Introduction
Colorectal cancer (CRC) has been recognized as one
of the most common human cancer types and the most common causes of
cancer-related death worldwide (1,2).
Although marked progress has been made in CRC diagnosis and
treatment in the past decades, the prognosis of CRC remains rather
dismal due to metastasis and recurrence (3). Therefore, further exploration of the
potential mechanisms underlying CRC development is urgently
required to identify novel diagnostic markers and therapeutic
targets for CRC.
Long noncoding RNA (lncRNA) is a class of
transcripts with >200 nucleotides in length (4,5).
Accumulating evidence has indicated that lncRNAs have crucial roles
in various biological processes (6-8).
Furthermore, emerging studies have suggested that lncRNAs have
important roles in the initiation and progression of human cancers
(9,10). Numerous studies have indicated that
certain lncRNAs are involved in the development of CRC (11,12).
Long intergenic ncRNA (LINC)01535 is a novel lncRNA that has been
implicated in the development of esophageal squamous cell cancer
(13), osteosarcoma (14) and cervical cancer (15). However, to date, the biological
functions and molecular mechanisms of LINC01535 in CRC have
remained elusive. In the present study, the expression of LINC01535
in CRC tissues and in paired adjacent normal tissues was evaluated,
and the association of LINC01535 expression with
clinicopathological indicators and prognosis of CRC patients was
further examined. In addition, the biological roles of LINC01535 in
CRC cell proliferation, invasion and cisplatin (DDP) sensitivity
in vitro and tumor growth in vivo were investigated.
Finally, the interaction between LINC01535 and microRNA
(miRNA/miR)-761 was examined as part of elucidating the molecular
mechanisms of LINC01535 in CRC in detail. To the best of our
knowledge, the present study was the first to indicate the
oncogenic role of LINC01535 in the development of CRC. These
results offer novel insight into the progression and
chemoresistance of CRC and may promote the development of novel
anticancer therapeutic strategies for CRC.
Materials and methods
Clinical samples
A total of 24 pairs of CRC tissues and matched
adjacent normal tissues were sourced from patients undergoing
resection surgery at the Affiliated Dongtai Hospital of Nantong
University (Dongtai, China) between January 2012 and September
2015. None of the patients received any chemotherapy or radiation
therapy prior to specimen collection. All of these participants
provided written informed consent prior to sample collection. The
samples from resection surgery were rapidly frozen and stored in
liquid nitrogen until required. This study was approved by the
Ethics and Research Committees of the Affiliated Dongtai Hospital
of Nantong University (Dongtai, China).
Cell culture
Fetal human cells (FHC) from normal colonic mucosa
and the human CRC cell lines HT29, LoVo, SW480 and HCT116 were
purchased from the Institute of Biochemistry and Cell Biology of
the Chinese Academy of Sciences. Cells were authenticated by short
tandem repeat profiling. The FHC cells were maintained in Ham's F12
medium (45%) (Gibco; Thermo Fisher Scientific, Inc.), Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.)
(45%), 25 mM HEPES, 10 ng/ml cholera toxin, 0.005 mg/ml
transferrin, 0.005 mg/ml insulin, 100 ng/Ml hydrocortisone, 20
ng/ml human recombinant EGF (PHG0311; Thermo Fisher Scientific,
Inc.) and fetal bovine serum (FBS, 10%) (HyClone; Cytiva). LoVo
cells were cultured in F-12K medium (Invitrogen; Thermo Fisher
Scientific, Inc.). HT29, HCT116 and SW480 cells were cultured in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS. All cells were maintained at 37˚C with 5% CO2
in a humidified incubator.
Constructs, synthesized oligos and
transfection
Short hairpin RNA (shRNA) targeting LINC01535
(sh-LINC01535), LINC01535 (overexpression vector), miR-761 mimics,
miR-761 inhibitors and their corresponding negative control (NC)
were purchased from GeneChem. All of the DNAs were inserted into
pcDNA3.1 (Shanghai GeneChem Co., Ltd.). Finally, Lipofectamine 3000
(Thermo Fisher Scientific, Inc.) was utilized to transfect the
oligonucleotides and constructs into the SW480 and HCT116 cells
according to the manufacturer's protocol. After 36-48 h of
transfection, the cells were ready for the following
experiments.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
Total RNA from CRC tissues and cells was extracted
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. A total of 2 µg RNA was
reverse transcribed into complementary DNA using the PrimeScript RT
Reagent kit (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol. qPCR was performed using SYBR-Green
Master Mix (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol on an ABI PRISM 7500 PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). GAPDH or U6
was used as a normalization control for mRNA and miRNA,
respectively. The relative expression levels of LINC01535 and
miR-761 were calculated via the 2-ΔΔCq method (16). The primer sequences are provided in
Table I.
 | Table IPrimer sequences for quantitative PCR
(5'-3'). |
Table I
Primer sequences for quantitative PCR
(5'-3').
| Gene | Forward primer | Reverse primer |
|---|
| LINC01535 |
GGGCGGCAGGTCACTGACAC |
GCCAGCAGCCGCTGGCTTAG |
| miR-761 |
ACAGCAGGCACAGAC |
GAGCAGGCTGGAGAA |
| GAPDH |
CCAGCCGAGCCACATCGCTC |
ATGAGCCCCAGCCTTCTCCAT |
| U6 |
CTCGCTTCGGCAGCACATATACT |
ACGCTTCACGAATTTGCGTGTC |
Cell proliferation assay
Cell proliferation was examined using Cell Counting
Kit-8 (CCK-8) and colony formation assays. For the CCK8 assay,
cells were seeded in 96-well plates at 3,000 cells/well. After 24,
48, 72 or 96 h of incubation, 10 µl CCK-8 stain (Dojindo Molecular
Technologies, Inc.) was added. After another 2 h of incubation, the
plates were washed using PBS and the absorbance was measured at 450
nm with a microplate reader (ELx800; Agilent Technologies,
Inc.).
Transfected DDP-resistant CRC cells (SW480/DDP and
HCT116/DDP) were cultured at 37˚C for 48 h prior to exposure to
different doses of DDP, and then the CCK-8 assay was performed. The
IC50 was calculated using a viability curve.
For the colony formation assay, cells were seeded in
a six-well plate at 1,000 cells/well and continuously cultured for
14 days, the medium was refreshed every 2-3 days. The colonies were
then fixed with 4% paraformaldehyde for 10 min and stained with 1%
crystal violet for 15 min at room temperature. Finally, the
colonies were counted manually under a light microscope (Olympus
Corp.) and images were captured.
Transwell invasion assay
Transwell invasion assays were performed to
determine the cell invasion potential using Transwell plates
(Corning, Inc.) that were coated with 50 µl of Matrigel (BD
Biosciences). In brief, 1x105 cells were suspended in
300 µl serum-free medium and added to the upper chamber, while 800
µl complete medium was placed in the lower chamber. After 24 h of
incubation, cells on the upper surface of the membrane were scraped
off. Cells on the lower side of the chamber were fixed with 100%
methanol for 15 min at room temperature and stained with 1% crystal
violet for 30 min at room temperature. The invading cells were
counted in at least five fields under a light microscope
(magnification, x200; Olympus Corp.) using ImageJ software
(National Institutes of Health).
