lipids of pathogenic Mycobacteria: Contributions to virulence and
host immune suppression. Transbound Emerg Dis. 56:255–268.
2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zheng RB, Jégouzo SAF, Joe M, Bai Y, Tran
HA, Shen K, Saupe J, Xia L, Ahmed MF, Liu YH, et al: Insights into
interactions of mycobacteria with the host innate immune system
from a novel array of synthetic mycobacterial glycans. ACS Chem
Biol. 12:2990–3002. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Mazurek J, Ignatowicz L, Kallenius G,
Svenson SB, Pawlowski A and Hamasur B: Divergent effects of
mycobacterial cell wall glycolipids on maturation and function of
human monocyte-derived dendritic cells. PLoS One.
7(e42515)2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang SP, Wu QH, Lei H, Zheng H, Zhou F,
Sun ZQ, Zhao JW, Yu XL and Zhang SL: Mannosylated structures of
mycobacterial lipoarabinomannans facilitate the maturation and
activation of dendritic cells. Cell Immunol. 335:85–92.
2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lahti M, Marttila R and Hallman M:
Surfactant protein C gene variation in the Finnish
population-association with perinatal respiratory disease. Eur J
Hum Genet. 12:312–320. 2004.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Puthothu B, Krueger M, Heinze J, Forster J
and Heinzmann A: Haplotypes of surfactant protein C are associated
with common paediatric lung diseases. Pediatr Allergy Immunol.
17:572–577. 2006.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wambach JA, Yang P, Wegner DJ, An P,
Hackett BP, Cole FS and Hamvas A: Surfactant protein-C promoter
variants associated with neonatal respiratory distress syndrome
reduce transcription. Pediatr Res. 68:216–220. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Peca D, Boldrini R, Johannson J, Shieh JT,
Citti A, Petrini S, Salerno T, Cazzato S, Testa R, Messina F, et
al: Clinical and ultrastructural spectrum of diffuse lung disease
associated with surfactant protein C mutations. Eur J Hum Genet.
23:1033–1041. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Fatahi N, Dalili H, Kalani M, Niknafs N,
Shariat M, Tavakkoly-Bazzaz J, Amini E, Esmaeilnia Shirvani T,
Hardani AK, Taheritafti R, et al: Association of SP-C gene codon
186 polymorphism (rs1124) and risk of RDS. J Matern Fetal Neonatal
Med. 30:2585–2589. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Liu J, Chen JH, Wang YQ, Nong GM, Zheng YJ
and Hao CL: Genetic variants in the surfactant protein C gene 218
Site are associated with pediatric interstitial lung disease: Seven
cases study. Zhonghua Er Ke Za Zhi. 57:21–26. 2019.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
18
|
Hsieh MH, Ou CY, Hsieh WY, Kao HF, Lee SW,
Wang JY and Wu LSH: Functional analysis of genetic variations in
surfactant protein d in mycobacterial infection and their
association with tuberculosis. Front Immunol.
9(1543)2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yang HY, Li H, Wang YG, Xu CY, Zhao YL, Ma
XG, Li XW and Chen H: Correlation analysis between single
nucleotide polymorphisms of pulmonary surfactant protein A gene and
pulmonary tuberculosis in the Han population in China. Int J Infect
Dis. 26:31–36. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Bai H, Wu Q, Hu X, Wu T, Song J, Liu T,
Meng Z, Lv M, Lu X, Chen X, et al: Clinical significance of
lnc-AC145676.2.1-6 and lnc-TGS1-1 and their variants in western
Chinese tuberculosis patients. Int J Infect Dis. 84:8–14.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhao Z, Zhang M, Ying J, Hu X, Zhang J,
Zhou Y, Zhou Y, Song X and Ying B: Significance of genetic
polymorphisms in long non-coding RNA AC079767.4 in tuberculosis
susceptibility and clinical phenotype in Western Chinese Han
population. Sci Rep. 7(965)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z,
He L and Shi Y: A partition-ligation-combination-subdivision EM
algorithm for haplotype inference with multiallelic markers: Update
of the SHEsis. (http://analysis.bio-x.cn).
Cell Res. 19:519–523. 2009.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang Y, Zhang MM, Huang WW, Wu SQ, Wang
MG, Tang XY, Sandford AJ and He JQ: Polymorphisms in toll-like
receptor 10 and tuberculosis susceptibility: Evidence from three
independent series. Front Immunol. 9(309)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bottazzi B, Doni A, Garlanda C and
Mantovani A: An integrated view of humoral innate immunity:
Pentraxins as a paradigm. Annu Rev Immunol. 28:157–183.
2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Verrall AJ, Schneider M, Alisjahbana B,
Apriani L, van Laarhoven A, Koeken VACM, van Dorp S, Diadani E,
Utama F, Hannaway RF, et al: Early clearance of Mycobacterium
tuberculosis is associated with increased innate immune responses.
J Infect Dis. 221:1342–1350. 2019.
|
|
26
|
Liu CH, Liu H and Ge B: Innate immunity in
tuberculosis: Host defense vs pathogen evasion. Cell Mol Immunol.
14:963–975. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhou KL, Li X, Zhang XL and Pan Q:
Mycobacterial Mannose-capped lipoarabinomannan: A modulator
bridging innate and adaptive immunity. Emerg Microbes Infect.
8:1168–1177. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang J, Zhao Z, Zhong H, Wu L, Zhou W,
Peng W, Hu X, Song J, Liu T, Wu Q, et al: Importance of common TLR2
genetic variants on clinical phenotypes and risk in tuberculosis
disease in a Western Chinese population. Infect Genet Evol.
60:173–180. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Moliva JI, Duncan MA, Olmo-Fontanez A,
Akhter A, Arnett E, Scordo JM, Ault R, Sasindran SJ, Azad AK,
Montoya MJ, et al: The lung mucosa environment in the elderly
increases host susceptibility to Mycobacterium tuberculosis
infection. J Infect Dis. 220:514–523. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Venosa A, Katzen J, Tomer Y, Kopp M, Jamil
S, Russo S, Mulugeta S and Beers M: Epithelial expression of an
interstitial lung disease-associated mutation in surfactant
protein-C modulates recruitment and activation of key myeloid cell
populations in mice. J Immunol. 202:2760–2771. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Xu Z and Taylor JA: SNPinfo: integrating
GWAS and candidate gene information into functional SNP selection
for genetic association studies. Nucleic Acids Res. 37:W600–605.
2009.PubMed/NCBI View Article : Google Scholar
|