Introduction
Oesophageal cancer (EC) is a common gastrointestinal
tumour that can be caused by complex genetic variants or an
unhealthy lifestyle (1,2). In 2016, 16,910 new cases of EC and
15,910 deaths caused by EC were recorded in the United States
(3). Current treatments for EC,
including surgical resection and radiotherapy, have greatly
improved the prognosis of patients with EC; however, due to delayed
diagnosis and frequent metastasis, the 5-year survival rate (at
41.7%) remains unsatisfactory (4).
Therefore, identifying novel therapeutic targets for EC is crucial
to improve clinical outcomes and allow for earlier diagnosis.
MicroRNAs (miRNAs/miRs) are highly conserved, small
(18-22 nucleotides) RNA molecules (5). Emerging evidence has demonstrated that
miRNAs serve vital roles in various biological processes, such as
cell differentiation, viability, apoptosis and the cell cycle, and
a number of miRNAs regulate the progression of various types of
cancer, such as lung cancer and colon cancer (6,7). The
correlation between miRNAs and tumours was first reported in
2002(8). Several miRNAs, such as
miR-206(9), miR-203a (10) and miR-543(11), have been reported to be involved in
the progression of EC. Previous studies have also demonstrated that
miRNAs, which can function as tumour suppressors or
cancer-promoting factors, could be useful for predicting cancer
prognosis (12-15).
The potential of miR-15b has been evaluated in neuroblastoma
(12), as well as in colorectal
(13) and gastric cancer (14). In addition, Wang et al
(15) demonstrated that miR-15b is
sponged by long intergenic non-protein coding RNA-regulator of
reprogramming, which promotes the development of oesophageal
squamous cell carcinoma (ESCC). Based on The Cancer Genome Atlas
database, a previous study reported that miR-15b is a novel
biomarker for ESCC (16). The
results of the aforementioned studies suggested that miR-15b may
serve as a potential biomarker in EC.
The PI3K/AKT signalling pathway serves an important
role in regulating various cellular processes, including cell
proliferation, apoptosis, the cell cycle and epithelial-mesenchymal
transition (17-19).
Increasing evidence suggests that the PI3K/AKT signalling pathway
also serves a critical role during the progression of EC (20,21).
Sheng et al (20) reported
that protease-activated receptor-2 increases EC cell migration and
invasion by activating the PI3K/AKT signalling pathway. In
addition, peroxiredoxin 1 knockdown suppresses EC cell
proliferation and induces apoptosis by inactivating the PI3K/AKT
signalling pathway (21). However,
whether the PI3K/AKT signalling pathway is related to the malignant
behaviours regulated by miR-15b in EC is not completely
understand.
Materials and methods
Study subjects
In the present study, 74 patients (<60, n=30;
≥60, n=44) with EC who had undergone surgery and 74 healthy
volunteers (<60, n=30; ≥60, n=44) who had undergone a physical
examination between January 2016 and December 2018 were recruited
in Shandong Cancer Hospital (Jinan, China). Both patients and
healthy volunteer groups comprised of 43 males and 31 females. The
age range of the 74 patients was 35 to 66 years. The age range of
74 healthy volunteers matched the age distribution of the patient
group. Patients with EC had not been treated with radiotherapy or
chemotherapy prior to surgery. Written informed consent was
obtained from all participants. The present study was approved by
the Ethics Committee of Shandong Cancer Hospital (approval no.
2017024).
Isolation of PBMCs and cell lines
Following a 12-h fast, peripheral blood was
collected into EDTA anticoagulant tubes. PBMCs were extracted using
Ficoll-Hypaque (450 x g, 20˚C for 20 min), washed by HBSS and
resuspended in PBS (1.0x106 cells/ml).
Human EC cell lines (EC109 and TE10) were obtained
from The Cell Bank of Type Culture Collection of the Chinese
Academy of Sciences and cultured in RPMI-1640 medium (Invitrogen;
Thermo Fisher Scientific, Inc.) containing 10% FBS (Invitrogen;
Thermo Fisher Scientific, Inc.). HEEpiCs were obtained from
ScienCell Research Laboratories, Inc. and cultured in EpiCM2 medium
containing 10% FBS, 5% EGF and 5% penicillin/streptomycin
(ScienCell Research Laboratories, Inc.). Cells were incubated at
37˚C in a humidified atmosphere containing 5% CO2.
Cell transfection
To adjust miR-15b expression levels in EC cells,
miR-15b mimics (forward: UAGCAGCACAUCAUGGUUUACA, reverse:
UAAACCAUGAUGUGCUGCUAUU), miR-15b inhibitor (forward:
UGUAAACCAUGAUGUGCUGCUA), and corresponding mimic negative control
(mimics NC, forward: UUCUCCGAACGUGUCACGUTT, reverse:
ACGUGACACGUUCGGAGAATT) and inhibitor negative control (inhibitor
NC, forward: CAGUACUUUUGUGUAGUACAA) were designed and synthesized
by Invitrogen; Thermo Fisher Scientific, Inc.. The non-interference
group was used as the blank control. Cells (2x106) were
transfected with the above agents (miR-15b mimics, mimics NC,
miR-15b inhibitor and inhibitor NC; 100 nM) for 48 h using the
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. To
determine the association between the PI3K/AKT signalling pathway
and miR-15b in EC, transfected EC cells (2x106) were
cultured in RPMI-1640 medium containing a PI3K/AKT signalling
pathway agonist, recilisib (10 µM; Sigma-Aldrich; Merck KGaA) at
37˚C for at least 48 h. The cells were then harvested to perform
the following experiments.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
After transfection for 24 h, total RNA was extracted
from cells using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). The concentration of total RNA was
measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher
Scientific, Inc.). Total RNA was reverse transcribed into cDNA at
42˚C for 45 min using the MiScript Reverse Transcription kit
(Qiagen GmbH). Subsequently, qPCR was performed using the MiScript
SYBR-Green PCR kit (Qiagen GmbH) and an ABI7500 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling
conditions were: 94˚C for 10 min, followed by 40 cycles at 94˚C for
10 sec, 60˚C for 20 sec and 72˚C for 1 min. The sequences of the
primers used in the present study are listed in Table I. Expression levels were quantified
using the 2-ΔΔCq method (22) and normalized to the internal
reference gene U6.