Luciferase reporter assay
The targeting interaction was predicted with a
bioinformatics tool, Targetscan version 7.2 (http://www.targetscan.org/vert_72/). The wild-type
(WT) and mutant-type (MUT) fragments of LINC01535 were amplified
and then inserted into a pGL3 vector (Promega Corporation) to
construct the luciferase reporter, referred to as LINC01535 WT and
LINC01535 MUT, respectively. MUT or WT fragments of LINC01535
containing miR-761 targeting site were synthesized and cloned into
a dual-luciferase reporter vector (pmirGLO; GenePharma Biotech
Corp.). Luciferase vectors and miR-761 mimics or miR-761 NC
together with Renilla plasmid were cotransfected into HCT116
cells by using Lipofectamine 3000. At 48 h after transfection, the
dual-luciferase assay (Promega Corp.) was used to examine the
Renilla and firefly luciferase activity following the
manufacturer's protocol. The luminescence activity of firefly
luciferase was normalized to that of Renilla luciferase.
RNA-binding protein
immunoprecipitation (RIP) assay
The Magna RIP RNA-Binding Protein
Immunoprecipitation Kit (EMD Millipore) was used for the RIP assay.
Cells were harvested and lysed, and lysis buffer containing
magnetic beads was incubated with human anti-Ago2 (argonaute 2)
antibody (cat. no. ab186733; 1:30; Abcam) to conjugate the antibody
to the magnetic beads. Subsequently, proteinase K (Sigma-Aldrich;
Merck KGaA) was added to digest the protein and the
immunoprecipitated RNAs were isolated using TRIzol reagent and
measured by RT-qPCR as aforementioned.
Tumor xenograft experiment
HCT116 cells (2x106) stably transfected
with sh-LINC01535 or sh-NC were subcutaneously injected
subcutaneously into the left flank of 6-week-old female nude mice
(n=5 mice per group; weight, ~20 g). For the animal study, humane
endpoints were utilized to prevent unnecessary animal suffering
according to the Guide for the Care and Use of laboratory Animals
from the National Institutes of Health. In brief, the tumor burden
was measured with calipers and animals were euthanized according to
guidelines before the tumor size reached the maximum permitted size
(tumor diameters remained <1 cm). Regardless of the tumor size,
mice were euthanized if tumors displayed ulceration, if there was
distension of covering tissues, or if severe body weight loss
occurred (consistent or rapid weight loss of 15% or greater
compared with their initial weight). Mice were also euthanized if
they displayed clinical signs necessitating immediate intervention.
The mice were then placed in a carbon dioxide (CO2)
euthanasia chamber (80 l; Shanghai Yuyan Instruments Co., Ltd.) and
sacrificed by excess CO2. The controlled flow rate of
CO2 was 20% of the volume of the euthanasia chamber per
minute. Once the animal lost consciousness, the flow rate was
increased to 100% of the euthanasia chamber volume per minute.
Death was confirmed by cardiac arrest and pupil enlargement. Animal
wellbeing was monitored every day and the tumor sizes were measured
every week. After 4 weeks, the mice were euthanized, the tumor
tissues were excised and weighed, and RT-qPCR was performed to
determine LINC01535 and miR-761 expression. The present study was
approved by the Ethics and Research Committees of the Affiliated
Dongtai Hospital of Nantong University (Dongtai, China) in
2015.
Statistical analysis
Values are expressed as the mean ± standard
deviation from at least three independent experiments. One-way
ANOVA followed by Tukey's post-hoc test or a two-tailed Student's
t-test was performed for comparisons among multiple or between two
groups, respectively. A paired t-test was used for the analysis of
tumor and adjacent non-tumor samples of the same individuals. The
correlation of the expression of genes was assessed by calculating
Pearson's correlation coefficient. P<0.05 was considered to
indicate statistical significance.
Results
LINC001535 is upregulated and miR-761
is downregulated in CRC tissues
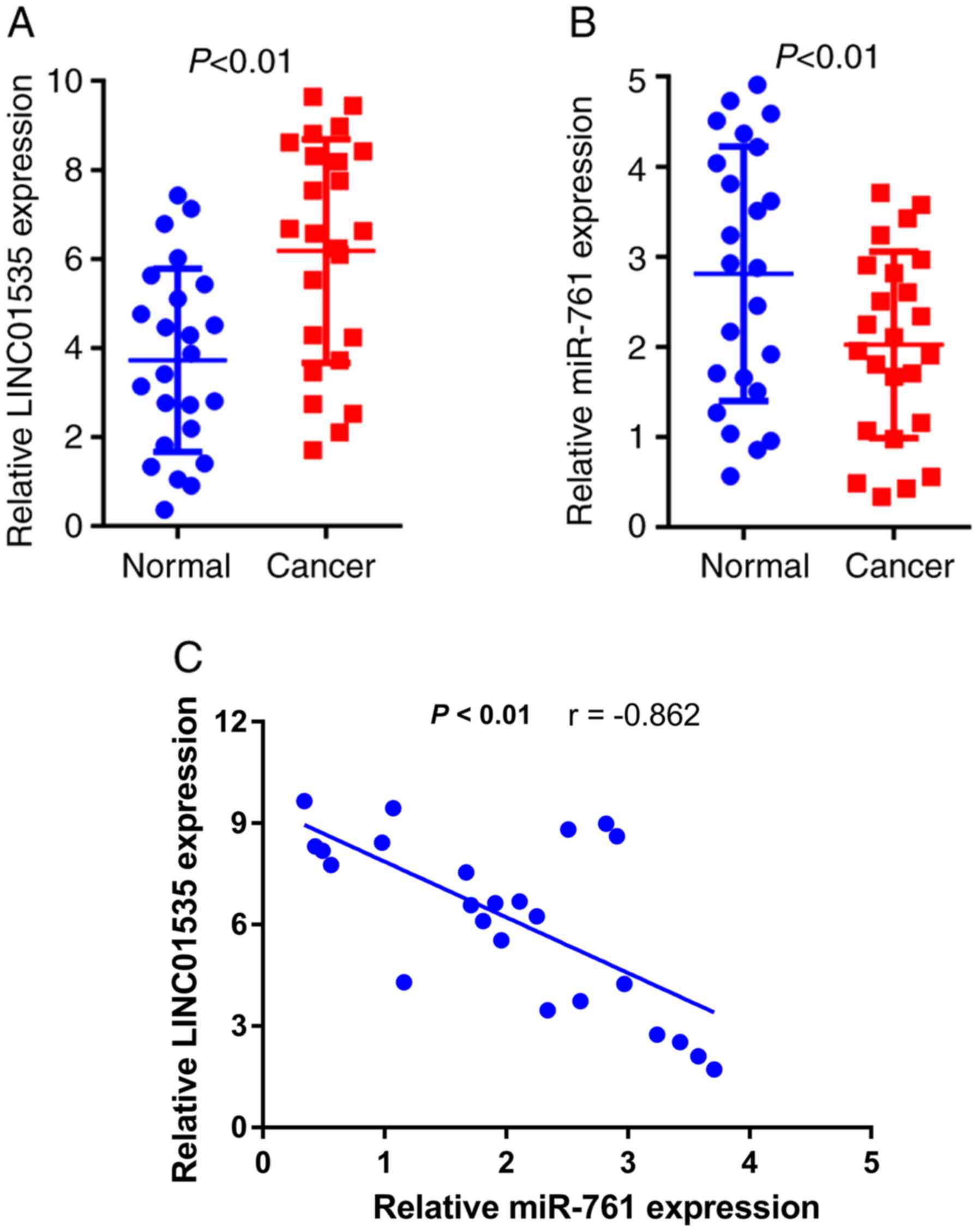
RT-qPCR analysis was performed to determine the
relative expression of LINC01535 and miR-761 in 24 pairs of CRC and
adjacent non-tumor tissues. The age of the patients ranged from
33-69 years, including 11 females and 13 males (data not shown).
