Introduction
Gastric cancer (GC) is one of the most common
malignant tumor types of the digestive tract in the world. Japan,
South Korea and China are regions with a high incidence of GC and
>600,000 new cases are registered each year in China, accounting
for 42% of newly registered cases in the world (1,2). The
mortality rate of patients with GC ranks among the top 3 of all
malignancies and has not exhibited any downward trend (3). Although early detection and early
treatment are the most effective measures to reduce the mortality
of GC, the detection rate of early GC is only 10% and the lack of
specific biomarkers for GC is a major obstacle (4).
Of the human genomic DNA (including introns and
exons), 75% is transcribed into RNA, but only 2% of the genome
encodes proteins and 98% of transcripts are non-coding RNAs
(5,6). A single-stranded RNA molecule with a
length of about 20-24 nucleotides is called a non-coding
single-stranded RNA molecule, and a non-coding RNA with a length of
>200 nucleotides is called long non-coding RNA (lncRNA)
(5,6). lncRNA was originally considered to be
‘junk’ RNA, but in recent years, research has indicated that lncRNA
has an important role in numerous biological processes such as dose
compensation effects, epigenetic regulation and control of the cell
cycle regulation and cell differentiation (7). In GC, increasing evidence has
indicated that certain lncRNAs are involved in the proliferation,
migration, invasion and evasion of apoptosis of GC cells, including
lncRNA LINC00978(8), lncRNA
ZFAS1(9) and lncRNA HAGLROS
(10). lncRNA HIT000218960 is a
newly discovered lncRNA and was reported to be highly expressed in
papillary thyroid cancer (11) and
GC (12). In addition, the
expression levels of HIT000218960 were associated with tumor size,
Tumor-Node-Metastasis (TNM) stage and lymph node metastasis in
patients with GC (12). However,
studies on lncRNA HIT000218960 in GC are rare. In the present
study, HIT000218960 expression was detected in GC tissues from 103
cases and normal gastric mucosal tissues from 62 subjects and its
prognostic value in patients with GC was assessed.
Materials and methods
Patients, tissues and grouping
Between January 2015 and June 2016, GC tissues from
103 cases and normal gastric mucosal tissues from 62 subjects were
collected for analysis in the present study. GC tissues were
collected from surgical removal of tumors and normal gastric
mucosal tissues were collected from gastric mucosal biopsies of
healthy individuals). The clinical data of the patients with GC are
provided in Table I, and the age
individuals providing normal gastric mucosal tissue was 55.2±13.8
years (38-72 years), with 25 males and 37 females. The inclusion
criteria were as follows: i) Diagnosed with GC, ii) complete 3-year
follow-up, iii) being informed of the content of the study and
voluntarily joining the research. The exclusion criteria were as
follows: i) Diagnosis with another malignant tumor type, severe
cardio-cerebral vascular disease or organ dysfunction, ii) a lack
of basic and 3-year follow-up records, iii) death from other
illnesses or an accident, iv) individuals who were pregnant,
lactating or diagnosed with chronic viral (HIV, hepatitis B and/or
C virus) or bacterial infections (M. tuberculosis), v)
individuals who underwent chemotherapy, immunotherapy or targeted
therapy prior to tissue acquisition. All patients were informed and
signed an informed consent form. All experiments involving human
tissues were approved and supervised by the Ethics Committee of the
970th Hospital of the PLA joint Logistics Support Force (Yantai,
China). In addition, patients were divided into groups based on
HIT000218960 expression and HMAG2 mRNA expression: The low HMGA2
expression group (n=53) consisted of patients with HMGA2 expression
levels ≤ median of the HMGA20 expression levels (2.505) and the
high HMGA2 expression group (n=50) consisted of patients with
HIT000218960 expression levels > median of the HMGA2
expression.
 | Table IHIT000218960 expression and clinical
data of patients with gastric cancer. |
Table I
HIT000218960 expression and clinical
data of patients with gastric cancer.
