Introduction
The with no lysine (WNK; including WNK1, WNK2, WNK3
and WNK4) kinases are serine/threonine kinases lacking an ATP-bound
lysine in the catalytic domain subtype II (1). WNK4 is highly expressed in the kidney,
where it regulates diverse ion transporters and channels, including
the Na-Cl co-transporter (NCC) (2-4).
WNK4 appears to act as a molecular switch that can balance NaCl
reabsorption and K+ secretion, thereby maintaining blood
pressure. A mutation in WNK4 (D561A) is responsible for
pseudohypoaldosteronism type II (PHA II) (5), which is characterized by familial
hyperkalemia and salt-sensitive hypertension (6,7). Our
previous study reported that polymorphism of WNK4 was associated
with hypertension (8). Therefore,
it has been demonstrated that WNK4 is involved in the
pathophysiological processes of fluid and electrolyte perturbations
and hypertension. Transcriptional regulation and post-translational
modification contribute toward the regulation of protein abundance
in tissues and cells. Our previous study described several
mechanisms of WNK4 transcriptional regulation (9-11),
but little is known regarding its post-translational
modification.
Ubiquitination is one of the most important
post-translational modifications. It has been proposed that WNK4 is
a substrate-recognition protein of the Cullin 3-based E3 ligase
complex, which targets WNK4 for ubiquitylation and degradation. It
has also been reported that WNK4 protein may be bound by Kelch-like
3 (KLHL3) and Kelch-like 2 (KLHL2) (12,13).
Furthermore, impaired KLHL3 was suggested to mediate the
ubiquitination of WNK4, thereby causing hypertension (14). This suggests that the ubiquitination
and degradation of WNK4 serves important roles in its function.
However, the exact site of ubiquitination in the WNK4 protein has
remained unclear. In addition to ubiquitination, phosphorylation is
also a normal post-translational modification used in eukaryotic
cells to adjust and control the protein activity and function.
Ubiquitination and phosphorylation are usually relatively
independent (13,15), but sometimes do affect each other.
For example, it has been proposed that during hypoxia,
phosphorylation is required for Na,K-ATPase ubiquitination
(16). However, it has remained
unclear whether there is an interaction between ubiquitination and
phosphorylation in WNK4.
It is well known that high salt levels are the
primary cause of high blood pressure (17,18);
and the present study reported that mice fed a high-salt diet have
increased blood pressure as well as increased sodium excretion
(19). The purpose of the present
study was to determine how high salt regulates WNK4 expression. The
aim of the present study was to identify the site of ubiquitination
in the WNK4 protein and to discover the mechanism by which high
salt levels increase WNK4 protein through crosstalk between the
ubiquitination and phosphorylation of WNK4.
Materials and methods
Animals
Experiments were performed on 12-18-week-old C57BL/6
male mice (provided by the Department of Laboratory Animal Science
of China Medical University) weighing 25-30 g. A total of 30 male
mice were maintained and housed in stainless steel, wire-bottomed
cages on a 12-h light/dark cycle at an ambient temperature of
23±2˚C and 60% humidity. The mice were fed a diet containing either
a normal salt level (0.4% NaCl) or a high level of salt (4% NaCl)
for 2, 4, 6 and 8 weeks, and were then euthanized with
CO2 at an air replacement rate of 20%/min at the same
age (20 weeks). Water and food were provided to all animals ad
libitum. All animal experimental protocols were approved by the
Ethics Committee of Shengjing Hospital of China Medical
University.
Cell culture and stimulation
HEK293 cells were provided by the Department of
Genetics, China Medical University and cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.), supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.), 100 µg/ml penicillin and 100 µg/ml
streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37˚C in a
humidified atmosphere with 5% CO2 in air. Subsequently,
the cells were seeded onto a 6-well plate and treated with NaCl (40
mM extra NaCl added to complete medium for 72 h) or with mannitol
(80 mM for 72 h) as a control. The medium was replaced every 2
days. Mycoplasma testing was performed on this cell line to ensure
that the cells had not been contaminated by mycoplasma.
Construct of mutant plasmid vectors
and transfections
Expression plasmids for HA-tagged human WNK4 were
established as previously described (8). The following primers were used to
construct the deleted plasmid vectors: K1010 forward,
5'-TGAAGAGGGACCGCAGCTTGTTGGGCGTTT-3' and reverse,
5'-CAAGCTGCGGTCCCTCTTCAGAGATGG GCG-3'; K1023 forward,
5'-GACTTCATCCGAACCG GCTGAGC CTCTTCC-3' and reverse, 5'-CAGCCGGT
TCGGATGAAGTCACTTGGAAAC-3'; K1092 forward,
5'-AGATGATGGGGAACCCCAAGTTGGGGGCAG-3' and reverse,
5'-CTTGGGGTTCCCCATCATCTCCTTCCT CCTCA-3'; and S1022 forward,
5'-TTGGGCGTTTCCA AGTGACTTCAAAGGAAC-3' and reverse, 5'-GGCTCA
GCCGGTTCCTTTGAAGTCACTTGG-3'
All constructs were verified by
sequencing
HEK293 cells (3x105 cells/6-cm dish) were
transfected with the indicated amount of expression constructs
using the jetPRIME® transfection reagent
(Polyplus-transfection®). Cell transfection was
performed at 37˚C for 6 h, the transfection regent was removed and
fresh DMEM medium was added. At 48 h following cell transfection,
cells were used for subsequent experimentation.