The results indicated that LINC01535 expression was significantly
upregulated (Fig. 1A), while
miR-761 levels were markedly downregulated (Fig. 1B) in CRC tissues compared with those
in the corresponding normal tissues. Furthermore, a significant
negative correlation between LINC01535 expression and miR-761
levels in CRC tissues was determined (Fig. 1C). These results suggested that
LINC01535 and miR-761 may be associated with CRC development.
Negative regulatory interaction of
LINC01535 and miR-761
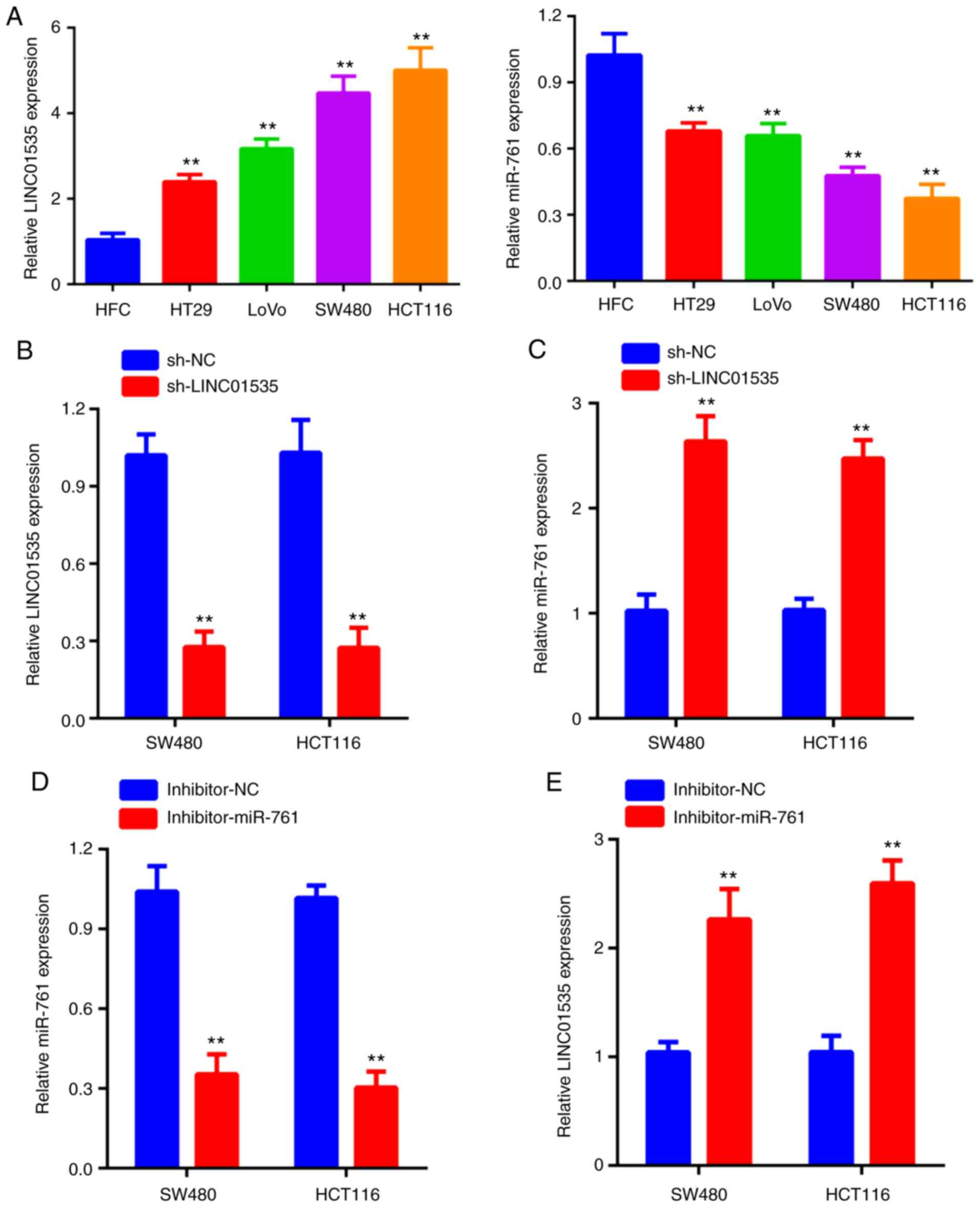
The result of the RT-qPCR analysis indicated that
LINC01535 expression was upregulated in CRC cell lines compared
with that in normal cells (human fetal cells; HFC), while the
expression of miR-671 was downregulated in CRC cell lines compared
with that in HFC (Fig. 2A). As
presented in Fig. 2B, LINC01535
expression was markedly decreased in two CRC cell lines following
transfection with sh-LINC01535. Of note, knockdown of LINC01535
significantly increased the expression of miR-761 (Fig. 2C). To further explore the
correlation between LINC01535 and miR-761, SW480 and HCT116 cells
were transfected with miR-761 inhibitor or corresponding control.
The reduced expression of miR-761 in the two cell lines was
verified by RT-qPCR (Fig. 2D).
Furthermore, it was revealed that inhibition of miR-761 obviously
promoted LINC01535 expression (Fig.
