Introduction
Long non-coding RNAs (lncRNAs) are characterized by
their length (>200 nucleotides), intron/exon structure, the
presence of a 3' untranslated region and termination region, and a
limited coding potential supported by the absence of open reading
frames (1). Biochemically, lncRNAs
are thought to mediate local gene expression as cis-regulatory
elements, affect transcription of multiple genes as
trans-regulatory elements and act as a scaffold for chromatin
structure maintenance (2,3). In terms of function, lncRNAs have been
reported to participate in numerous biological processes, including
X chromosome inactivation, genomic imprinting, cell cycle
regulation and the regulation of stem cell pluripotency (4,5).
Additionally, the molecular functions of lncRNAs have been
highlighted to have roles in various diseases, particularly those
relevant to endocrinology, reproduction, metabolism, immunology,
neurobiology, muscle biology and cancer (6-9).
Several studies have investigated the association
between neonatal lung diseases and lncRNAs. Cheng et al
(10) reported 9 lncRNAs that were
potentially associated with bronchopulmonary dysplasia (BPD) and
these data may provide novel insight into the biological roles of
lncRNAs in the pathogenesis of BPD. Numerous lncRNAs are
significantly differentially expressed in various lung diseases.
For instance, metastasis-associated lung adenocarcinoma transcript
1 (MALAT1) has been reported to have an important role in lung
cancer progression (11-13).
Although the loss of MALAT1 does not affect lung development
(14), a previous study indicated
that the upregulation of MALAT1 may protect preterm infants with
BPD by inhibiting apoptosis (15).
Deletions of chromosomes encompassing other lncRNAs may cause
lethal lung developmental disorders (16). Szafranski et al (17) demonstrated that deletion of a small
non-coding methylated region at 16q24.1, included in lncRNA genes,
caused the lethal lung developmental disorder alveolar capillary
dysplasia with misalignment of pulmonary veins with
parent-of-origin effects in a human model. These data indicated
that lncRNAs may regulate fetal lung development.
As the survival rate increases among preterm
infants, respiratory distress syndrome (RDS) and BPD are becoming
more and more common (18). One
widely accepted cause of BPD is insufficient fetal lung development
during pregnancy (19).
Additionally, a lack of a pulmonary surfactant synthesized by type
II alveolar epithelial cells is acknowledged as a major cause of
RDS (20). Consequently, there may
be a close association between neonatal respiratory diseases and
lung development.
Fetal lung development is a complex and continuous
process. The development of lung structure includes the embryonic,
fetal and postnatal stages (21).
Fetal embryonic lung development may be divided into the
pseudo-glandular period (7-16 weeks of gestation), canalicular
period (16-25 weeks of gestation) and terminal saccular period (25
weeks to full term gestation) (22). Different stages of lung development
have different characteristics (23). During the terminal saccular period,
with the appearance of alveolar septum, capillaries, elastic fibers
and collagen fibers, terminal vesicles become alveolar (21). Alveolar maturation begins at 30
weeks of gestation. Lung potential gas volume and surface area
increase from 25 weeks of gestation to full term (24). Increased alveoli, lung volume and
surface area provide the anatomical potential for gas exchange and,
therefore, provide the basis for fetal survival following birth
(25-27).
In the present study, three periods of fetal lung development,
divided by gestational week, were investigated through the
examination of morphological and lncRNA expression changes in three
groups of fetal lung tissue samples. An Affymetrix Human GeneChip
was employed to assess the differential expression of lncRNAs
between three phases of fetal lung development. Bioinformatics
methods were also used to analyze the potential functions and
pathways associated with the protein-coding genes associated with
the differentially expressed lncRNA. These data may provide a
theoretical basis for the prevention and treatment of neonatal lung
developmental diseases, including RDS and BPD.
Materials and methods
Patients and samples
The present study protocol was approved by the
Ethics Committee of Nanjing Maternal and Child Health Care
Hospital, Nanjing, China [approval no. (2014) 74]. All of the
patients included provided written informed consent to participate
in the current study. Abortion was most commonly induced due to
personal or social factors, rather than due to congenital
problems.
The inclusion criteria were as follows: i)
Gestational age of the fetus at the time-point of abortion was 7-28
weeks; ii) fetal abortion was performed due to personal or social
factors; iii) pregnant females were aged 20-35 years; and iv)
pregnant females provided written informed consent.
The exclusion criteria were as follows: i) Pregnant
females with a history of hypertension or diabetes, or kidney,
heart, connective tissue or autoimmune diseases; ii) pregnant
females with a known history of exposure to radioactivity, toxic
substances or drugs; iii) maternal use of glucocorticoids (e.g.,
dexamethasone, prednisone or beclomethasone) prior to abortion; iv)
pregnant females with signs of infection, including positive
amniotic fluid culture, increased C-reactive protein or
procalcitonin; and v) previously detected chromosomal abnormalities
or congenital malformations of the fetus.
Abortion procedure
Procedures took place between August 2014 and
February 2015 at Nanjing Maternity and Child Health Care Hospital.
Physicians explained the medical method and possible adverse
reactions of abortion to the pregnant females who then provided
voluntary written informed consent. Females at 8-13 weeks of
gestation took mifepristone (200 mg orally), then misoprostol (400
µg sublingually) followed by mifepristone (200 mg orally) after
24-48 h. If the abortion was not complete, repeated misoprostol was
taken sublingually every 3 h and up to 4 doses were administered
until complete abortion. For females at 14-28 weeks of gestation,
amniocentesis was performed prior to the intra-amniotic injection
of 0.5% rivanol solution (100 mg) to induce contractions and
initiate labor. The pregnant females took mifepristone (25 mg
orally), which was used to soften the cervix (28-30).
Fetal right-lung tissue samples were collected within 2 h of labor.
Fetal lungs were isolated and were divided into three groups
according to the fetal gestational age. These groups were termed S1
(embryonic week, 7-16), S2 (embryonic week, 16-25) and S3
(embryonic week, 25-28). A total of 10 samples were collected at S1
(mean age, 27.2±1.56 years; mean fetal gestational age, 13.57±0.73
weeks) 14 samples at S2 (mean age, 25.5±0.84 years; mean fetal
gestational age, 20.73±0.72) and 12 samples at S3 (mean age,
25.92±1.13 years; mean fetal gestational age, 26.75±0.23 weeks).
The median overall gestational age was 21.35 weeks with a range of
15.58-26.18 weeks.