 | Table IPrimers used for reverse
transcription-quantitative PCR. |
Table I
Primers used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5'→3') |
|---|
| MircoRNA-15b | F:
TAGCAGCACATCATGGTTTACA |
| | R:
TGCGTGTCGTGGAGTC |
| U6 | F:
GCTTCGGCAGCACATATACTAAAAT |
| | R:
CGCTTCACGAATTTGCGTGTCAT |
Protein isolation and western
blotting
At 48 h post-transfection, total protein was
extracted from the cells using RIPA lysis buffer (Beyotime
Institute of Biotechnology) and denatured by incubation in 5X
loading buffer (Invitrogen; Thermo Fisher Scientific, Inc.) at 95˚C
for 10 min. BCA Protein Assay kit (Abcam) was used to detect the
protein concentrations. Protein (20 µg per lane) was separated via
SDS-PAGE (15%) and transferred to PVDF membranes, which were
blocked with 5% skimmed milk for 2 h at 25˚C. Subsequently, the
membranes were incubated overnight at 4˚C with primary antibodies
targeted against the following: Bcl-2 (1:1,000; cat. no. AF6139;
Affinity Biosciences), Bax (1:1,000; cat. no. AF0120; Affinity
Biosciences), PI3K (1:1,000; cat. no. AF6241; Affinity
Biosciences), phosphorylated (p)-PI3K (1:1,000; cat. no. AF3242;
Affinity Biosciences), AKT (1:1,000; cat. no. 4685; Cell Signaling
Technology, Inc.) and p-AKT (1:2,000; cat. no. 4060; Cell Signaling
Technology, Inc.). After washing three times with TBST (Tween-20;
0.05%) for 5 min, the membranes were incubated with horseradish
peroxidase-conjugated secondary antibody anti-mice immunoglobulin G
(1:5,000; cat. no. 14708; Cell Signaling Technology, Inc.) for 1 h
at 25˚C. GAPDH (1:1,000; cat. no. MA5-15738; Thermo Fisher
Scientific, Inc.) was used as the internal reference. The membranes
were developed using Chemiluminescence reagents (Thermo Fisher
Scientific, Inc.) with a Gel-Pro analyzer (version 4.0; Media
Cybernetics, Inc.).
MTT assay
The effect of miR-15b on EC cell viability was
assessed by performing the MTT assay. At 24 h post-transfection,
cells were collected and seeded (2x103 cells/well) into
96-well plates. At the indicated time point, 20 µl MTT reagent (5
mg/ml; Sigma-Aldrich; Merck KGaA) was added to each well and
incubated at 37˚C with 5% CO2 for 4 h. DMSO (150 µl;
Sigma-Aldrich; Merck KGaA) was added to each well and the optical
density at 450 nm was measured using an ELX800 microplate reader
(Perkin Elmer, Inc.).
Flow cytometry analysis
The early apoptosis of EC cells was evaluated using
the Annexin V-PI apoptosis detection kit (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Briefly, 1-5x105 cells were resuspended in 500 µl
binding buffer and stained with 5 µl Annexin V-EGFP and PI at 4˚C
for 15 min in the dark. Subsequently, 400 µl binding buffer was
added to the cells and apoptosis was assessed using a FACScan flow
cytometer (version 2.0; Becton, Dickinson and Company).
Transwell invasion and migration
assays
Cell invasion was assessed using Transwell chambers
(Corning, Inc.) pre-coated (at 37˚C for 30 min) with
Matrigel® (BD Biosciences). Briefly, transfected EC
cells were resuspended in serum-free medium and 200 µl cell
suspension (1x105 cells) was placed in the upper
chamber. RPMI-1640 medium containing 10% FBS (600 µl) was added to
the lower chamber. Following incubation for 24 h at 37˚C with 5%
CO2, cells on the upper surface were removed using a
cotton swab. Invading or migratory cells were fixed at room
temperature for 20 min with 3.7% formaldehyde and stained with 0.1%
crystal violet (at 37˚C for 15 min). Stained cells were observed in
five randomly selected fields using a light microscope
(magnification, x200). To assess migration, the Transwell chambers
were not pre-coated with Matrigel® and the cell density
in the upper chamber was 3x104 cells/200 µl.
Statistical analysis
Data are presented as the mean ± SD. Statistical
analyses were performed using GraphPad Prism software (version 7.0;
GraphPad Software, Inc.). The Student's t-test was used to analyse
differences between two groups. One-way ANOVA followed by Tukey's
post hoc test was used to analyse differences among multiple
groups. The association between miR-15b expression levels and
clinical data of patients with EC were analysed using the
χ2 test. P<0.05 was considered to indicate a
statistically significant difference. The diagnostic analysis was
performed through ROC curve analysis with healthy controls as true
negative cases and EC patients as true positive cases. Each
experiment was performed at least three times.
Results
miR-15b expression levels are
decreased in PBMCs isolated from patients with EC
To explore the biological function of miR-15b in EC,
RT-qPCR was performed to measure miR-15b expression levels in PBMCs
isolated from patients with EC and healthy volunteers. The results
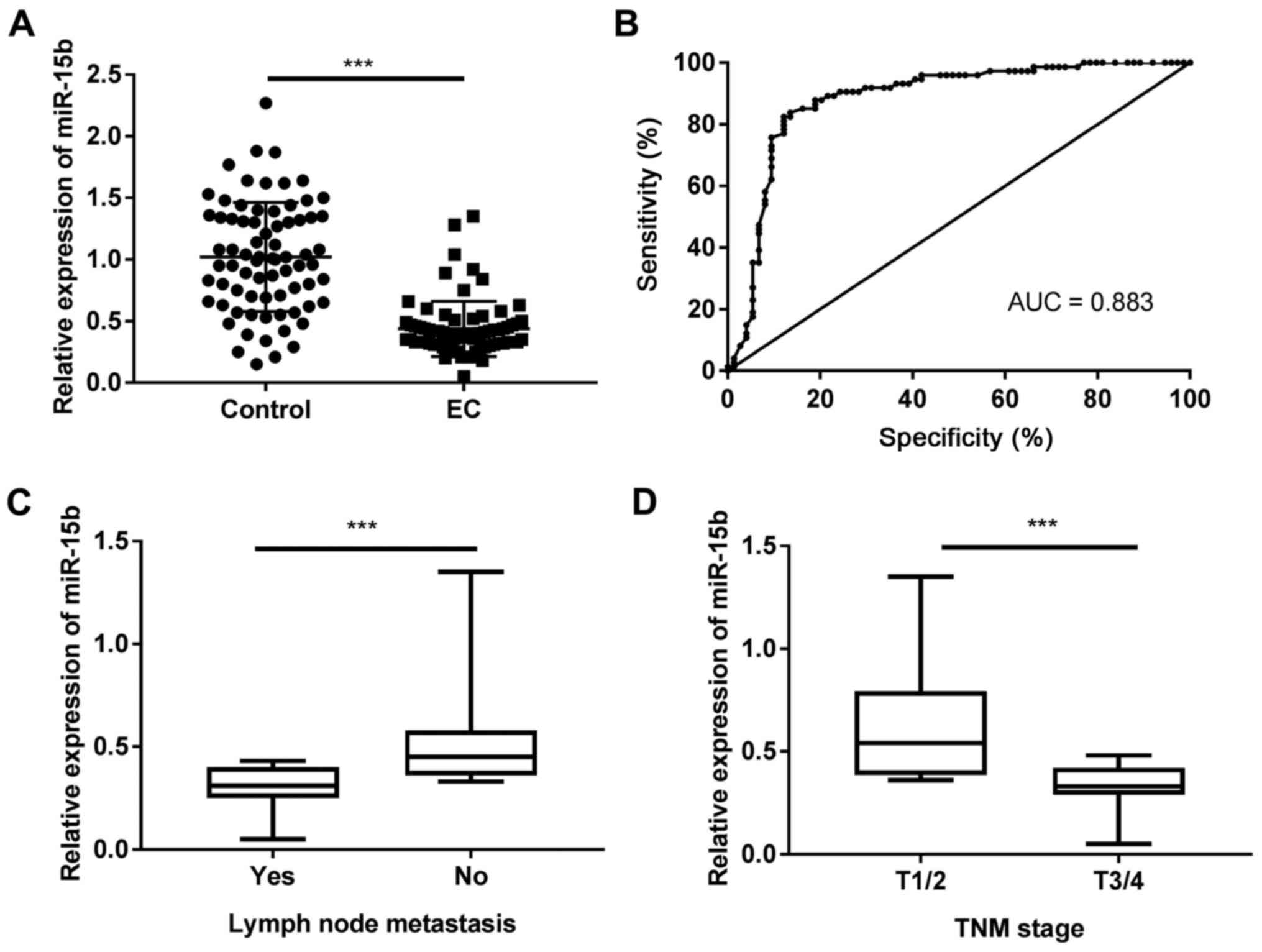
indicated that miR-15b expression levels were significantly lower
in patients with EC compared with healthy volunteers (P<0.001;
Fig. 1A). The receiver operating
characteristic curve revealed that the area under the curve of the
EC and control groups distinguished by miR-15b was 0.883 with a
cut-off value of 0.525, 82.43% sensitivity and 87.84% specificity
(Fig. 1B). Patients with EC were
then divided into two groups (high and low expression) according to
the median miR-15b expression level (0.46). The association between
miR-15b expression and various clinical parameters was determined
and the results are presented in Table
II. miR-15b expression was associated with tumour size, lymph
node metastasis, TNM stage (23),
fibrous membrane invasion and histologic grade (P<0.05).