Co-immunoprecipitation (Co-IP) and
western blotting (WB)
The renal samples were immediately frozen at -80˚C
after being removed from the mice. A total of 50 mg kidney tissue
was obtained and cut into pieces, followed by washing with 1 ml
PBS. Renal protein samples were prepared by homogenizing the frozen
tissues in RIPA lysis buffer (cat. no. P0013B; Beyotime
Biotechnology, Inc.) containing protease inhibitors (cat. no.
5871S; Cell Signaling Technology, Inc.). The cell proteins were
prepared in lysis buffer containing protease and phosphatase
inhibitors. The protein concentration was determined using the
Bradford method with 5% BSA as the washing reagent. For IP, 500 ug
protein samples were pre-incubated with a primary antibody against
ubiquitin (Ub; cat. no. 3933; dilution 1:1,000; Cell Signaling
Technology, Inc.), HA-tag (cat. no. 26183; dilution 1:1,000; Thermo
Fisher Scientific, Inc.) and Phospho-(Ser/Thr) (P-S/T; cat. no.
9631S; dilution 1:1,000; Cell Signaling Technology, Inc.) by
rotating (8 x g) at 4˚C overnight, followed by the addition of 20
µl protein A/G PLUS-Agarose (cat. no. sc-2003; Santa Cruz
Biotechnology, Inc.; ) and a further incubation with rotation (8 x
g, 37˚C) for 1 h. A total of 500 µg of each protein sample in 20 µl
protein A/G beads was collected and washed with lysis buffer three
times. Immunoprecipitates were resolved by 8% SDS-polyacrylamide
gel electrophoresis (PAGE) and analyzed by western blotting. For
western blot analysis, 50 µg protein samples were subjected to 8 or
10% SDS-PAGE and transferred onto polyvinylidene difluoride
membranes (BioRad Laboratories, Inc.). The membranes were blocked
with 5% dry skimmed milk in TBS containing 0.1% Tween-20 (in
phosphorylation experiments, the membranes were blocked with 5% BSA
buffer in TBS containing 0.1% Tween-20) and incubated with primary
antibodies against WNK4 (cat. no. 5713; dilution 1:1,000; Cell
Signaling Technology, Inc.), Phospho-(Ser/Thr) (P-S/T; cat. no.
9631S; dilution 1:1,000; Cell Signaling Technology, Inc.),
ubiquitin (Ub, cat. no. 3933; dilution 1:1,000; Cell Signaling
Technology, Inc.), HA-tag (cat. no. 26183; dilution 1:1,000; Thermo
Fisher Scientific, Inc.) and GAPDH (cat. no. 60004-1-Ig; dilution
1:10,000; Proteintech Group, Inc.), prior to being incubated with
horseradish peroxidase-conjugated secondary antibodies, goat
anti-rabbit IgG-HRP (cat. no. sc-2004; dilution 1:5,000; Santa Cruz
Biotechnology, Inc.) and goat anti-mouse IgG-HRP (cat. no. sc-2005;
dilution 1:5,000; Santa Cruz Biotechnology, Inc.) for 1 h at room
temperature. The results were visualized using an enhanced
chemiluminescence kit obtained from Thermo Fisher Scientific, Inc.
Protein levels were normalized using ImageJ software (version
1.8.0; National Institutes of Health).
Forecasting
The UbPred database (www.ubpred.org) was used to identify the
ubiquitination sites in the WNK4 protein. Potential phosphorylation
sites of WNK4 were predicted with Netphos2.0 (www.cbs.dtu.dk/services/NetPhos/) and
Musite software (Version 1.0; Digital Biology Laboratory,
Inc.).