The right lungs were washed with PBS and cut into
several parts (2x4x6 mm), all of which were kept for histological
examination. A 5 mg sample of each of the lungs was cut into small
pieces in homogenization buffer (Trevigen, Inc.). The lungs were
homogenized with a Sonifier (Branson Ultrasonics Corporation) with
an amplitude of 14 microns for 10 sec. The cell supernatant was
obtained by centrifugation at 12,000 x g for 15 min at 4˚C. Total
RNA from the right lungs was isolated from the supernatant using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. RNA quality and
quantity were measured on a NanoDrop 2000 spectrophotometer (Thermo
Fisher Scientific, Inc.). A total of 1 µg of total RNA was taken
from each sample and subjected to 1.5% agarose gel electrophoresis
(120 V) for 15 min to determine the integrity of 28 and 18s
ribosomal RNA under a gel imager (Bio-Rad Laboratories, Inc.) and
to ensure that there were no residual RNA enzymes.
Histology
Lung tissues were fixed with 4% buffered
paraformaldehyde at 4˚C overnight, dehydrated [50% ethanol for 2 h,
70% ethanol for 2 h, 85% ethanol for 2 h, 95% ethanol for 2 h,
anhydrous ethanol for 1.5 h and anhydrous ethanol (fresh
configuration) II for 1.5 h] and embedded in paraffin. Sections
with a thickness of 3-4 µm were prepared for H&E staining and
immunohistochemistry. Sections were dewaxed with xylene and
rehydrated in a graded series of ethanol/water solutions. The
sections were then stained with hematoxylin for 5 min,
differentiated with 1% ethanol hydrochloride for 3 sec and
transferred to eosin solution for 2 min. All procedures were
performed at room temperature. The sections were then dehydrated
and mounted. A total of three sections were randomly selected from
each sample and a total of 108 H&E-stained sections were taken
for image analysis, which was performed under a light microscope
(BX51; Olympus Corporation) at magnifications of x200 and x400 to
observe changes in fetal lung development between groups S1, S2 and
S3.
Affymetrix Human GeneChip
analysis
The GeneChip® Human Transcriptome Array
2.0 (Affymetrix Inc.) serves as an advanced and comprehensive gene
expression profiling tool for whole-transcript coverage available
on any microarray platform (31).
Probes are distributed across the full length of a gene, including
specific probes covering splice junctions, providing a more
complete and accurate picture of overall gene expression with
additional capacity for transcript isoform analysis. In brief,
following the extraction of total RNA, 10 µg of RNA was used to
synthesize double-stranded complementary (c)DNA using an Ambion WT
expression kit (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The cDNA was then
fragmented and labelled with the Affymetrix GeneChip WT terminal
labelling kit (Affymetrix Inc.), according to the manufacturer's
protocol. The fragmented cDNA was hybridized using the GeneChip
hybridization, wash and stain kit (Affymetrix Inc.), according to
the manufacturer's protocol. Hybridization was performed at 65˚C
with rotation for 16 h in an Affymetrix GeneChip Hybridization Oven
645 (Affymetrix Inc.). The GeneChip arrays were washed and stained
on an Affymetrix Fluidics Station 450 (Affymetrix Inc.), followed
by scanning on a GeneChip Scanner 3000 (Affymetrix Inc.). The
microarray analysis was performed by Genminix Informatics Co.,
Ltd.
Reverse transcription-quantitative PCR
(RT-qPCR)
A total of 0.1 g of lung tissue was homogenized in a
homogenizer (Kinematica AG). Total RNA was isolated from fetal
lungs using TRIzol® reagent. cDNA was synthesized with
the Transcriptor First Strand cDNA Synthesis kit (Roche Diagnostics
Co., Ltd.), according to the manufacturer's protocol. An aliquot of
1 µg total RNA was added to each reaction mixture. RT-qPCR was
performed on an ABI 7500 thermal cycler (Applied Biosystems; Thermo
Fisher Scientific, Inc.) with SYBR Green (Roche Diagnostics Co.,
Ltd.). The thermocycling conditions were as follows: 95˚C for 5
min, followed by 40 cycles of 95˚C for 20 sec and 55˚C for 20 sec.
At the end of each run, a melting curve analysis was performed at
72˚C to monitor primer dimers and formation of non-specific
products. Relative quantification of gene expression in multiple
samples was achieved by normalization to the expression of an
endogenous control gene, GAPDH. The relative expression levels were
calculated by the 2-∆∆Cq method
(32). Primer sequences are listed
in Table I.
 | Table IReverse transcription-quantitative
PCR primers. |
Table I
Reverse transcription-quantitative
PCR primers.
| Gene | Primer sequence
(5'-3') |
|---|
| n387037 | Forward
TGGAAATTGGAAGAGCACAA |
| | Reverse
TGTATGAGGGTGCATGGAAA |
| n340848 | Forward
TTACAAGCTCCATCAGCACAG |
| | Reverse
TCCACCTGTTCATTGGTTCA |
|
ENST00000445168 | Forward
GGTGGCAGAGCTAGAACTCG |
| | Reverse
GGGTAAGCCTCGTGTACCAA |
| n336823 | Forward
TTGTGGGCCTCCTCATATTT |
| | Reverse
GAAGTTTGTCCACCGCAAAG |
| GAPDH | Forward
AACTTTGGCATTGTGGAAGG |
| | Reverse
GGATGCAGGGATGATGTTCT |
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analyses
The GO project offers a controlled vocabulary to
label gene and gene product attributes in any organism (geneontology.org; date of access, May 2015).
Categories covered by GO analysis include biological process,
cellular component and molecular function. GO analysis provides an
interpretation of the relevance of genes differentially expressed
between the groups by suggesting possible functions of the genes
and functions associated with the genes. Fisher's exact test and
the χ2 test were performed to calculate the P-value and
false discovery rate of each GO term function. The input used in
the bioinformatics analysis was the crossover genes of
differentially expressed lncRNA-associated genes and differential
mRNA genes co-expressed with lncRNA that were screened in the
lncRNA expression profiling results. The criterion for screening
differentially expressed genes for statistical significance was
P<0.05, thus screening out the significant functions exerted by
the differentially expressed genes.
KEGG (kegg.jp/kegg/pathway.html) pathway analysis is a
functional analysis tool, mapping a set of genes that may be
associated with a certain lncRNA to potential pathways they are
accumulated in. Fisher's exact test and χ2 test were
used to identify differentially expressed genes. P<0.05 was used
to screen and obtain significantly associated pathways. The
probability of enrichment of a differentially expressed gene set in
a term entry was represented by an enrichment score (EC), with a
higher EC indicating a higher significance of the entry. The EC was
calculated as the negative base 10 log of the P-value.