2E). Taken together, these results indicated the negative
regulation between LINC01535 and miR-761.
miR-761 is a direct target of
LINC01535
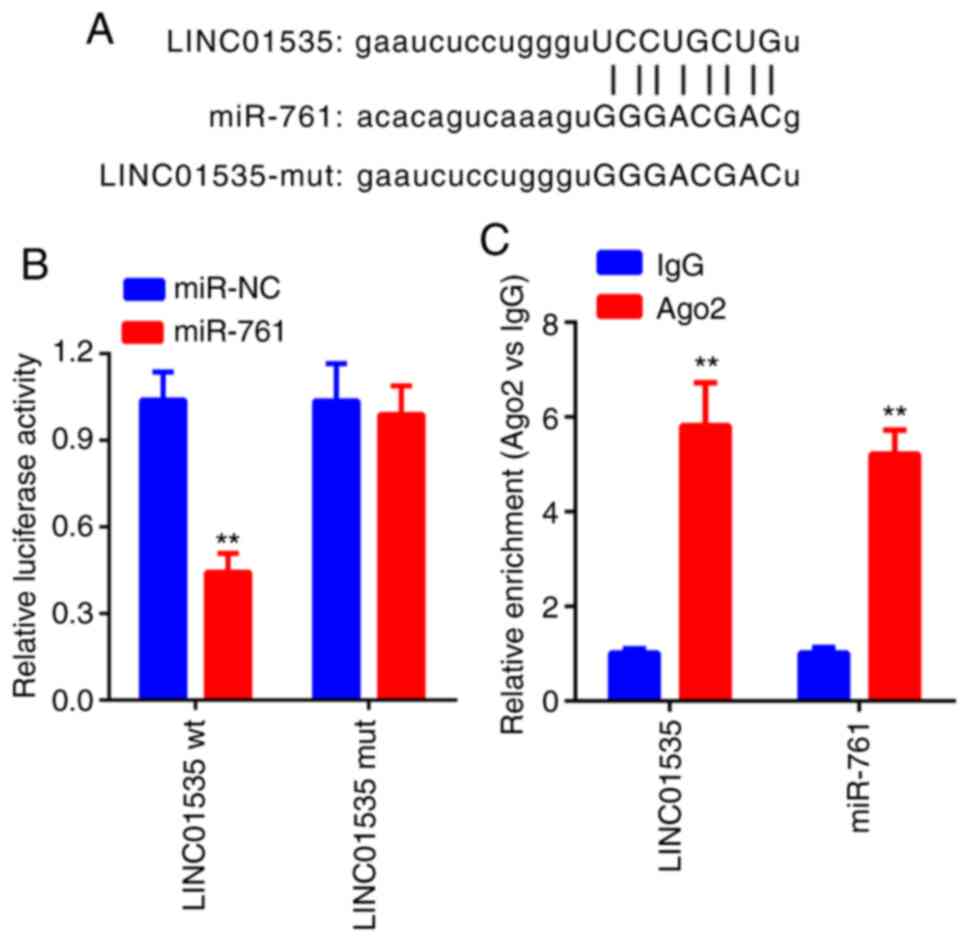
A dual-luciferase reporter assay was performed to
further determine whether miR-761 is a direct target of LINC01535.
The result indicated that miR-761 mimics obviously decreased the
luciferase activity in the cells transfected with LINC01535-wt but
not in the cells transfected with LINC01535-mut in comparison to
the NC group (Fig. 3A and B). It is widely acknowledged that miRNA
functions through RNA-induced silencing complex (RISC) regulation.
Ago2, a key component of RISC, exerts crucial roles in RNA cleavage
(17). Thus, an RIP assay was
performed to determine whether miR-28 regulates LINC01535 via RISC
formation. The result indicated that, compared to the NC (IgG),
LINC01535 was preferentially enriched in anti-Ago2 antibody-treated
beads (Fig. 3C). Collectively,
these results indicated that LINC01535 directly binds to
miR-761.
Effects of LINC01535 and miR-761 on
CRC cell proliferation and invasion
The biological effects of LINC01535 and miR-761 on
CRC cell proliferation were then explored. The results of the CCK-8
assay suggested that knockdown of LINC01535 significantly inhibited
the proliferation of SW480 and HCT16 cells but the inhibitory
effect was abrogated when the cells were co-transfected with
sh-LINC01535 and the miR-761 inhibitor (Fig. 4A). Similarly, the colony formation
assay further confirmed that knockdown of LINC01535 significantly
inhibited the proliferation of SW480 and HCT16 cells but the
inhibitory effect was reversed when the cells were co-transfected
with sh-LINC01535 and the miR-761 inhibitor (Fig. 4B). It was also examined whether
LINC01535 and miR-761 affected CRC-cell invasion. The Transwell
invasion assay suggested that silencing of LINC01535 suppressed the