Statistical analysis
Data are expressed as the mean ± standard error of
the mean of three independent experiments. Bar graphs were
constructed using GraphPad Prism 5 (Prism 5 for Windows; GraphPad
Software, Inc.). Student's unpaired two-tailed test or one-way
analysis of variance followed by Dunnett's test was used to assess
the statistical significance of differences among different groups
using the statistical software SPSS (version 17.0 for Windows;
SPSS, Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
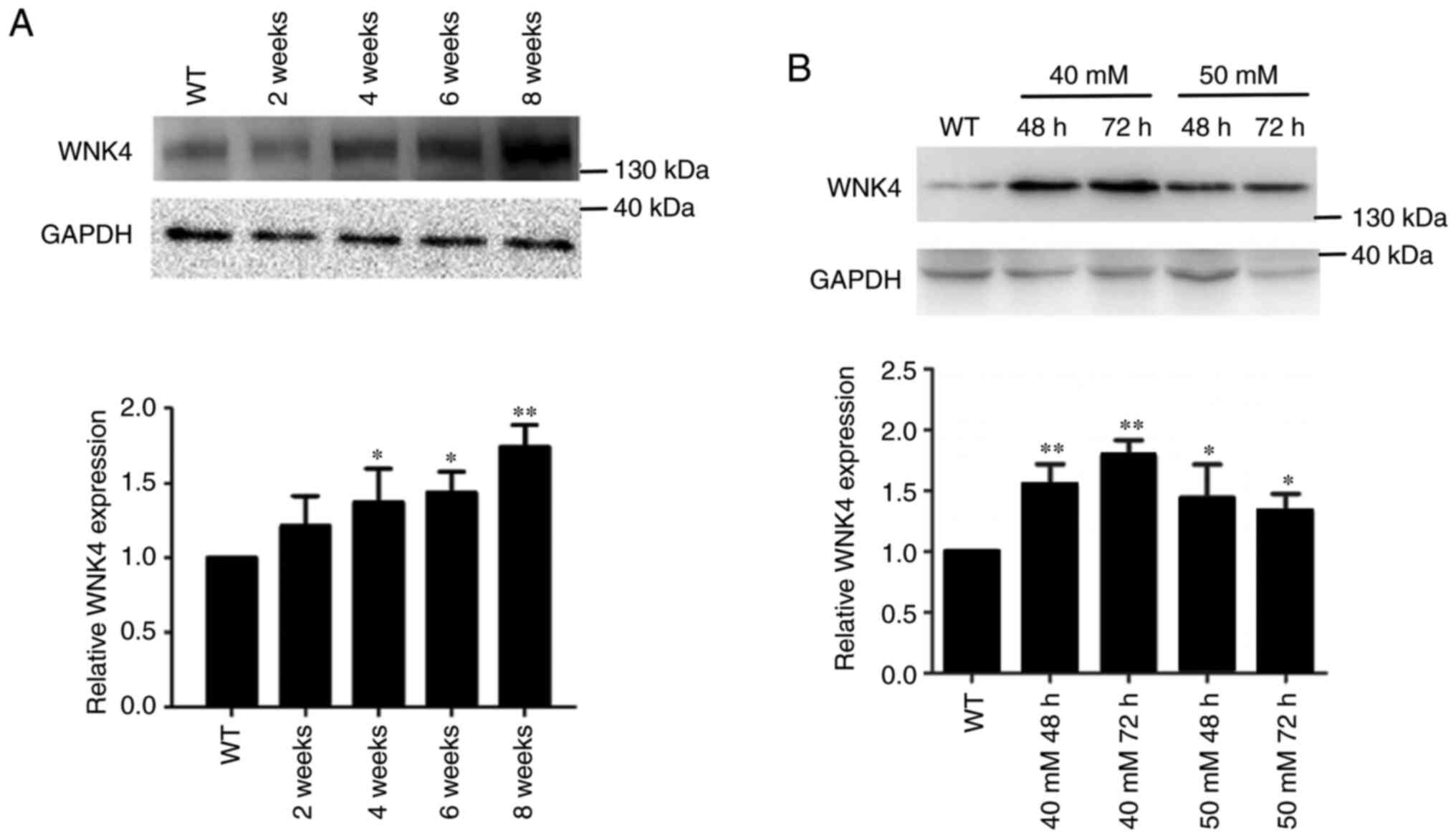
High salt increases the expression of
WNK4
Mice were fed a high-salt diet for 2, 4, 6 or 8
weeks, after which the expression of the WNK4 protein in the kidney
was determined by western blotting. The abundance of WNK4 in the
kidneys of high-salt-fed mice was significantly higher than that in
the kidney of wild-type (WT) mice (Fig.
1A). Since the level of WNK4 increased with increasing duration
of high-salt intake, samples were selected after 8 weeks of feeding
for the follow-up experiments. Similarly, as shown in Fig. 1B, WNK4 was upregulated in HEK293
cells following salt stimulation under different conditions (40 or
50 mM for either 48 or 72 h). As the increase in WNK4 protein was
most pronounced at 40 mM for 72 h, these conditions were used in
the subsequent experiments.