Moreover, patients with EC with lymph node metastasis displayed
significantly lower miR-15b expression levels compared with
patients without lymph node metastasis (P<0.001; Fig. 1C). In addition, miR-15b expression
levels were significantly lower in patients with T3/4 stage EC
compared with patients with T1/2 stage EC (P<0.001; Fig. 1D).
 | Table IIAssociation between microRNA-15b
expression and clinical features of patients with oesophageal
cancer. |
Table II
Association between microRNA-15b
expression and clinical features of patients with oesophageal
cancer.
| | MicroRNA-15b
expression | |
|---|
| Variable | Total | Low | High | P-value |
|---|
| Age | | | | 0.344 |
|
<60 | 30 | 13 | 17 | |
|
≥60 | 44 | 24 | 20 | |
| Gender | | | | 0.48 |
|
Male | 43 | 20 | 23 | |
|
Female | 31 | 17 | 14 | |
| Tumour size | | | | 0.013a |
|
<2
cm | 24 | 7 | 17 | |
|
≥2 cm | 50 | 30 | 20 | |
| Lymph node
metastasis | | | | 0.034a |
|
Yes | 31 | 20 | 11 | |
|
No | 43 | 17 | 26 | |
| TNM stage | | | | 0.027a |
|
T1/2 | 25 | 8 | 17 | |
|
T3/4 | 49 | 29 | 20 | |
| Fibrous membrane
invasion | | | | 0.008b |
|
Yes | 47 | 29 | 18 | |
|
No | 27 | 8 | 19 | |
| Histologic
grade | | | | 0.049a |
|
Well | 26 | 9 | 17 | |
|
Modest | 30 | 15 | 15 | |
|
Poor | 18 | 13 | 5 | |
miR-15b reduces EC cell viability and
induces apoptosis
To determine the function of miR-15b in EC, the role
of miR-15b in EC cell viability and apoptosis was assessed. First,
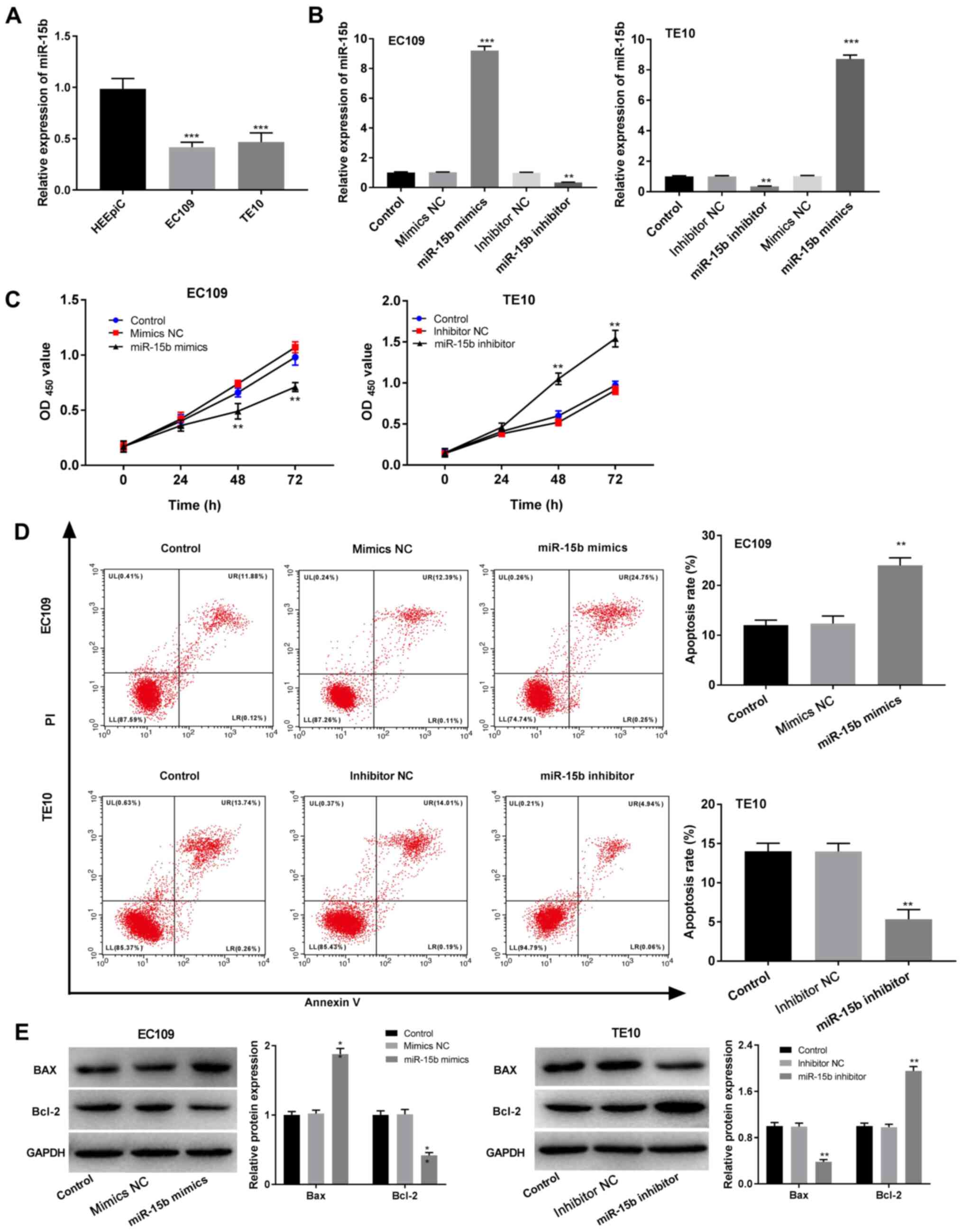
miR-15b expression levels in HEEpiC and EC cells (EC109 and TE10)
were measured via RT-qPCR. Compared with HEEpiC cells, EC109 and
TE10 cells expressed significantly lower levels of miR-15b
(P<0.001; Fig. 2A). After
transfection of miR-15b mimics/NC and miR-15b inhibitor/NC into