invasive ability of CRC cells. Similar to the above, this effect
was also abrogated when sh-LINC01535 and the miR-761 inhibitor were
co-transfected (Fig. 4C).
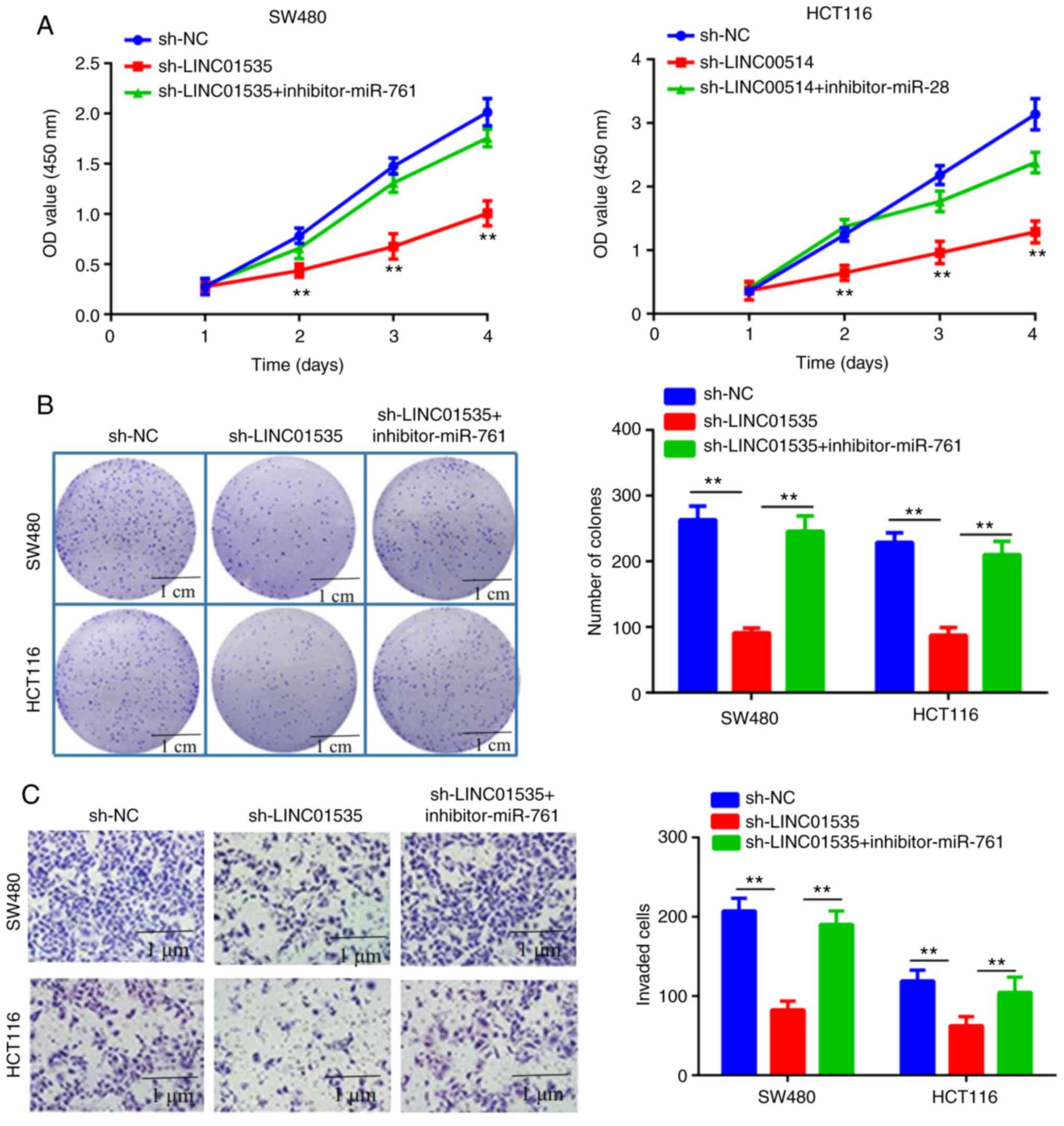 | Figure 4Roles of LINC01535 and miR-761 in
colorectal cancer cell proliferation and invasion. (A) A Cell
Counting Kit-8 cell viability assay was used to evaluate the
proliferation in the sh-NC, sh-LINC01535 and sh-LINC01535+miR-761
inhibitor groups from days 1-4 after inoculation. (B) Colony
formation assays were used to evaluate cell migration in the sh-NC,
sh-LINC01535 and sh-LINC01535+miR-761 inhibitor groups (scale bars,
1 cm). (C) Transwell assays were used to evaluate the invasion
ability of sh-NC, sh-LINC01535 and sh-LINC01535+miR-761 inhibitor
groups (scale bars, 1 µm). **P<0.01 vs. NC. miR,
microRNA; LINC, long intergenic non-coding RNA; sh-LINC01535, short
hairpin RNA targeting LINC01535; NC, negative control; OD, optical
density. |
LINC01535 knockdown enhances DDP
sensitivity of CRC cells
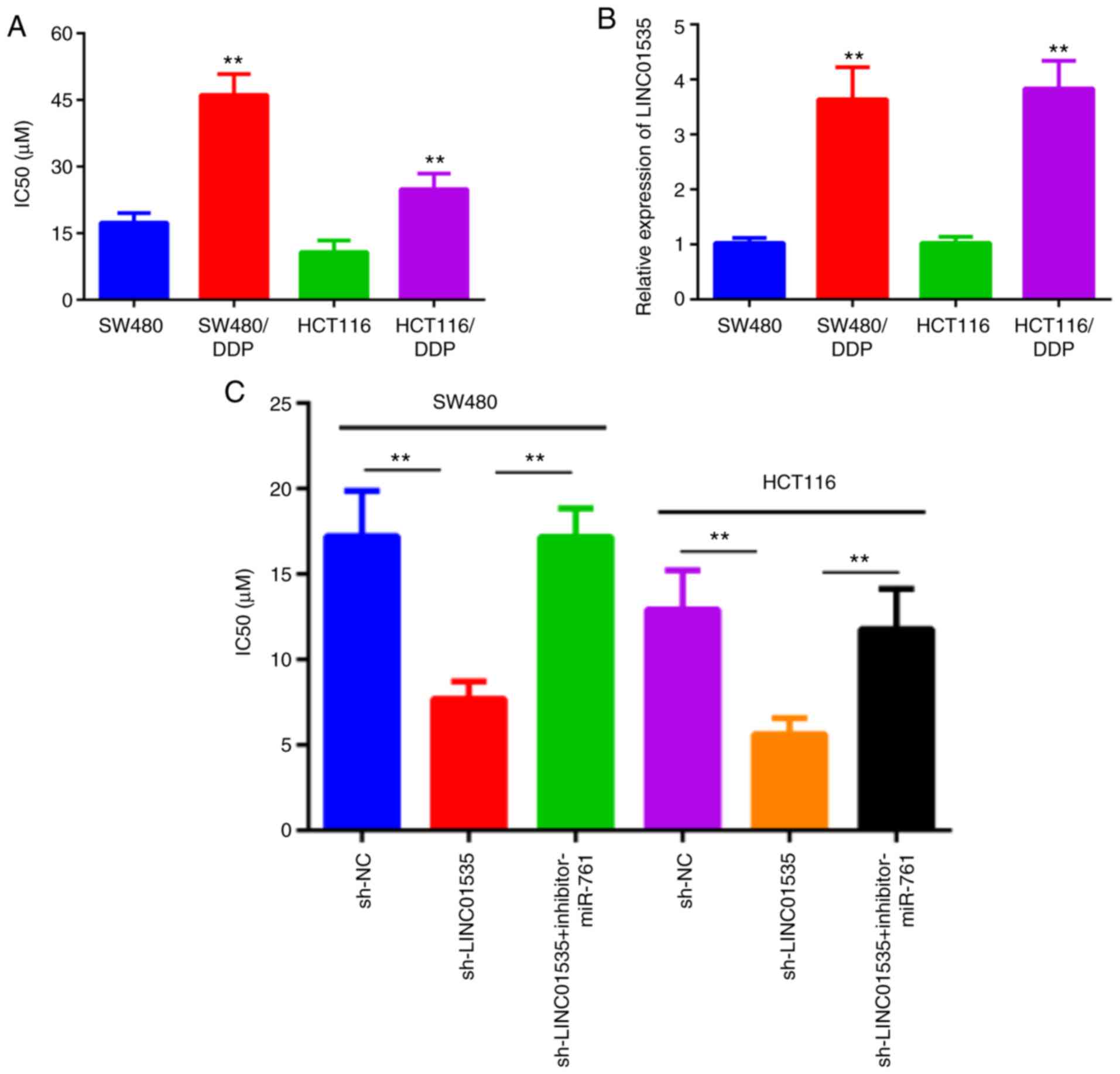
To further investigate the effect of LINC01535 and
miR-761 on the DDP sensitivity of CRC cells, two DDP-resistant CRC
cell lines were established (SW480/DDP and HCT116/DDP). For
SW480/DDP and HCT116/DDP cells, higher IC50 values than
those in the corresponding parental cell lines SW480 and HCT116
were obtained, indicating an increased resistance to DDP (Fig. 5A). Furthermore, RT-qPCR was used to
measure LINC01535 expression in parental and DDP-resistant CRC
cells. Significantly increased LINC01535 expression was observed in
SW480/DDP and HCT116/DDP cells (Fig.
5B). Furthermore, LINC01535 knockdown in SW480 and HCT116 cells
resulted in significantly lower IC50 values than those
for the control group, and this effect was reversed by miR-761
inhibitor (Fig. 5C). Taken
together, these results suggested that LINC01535 induces DDP
resistance in CRC cells.