| | HIT000218960
expression | |
|---|
| Item | Cases (n) | Low (n=56) | High (n=47) | χ2 | P-value |
|---|
| Sex | | | | 0.920 | 0.337 |
|
Female | 65 | 33 | 32 | | |
|
Male | 38 | 23 | 15 | | |
| Age (years) | | | | 0.690 | 0.406 |
|
≥60 | 59 | 30 | 29 | | |
|
<60 | 44 | 26 | 18 | | |
| Tumor diameter
(cm) | | | | 6.756 | 0.009 |
|
≥3 | 65 | 29 | 36 | | |
|
<3 | 38 | 27 | 11 | | |
| TNM stage | | | | 10.914 | 0.004 |
|
II | 41 | 29 | 12 | | |
|
III | 48 | 24 | 24 | | |
|
IV | 14 | 3 | 11 | | |
| Histological
grade | | | | 6.336 | 0.042 |
|
I | 28 | 19 | 9 | | |
|
II | 58 | 32 | 26 | | |
|
III | 17 | 5 | 12 | | |
| Lymph node
metastasis (n) | | | | 13.195 | 0.004 |
|
0 | 18 | 15 | 3 | | |
|
1-2 | 20 | 13 | 7 | | |
|
3-6 | 28 | 15 | 13 | | |
|
≥7 | 37 | 13 | 25 | | |
| HMGA2
expression | | | | 27.234 | <0.001 |
|
Low | 53 | 42 | 11 | | |
|
High | 50 | 14 | 36 | | |
Reverse transcription-quantitative
(RT-q)PCR analysis
Total RNA was extracted fµrom the tissues using
RNAiso plus (cat. no. 9108; Takara Bio, Inc.). After preparing the
complementary DNA using a PrimeScript RT reagent kit with gDNA
eraser (cat. no. RR047A; Takara Bio, Inc.), 20 µl of a qPCR mixture
were prepared using GoTaq qPCR Master Mix (cat. no. A6001; Promega,
Corp.) according to the manufacturers' protocols for both RT and
PCR using Touch system (CFX384; Bio-Rad Laboratories, Inc.) and the
thermocycling conditions were showed as follows: 95˚C of 2 min,
(95˚C of 5 sec and 65C of 15 sec) for 40 cycles. The primer
sequences were as follows: β-actin forward, 5'-AGCCCATCCTTCGAGTA
CAAA-3' and reverse, 5'-TCTTGGTGCGATAACTGGTGG-3'; HIT000218960
forward, 5'-CCACCTACCCATCTGAC TTTG-3' and reverse,
5'-CCACTATTTCCCACTGCCTT-3'; high mobility group AT-hook 2 (HMGA2)
forward, 5'-ACCCA GGGGAAGACCCAAA-3' and reverse, 5'-CCTCTTGGCC
GTTTTTCTCCA-3'. The relative expression of HIT000218960 and HMGA2
mRNA were calculated with the 2-∆∆Cq method (13) and β-actin was used as a housekeeping
gene.
Immunohistochemistry
Paraffin sections that were 4-µm thick of preserved
GC tumor tissue and normal gastric mucosal tissues were subjected
to immunohistochemistry for the detection of the expression of
HMGA2 protein. HMGA2 staining was performed in accordance with the
instructions of the VECTASTAIN® Elite® ABC
Kit (Vector Laboratories, Inc.). Anti-HMGA2 antibody (1:500
dilution; cat. no. ab207301; Abcam) was selected as the primary
antibody and goat Anti-Rabbit IgG H&L (HRP) (1:2,000 dilution;
cat. no. ab6702; Abcam) was selected as the secondary antibody.
Samples were incubated with primary antibody overnight at 4˚C and
with secondary antibody for 2 h at 37˚C, and then the slices were
soaked in hematoxylin and stained for 30 sec at room temperature.
Finally, slices were analyzed using a Leica TCS SP5 microscope
(Leica Microsystems) with the LAS AF Lite 4.0 image browser
software (Leica Microsystems) and the HMGA2-positive cells were
recorded.
Statistical analysis
GraphPad Prism 5 software was used for data
analysis. Differences between two groups were analyzed using the
Student's t-test or the χ2 test, and differences between
multiple groups were assessed using one-way ANOVA with Tukey's test
as the post-hoc test. The log-rank (Mantel-Cox) test was used to
compare the survival of patients with GC. The Pearson correlation
coefficient was used to measure the correlation between two
variables in the cohort. The Cox regression model was employed for
analyzing factors that affect the survival of patients with GC.
P<0.05 was considered to indicate statistical significance.