EC109 and TE10 cells, transfection efficiency was assessed via
RT-qPCR. The results indicated that miR-15b mimics significantly
increased miR-15b expression in contrast to the mimics NC group,
whereas miR-15b inhibitor significantly decreased miR-15b
expression in comparison to the inhibitor NC group (P<0.01;
Fig. 2B). The overexpression and
knockdown cell lines were then used in further experiments to
assess the function of miR-15b in EC. The MTT assay results
revealed that miR-15b overexpression significantly inhibited EC109
cell viability in contrast to the mimics NC group, whereas miR-15b
knockdown significantly enhanced TE10 cell viability in comparison
to the inhibitor NC group (P<0.01; Fig. 2C). Flow cytometry analysis suggested
that miR-15b overexpression significantly promoted EC109 cell
apoptosis, increased Bax expression and decreased Bcl-2 expression
in contrast to the mimics NC group, whereas miR-15b knockdown
displayed the opposite effects on TE10 cells (P<0.01; Fig. 2D and E).
miR-15b reduces EC cell migration and
invasion
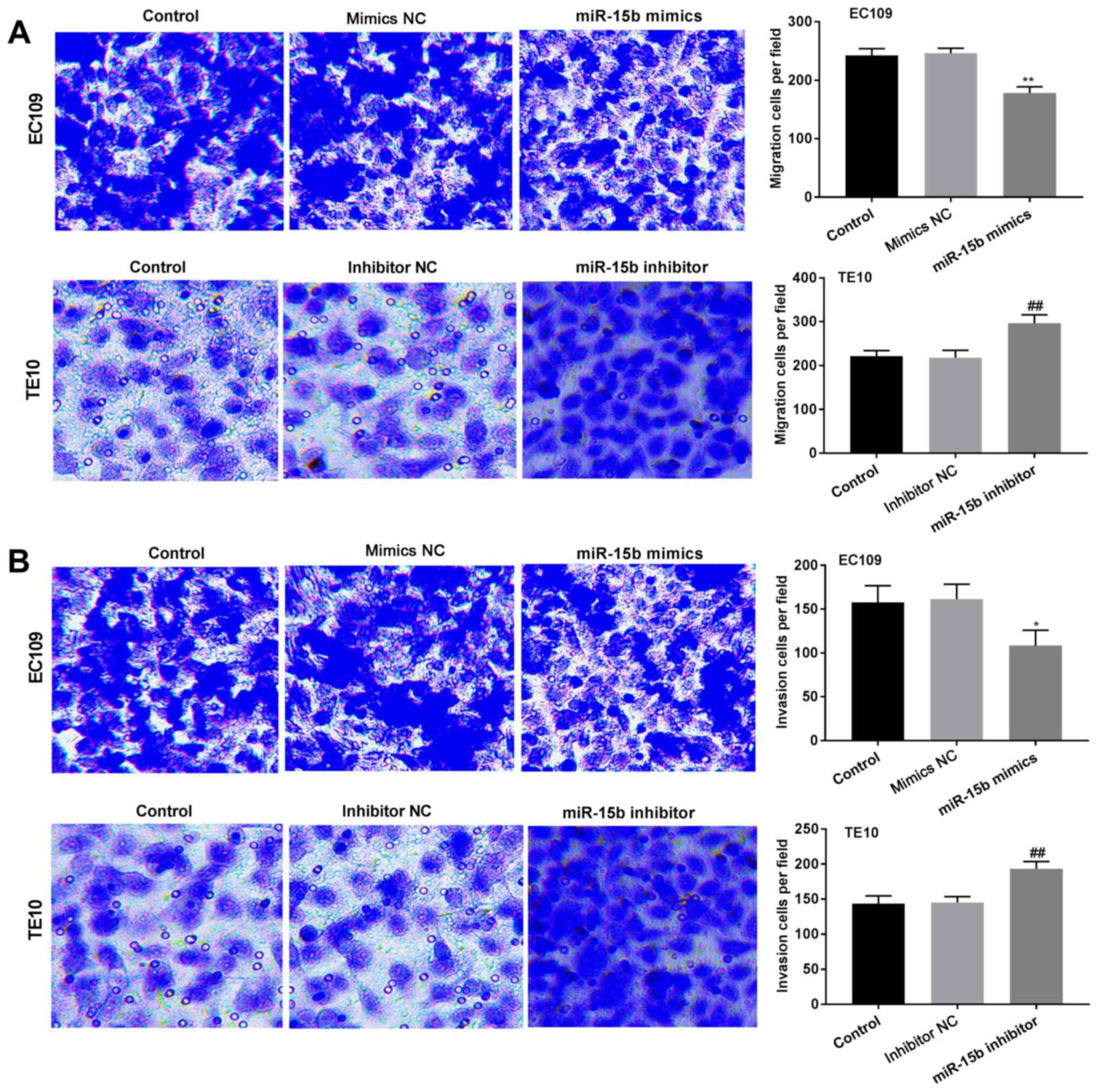
Transwell assays were performed to assess the
effects of miR-15b on EC cell migration and invasion. miR-15b
overexpression significantly decreased EC109 cell migration and
invasion (P<0.05), whereas miR-15b knockdown significantly
increased TE10 cell migration and invasion (P<0.01) compared
with the corresponding NC cells (Fig.
3).
miR-15b inhibits the PI3K/AKT
signalling pathway in EC
A previous study demonstrated that miR-15b
upregulation suppressed the PI3K/AKT signalling pathway in ovarian
cancer (23). In addition,
activation of the PI3K/AKT signalling pathway increased cancer cell
viability and inhibited tumour cell death (24,25).