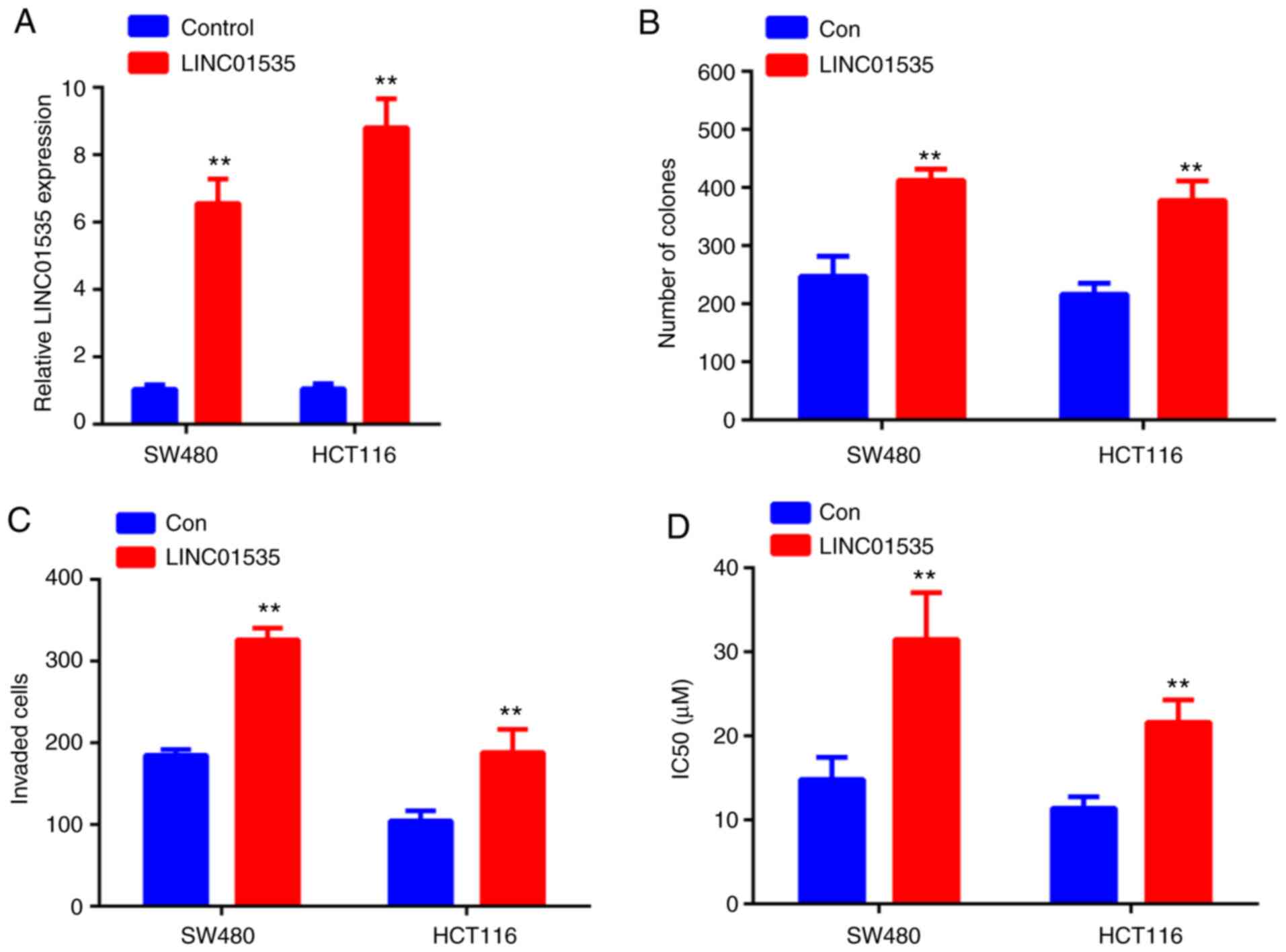
Overexpression of LINC01535 promotes
CRC cell proliferation, invasion and decreases DDP sensitivity of
CRC cells
As demonstrated in Fig.
6A, following transfection with LINC01535 overexpression
vector, LINC01535 levels markedly increased in both cell lines
compared with those in the negative control group. Furthermore,
overexpression of LINC01535 enhanced the colony forming ability of
SW480 and HCT116 cells (Figs. 6B
and S1), as well as their invasive
capacity (Figs. 6C and S1). In addition, overexpression of
LINC01535 in SW480 and HCT116 cells resulted in significantly
higher IC50 values for DDP than those for the control
group, further suggesting that LINC01535 induces DDP resistance in
CRC cells.
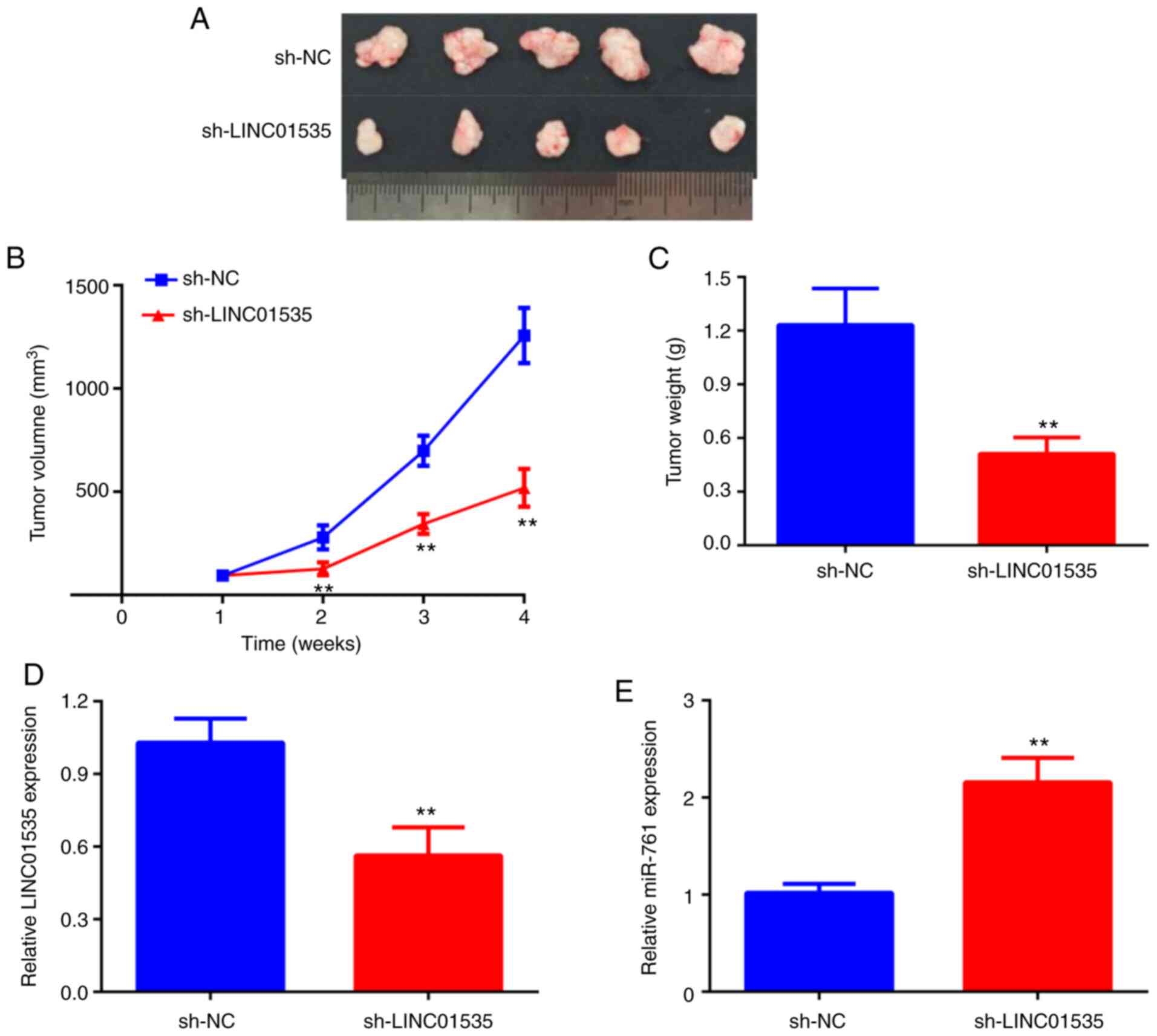
LINC01535 silencing inhibits CRC tumor
growth in vivo
To further verify the in vitro results, a
subcutaneous xenograft tumor model was established by injecting
HCT116 cells stably transfected with sh-NC or sh-LINC01535 into
nude mice. Consistent with the in vitro findings, the in
vivo study demonstrated that the volume and weight of tumors in
the sh-LINC01535 group were significantly reduced compared with
those in the sh-NC group (Fig.