Statistical analysis
All quantitative data are expressed as the mean ±
standard error of the mean. All experiments were repeated ≥3 times
with similar results. Data were analyzed using a SPSS statistical
package (version no. 17.0; SPSS, Inc.) and the results of the
RT-qPCR were evaluated by one-way ANOVA with the
Student-Newman-Keuls post-hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
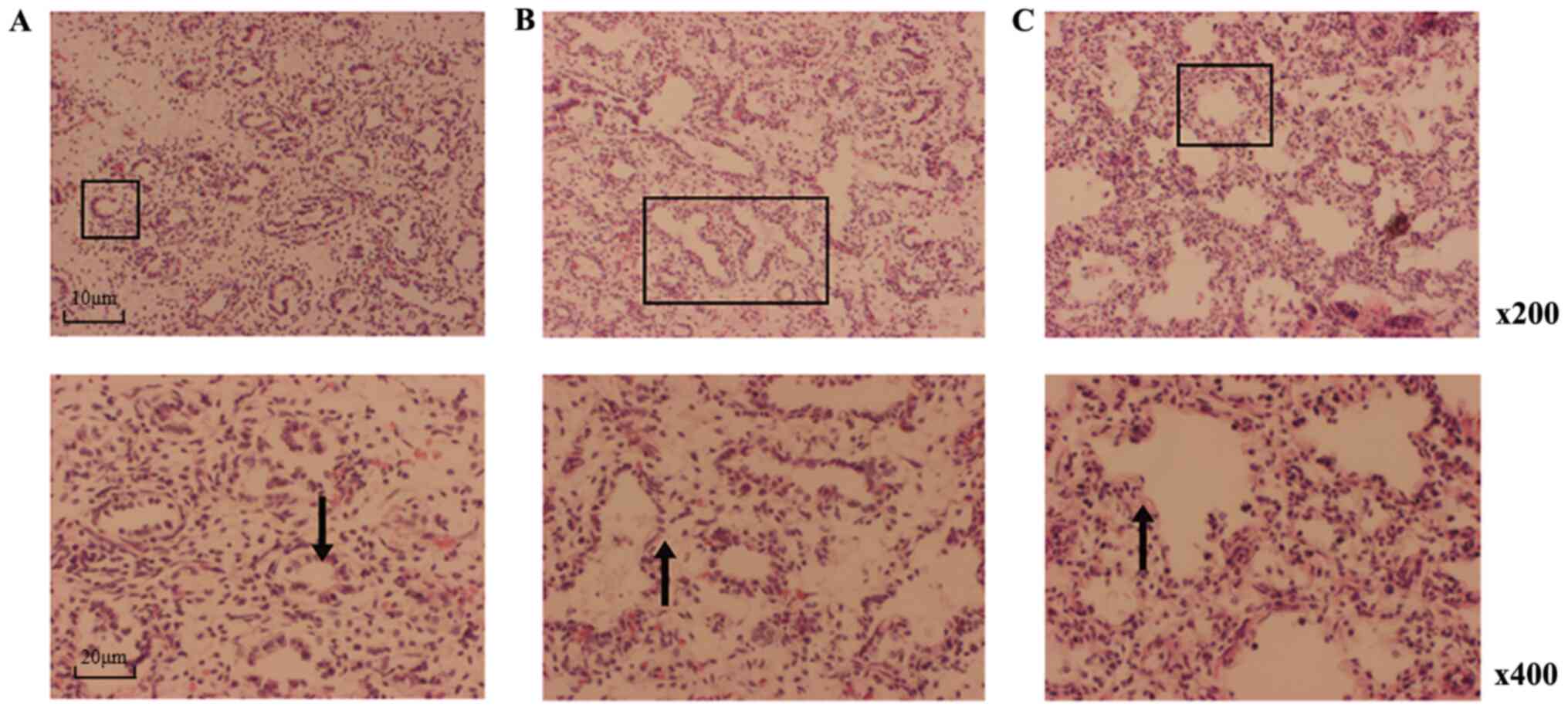
Histology
In the S1 group, the bronchial tree extended to
numerous bronchioles, which were composed of epithelial cells. In a
single layer of cubic cells, cells were tall, columnar and arranged
in rings. In the S2 group, terminal bronchioles branched out into
greater numbers of respiratory bronchioles and cubic epithelial
cells appeared short and columnar compared with S1. A dilated
alveolar lumen, increased alveolar septa, thinner interstitium and
hyperplasia of capillaries were observed in the fetal lungs in the
S2 and S3 groups. Alveolar sacs formed in the S3 group and
pulmonary alveoli took shape. In S3, a greater number of alveolar
septa appeared compared with the S1 and S2 groups, and the
interstitium was thinner compared with the S1 and S2 groups. In S3,
epithelial cells had a cubic or flat shape (Fig. 1).
lncRNA microarray profiles
Affymetrix Human GeneChip was utilized to determine
the expression spectrum of lncRNAs during fetal lung development.
As a result, 687 lncRNAs were indicated to be differentially
expressed among the three groups (S1, S2 and S3) of fetal lung
tissue samples. According to these data, there were 34 upregulated
lncRNAs and 12 downregulated lncRNAs that were significantly
differentially expressed among all combinations of S1 vs. S2 vs. S3
(fold-change ≥1.5). Among the 687 differentially expressed lncRNAs,
39 downregulated and 202 upregulated lncRNAs were identified in the
S2 vs. S1 comparison (fold-change >2). Furthermore, 24 lncRNAs
were downregulated and 78 upregulated in the S3 vs. S2 comparison
(fold-change >2) and 77 downregulated and 535 upregulated
lncRNAs were identified in the S3 vs. S1 comparison (fold-change
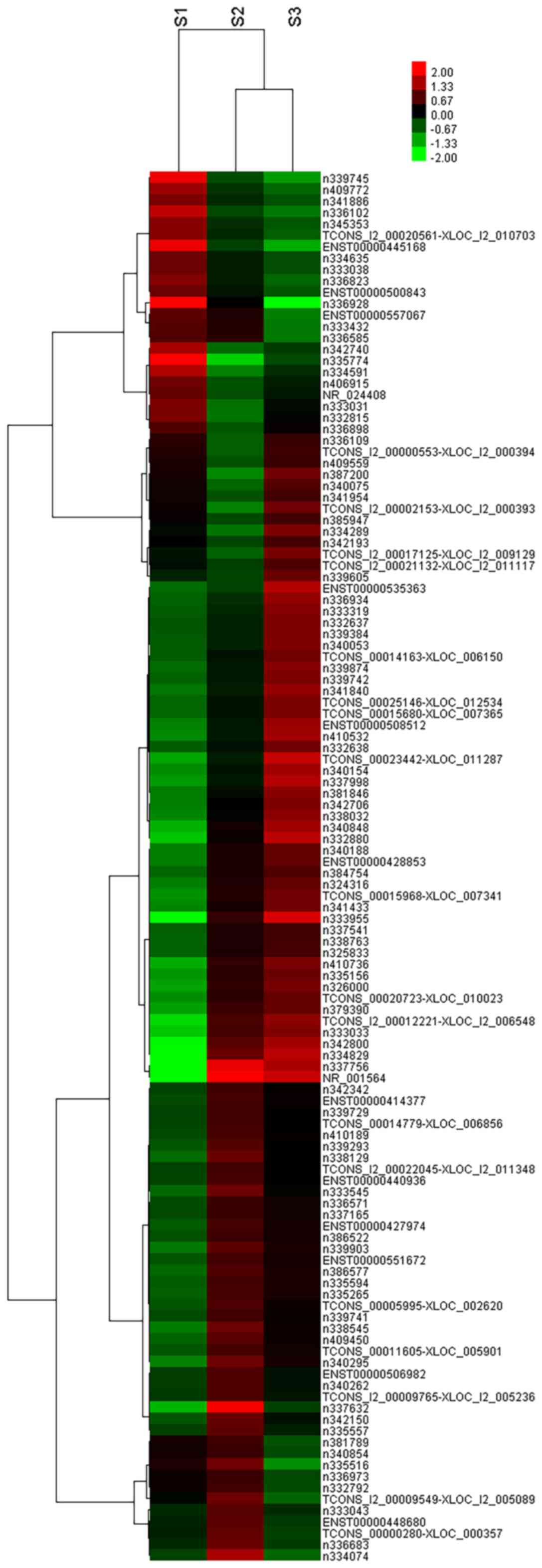
>2; Tables II and III). Hierarchical clustering was
performed in order to display distinguishable lncRNA expression
profiles among the groups. Taken together, these data were
consistent with the notion that different lncRNAs may be involved
in the different phases of lung development (Fig. 2).