Results
HIT000218960 is highly expressed in GC
tissues
Normal tissues from a total of 62 subjects and GC
tissues from 103 cases were collected and used to detect
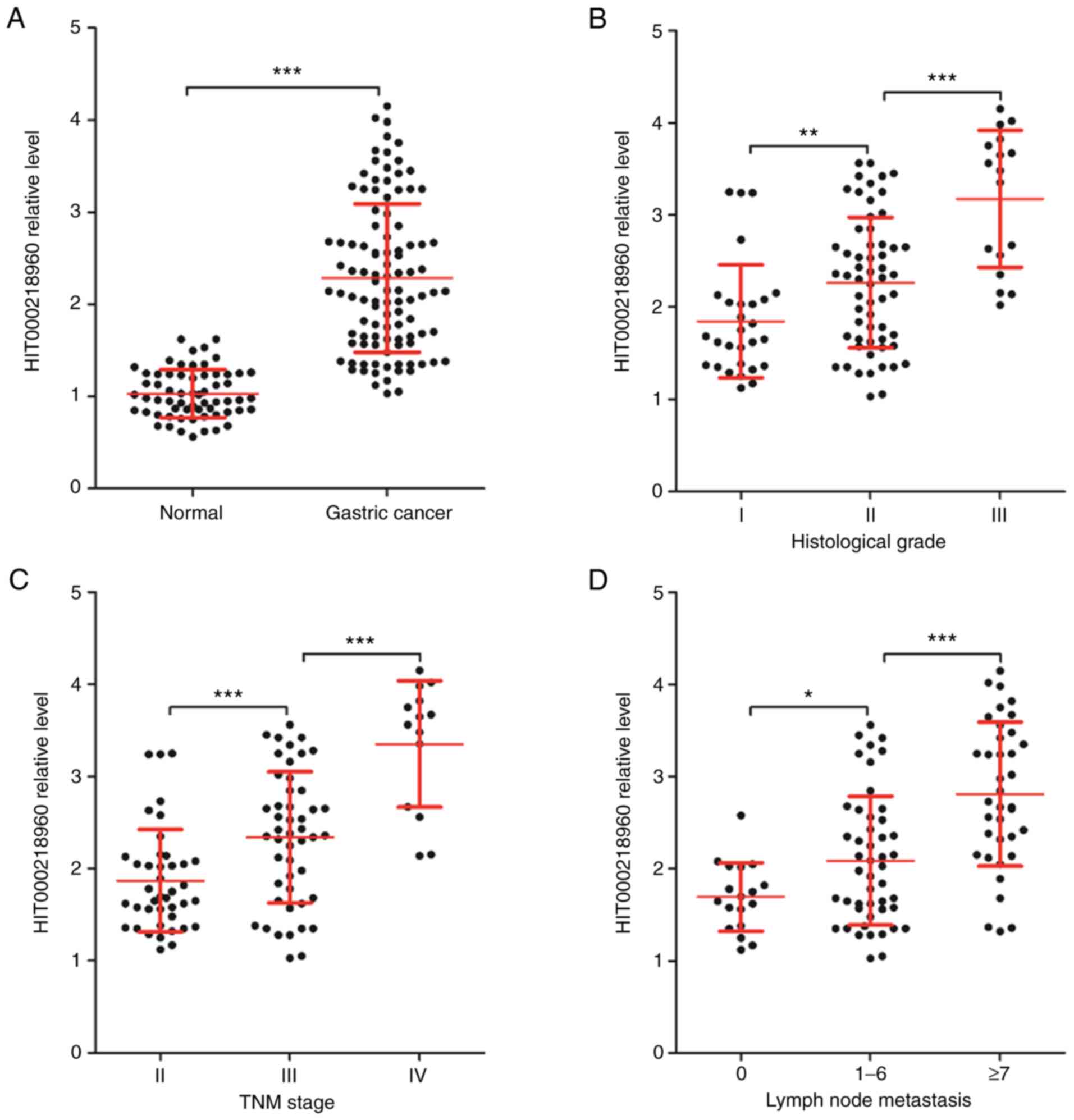
HIT000218960 expression by RT-qPCR. As presented in Fig. 1A, the expression of HIT000218960 in
GC tissues was significantly higher than that in normal tissues.
Next, HIT000218960 expression was compared among different
subgroups of patients with GC based on clinical features and
significant differences were obtained among patients with different
histological grades (Fig. 1B), TNM
stages (Fig. 1C) and numbers of
lymph nodes with metastasis (Fig.
1D). The expression of HIT000218960 was lowest in 28 cases of
histological grade I and in 17 cases of histological grade III, it
was the highest (Fig. 1B).
Furthermore, in 41 cases of TNM stage-II, 48 cases of stage-III and
14 cases of stage-IV GC, HIT000218960 expression increased
gradually with the TNM stage (Fig.