To determine whether the effects of miR-15b in EC were associated
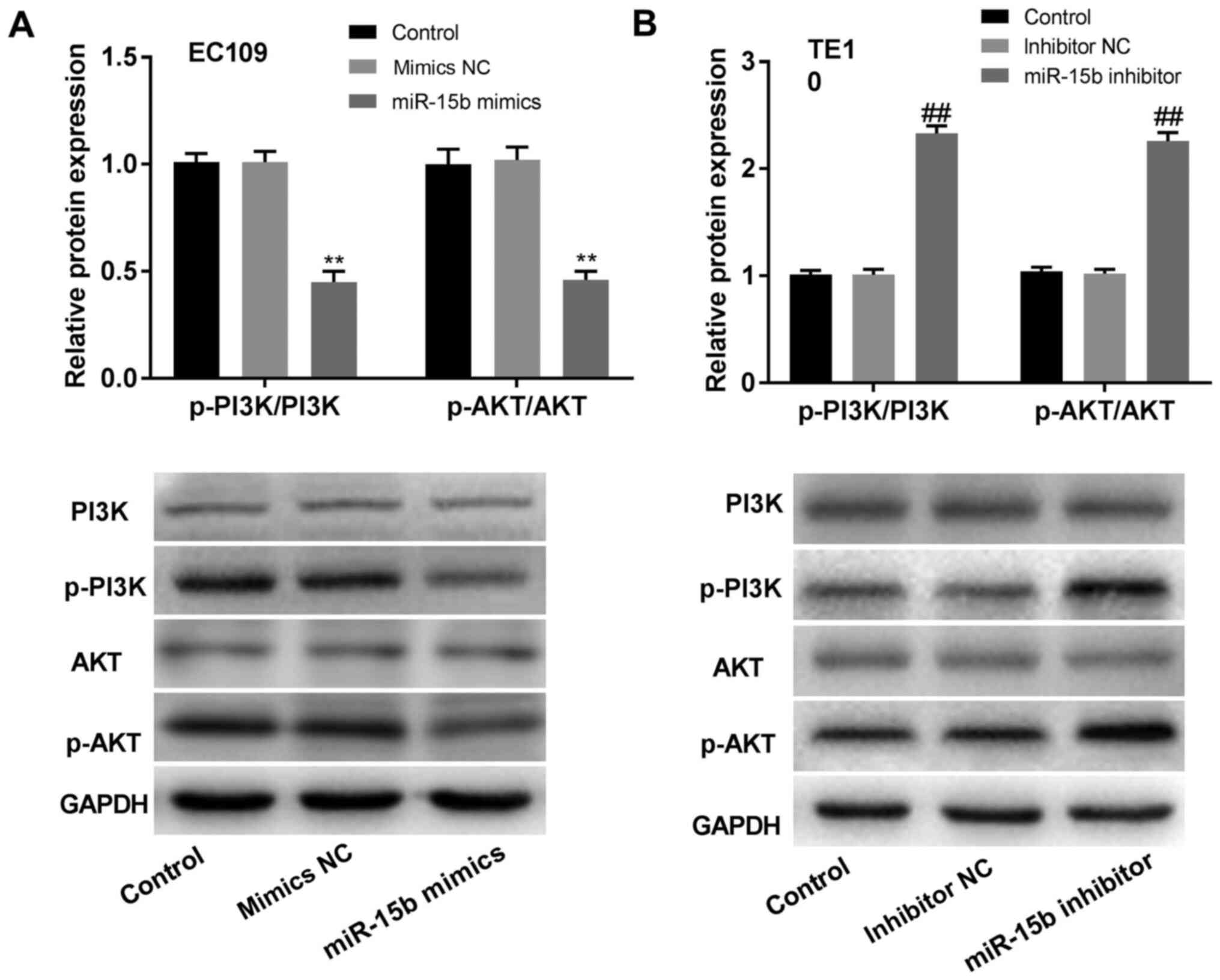
with the PI3K/AKT signalling pathway, western blotting was
performed. The results indicated that the expression levels of
p-PI3K and p-AKT were significantly reduced in
miR-15b-overexpression EC109 cells compared to those in the mimics
NC EC109 cells, but significantly increased in miR-15b-knockdown
TE10 cells compared with the inhibitor NC TE10 cells (P<0.01;
Fig. 4A and B). There were no significant differences
in the expression levels of PI3K and AKT between
miR-15b-overexpression/knockdown cells and the corresponding NC
cells (P>0.05; Fig. 4A and
B).
miR-15b suppresses the malignant
behaviours of EC cells by regulating the PI3K/AKT signalling
pathway
To further explore the relationship between miR-15b
and the PI3K/AKT signalling pathway during EC progression, TE10
cells were treated with recilisib, a PI3K/AKT signalling pathway
agonist. Recilisib treatment significantly increased the ratios of
p-PI3K/PI3K and p-AKT/AKT in TE10 cells compared with untreated
controls (P<0.01; Fig. 5A).
However, miR-15b mimic significantly reduced recilisib-induced
increases in p-PI3K/PI3K and p-AKT/AKT ratios in TE10 cells
compared with the mimics NC + recilisib group (P<0.01; Fig. 5A). Functional in vitro
experiments suggested that recilisib significantly promoted TE10
cell viability, migration and invasion, and attenuated TE10 cell
apoptosis compared with the control group (P<0.01; Fig. 5B-E). However, miR-15b mimic blocked
recilisib-induced cell viability, migration and invasion, and also
inhibited the inhibitory effects of recilisib on TE10 cell
apoptosis (P<0.01; Fig. 5B-E).
Compared with the control group, recilisib treatment significantly
decreased Bax expression and increased Bcl-2 expression
(P<0.01), which were reversed by miR-15b mimic (P<0.01;
Fig. 5F).