 | Table IIDownregulated differentially
expressed lncRNAs between the three stages of fetal lung
development. |
Table II
Downregulated differentially
expressed lncRNAs between the three stages of fetal lung
development.
| A, S2 vs. S1 | |
|---|
| lncRNA | Fold-change |
|---|
| n335774 | 0.06935588 |
| n339745 | 0.182483003 |
| n334591 | 0.186404811 |
|
ENST00000445168 | 0.194416962 |
| n342740 | 0.223988722 |
| n336928 | 0.225022474 |
| n336102 | 0.234549609 |
| n335785 | 0.254695963 |
| n333031 | 0.272814078 |
| n332815 | 0.275703789 |
|
OTTHUMT00000312398 | 0.30750654 |
| n409772 | 0.320872488 |
| n338599 | 0.34498774 |
| n334031 | 0.346874815 |
| n406915 | 0.347614219 |
| n345353 | 0.365559716 |
|
ENST00000559553 | 0.370347627 |
| NR_024408 | 0.371285274 |
| n333438 | 0.376855146 |
|
TCONS_l2_00020561-XLOC_l2_010703 | 0.381374424 |
| B, S3 vs. S2 |
| lncRNA | Fold-change |
| n337632 | 0.186974574 |
| n336928 | 0.221248411 |
| n334074 | 0.231631923 |
| n335516 | 0.250004679 |
|
TCONS_l2_00009549-XLOC_l2_005089 | 0.322783111 |
| n336841 | 0.35573011 |
| n410723 | 0.39944174 |
| n336683 | 0.412094182 |
| n333958 | 0.414088201 |
| TCONS_00000280
XLOC_000357 | 0.415518323 |
| n336585 | 0.417651927 |
| n333432 | 0.424395128 |
| n408121 | 0.425823065 |
|
ENST00000557067 | 0.428630673 |
|
ENST00000448680 | 0.4427573 |
| n381789 | 0.457561636 |
| n332792 | 0.469538382 |
| n333380 | 0.482586011 |
| n340854 | 0.486920451 |
| n335620 | 0.488461581 |
| C, S3 vs. S1 |
| lncRNA | Fold-change |
| n336928 | 0.049785865 |
|
ENST00000445168 | 0.108667618 |
| n339745 | 0.117414297 |
| n335774 | 0.144621906 |
| n336102 | 0.179262671 |
| n409772 | 0.243588174 |
| n338817 | 0.26669321 |
| n336823 | 0.27863222 |
| n335785 | 0.281131886 |
|
TCONS_l2_00020561-XLOC_l2_010703 | 0.283204481 |
| n342740 | 0.288809679 |
| n345353 | 0.295015569 |
| n334591 | 0.295785862 |
|
ENST00000557067 | 0.303030728 |
| n336841 | 0.322103669 |
| n333432 | 0.322647779 |
| n341886 | 0.322920291 |
| n336585 | 0.333848393 |
| NR_039890 | 0.343190138 |
|
ENST00000500843 | 0.344062843 |
 | Table IIIUpregulated differentially expressed
lncRNAs between the 3 stages of fetal lung development. |
Table III
Upregulated differentially expressed
lncRNAs between the 3 stages of fetal lung development.
| A, S2 vs. S1 |
|---|
| lncRNA | Fold-change |
|---|
| NR_001564 | 47.08876564 |
| n337756 | 32.68860714 |
| n337632 | 10.19552386 |
| n339163 | 9.001615862 |
| NR_003349 | 8.523677449 |
| n334829 | 8.462693991 |
| NR_003347 | 8.142292223 |
| NR_003298 | 7.369409383 |
| NR_003314 | 7.246287266 |
| NR_003303 | 6.652354265 |
| NR_003355 | 6.62124723 |
| NR_003359 | 6.399445927 |
| n342800 | 6.291303669 |
| NR_003297 | 6.236367731 |
| n333955 | 5.766172512 |
| NR_001291 | 5.567478185 |
| NR_003308 | 5.567478185 |
| NR_003348 | 5.40086692 |
| NR_002974 | 5.268747348 |
| NR_002581 | 5.046048245 |
| B, S3 vs. S2 |
| lncRNA | Fold-change |
| n386326 | 4.555394507 |
| NR_029493 | 4.4777873 |
| NR_024065 | 4.340466177 |
| n382996 | 4.289896973 |
| NR_036677 | 4.269902123 |
| NR_026703 | 3.970979739 |
| n408293 | 3.920158536 |
| n387200 | 3.917295604 |
|
ENST00000535363 | 3.798942827 |
|
TCONS_l2_00002153-XLOC_l2_000393 | 3.759703782 |
| n334289 | 3.748358652 |
|
ENST00000459059 | 3.538294177 |
| n340146 | 3.474537616 |
|
TCONS_00023442-XLOC_011287 | 3.453784337 |
|
ENST00000379816 | 3.283596831 |
|
TCONS_l2_00017125-XLOC_l2_009129 | 3.263300854 |
| NR_004407 | 3.159558548 |
| NR_028502 | 3.103420093 |
| NR_002581 | 3.063370788 |
| n337998 | 3.052347039 |
| C, S3 vs. S1 |
| lncRNA | Fold-change |
| NR_001564 | 35.47473495 |
| n337756 | 23.19649621 |
| NR_002581 | 15.45791679 |
| n333955 | 14.17277149 |
| n334829 | 13.39219004 |
| NR_026703 | 10.89074695 |
| n339163 | 10.31981355 |
| n342800 | 9.393546857 |
| NR_004407 | 8.566180043 |
| n332880 | 8.210340188 |
|
TCONS_00023442-XLOC_011287 | 7.217168811 |
|
TCONS_l2_00012221-XLOC_l2_006548 | 7.062979945 |
| NR_002974 | 6.958339161 |
| NR_003349 | 6.388459125 |
| n340848 | 6.261155795 |
| n337998 | 6.218717152 |
| n333033 | 6.04561874 |
| NR_003347 | 5.995399957 |
| NR_002977 | 5.681622584 |
| NR_003314 | 5.591673021 |
As certain lncRNAs exhibited more significant
fold-changes among the three groups, 4 lncRNAs were selected based
on these data, including two downregulated lncRNAs (n336823 and
ENST00000445168; Table IV) and two
upregulated lncRNAs (n3408848 and n387037; Table V).