7A-C). Furthermore, in the tumors of the sh-LINC01535 group,
the expression of LINC01535 was decreased, while miR-761 expression
was increased compared with that in the sh-NC group (Fig. 7D and E). Therefore, it was concluded that
LINC01535 knockdown inhibited CRC tumor growth in vivo.
Discussion
Tumor cell proliferation, invasion and
chemoresistance are important types of aggressive behavior of human
cancers (18,19). A large number of studies have
revealed that lncRNAs may function as promoters or inhibitors of
the proliferation, invasion and chemoresistance of human cancer
cells (20,21). It has been widely acknowledged that
numerous lncRNAs are involved in the progression of CRC. For
instance, LINC00963 promotes cell proliferation and migration via
the miR-124-3p/frizzled class receptor 4 signaling pathway in CRC
(22). LncRNA UCID was reported to
promote CRC cell migration and invasion by sponging miR-152-3P and
activation of the Wnt/β-catenin signaling pathway (23). LncRNA heart and neural crest
derivatives expressed 2-antisense 1 reduced 5-fluorouracil
resistance of CRC cells through regulating the miR-20a/programmed
cell death 4 axis (24). However,
the exact functions and underlying mechanisms of the majority of
lncRNAs in CRC have remained to be elucidated.
LINC01535, a newly identified lncRNA, was reported
to promote cervical cancer cell proliferation, migration and
invasion in vitro and tumor growth in vivo via
modulating the miR-214/enhancer of zeste 2 polycomb repressive
complex 2 subunit feedback loop (15). LINC01535 was also reported to
facilitate cell proliferation and inhibit apoptosis in esophageal
squamous cell carcinoma through activating the JAK/STAT3 signaling
pathway (13). In addition,
LINC01535 was indicated to promote osteosarcoma progression via
regulating the miR-214-3p/potassium voltage-gated channel subfamily
C member 4 axis (14). In the
present study, it was determined that the expression of LINC01535
was significantly increased in CRC tissues and cell lines.
Considering the relatively small number of included patients, the
association between LINC001535 expression and the
clinicopathological characteristics of patients with CRC was not
explored. Furthermore, an in vitro functional study
demonstrated that silencing of LINC01535 suppressed cell
proliferation and invasion. In addition, the in vivo results
confirmed that LINC01535 knockdown inhibited CRC growth in a
xenograft model. DDP is a classic and effective cell cycle
non-specific anti-cancer drug, which has been widely used to treat
a variety of solid malignancies, including CRC (25,26).
Patients with CRC frequently display good initial responses to
DDP-based chemotherapy; however, DDP resistance usually occurs.
Therefore, enhancing DDP sensitivity in DDP-resistant CRC cells is
extremely important for the treatment of CRC (27). The present study provides the first
evidence, to the best of our knowledge, that LINC01535 knockdown
enhanced DDP sensitivity in DDP-resistant CRC cells.
Accumulating studies suggested that lncRNAs exert
their functions through binding with miRNAs as competing endogenous
(ce)RNAs (28). For instance,
LINC02418 upregulated maternal embryonic leucine zipper kinase
expression by acting as a ceRNA in CRC (29). LncRNA HOX transcript antisense RNA
knockdown enhanced radiosensitivity through regulating the
miR-93/autophagy related 12 axis in CRC (30). In the present study, an online
database was used to identify potential target miRNAs of LINC01535
and miR-761 was selected. Previous studies have established the
inhibitory function of miR-761 in various human cancer types,
including colorectal cancer (31),
gastric cancer (32) and ovarian
cancer (33). In
LINC01535-knockdown CRC cells, the expression of miR-761 was
significantly increased. In addition, the expression of miR-761 was
negatively associated with LINC01535 expression in CRC tissues. The
results of the luciferase reporter and RIP assay further confirmed
that LINC01535 directly bound to miR-761. Finally, the results of
the functional rescue assay indicated that LINC01535 knockdown
inhibited CRC cell proliferation and invasion and enhanced DDP
sensitivity through regulating miR-761. Additionally, there are
some limitations that should be addressed. Firstly, in the animal
experiments, DDP could have been used in some additional groups to
determine the effect of LINC01535 on DDP resistance in vivo.
Furthermore, in a future study, the sample size should be increased
to evaluate the association of LINC01535 and miR-761 with
clinicopathological characteristics and their prognostic value.
In conclusion, the present results indicated that
LINC01535 expression is upregulated in CRC tissues and cell lines.
Furthermore, LINC01535 promoted CRC proliferation, invasion and DDP
resistance, partly through regulating miR-761. These results
demonstrated that LINC01535 may be an important oncogenic factor in
the development of CRC and may be a promising therapeutic target
for CRC.
Supplementary Material
Overexpression of LINC01535 promotes
CRC cell proliferation (left; scale bar, 1 cm) and invasion (right;
scale bar, 1 μm). LINC, long intergenic non-coding RNA.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WC conceived and designed the study, and drafted the
first draft of the manuscript. CZ and WC confirm the authenticity
of all the raw data WC, CZ, QJ and LC conducted the experiments.
CZ, LC and QJ analyzed and collated the results. All authors
reviewed, critiqued and agreed to the final submission of the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from every
patient and the study was approved by The Ethics and Research
Committees of the Affiliated Dongtai Hospital of Nantong University
(Dongtai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang Y, Huang W, Yuan Y, Li J, Wu J, Yu
J, He Y, Wei Z and Zhang C: Long non-coding RNA H19 promotes
colorectal cancer metastasis via binding to hnRNPA2B1. J Exp Clin
Cancer Res. 39(141)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Liang L, Chen Y, Yu Y, Pan W, Cui Y, Xu X,
Peng K, Liu M, Rashid K, Hou Y and Liu T: SLC25A18 has prognostic
value in colorectal cancer and represses Warburg effect and cell
proliferation via Wnt signaling. Am J Cancer Res. 10:1548–1567.