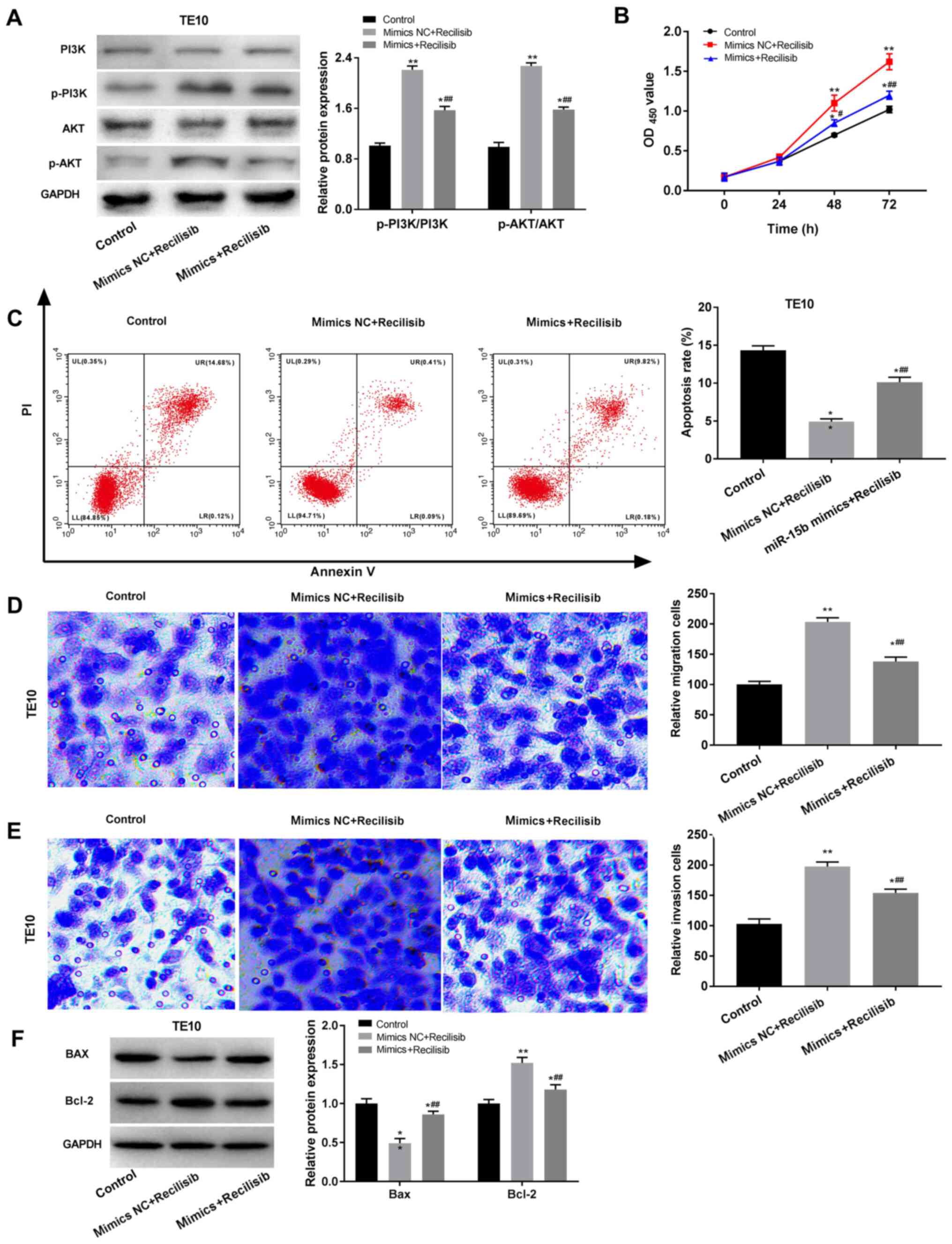 | Figure 5miR-15b suppresses oesophageal cancer
cell malignant behaviours by mediating the PI3K/AKT signalling
pathway. (A) Western blotting was performed to measure the
expression levels of PI3K/AKT signalling pathway-related proteins.
TE10 cell. (B) viability, (C) apoptosis, (D) migration and (E)
invasion (magnification, x200) were detected by performing MTT,
flow cytometry and Transwell assays, respectively. (F) Western
blotting was performed to measure the expression levels of Bax and
Bcl-2 in TE10 cells. *P<0.05, **P<0.01
vs. control; ##P<0.01 vs. mimics NC + Recilisib. miR,
microRNA; p, phosphorylated; NC, negative control; OD, optical
density; PI, propidium iodide. |
Discussion
EC is one of the most serious digestive
malignancies, resulting in unfavourable prognosis due to tumour
metastasis and recurrence (24,25).
The present study suggested that miR-15b expression levels were
significantly decreased in PBMCs isolated from patients with EC
compared with healthy volunteers. In addition, miR-15b expression
was significantly associated with tumour size, lymph node
metastasis, TNM stage, fibrous membrane invasion and histologic
grade in patients with EC. Li et al (16) identified miR-15b as a potential
biomarker for ESCC, and demonstrated that miR-15b was correlated
with tumour histological grade, TNM stage and overall survival.
miR-15b was also reported to be related to major clinical features
in other types of cancer, such as glioblastoma and cervical
carcinoma (26,27). The functional in vitro
experiments conducted in the present study suggested that miR-15b
overexpression inhibited EC cell viability, migration and invasion,
and promoted EC cell apoptosis compared with the corresponding NC
group. Moreover, the results indicated that the effects of miR-15b
were associated with the PI3K/AKT signalling pathway.
miRNAs have received increasing attention as
potential therapeutic targets for various tumours, including EC
(9,10,11,28).
miR-15b is an important RNA molecule that is located on chromosome
3q25(29). miR-15b levels in PBMCs
are correlated with baseline blood glucose concentrations and might
serve as a useful indicator of diabetes (30). A previous study demonstrated that
miR-15b displays an inhibitory effect on glioblastoma development
by inhibiting cell proliferation, cell cycle arrest and invasion
(31). In addition, miR-15b
upregulation significantly reduces neuroblastoma cell
proliferation, migration and invasion by directly targeting MYCN
proto-oncogene, bHLH transcription factor (12). Similarly, miR-15b was reported to
function as a tumour suppressor by reducing cellular malignant
behaviours in osteosarcoma (32)
and oral tongue squamous cell cancer (33). Lu et al (34) demonstrated that miR-15b inhibits
thyroid cancer cell proliferation, migration and invasion by
regulating Bcl-2. By contrast, miR-15b displays cancer-promoting
effects in other types of cancer. For example, miR-15b upregulation
improved resistance to sunitinib, and promoted cell survival and
invasion in renal cell carcinoma (35). Moreover, miR-15b-5p serves an
oncogenic role in hepatocarcinogenesis by mediating multiple
complex regulatory pathways (36).
Therefore, it was hypothesized that the function of miR-15b may
vary according to the type of cancer. In the present study, miR-15b
overexpression reduced EC cell viability, invasion and migration,
and promoted EC cell apoptosis compared with the corresponding NC
group, which suggested that miR-15b might serve a suppressive role
in EC.
Accumulating evidence suggests that the PI3K/AKT
signalling pathway is associated with the development of EC
(10,37). Activation of the PI3K/AKT signalling
pathway can increase cancer cell viability and inhibit tumour cell
death (38,39). A number of miRNAs are involved in
activating the PI3K/AKT signalling pathway. For example, Zheng
et al (40) reported that
miR-145 could promote apoptosis by suppressing the PI3K/AKT
signalling pathway in ESCC. The aggressive phenotype of EC is
stimulated by miR-508 via activation of the PI3K/AKT signalling
pathway (41). In addition, Pan
et al (42) demonstrated
that miR-205 can promote radioresistance by regulating the PI3K/AKT
signalling pathway in ESCC, which affects clinical outcomes.
miR-15b overexpression alleviated ovarian cancer by inhibiting the
PI3K/AKT signalling pathway (43).
By contrast, miR-15b participates in the development of peripheral
arterial disease by inactivating the PI3K/AKT signalling pathway
(44). Therefore, it was speculated
that the effects of miR-15b in EC may be related to the PI3K/AKT
signalling pathway. Mechanistically, AKT and PI3K serve vital roles
in PI3K/AKT/mTOR signalling (45)
as PI3K binds stably to the pleckstrin homology domain of AKT,
partially activating it (46). The
results of the present study suggested that miR-15b may block
activation of the PI3K/AKT signalling pathway in EC cells by
reducing p-PI3K and p-AKT expression levels. Some compounds, such
as PI3K activators or inhibitors, have been widely used to explore
the crosstalk between the PI3K/AKT signalling pathway and various
miRNAs in tumorigenesis (47,48).