 | Table IVSpecific fold-changes of ≥1.5
consistently upregulated lncRNAs identified following
screening. |
Table IV
Specific fold-changes of ≥1.5
consistently upregulated lncRNAs identified following
screening.
| Comparison | n34048 | n387037 |
|---|
| S2 vs. S1 | 2.905918 | 1.571261 |
| S3 vs. S2 | 2.154622 | 1.509102 |
| S3 vs. S1 | 6.261156 | 2.371193 |
 | Table VSpecific fold-changes of ≥1.5
consistently downregulated lncRNAs identified following
screening. |
Table V
Specific fold-changes of ≥1.5
consistently downregulated lncRNAs identified following
screening.
| Fold-change | n336823 |
ENST00000445168 |
|---|
| S2 vs. S1 | 0.414761 | 0.194416 |
| S3 vs. S2 | 0.471789 | 0.458941 |
| S3 vs. S1 | 0.278632 | 0.108668 |
RT-qPCR
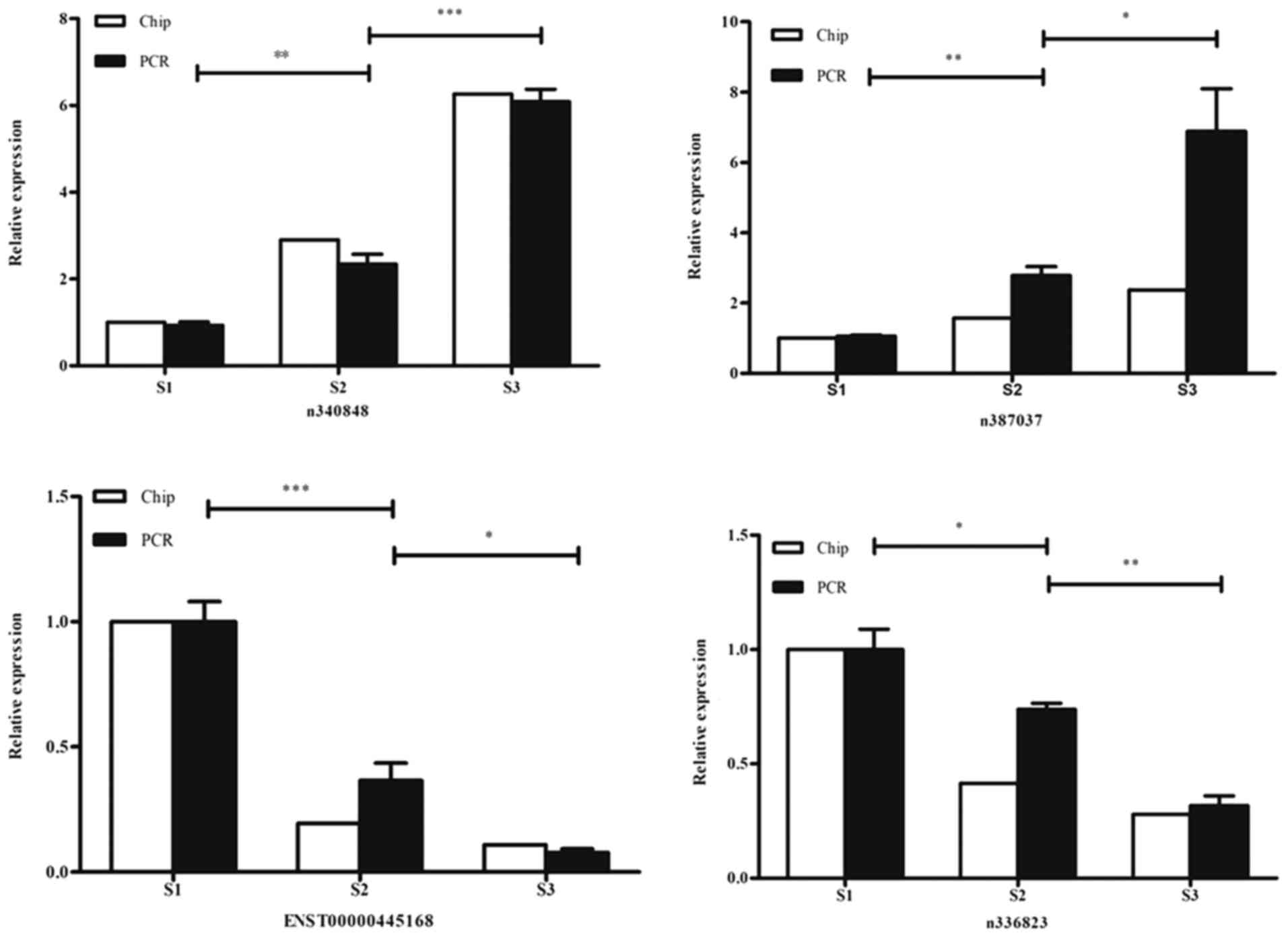
The expression levels of the selected lncRNAs were
confirmed by RT-qPCR. Among these differentially expressed lncRNAs,
n340848 and n387037 were indicated to be continuously increased in
expression with progression through the three phases of lung
development, while the expression levels of ENST00000445168 and
n336823 were reduced with progression. These results were
consistent with the GeneChip data obtained. The relative trends in
expression of these lncRNAs are presented in Fig. 3.