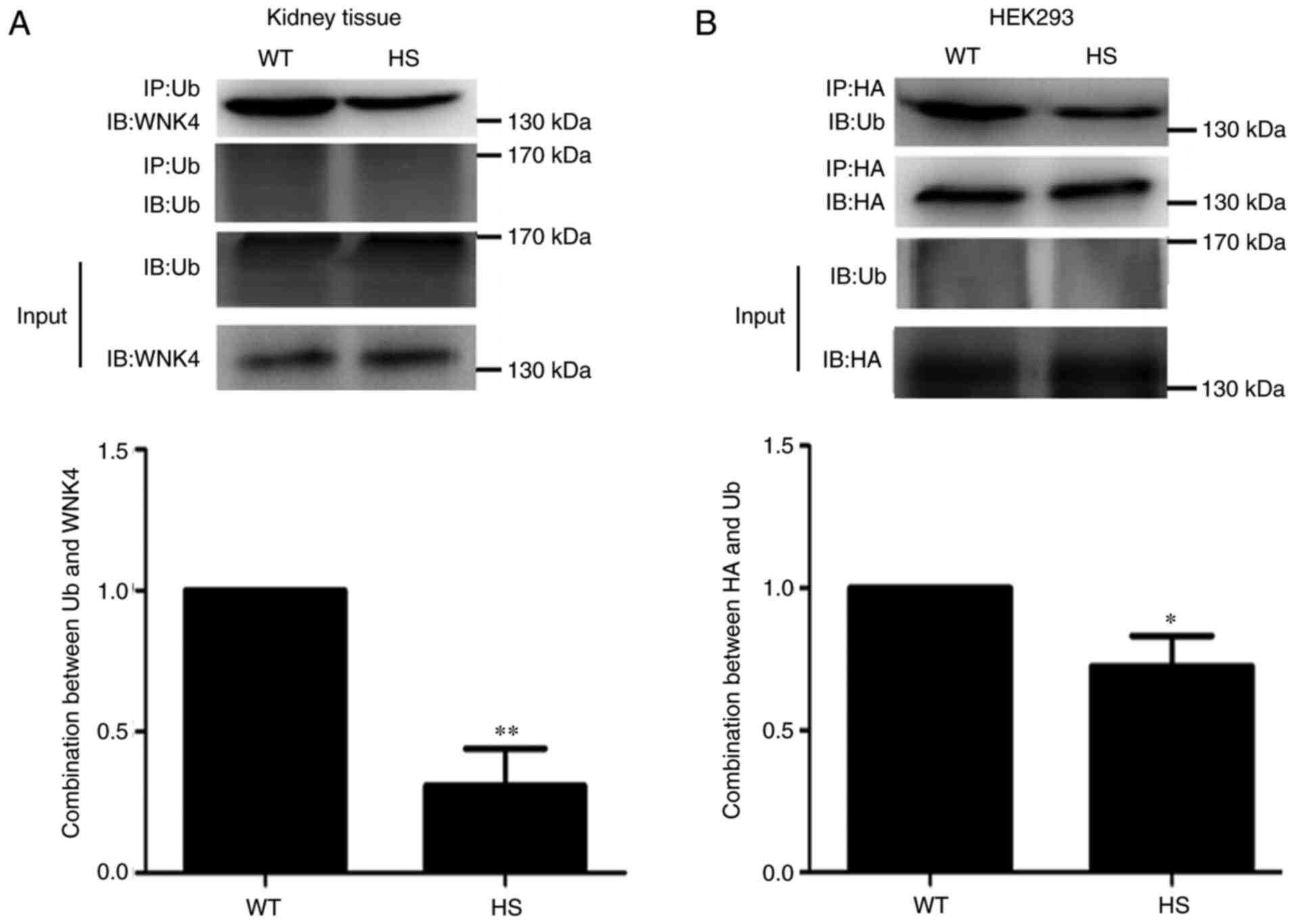
High-salt conditions reduces WNK4
ubiquitination
To determine the ubiquitination of WNK4 under
high-salt conditions, Co-IP was used to detect the interactions
between WNK4 and Ub. As shown in Fig.
2A, in high-salt-fed mice, the level of binding of Ub to WNK4
was significantly lower than that in WT mice. In addition, in
HEK293 cells transfected with the HA-tagged WNK4 expression vector,
the binding of Ub to WNK4 was lower under high-salt simulation than
in the control cells (Fig. 2B).
These findings suggested that the increased abundance of WNK4
induced by high salt was caused, at least in part, by a decrease in
WNK4 ubiquitination and degradation.
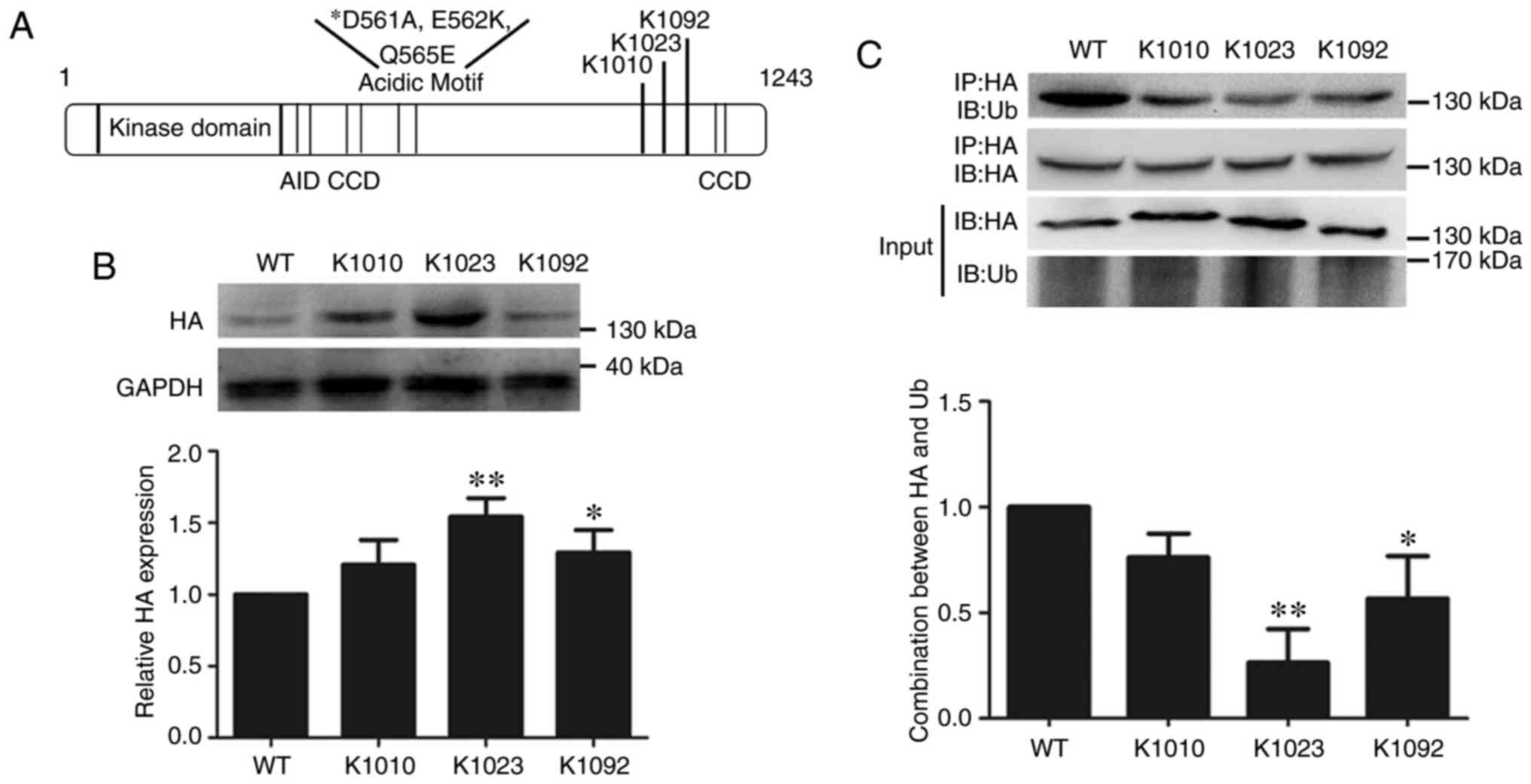
Identification of the ubiquitination
sites in the WNK4 protein
When attempting to determine the ubiquitination
sites in the WNK4 protein, three lysine residues (K1010, K1023 and
K1092) with high confidence scores were identified using UbPred
software, a sketch map of which is shown in Fig. 3A. Plasmids containing WNK4 with
mutations in each lysine residue (K1010, K1023 and K1092) were
constructed. WNK4 protein expression levels were greatly increased
when the K1023 lysine was deleted, but only slightly or not
increased when the K1092 or K1010 residue was deleted, compared
with the level for WT WNK4 (Fig.
3B). Next, Co-IP was used to determine the ability of mutated
WNK4 proteins to bind Ub, and the three mutated forms of WNK4 all
exhibited decreased binding to Ub, compared with that of WT WNK4,
particularly when the K1023 site was mutated (Fig. 3C). These results demonstrated that
the K1023 site was the most important for ubiquitination of the
WNK4 protein.
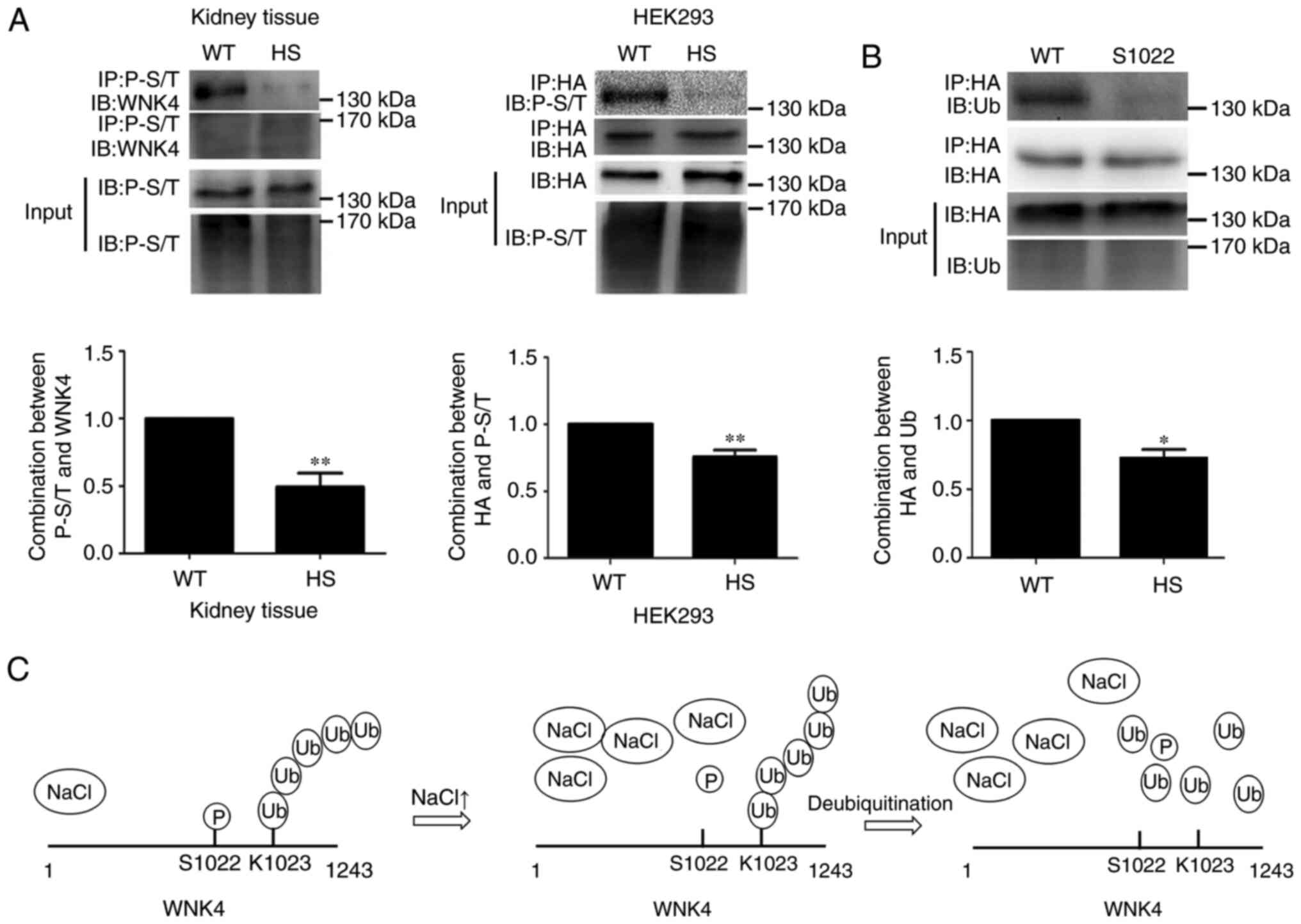