1C). According to the number of lymph nodes with metastases,
the 103 cases of patients with GC were divided into 3 groups with 0
(n=18), 1-6 (n=48) and ≥7 (n=37) lymph nodes with metastases. As
presented in Fig. 1D, HIT000218960
expression increased gradually with the number of lymph nodes with
metastases in patients with GC.
HIT000218960 expression is related to
HMGA2
HMGA2 is an oncogene for GC and has been indicated
to be elevated in GC tissues (14,15).
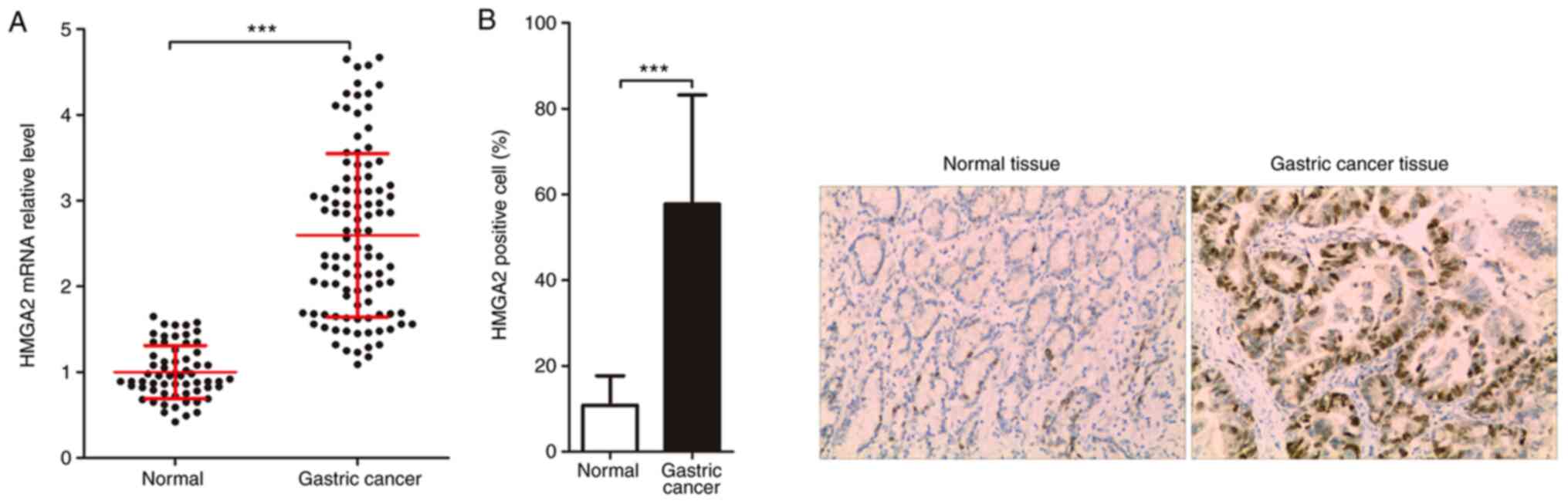
In the present study, the expression of HMGA2 was first detected in
normal tissues and GC tissues and the correlation between
HIT000218960 expression and HMGA2 expression was then analyzed to
assess the significance of HIT000218960 expression in GC tissues.
The data of the RT-qPCR analysis suggested that the expression of
HMGA2 mRNA in GC tissues was significantly higher than that in
normal tissues (Fig. 2A).
Similarly, the results of the immunohistochemistry analysis
indicated that the expression of HMGA2 protein in GC tissues was
also significantly higher than that in normal tissues (Fig. 2B). Of note, the correlation between
HIT000218960 expression and HMGA2 mRNA expression in GC tissues was
analyzed and it was revealed that there was a positive correlation
between HIT000218960 expression and HMGA2 mRNA expression in GC
tissues (Fig. 2C).
Clinical significance of HIT000218960
expression
To further evaluate the clinical significance of
HIT000218960 expression in patients with GC, the 103 cases of GC
were divided into 2 groups based on HIT000218960 expression, namely
the low HIT000218960 expression group (n=56; HIT000218960
expression levels ≤ median of the HIT000218960 expression levels in
103 the GC patients) and the high HIT000218960 expression group
(n=47; HIT000218960 expression levels > median of the
HIT000218960 expression levels in the 103 GC patients). At the same
time, GC patients also were divided into 2 groups based on HMGA2
mRNA and HIT000218960 expression. As presented in Table I, the expression of HIT000218960 in
the 103 cases of GC was not significantly associated with sex
(P=0.337) or age (P=0.406), while it was significantly related to
tumor diameter (P=0.009), TNM stage (P=0.004), histological grade
(P=0.042), the number of lymph nodes with metastasis (P=0.004) and
HMGA2 expression (P<0.001).