The PI3K/AKT pathway activator recilisib was used in the present
study, and the results indicated that recilisib increased cell
viability, migration and invasion, and promoted apoptosis in TE10
cells. In addition, recilisib reversed the inhibitory effects of
miR-15b overexpression.
In conclusion, in the present study, miR-15b
expression levels were significantly lower in PBMCs isolated from
patients with EC compared with PBMCs isolated from healthy
controls. In addition, miR-15b expression was associated with
various clinicopathological characteristics in patients with EC.
Functional in vitro experiments further indicated that
miR-15b may function as a suppressive factor in EC by inhibiting
the PI3K/AKT signalling pathway. In summary, the present study
improved the current understanding of the mechanism underlying EC
and suggested potential novel therapeutic targets for EC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL made substantial contributions to the conception
and design of the study. JL, HX, NW and MS made substantial
contributions to the acquisition, analysis and interpretation of
data, as well as the drafting and revision of the manuscript. All
authors agreed to be accountable for the work, read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
participants. The present study was approved by the Ethics
Committee of Shandong Cancer Hospital (Jinan, China; approval no.
2017024).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jia X, Liu P, Zhang M, Feng T, Tang H,
Tang Z, Zhao H and Jin T: Genetic variants at 6p21, 10q23, 16q21
and 22q12 are associated with esophageal cancer risk in a Chinese
Han population. Int J Clin Exp Med. 8:19381–19387. 2015.PubMed/NCBI
|
|
2
|
Oze I, Matsuo K, Wakai K, Nagata C, Mizoue
T, Tanaka K, Tsuji I, Sasazuki S, Inoue M and Tsugane S: Research
Group for the Development and Evaluation of Cancer Prevention
Strategies in Japan. Alcohol drinking and esophageal cancer risk:
An evaluation based on a systematic review of epidemiologic
evidence among the Japanese population. Jpn J Clin Oncol.
41:677–692. 2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Short MW, Burgers KG and Fry VT:
Esophageal cancer. Am Fam Physician. 95:22–28. 2017.PubMed/NCBI
|
|
4
|
Yang C, Shen S, Zheng X, Ye K, Sun Y, Lu Y
and Ge H: Long noncoding RNA HAGLR acts as a microRNA-143-5p sponge
to regulate epithelial-mesenchymal transition and metastatic
potential in esophageal cancer by regulating LAMP3. FASEB J.
33:10490–10504. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lowery AJ, Miller N, McNeill RE and Kerin
MJ: MicroRNAs as prognostic indicators and therapeutic targets:
Potential effect on breast cancer management. Clin Cancer Res.
14:360–365. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Guo L and Lu Z: The fate of miRNA* strand
through evolutionary analysis: Implication for degradation as
merely carrier strand or potential regulatory molecule? PLoS One.
5(e11387)2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531.
2004.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Du G, Zhou J, Cheng L, Ma X, Gui Y and Tan
B: High expression of miR-206 predicts adverse outcomes: A
potential therapeutic target for esophageal cancer. Comb Chem High
Throughput Screen. 22:599–611. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang L, Zhang Z, Yu X, Li Q, Wang Q, Chang
A, Huang X, Han X, Song Y, Hu J, et al: SOX9/miR-203a axis drives
PI3K/AKT signaling to promote esophageal cancer progression. Cancer
Lett. 468:14–26. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang L, Chen J, Wang L, Chen L, Du Z, Zhu
L, Cui M, Zhang M and Song L: Linc-PINT acted as a tumor suppressor
by sponging miR-543 and miR-576-5p in esophageal cancer. J Cell
Biochem. 120:19345–19357. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chava S, Reynolds CP, Pathania AS,
Gorantla S, Poluektova LY, Coulter DW, Gupta SC, Pandey MK and
Challagundla KB: MiR-15a-5p, miR-15b-5p, and miR-16-5p inhibit
tumor progression by directly targeting MYCN in neuroblastoma. Mol
Oncol. 14:180–196. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Marcuello M, Duran-Sanchon S, Moreno L,
Lozano JJ, Bujanda L, Castells A and Gironella M: Analysis of A
6-Mirna signature in serum from colorectal cancer screening
participants as non-invasive biomarkers for advanced adenoma and
colorectal cancer detection. Cancers (Basel).
11(1542)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yuan C, Zhang Y, Tu W and Guo Y:
Integrated miRNA profiling and bioinformatics analyses reveal
upregulated miRNAs in gastric cancer. Oncol Lett. 18:1979–1988.
2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang L, Yu X, Zhang Z, Pang L, Xu J, Jiang
J, Liang W, Chai Y, Hou J and Li F: Linc-ROR promotes esophageal
squamous cell carcinoma progression through the derepression of
SOX9. J Exp Clin Cancer Res. 36(182)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li CY, Zhang WW, Xiang JL, Wang XH, Li J
and Wang JL: Identification of microRNAs as novel biomarkers for
esophageal squamous cell carcinoma: A study based on the cancer
genome atlas (TCGA) and bioinformatics. Chin Med J (Engl).
132:2213–2222. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Liu Z, Chen M, Xie LK, Liu T, Zou ZW, Li
Y, Chen P, Peng X, Ma C, Zhang WJ and Li PD: CLCA4 inhibits cell
proliferation and invasion of hepatocellular carcinoma by
suppressing epithelial-mesenchymal transition via PI3K/AKT
signaling. Aging (Albany NY). 10:2570–2584. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yang J, Zhang L, Jiang Z, Ge C, Zhao F,
Jiang J, Tian H, Chen T, Xie H, Cui Y, et al: TCF12 promotes the
tumorigenesis and metastasis of hepatocellular carcinoma via
upregulation of CXCR4 expression. Theranostics. 9:5810–5827.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhang F, Li K, Yao X, Wang H, Li W, Wu J,
Li M, Zhou R, Xu L and Zhao L: A miR-567-PIK3AP1-PI3K/AKT-c-Myc
feedback loop regulates tumour growth and chemoresistance in
gastric cancer. EBioMedicine. 44:311–321. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sheng J, Deng X, Zhang Q, Liu H, Wang N,
Liu Z, Dai E and Deng Q: PAR-2 promotes invasion and migration of
esophageal cancer cells by activating MEK/ERK and PI3K/Akt
signaling pathway. Int J Clin Exp Pathol. 12:787–797.
2019.PubMed/NCBI
|
|
21
|
Song Y, Liu H, Cui C, Peng X, Wang C, Tian
X and Li W: Silencing of peroxiredoxin 1 inhibits the proliferation
of esophageal cancer cells and promotes apoptosis by inhibiting the
activity of the PI3K/AKT pathway. Cancer Manag Res. 11:10883–10890.