Phosphorylation of WNK4 is involved in
its ubiquitination
It is known that crosstalk can occur between
phosphorylation and ubiquitination under certain conditions
(16); therefore, we hypothesized
that phosphorylation of WNK4 may contribute toward its
ubiquitination under high-salt conditions. WNK4 phosphorylation was
detected by Co-IP, immunoprecipitated with anti-phosphorylated
serine or threonine antibody (P-S/T) and blotted with an
anti-HA-tag antibody. As shown in Fig.
4A, in high-salt-fed mice, WNK4 phosphorylation was
significantly decreased following high-salt stimulation, compared
with the level in unstimulated kidney; in HEK293 cells transfected
with HA-tagged WNK4 expression vector, the phosphorylation level of
WNK4 was also lower under high-salt simulation than in unstimulated
HEK293 cells. Potential phosphorylation sites of WNK4 were
predicted using Netphos2.0 and Musite software. Next, the putative
phosphorylation site, S1022, was selected to construct a
site-mutated WNK4 protein, as this site is located proximal to the
K1023 ubiquitination site. Using Co-IP, it was found that the
binding of Ub to the WNK4-S1022 mutant was significantly lower than
that to wild-type WNK4 (Fig. 4B),
implying that the phosphorylation of WNK4 at position S1022 affects
the binding between WNK4 and Ub. These findings demonstrated that
the decreased phosphorylation of WNK4 under high-salt conditions
may result in its decreased ubiquitination and this process is
shown in Fig. 4C.
Discussion
The present study identified that WNK4 protein
expression was increased under high-salt conditions in mice and
in vitro, which was consistent with a previous report
(20). Furthermore, it was
demonstrated that high salt downregulated phosphorylation at the