HIT000218960 is associated with the
prognosis of patients with GC
Survival prognosis is the best indicator for tumor
patients to obtain treatment benefits and thus, the prognostic
significance of HIT000218960 expression in GC patients was assessed
by analyzing factors affecting prognosis. Following univariate
logistic regression analysis, the results of the multivariate Cox
regression analysis indicated that TNM stage (hazard ratio =2.658,
95% CI=0.891-3.254), histological grade (OR=1.026, 95%
CI=0.658-1.627), lymph node metastasis (OR=2.232, 95%
CI=1.241-3.654), HIT000218960 expression (OR=1.567, 95%
CI=1.118-3.567) and HMGA2 expression (OR=2.892, 95% CI=1.387-4.125)
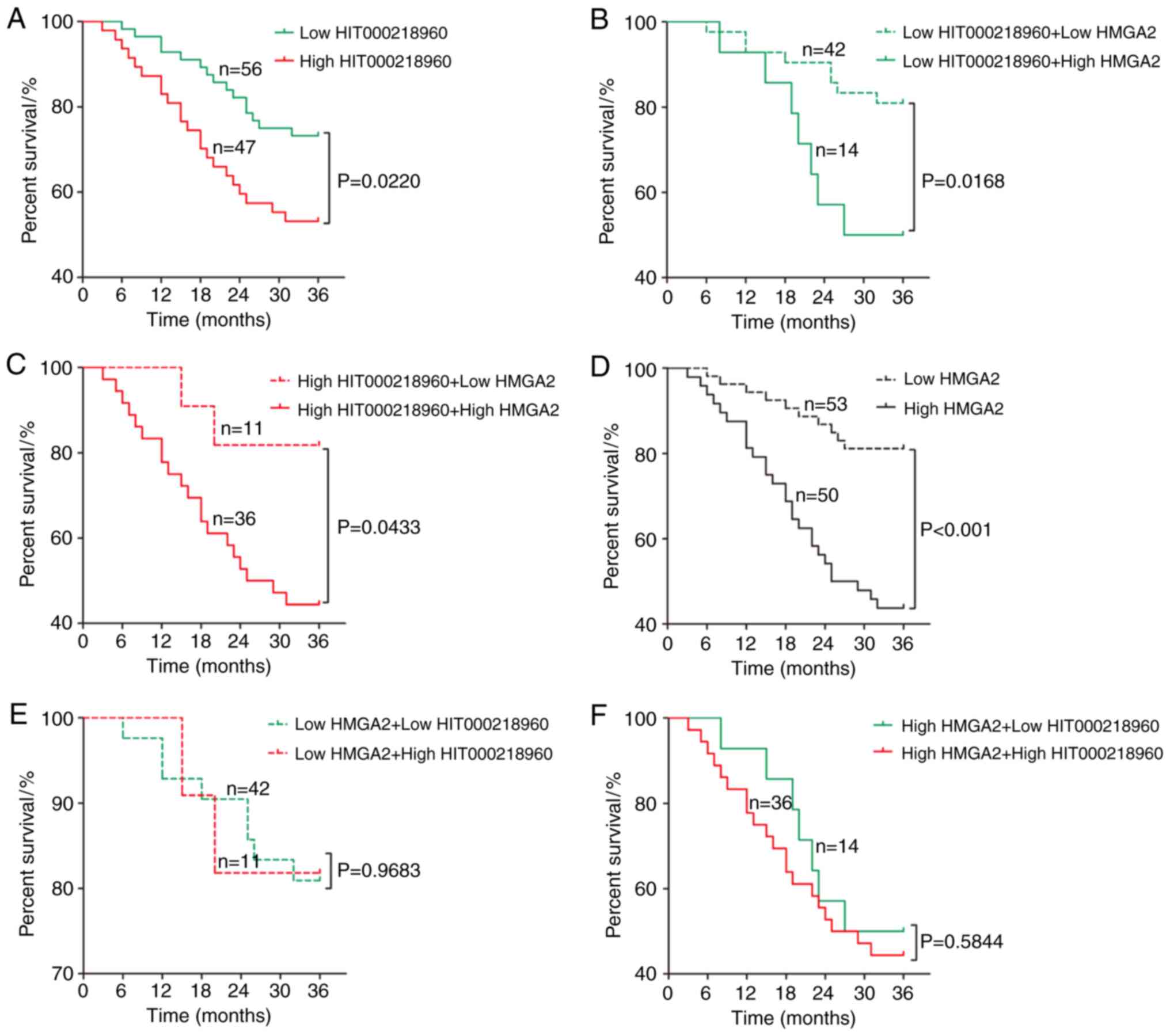
were factors affecting the prognosis of patients with GC (Table II). Next, the 3-year survival of
patients with GC with different HIT000218960 and HMGA2 expression
was compared. As presented in Fig.