GO analysis and KEGG pathway
analysis
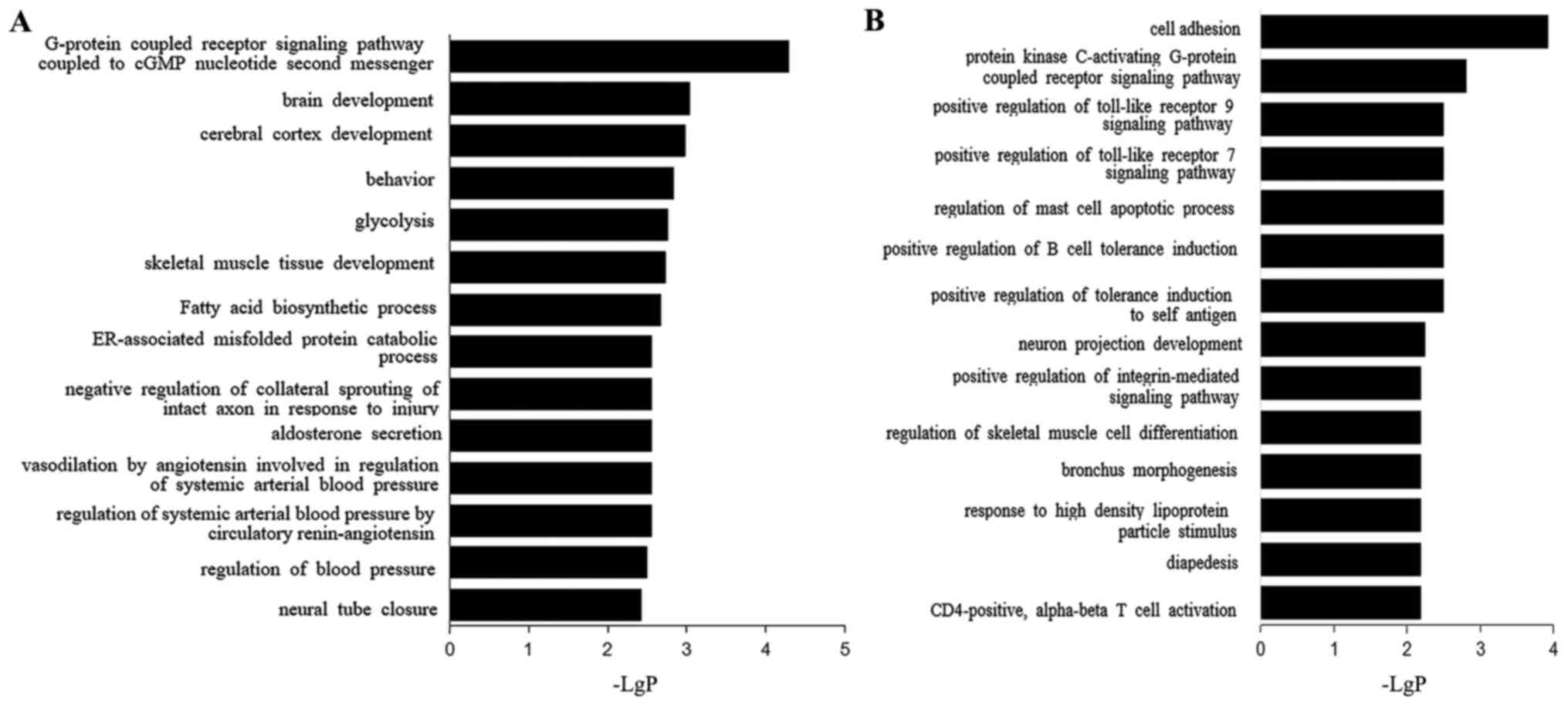
Upregulated transcripts were indicated to be
associated with the GO terms cell adhesion, hydrogen peroxide
decomposition and protein kinase C of a G protein-coupled receptor
signaling pathway (Fig. 4A). The
top three biological process terms associated with the
downregulated transcripts were G protein-coupled receptor signaling
pathway coupled to the cyclic guanosine monophosphate nucleotide
second messenger, brain development and cerebral cortex development
(Fig. 4A).
Additionally, KEGG enrichment analysis was performed
to investigate the possible roles of the lncRNA-associated
protein-coding genes. The most significant pathways enriched in the
set of upregulated protein-coding genes included cell adhesion
molecules, as well as adherens junction and glyceride metabolism
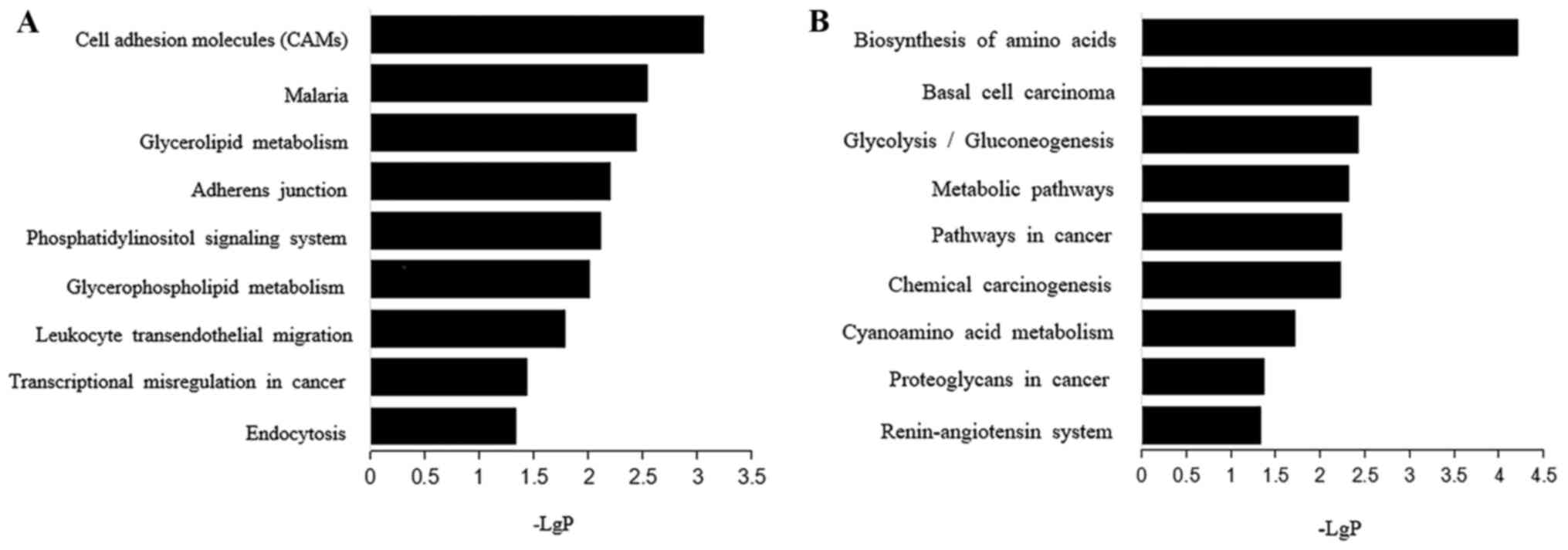
(Fig. 5A). Biosynthesis of an amino
acid, basal cell carcinoma and glycolysis/gluconeogenesis were the
most important pathways enriched in the set of down-regulated genes
(Fig. 5B).