2020.PubMed/NCBI
|
|
3
|
Li Y, Liu J, Xiao Q, Tian R, Zhou Z, Gan
Y, Li Y, Shu G and Yin G: EN2 as an oncogene promotes tumor
progression via regulating CCL20 in colorectal cancer. Cell Death
Dis. 11(604)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Liu Y, Sun H, Makabel B, Cui Q, Li J, Su
C, Ashby CR Jr, Chen Z and Zhang J: The targeting of non-coding
RNAs by curcumin: Facts and hopes for cancer therapy (review).
Oncol Rep. 42:20–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Grillone K, Riillo C, Scionti F, Rocca R,
Tradigo G, Guzzi PH, Alcaro S, Di Martino MT, Tagliaferri P and
Tassone P: Non-coding RNAs in cancer: Platforms and strategies for
investigating the genomic ‘dark matter’. J Exp Clin Cancer Res.
39(117)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Dong L, Zhu K, Chen M, Li D, Jiang C and
Chen L: Long non-coding RNA GACAT3 promotes liver cancer
progression by regulating the proliferation, apoptosis and
migration of tumor cells. Exp Ther Med. 19:3377–3383.
2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zong Y, Zhang Y, Hou D, Xu J, Cui F, Qin Y
and Sun X: The lncRNA XIST promotes the progression of breast
cancer by sponging miR-125b-5p to modulate NLRC5. Am J Transl Res.
12:3501–3511. 2020.PubMed/NCBI
|
|
8
|
Huo W, Qi F and Wang K: Long noncoding RNA
BCYRN1 promotes prostate cancer progression via elevation of
HDAC11. Oncol Rep. 44:1233–1245. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Han Q, Li J, Xiong J and Song Z: Long
noncoding RNA LINC00514 accelerates pancreatic cancer progression
by acting as a ceRNA of miR-28-5p to upregulate Rap1b expression. J
Exp Clin Cancer Res. 39(151)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Song Z, Zhang X, Lin Y, Wei Y, Liang S and
Dong C: LINC01133 inhibits breast cancer invasion and metastasis by
negatively regulating SOX4 expression through EZH2. J Cell Mol Med.
23:7554–7565. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lai F, Deng W, Fu C, Wu P, Cao M and Tan
S: Long non-coding RNA SNHG6 increases JAK2 expression by targeting
the miR-181 family to promote colorectal cancer cell proliferation.
J Gene Med. 22(e3262)2020.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Chen B, Dragomir MP, Fabris L, Bayraktar
R, Knutsen E, Liu X, Tang C, Li Y, Shimura T, Ivkovic TC, et al:
The long noncoding RNA CCAT2 induces chromosomal instability
through BOP1-AURKB signaling. Gastroenterology. 159:2146–2162.e33.
2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Fang Y, Zhang S, Yin J, Shen YX, Wang H,
Chen XS and Tang H: LINC01535 promotes proliferation and inhibits
apoptosis in esophageal squamous cell cancer by activating the
JAK/STAT3 pathway. Eur Rev Med Pharmacol Sci. 24:3694–3700.
2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yao X, Wu L, Gu Z and Li J: LINC01535
promotes the development of osteosarcoma through modulating
miR-214-3p/KCNC4 axis. Cancer Manag Res. 12:5575–5585.
2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Song H, Liu Y, Jin X, Liu Y, Yang Y, Li L,
Wang X and Li G: Long non-coding RNA LINC01535 promotes cervical
cancer progression via targeting the miR-214/EZH2 feedback loop. J
Cell Mol Med. 23:6098–6111. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Chen H, Xu Z and Liu D: Small non-coding
RNA and colorectal cancer. J Cell Mol Med. 23:3050–3057.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xu Q, Deng F, Qin Y, Zhao Z, Wu Z, Xing Z,
Ji A and Wang QJ: Long non-coding RNA regulation of
epithelial-mesenchymal transition in cancer metastasis. Cell Death
Dis. 7(e2254)2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang H, Luan H, Zhan T, Liu X, Song J and
Dai H: Long non-coding RNA LINC00707 acts as a competing endogenous
RNA to enhance cell proliferation in colorectal cancer. Exp Ther
Med. 19:1439–1447. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liu B, Pan S, Xiao Y, Liu Q, Xu J and Jia
L: LINC01296/miR-26a/GALNT3 axis contributes to colorectal cancer
progression by regulating O-glycosylated MUC1 via PI3K/AKT pathway.
J Exp Clin Cancer Res. 37(316)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zheng K and Zhang TK: LncRNA LINC00963
promotes proliferation and migration through the miR-124-3p/FZD4
pathway in colorectal cancer. Eur Rev Med Pharmacol Sci.
24:7634–7644. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sun LB, Zhao SF, Zhu JJ, Han Y and Shan
TD: Long noncoding RNA UCID sponges miR-152-3p to promote
colorectal cancer cell migration and invasion via the Wnt/β-catenin
signaling pathway. Oncol Rep. 44:1194–1205. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Jiang Z, Li L, Hou Z, Liu W, Wang H, Zhou
T, Li Y and Chen S: LncRNA HAND2-AS1 inhibits 5-fluorouracil
resistance by modulating miR-20a/PDCD4 axis in colorectal cancer.
Cell Signal. 66(109483)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wan X, Wang C, Huang Z, Zhou D, Xiang S,
Qi Q, Chen X, Arbely E, Liu CY, Du P and Yu W: Cisplatin inhibits
SIRT3-deacetylation MTHFD2 to disturb cellular redox balance in
colorectal cancer cell. Cell Death Dis. 11(649)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang P, Zhao S, Lu X, Shi Z, Liu H and
Zhu B: Metformin enhances the sensitivity of colorectal cancer
cells to cisplatin through ROS-mediated PI3K/Akt signaling pathway.
Gene. 745(144623)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang Z, Zhang Y, Qin X, Wang Y and Fu J:
FGF9 promotes cisplatin resistance in colorectal cancer via
regulation of Wnt/β-catenin signaling pathway. Exp Ther Med.
19:1711–1718. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ala U: Competing endogenous RNAs,
non-coding RNAs and diseases: An intertwined story. Cells.
9(1574)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhao Y, Du T, Du L, Li P, Li J, Duan W,
Wang Y and Wang C: Long noncoding RNA LINC02418 regulates MELK
expression by acting as a ceRNA and may serve as a diagnostic
marker for colorectal cancer. Cell Death Dis.
10(568)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Liu Y, Chen X, Chen X, Liu J, Gu H, Fan R
and Ge H: Long non-coding RNA HOTAIR knockdown enhances
radiosensitivity through regulating microRNA-93/ATG12 axis in
colorectal cancer. Cell Death Dis. 11(175)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Xiong W, Yang S, Zhang W, Chen Y and Wang
F: miR-761 inhibits colorectal cancer cell proliferation and
invasion through targeting HDAC1. Pharmazie. 74:111–114.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhang Q, Sui Y and Sui X: MicroRNA-761
inhibits the metastasis of gastric cancer by negatively regulating
Ras and Rab interactor 1. Oncol Lett. 18:3097–3103. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Shi C and Zhang Z: miR-761 inhibits tumor
progression by targeting MSI1 in ovarian carcinoma. Tumour Biol.
37:5437–5443. 2016.PubMed/NCBI View Article : Google Scholar
|