3, the 3-year survival of patients with GC with low
HIT000218960 expression was significantly higher than that in
patients with high HIT000218960 expression (Fig. 3A). Among the 56 cases of GC with low
HIT000218960 expression, the 3-year survival of patients with low
HMGA2 expression was significantly higher than that of patients
with high HMGA2 expression (Fig.
3B), and similar results were also obtained for the 47 cases of
GC with high HIT000218960 expression (Fig. 3C). Similarly, the 3-year survival of
patients with GC and low HMGA2 expression was significantly higher
than that of patients with high HMGA2 expression (Fig. 3D). However, there was no significant
difference in survival between GC patients with low and high
HIT000218960 expression, whether in the group of 53 cases with GC
low HMGA2 expression (Fig. 3E) or
among the 50 cases of GC with high HMGA2 expression (Fig. 3F).
 | Table IIUnivariate and multivariate Cox
regression analysis for the prognosis of patients with gastric
cancer. |
Table II
Univariate and multivariate Cox
regression analysis for the prognosis of patients with gastric
cancer.
| | Univariate
analysis | Multivariate
analysis |
|---|
| Clinical
parameter | OR | 95% CI | P-value | OR | 95% CI | P-value |
|---|
| Sex (males vs
females) | 1.032 | 0.569-2.031 | 0.632 | - | - | - |
| Age (≥60 vs <60
) | 0.628 | 0.235-1.351 | 0.289 | - | - | - |
| Tumor diameter, ≥3
vs <3 | 2.325 | 1.256-3.541 | 0.068 | - | - | - |
| TNM stage, II vs
III+IV | 3.021 | 1.568-4.025 | <0.001 | 2.658 | 0.891-3.254 | <0.001 |
| Histological grade,
I vs II+III | 2.541 | 1.335-2.987 | 0.021 | 1.026 | 0.658-1.627 | 0.038 |
| Lymph node
metastasis, yes vs no | 1.524 | 0.985-3.127 | 0.005 | 2.232 | 1.241-3.654 | 0.002 |
| HIT000218960
expression, low vs high | 1.023 | 0.569-1.352 | 0.038 | 1.567 | 1.118-3.567 | 0.042 |
| HMGA2 expression,
low vs high | 1.953 | 1.055-3.246 | <0.001 | 2.892 | 1.387-4.125 | 0.003 |
Discussion
According to data released by the China National
Cancer Center (3), the morbidity
and mortality of GC rank second among all malignancies in males and
679,000 new cases and 60,000 deaths are reported each year. Of
note, most patients with GC are already at an advanced stage at the
time-point of diagnosis, and thus, the 5-year survival for patients
with GC is low, e.g. the 5-year overall survival rate of patients
with GC treated only by surgery is ~20% and 30-50% in patients
treated by surgery and adjuvant therapy (16,17).
Therefore, it may be worthwhile to identify biomarkers that may be
used to diagnose and evaluate the prognosis of patients with GC. In
the present study, it was not only determined that HIT000218960 was
highly expressed in GC tissue, but also that HIT000218960
expression increased with the degree of GC differentiation,
clinical stage and number of lymph nodes with metastases. A recent
similar study suggested that HIT000218960 was highly expressed in
GC as compared to normal tissues adjacent to GC tissues (12). Furthermore, a previous study
suggested that HIT000218960 was highly expressed in papillary
thyroid cancer tissues and promoted the proliferation and
metastasis of papillary thyroid cancer cells in vitro
(11). Of note, these two studies
all indicated that the function of HIT000218960 in cancer cells was
related to HMGA2.