Discussion
In the present study, 687 lncRNAs were identified to
be differentially expressed among three groups (embryonic periods
S1, S2 and S3) of human fetal lung tissue samples. The results
revealed 34 upregulated lncRNAs and 12 downregulated lncRNAs, which
were significantly differentially expressed among all combinations
of S1 vs. S2 vs. S3 (fold-change ≥1.5; P<0.05). Among these
differentially expressed lncRNAs, n340848, n387037, n336823 and
ENST00000445168 were then validated by RT-qPCR. These results were
consistent with the GeneChip results. GO enrichment analysis
revealed that the majority of the GO terms associated with these
genes belonged to the biological process category. The fact that
one lncRNA is able to target numerous genes suggests that lncRNAs
may be involved in a series of biological processes.
Among the four lncRNAs selected, lncRNA n340848 is
located on chromosome 6, overlapping with a gene called neural
precursor cell expressed developmentally down-regulated 9 (NEDD9).
The highest levels of NEDD9 mRNA and protein have been detected in
lungs and kidneys (33). NEDD9 has
been reported to act as a scaffold protein and is part of the
Crk-associated substrate family, which regulates protein complex
control of cell invasion and differentiation (34). n387037 is a 1,342-bp lncRNA with a
genomic overlap with the platelet and endothelial cell adhesion
molecule 1 (PECAM1) gene (5).
PECAM1 expression has been reported in almost all tissues and is
expressed at the highest levels in the placenta, lungs and fat
tissues (35). It has been
indicated as a novel therapeutic target in neonatal respiratory
distress syndrome and ventilator-induced lung injury (36). LncRNA n336823 is 26,063 bp long and
overlaps with patched 1, which encodes a member of the patched
family of proteins and a component of the hedgehog (HH) signaling
pathway (37). HH signaling is
crucial for embryonic development and tumorigenesis (38). Numerous studies have indicated that
activation of the HH signaling pathway is associated with the
progression of multiple solid tumors, including lung cancer
(1,39).
lncRNA ENST0000445168 (also known as 02038-202),
encoded on chromosome 3, is an intergenic lncRNA, whose
transcription occurs entirely within the genomic interval between
two adjacent protein-coding genes (40,41).
These results indicated that the four lncRNAs mentioned above may
take part in the etiology and pathogenesis of disorders of neonatal
lung development; however, further research is required for
confirmation.
Numerous studies have indicated that various lncRNAs
are involved in different biological events (38,42,43).
lncRNA NR_033925, also known as forkhead box (FOX)F1-AS1 or
FOXF1-adjacent non-coding developmental regulatory RNA, is encoded
upstream of FOXF1(44). It is
highly expressed in human lungs and has an important effect on the
development of the heart and gastrointestinal tract (45). lncRNA n409380, associated with the
FOXP2 gene, promotes embryonic development and the proliferation of
lung epithelial cells (46). lncRNA
n335087 and lncRNA n339275 overlap with gene TGF-β receptor 2 and
are involved in lung development-associated tracheal morphogenesis
and leukocytic protein phosphorylation (17).
Certain studies have addressed the specific
expression patterns or function of lncRNAs in lung development.
Herriges et al (47)
screened 363 lncRNAs in the lung and foregut endoderm and indicated
that they were spatially associated with transcription factors
across the genome. It has been reported that lncRNAs in the lungs
are located near genes of transcription factors, including NK2
homeobox 1 (NKX2.1), GATA binding protein 6, FOXA2 and FOXF1, which
are essential in foregut and lung development (48,49).
One of these lncRNAs, NKX2.1-associated non-coding intergenic RNA
(NANCI), performs an important function in lung development by
acting upstream of the critical transcription factor NKX2.1 and
downstream of Wnt/β-catenin signaling to regulate lung endoderm
gene expression and morphogenesis (16). Furthermore, a previous study
identified a feedback loop within the NANCI-NKX2.1 gene duplex to
explain how this subset acts as a rheostat to buffer the expression
of neighboring transcription factor genes, to maintain
tissue-specific cellular identity during development and postnatal
homeostasis (47).
In conclusion, the results of the present study
indicated that the lncRNA expression profile varies among different
phases of fetal lung development. These results provided certain
indications of the roles of lncRNAs in human fetal lung
development. In the future, functional verification, target gene
prediction and validation of the identified lncRNAs will be
performed in vitro and in vivo to provide novel
insight into the pathogenesis of neonatal pulmonary diseases.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 781270725).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JS, ZB, YY, XGZ and XYZ designed the experiments.
JS, ZB, WZ, CM, YS and QK performed the experiments, collected
data, generated the figures and interpreted the results. JS, ZB,
XGZ and XYZ wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
All procedures were approved by the Ethics Committee
of Nanjing Maternal and Child Health Hospital, Nanjing, China
[approval no. (2014) 74] and all patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Schmitz SU, Grote P and Herrmann BG:
Mechanisms of long noncoding RNA function in development and
disease. Cell Mol Life Sci. 73:2491–2509. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46.
2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Wang C, Wang L, Ding Y, Lu X, Zhang G,
Yang J, Zheng H, Wang H, Jiang Y and Xu L: LncRNA structural
characteristics in epigenetic regulation. Int J Mol Sci.
18(2659)2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wangyang Z, Daolin J, Yi X, Zhenglong L,
Lining H, Yunfu C and Xingming J: NcRNAs and Cholangiocarcinoma. J
Cancer. 9:100–107. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Jiang W, Liu Y, Liu R, Zhang K and Zhang
Y: The lncRNA DEANR1 facilitates human endoderm differentiation by
activating FOXA2 expression. Cell Rep. 11:137–148. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang P, Lu S and Mao H, Bai Y, Ma T, Cheng
Z, Zhang H, Jin Q, Zhao J and Mao H: Identification of biomarkers
for the detection of early stage lung adenocarcinoma by microarray
profiling of long noncoding RNAs. Lung Cancer. 88:147–153.
2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cheng HR, He SR, Wu BQ, Li DC, Hu TY, Chen
L and Deng ZH: Deep Illumina sequencing reveals differential
expression of long non-coding RNAs in hyperoxia induced
bronchopulmonary dysplasia in a rat model. Am J Transl Res.
9:5696–5707. 2017.PubMed/NCBI
|
|
11
|
Schmidt LH, Spieker T, Koschmieder S,
Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D,
Marra A, et al: The long noncoding MALAT-1 RNA indicates a poor
prognosis in non-small cell lung cancer and induces migration and
tumor growth. J Thorac Oncol. 6:1984–1992. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xu C, Yang M, Tian J, Wang X and Li Z:
MALAT-1: A long non-coding RNA and its important 3' end functional
motif in colorectal cancer metastasis. Int J Oncol. 39:169–175.