S1022 site, which in turn decreased the ubiquitination of WNK4
protein, leading to a decrease in its degradation and thus an
increase in its abundance.
The WNK4 protein possesses a kinase domain at its
N-terminus, which is abutted by an auto-inhibitory domain and then
two putative coiled-coil domains (21). After the first putative coiled-coil
domain, there is a region called the ‘acidic domain̓, which binds
to an E3 ligase complex, including Cullin3-KLHL3 and Cullin3-KLHL2
(22,23). Next, WNK4 protein connects to
ubiquitin molecules near specific lysine residues. Finally, the
ubiquitinated WNK4 protein is degraded by proteasomes. Numerous
studies have reported that mutant KLHL3 and Cullin3 molecules may
decrease the ubiquitination of WNK4 (24,25).
In addition, WNK4 mutations (E562K, Q565E and D561A) (5,13,26)
have been reported to inhibit the binding of WNK4 to KLHL3;
however, the exact site of ubiquitination in the WNK4 protein
remained unclear. In the present study, UbPred software was used to
predict three ubiquitination sites with high confidence (K1010,
K1023 and K1092) and constructed the respective mutant WNK4
expression plasmids. These experiments demonstrated that deletion
of the K1023 site resulted in increased protein levels of WNK4
associated with decreased ubiquitin binding, compared with that for
wild-type WNK4. Therefore, the present study was the first to
identify an important ubiquitination site for the WNK4 protein at
K1023.
In addition to ubiquitination, phosphorylation is
another important post-translational modification affecting protein
activity and function. Protein phosphorylation may occur at
multiple amino acids, but mostly at serine or threonine (27), and it has been reported that high
salt may decrease p38 and SPAK phosphorylation (28,29).