HMGA2 has been recognized as a novel oncogene and
HMGA2 protein is a non-histone chromatin-related protein that may
change its structure by binding to chromatin or directly interact
with related proteins to regulate the transcription of other genes
to then induce tumor formation (18,19).
HMGA2 was reported to be involved in numerous aspects of the
biological characteristics of GC, such as proliferation (14), epithelial-mesenchymal transition
(15,20), migration and invasion (21,22),
chemical resistance (23), tumor
angiogenesis (21,24) and tumor stem cell characteristics
(25). In the present study, HMGA2
mRNA expression was detected using RT-qPCR and HMGA2 protein
expression using immunohistochemistry, and it was determined that
HMGA2 was highly expressed in GC tissues, which was consistent with
previous studies (15,20). Furthermore, the expression of
HIT000218960 was positively correlated with the expression of HMGA2
mRNA in GC tissues. This suggested that HIT000218960 may be an
oncogene in GC.
To further explore the significance of HIT000218960
expression in patients with GC, the association between
HIT000218960 expression and clinical features of GC was explored.
As expected, HIT000218960 was significantly associated with
clinical stage, tumor size, lymph node metastasis and HMGA2
expression in GC, and HIT000218960 was a factor affecting the
prognosis of patients with GC. Furthermore, the effect of HMGA2
expression on the 3-year survival of patients with GC in
HIT000218960 low or high expression was compared and significant
differences were obtained, that was patients with GC with lower
HIT000218960 or HMGA2 expression had more favorable 3-year
survival. However, insignificant differences were obtained when
comparing the effect of HIT000218960 expression on the 3-year
survival of patients with GC with HMGA2 low or high expression
individually.
As an lncRNA, HIT000218960 does not directly
regulate biological processes of cells and it may only have a
biological function by directly interacting with the mRNA encoding
a protein or by indirectly regulating the expression of a protein
by binding to the microRNA (26,27).
Previous studies indicated that HIT000218960 promoted HMGA2 protein
expression in GC and papillary thyroid cancer (11,12).
Therefore, HIT000218960 may affect the biological characteristics
of GC cells through HMGA2 and then affect the progression and
prognosis of patients with GC. Overall, the present study suggested
that HIT000218960 was highly expressed and was related to poor
prognosis in patients with GC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YC, LB and KD conceived and designed the study. LB
and KD collected data, drafted the manuscript and critically
revised the manuscript for the important intellectual content. DT,
XS and SW analyzed and interpreted the experimental data. YC and LB
confirm the authenticity of all the raw data. All authors read and
approved the final version of the study.
Ethics approval and consent to
participate
The present study was performed with the approval of
the Ethics Committee of The 970th Hospital of the PLA joint
Logistics Support Force (Yantai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386.
2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bertuccio P, Chatenoud L, Levi F, Praud D,
Ferlay J, Negri E, Malvezzi M and La Vecchia C: Recent patterns in
gastric cancer: A global overview. Int J Cancer. 125:666–673.
2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen M, Wang W, Ma J, Ye P and Wang K:
High glucose induces mitochondrial dysfunction and apoptosis in
human retinal pigment epithelium cells via promoting SOCS1 and
Fas/FasL signaling. Cytokine. 78:94–102. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Shiraishi N, Yasuda K and Kitano S:
Laparoscopic gastrectomy with lymph node dissection for gastric
cancer. Gastric Cancer. 9:167–176. 2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y,
Li Z, Bu D, Sun N, Zhang MQ, et al: NONCODE 2016: An informative
and valuable data source of long non-coding RNAs. Nucleic Acids
Res. 44D1:D203–D208. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ulitsky I: Evolution to the rescue: Using
comparative genomics to understand long non-coding RNAs. Nat Rev
Genet. 17:601–614. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Fu M, Huang Z, Zang X, Pan L, Liang W,
Chen J, Qian H, Xu W, Jiang P and Zhang X: Long noncoding RNA
LINC00978 promotes cancer growth and acts as a diagnostic biomarker
in gastric cancer. Cell Prolif. 51(e12425)2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pan L, Liang W, Fu M, Huang ZH, Li X,
Zhang W, Zhang P, Qian H, Jiang PC, Xu WR, et al: Exosomes-mediated
transfer of long noncoding RNA ZFAS1 promotes gastric cancer
progression. J Cancer Res Clin Oncol. 143:991–1004. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen J-F, Wu P, Xia R, Yang J, Huo XY, Gu
DY, Tang CJ, De W and Yang F: STAT3-induced lncRNA HAGLROS
overexpression contributes to the malignant progression of gastric
cancer cells via mTOR signal-mediated inhibition of autophagy. Mol
Cancer. 17(6)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li T, Yang XD, Ye CX, Shen ZL, Yang Y,
Wang B, Guo P, Gao ZD, Ye YJ, Jiang KW, et al: Long noncoding RNA
HIT000218960 promotes papillary thyroid cancer oncogenesis and
tumor progression by upregulating the expression of high mobility
group AT-hook 2 (HMGA2) gene. Cell Cycle. 16:224–231.