2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang J, Zhou Y, Jiang K, Shen Z, Ye Y and
Wang S: Evaluation of the seventh AJCC TNM staging system for
gastric cancer: A meta-analysis of cohort studies. Tumour Biol.
35:8525–8532. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shimada A, Tsushima T, Tsubosa Y, Booka E,
Takebayashi K, Niihara M, Isaka M, Ohde Y, Machida N, Onozawa Y, et
al: Validity of surgical resection for lymph node or pulmonary
recurrence of esophageal cancer after definitive treatment. World J
Surg. 43:1286–1293. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Steffen T, Dietrich D, Schnider A,
Kettelhack C, Huber O, Marti WR, Furrer M, Gloor B, Schiesser M,
Thierstein S, et al: Recurrence patterns and long-term results
after induction chemotherapy, chemoradiotherapy, and curative
surgery in patients with locally advanced esophageal cancer. Ann
Surg. 269:83–87. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Matos B, Bostjancic E, Matjasic A, Popovic
M and Glavac D: Dynamic expression of 11 miRNAs in 83 consecutive
primary and corresponding recurrent glioblastoma: Correlation to
treatment, time to recurrence, overall survival and MGMT
methylation status. Radiol Oncol. 52:422–432. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wen F, Xu JZ and Wang XR: Increased
expression of miR-15b is associated with clinicopathological
features and poor prognosis in cervical carcinoma. Arch Gynecol
Obstet. 295:743–749. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liu Z, Hu G, Zhao Y, Xiao Z, Yan M and Ren
M: Silence of cZNF292 suppresses the growth, migration, and
invasion of human esophageal cancer Eca-109 cells via upregulating
miR-206. J Cell Biochem. 121:2354–2362. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lovat F, Fassan M, Sacchi D, Ranganathan
P, Palamarchuk A, Bill M, Karunasiri M, Gasparini P, Nigita G,
Distefano R, et al: Knockout of both miR-15/16 loci induces acute
myeloid leukemia. Proc Natl Acad Sci USA. 115:13069–13074.
2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gottmann P, Ouni M, Saussenthaler S, Roos
J, Stirm L, Jähnert M, Kamitz A, Hallahan N, Jonas W, Fritsche A,
et al: A computational biology approach of a genome-wide screen
connected miRNAs to obesity and type 2 diabetes. Mol Metab.
11:145–159. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wang J, Liu H, Tian L, Wang F, Han L,
Zhang W and Bai YA: MiR-15b inhibits the progression of
glioblastoma cells through targeting insulin-like growth factor
receptor 1. Horm Cancer. 8:49–57. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Weng Y, Shen Y, He Y, Pan X, Xu J, Jiang
Y, Zhang Q, Wang S, Kong F, Zhao S, et al: The miR-15b-5p/PDK4 axis
regulates osteosarcoma proliferation through modulation of the
Warburg effect. Biochem Biophys Res Commun. 503:2749–2757.
2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Xin QL, Deng CL, Chen X, Wang J, Wang SB,
Wang W, Deng F, Zhang B, Xiao G and Zhang LK: Quantitative
proteomic analysis of mosquito C6/36 cells reveals host proteins
involved in Zika virus infection. J Virol. 91:e00554–17.
2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Lu Z, Wu Z, Hu J, Wei W, Ma B and Wen D:
MicroRNA-15 regulates the proliferation, migration and invasion of
thyroid cancer cells by targeting Bcl-2. J BUON. 24:2114–2119.
2019.PubMed/NCBI
|
|
35
|
Lu L, Li Y, Wen H and Feng C:
Overexpression of miR-15b promotes resistance to sunitinib in renal
cell carcinoma. J Cancer. 10:3389–3396. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Pan WY, Zeng JH, Wen DY, Wang JY, Wang PP,
Chen G and Feng ZB: Oncogenic value of microRNA-15b-5p in
hepatocellular carcinoma and a bioinformatics investigation. Oncol
Lett. 17:1695–1713. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Shi N, Yu H and Chen T: Inhibition of
esophageal cancer growth through the suppression of PI3K/AKT/mTOR
signaling pathway. Onco Targets Ther. 12:7637–7647. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yang SX, Polley E and Lipkowitz S: New
insights on PI3K/AKT pathway alterations and clinical outcomes in
breast cancer. Cancer Treat Rev. 45:87–96. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Janku F, Yap TA and Meric-Bernstam F:
Targeting the PI3K pathway in cancer: Are we making headway? Nat
Rev Clin Oncol. 15:273–291. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zheng TL, Li DP, He ZF and Zhao S: miR-145
sensitizes esophageal squamous cell carcinoma to cisplatin through
directly inhibiting PI3K/AKT signaling pathway. Cancer Cell Int.
19(250)2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Lin C, Liu A, Zhu J, Zhang X, Wu G, Ren P,
Wu J, Li M, Li J and Song L: miR-508 sustains phosphoinositide
signalling and promotes aggressive phenotype of oesophageal
squamous cell carcinoma. Nat Commun. 5(4620)2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Pan F, Mao H, Bu F, Tong X, Li J, Zhang S,
Liu X, Wang L, Wu L, Chen R, et al: Sp1-mediated transcriptional
activation of miR-205 promotes radioresistance in esophageal
squamous cell carcinoma. Oncotarget. 8:5735–5752. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Li GC, Qin XL, Song HH, Li YN, Qiu YY, Cui
SC, Wang YS, Wang H and Gong JL: Upregulated microRNA-15b
alleviates ovarian cancer through inhitbition of the PI3K/Akt
pathway by targeting LPAR3. J Cell Physiol. 234:22331–22342.
2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Sun Y, Gao Y, Song T, Yu C, Nie Z and Wang
X: MicroRNA-15b participates in the development of peripheral
arterial disease by modulating the growth of vascular smooth muscle
cells. Exp Ther Med. 18:77–84. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Park J, Feng J, Li Y, Hammarsten O, Brazil
DP and Hemmings BA: DNA-dependent protein kinase-mediated
phosphorylation of protein kinase B requires a specific recognition
sequence in the C-terminal hydrophobic motif. J Biol Chem.
284:6169–6174. 2009.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Szymonowicz K, Oeck S, Malewicz NM and
Jendrossek V: New insights into protein kinase B/Akt signaling:
Role of localized Akt activation and compartment-specific target
proteins for the cellular radiation response. Cancers (Basel).
10(78)2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Porta C, Paglino C and Mosca A: Targeting
PI3K/Akt/mTOR Signaling in cancer. Front Oncol.
4(64)2014.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Hu H, Wang F, Wang M, Liu Y, Wu H, Chen X
and Lin Q: FAM83A is amplified and promotes tumorigenicity in
non-small cell lung cancer via ERK and PI3K/Akt/mTOR pathways. Int
J Med Sci. 17:807–814. 2020.PubMed/NCBI View Article : Google Scholar
|