2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang B, Arun G, Mao YS, Lazar Z, Hung G,
Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C and Spector DL:
The lncRNA Malat1 is dispensable for mouse development but its
transcription plays a cis-regulatory role in the adult. Cell Rep.
2:111–123. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Cai C, Qiu J, Qiu G, Chen Y, Song Z, Li J
and Gong X: Long non-coding RNA MALAT1 protects preterm infants
with bronchopulmonary dysplasia by inhibiting cell apoptosis. BMC
Pulm Med. 17(199)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Herriges MJ, Swarr DT, Morley MP, Rathi
KS, Peng T, Stewart KM and Morrisey EE: Long noncoding RNAs are
spatially correlated with transcription factors and regulate lung
development. Genes Dev. 28:1363–1379. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Szafranski P, Dharmadhikari AV, Brosens E,
Gurha P, Kolodziejska KE, Zhishuo O, Dittwald P, Majewski T, Mohan
KN, Chen B, et al: Small noncoding differentially methylated
copy-number variants, including lncRNA genes, cause a lethal lung
developmental disorder. Genome Res. 23:23–33. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Moss TJ: Respiratory consequences of
preterm birth. Clin Exp Pharmacol Physiol. 33:280–284.
2006.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Pierro M, Ciarmoli E and Thébaud B:
Bronchopulmonary dysplasia and chronic lung disease: Stem cell
therapy. Clin Perinatol. 42:889–910. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kumar A and Bhatnagar V: Respiratory
distress in neonates. Indian J Pediatr. 72:425–428. 2005.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Schittny JC: Development of the lung. Cell
Tissue Res. 367:427–444. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Burri PH: Fetal and postnatal development
of the lung. Annu Rev Physiol. 46:617–628. 1984.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Copland I and Post M: Lung development and
fetal lung growth. Paediatr Respir Rev. 5 (Suppl A):S259–S264.
2004.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Roth-Kleiner M and Post M: Similarities
and dissimilarities of branching and septation during lung
development. Pediatr Pulmonol. 40:113–134. 2005.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Burri PH: Structural aspects of postnatal
lung development-alveolar formation and growth. Biol Neonate.
89:313–322. 2006.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zoetis T and Hurtt ME: Species comparison
of lung development. Birth Defects Res B Dev Reprod Toxicol.
68:121–124. 2003.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hislop AA: Airway and blood vessel
interaction during lung development. J Anat. 201:325–334.
2002.PubMed/NCBI View Article : Google Scholar
|
|
28
|
WHO 2018: Medical management of abortion:
Available online: https://www.who.int/reproductivehealth/publications/medicalmanagement-abortion/en/.
|
|
29
|
Chen QJ, Hou SP, Meads C, Huang YM, Hong
QQ, Zhu HP and Cheng LN: EBM-CONNECT Collaboration: Mifepristone in
combination with prostaglandins for termination of 10-16 weeks'
gestation: A systematic review. Eur J Obstet Gynecol Reprod Biol.
159:247–254. 2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
ACOG Practice Bulletin No. 135:
Second-trimester abortion. Obstet Gynecol 121: 1394-1406, 2013.
|
|
31
|
Young RA: Biomedical discovery with DNA
arrays. Cell. 102:9–15. 2000.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Duff MO, Olson S, Wei X, Garrett SC, Osman
A, Bolisetty M, Plocik A, Celniker SE and Graveley BR: Genome-wide
identification of zero nucleotide recursive splicing in Drosophila.
Nature. 521:376–379. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chang JX, Gao F, Zhao GQ and Zhang GJ:
Expression and clinical significance of NEDD9 in lung tissues. Med
Oncol. 29:2654–2660. 2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gumina RJ, Kirschbaum NE, Rao PN, van
Tuinen P and Newman PJ: The human PECAM1 gene maps to 17q23.
Genomics. 34:229–232. 1996.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Villar J, Zhang H and Slutsky AS: Lung
repair and regeneration in ARDS: Role of PECAM1 and Wnt Signaling.
Chest. 155:587–594. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Jia Y, Wang Y and Xie J: The Hedgehog
pathway: Role in cell differentiation, polarity and proliferation.
Arch Toxicol. 89:179–191. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
McMillan R and Matsui W: Molecular
pathways: The hedgehog signaling pathway in cancer. Clin Cancer
Res. 18:4883–4888. 2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Bu D, Yu K, Sun S, Xie C, Skogerbø G, Miao
R, Xiao H, Liao Q, Luo H, Zhao G, et al: NONCODE v3.0: Integrative
annotation of long noncoding RNAs. Nucleic Acids Res. 40 (Database
Issue):D210–D215. 2012.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672.
2009.PubMed/NCBI View Article : Google Scholar
|
|
42
|
McDonel P and Guttman M: Approaches for
understanding the mechanisms of long noncoding RNA regulation of
gene expression. Cold Spring Harb Perspect Biol.
11(a032151)2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wu H, Yang L and Chen LL: The diversity of
long noncoding RNAs and their generation. Trends Genet. 33:540–552.
2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Schnatwinkel C and Niswander L: Nubp1 is
required for lung branching morphogenesis and distal progenitor
cell survival in mice. PLoS One. 7(e44871)2012.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Sauvageau M, Goff LA, Lodato S, Bonev B,
Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E,
Spence M, et al: Multiple knockout mouse models reveal lincRNAs are
required for life and brain development. Elife.
2(e01749)2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Mahlapuu M, Enerbäck S and Carlsson P:
Haploinsufficiency of the forkhead gene Foxf1, a target for sonic
hedgehog signaling, causes lung and foregut malformations.
Development. 128:2397–2406. 2001.PubMed/NCBI
|
|
47
|
Herriges MJ, Tischfield DJ, Cui Z, Morley
MP, Han Y, Babu A, Li S, Lu M, Cendan I, Garcia BA, et al: The
NANCI-Nkx2.1 gene duplex buffers Nkx2.1 expression to maintain lung
development and homeostasis. Genes Dev. 31:889–903. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Goss AM, Tian Y, Tsukiyama T, Cohen ED,
Zhou D, Lu MM, Yamaguchi TP and Morrisey EE: Wnt2/2b and
beta-catenin signaling are necessary and sufficient to specify lung
progenitors in the foregut. Dev Cell. 17:290–298. 2009.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Yang H, Lu MM, Zhang L, Whitsett JA and
Morrisey EE: GATA6 regulates differentiation of distal lung
epithelium. Development. 129:2233–2246. 2002.PubMed/NCBI
|