Recent evidence has suggested that there are three primary ways in
which phosphorylation regulates the ubiquitination of proteins. To
begin with, phosphorylation may positively and negatively regulate
the activity of the E3 ligase responsible for Ub transfer.
Additionally, phosphorylation promotes the identification of E3
ligase by producing a phosphodegron. Furthermore, phosphorylation
may influence ubiquitination by regulating the substrate/ligase
interaction at the level of subcellular compartmentalization
(30,31). The present study reported that the
phosphorylation of WNK4 was significantly decreased in HEK293 cells
following high-salt stimulation, compared with that in unstimulated
HEK293 cells, which is similar to reports suggesting that high salt
decreases SPAK phosphorylation (29). We hypothesized that the
phosphorylation of WNK4 contributes toward the process of WNK4
ubiquitination during high-salt stimulation. In certain proteins,
the ubiquitination and phosphorylation sites are thought to be
adjacent to each other (16,31,32).
Therefore, using the phosphorylation sites predicted by Musite and
Netphos2.0 software, the phosphorylation site that was nearest to
the K1023 ubiquitination site was selected to be mutated. It was
observed that WNK4-S1022 exhibited decreased binding to Ub,
suggesting that phosphorylation of S1022 was required for the
ubiquitination of WNK4. Although the results of the present study
demonstrated that high-salt conditions may decrease phosphorylation
of the WNK4 protein, it is not possible to determine whether this
was due to a decreased phosphorylation or increased
dephosphorylation. Further studies are required to investigate
whether phosphorylation or dephosphorylation involved in this
process.
In conclusion, the results of the present study
demonstrated that high-salt conditions would result in increased
expression of WNK4 protein. The mechanism behind this may involve
high salt inhibiting the ubiquitination of the WNK4 protein by
decreasing its phosphorylation, which then decreases the
degradation of WNK4 and eventually results in an increases level of
it. These findings suggested that salt-sensitive hypertension may
be successfully addressed through the targeted treatment of WNK4
protein ubiquitination or pharmacological inhibition of the WNK4
protein.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Key
Special Projects of China (grant nos. 2016YFC1000700 and
2016YFC1000702), the 345 Talent Project (grant no. M0298) and the
Free Researcher Fund (grant no. ME57) of Shengjing Hospital.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ, GL and YZ designed the research and revised the
manuscript. XZ, GL, YZ and JT performed the experiments and drafted
the manuscript. JT and SL performed data analysis. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Institutional Animal Care and Use Committee of Shengjing Hospital
and were conducted in strict accordance with the National
Institutes of Health guidelines for the use of experimental
animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shekarabi M, Zhang J, Khanna AR, Ellison
DH, Delpire E and Kahle KT: WNK kinase signaling in ion homeostasis
and human disease. Cell Metab. 25:285–299. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Subramanya AR, Yang CL, McCormick JA and
Ellison DH: WNK kinases regulate sodium chloride and potassium
transport by the aldosterone-sensitive distal nephron. Kidney Int.
70:630–634. 2006.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Tang BL: (WNK)ing at death: With-no-lysine
(Wnk) kinases in neuropathies and neuronal survival. Brain Res
Bull. 125:92–98. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hadchouel J, Ellison DH and Gamba G:
Regulation of renal rlectrolyte transport by WNK and SPAK-OSR1
kinases. Annu Rev Physiol. 78:367–389. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chu PY, Cheng CJ, Wu YC, Fang YW, Chau T,
Uchida S, Sasaki S, Yang SS and Lin SH: SPAK deficiency corrects
pseudohypoaldosteronism II caused by WNK4 mutation. PLoS One.
8(e72969)2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gordon RD: Syndrome of hypertension and
hyperkalemia with normal glomerular filtration rate. Hypertension.
8:93–102. 1986.PubMed/NCBI View Article : Google Scholar
|
|
7
|
O'Shaughnessy KM: Gordon Syndrome: A
continuing story. Pediatr Nephrol. 30:1903–1908. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sun Z, Li Y, Lu J, Ding Q, Liang Y, Shi J,
Li-Ling J and Zhao Y: Association of Ala589Ser polymorphism of WNK4
gene with essential hypertension in a high-risk Chinese population.
J Physiol Sci. 59:81–86. 2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li C, Li Y, Li Y, Liu H, Sun Z, Lu J and
Zhao Y: Glucocorticoid repression of human with-no-lysine (K)
kinase-4 gene expression is mediated by the negative response
elements in the promoter. Mol Endocrinol. 40:3–12. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li M, Zhao Y, Li Y, Li C, Chen F, Mao J
and Zhang Y: Upregulation of human with-no-lysine kinase-4 gene
expression by GATA-1 acetylation. Int J Biochem Cell Biol.
41:872–878. 2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang Y, Li C, Li W and Zhao Y: Estrogen
regulation of human with-no-lysine (K) kinase-4 gene expression
involves AP-1 transcription factor. Mol Cell Endocrinol.
332:140–148. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Schumacher FR, Sorrell FJ, Alessi DR,
Bullock AN and Kurz T: Structural and biochemical characterization
of the KLHL3–WNK kinase interaction important in blood pressure
regulation. Biochem J. 460:237–246. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Takahashi D, Mori T, Wakabayashi M, Mori
Y, Susa K, Zeniya M, Sohara E, Rai T, Sasaki S and Uchida S: KLHL2
Interacts with and ubiquitinates WNK kinases. Biochem Biophys Res
Commun. 437:457–462. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Susa K, Sohara E, Rai T, Zeniya M, Mori Y,
Mori T, Chiga M, Nomura N, Nishida H, Takahashi D, et al: Impaired
degradation of WNK1 and WNK4 kinases causes PHAII in mutant KLHL3
knock-in mice. Hum Mol Genet. 23:5052–5060. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ardito F, Giuliani M, Perrone D, Troiano G
and Lo Muzio L: The crucial role of protein phosphorylation in cell
signaling and its use as targeted therapy (Review). Int J Mol Med.
40:271–280. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Dada LA, Welch LC, Zhou G, Ben-Saadon R,
Ciechanover A and Sznajder JI: Phosphorylation and ubiquitination
are necessary for Na,K-ATPase endocytosis during hypoxia. Cell
Signal. 19:1893–1898. 2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Pilic L, Pedlar CR and Mavrommatis Y:
Salt-sensitive hypertension: Mechanisms and effects of dietary and
other lifestyle factors. Nutr Rev. 74:645–658. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Friso S, Carvajal CA, Fardella CE and
Olivieri O: Epigenetics and arterial hypertension: The challenge of
emerging evidence. Transl Res. 165:154–165. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wu J, Liu X, Lai G, Yang X, Wang L and
Zhao Y: Synergistical effect of 20-HETE and high salt on NKCC2
protein and blood pressure via ubiquitin-proteasome pathway. Hum
Genet. 132:179–187. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lai L, Feng X, Liu D, Chen J, Zhang Y, Niu
B, Gu Y and Cai H: Dietary salt modulates the sodium chloride
cotransporter expression likely through an aldosterone-mediated
WNK4-ERK1/2 signaling pathway. Pflugers Arch. 463:477–485.
2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang Z, Subramanya AR, Satlin LM,
Pastor-Soler NM, Carattino MD and Kleyman TR: Regulation of
large-conductance Ca2+-activated K+ channels
by WNK4 kinase. Am J Physiol Cell Physiol. 305:C846–C853.
2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kliuk-Ben Bassat O, Carmon V, Hanukoglu A,
Ganon L, Massalha E, Holtzman EJ, Farfel Z and Mayan H: Familial
hyperkalemia and hypertension (FHHt) and KLHL3: Description of a
family with a new recessive mutation (S553L) compared to a family
with a dominant mutation, Q309R, with analysis of urinary sodium
chloride cotransporter. Nephron. 137:77–84. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kasagi Y, Takahashi D, Aida T, Nishida H,
Nomura N, Zeniya M, Mori T, Sasaki E, Ando F, Rai T, et al:
Impaired degradation of medullary WNK4 in the kidneys of KLHL2
knockout mice. Biochem Biophys Res Commun. 487:368–374.
2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Susa K, Sohara E, Takahashi D, Okado T,
Rai T and Uchida S: WNK4 is indispensable for the pathogenesis of
pseudohypoaldosteronism type II caused by mutant KLHL3. Biochem
Biophys Res Commun. 491:727–732. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ibeawuchi SR, Agbor LN, Quelle FW and
Sigmund CD: Hypertension- causing mutations in Cullin3 protein
impair RhoA protein ubiquitination and augment the association with
substrate adaptors. J Biol Chem. 290:19208–19217. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ohta A, Schumacher FR, Mehellou Y, Johnson
C, Knebel A, Macartney TJ, Wood NT, Alessi DR and Kurz T: The
CUL3-KLHL3 E3 ligase complex mutated in Gordon's hypertension
syndrome interacts with and ubiquitylates WNK isoforms:
Disease-causing mutations in KLHL3 and WNK4 disrupt interaction.
Biochem J. 451:111–122. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Lee HK: Synaptic plasticity and
phosphorylation. Pharmacol Ther. 112:810–832. 2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Dinarello CA: Hyperosmolar sodium
chloride, p38 mitogen activated protein and cytokine-mediated
inflammation. Semin Dial. 22:256–259. 2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zeniya M, Sohara E, Kita S, Iwamoto T,
Susa K, Mori T, Oi K, Chiga M, Takahashi D, Yang SS, et al: Dietary
salt intake regulates WNK3-SPAK-NKCC1 phosphorylation cascade in
mouse aorta through angiotensin II. Hypertension. 62:872–878.
2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hunter T: The age of crosstalk:
Phosphorylation, ubiquitination, and beyond. Mol Cell. 28:730–738.
2007.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Filipčík P, Curry JR and Mace PD: When
worlds collide-mechanisms at the interface between phosphorylation
and ubiquitination. J Mol Biol. 429:1097–1113. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kelm KB, Huyer G, Huang JC and Michaelis
S: The internalization of yeast Ste6p follows an ordered series of
events involving phosphorylation, ubiquitination, recognition and
endocytosis. Traffic. 5:165–180. 2004.PubMed/NCBI View Article : Google Scholar
|