2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sun L, Yu J, Wang P, Shen M and Ruan S:
HIT000218960 promotes gastric cancer cell proliferation and
migration through upregulation of HMGA2 expression. Oncol Lett.
17:4957–4963. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Livak K J and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li W, Li J, Mu H, Guo M and Deng H:
miR-503 suppresses cell proliferation and invasion of gastric
cancer by targeting HMGA2 and inactivating WNT signaling pathway.
Cancer Cell Int. 19(164)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Dong J, Wang R, Ren G, Li X, Wang J, Sun
Y, Liang J, Nie Y, Wu K, Feng B, et al: HMGA2-FOXL2 Axis Regulates
Metastases and Epithelial-to-Mesenchymal Transition of
Chemoresistant Gastric Cancer. Clin Cancer Res. 23:3461–3473.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jiang L, Yang KH, Guan QL, Chen Y, Zhao P
and Tian JH: Survival benefit of neoadjuvant chemotherapy for
resectable cancer of the gastric and gastroesophageal junction: A
meta-analysis. J Clin Gastroenterol. 49:387–394. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Katai H, Ishikawa T, Akazawa K, Isobe Y,
Miyashiro I, Oda I, Tsujitani S, Ono H, Tanabe S, Fukagawa T, et
al: Registration Committee of the Japanese Gastric Cancer
Association: Five-year survival analysis of surgically resected
gastric cancer cases in Japan: A retrospective analysis of more
than 100,000 patients from the nationwide registry of the Japanese
Gastric Cancer Association (2001-2007). Gastric Cancer. 21:144–154.
2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang S, Mo Q and Wang X: Oncological role
of HMGA2 (Review). Int J Oncol. 55:775–788. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ahmed SM, Ramani PD, Wong SQR, Zhao X,
Ivanyi-Nagy R, Leong TC, Chua C, Li Z, Hentze H, Tan IB, et al: The
chromatin structuring protein HMGA2 influences human subtelomere
stability and cancer chemosensitivity. PLoS One.
14(e0215696)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhao X-P, Zhang H, Jiao J-Y, Tang D-X, Wu
YL and Pan C-B: Overexpression of HMGA2 promotes tongue cancer
metastasis through EMT pathway. J Transl Med. 14(26)2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sun J, Sun B, Zhu D, Zhao X, Zhang Y, Dong
X, Che N, Li J, Liu F, Zhao N, et al: HMGA2 regulates CD44
expression to promote gastric cancer cell motility and sphere
formation. Am J Cancer Res. 7:260–274. 2017.PubMed/NCBI
|
|
22
|
Wang H, Jiang Z, Chen H, Wu X, Xiang J and
Peng J: MicroRNA-495 inhibits gastric cancer cell migration and
invasion possibly via targeting high mobility group AT-Hook 2
(HMGA2). Med Sci Monit. 23:640–648. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yang X, Zhao Q, Yin H, Lei X and Gan R:
miR-33b-5p sensitizes gastric cancer cells to chemotherapy drugs
via inhibiting HMGA2 expression. J Drug Target. 25:653–660.
2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lee MY, da Silva B, Ramirez DC and Maki
RG: Novel HMGA2-YAP1 fusion gene in aggressive angiomyxoma. BMJ
Case Rep. 12(e227475)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li W, Wang Z, Zha L, Kong D, Liao G and Li
H: HMGA2 regulates epithelial-mesenchymal transition and the
acquisition of tumor stem cell properties through TWIST1 in gastric
cancer. Oncol Rep. 37:185–192. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Engreitz JM, Haines JE, Perez EM, Munson
G, Chen J, Kane M, McDonel PE, Guttman M and Lander ES: Local
regulation of gene expression by lncRNA promoters, transcription
and splicing. Nature. 539:452–455. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ferrè F, Colantoni A and Helmer-Citterich
M: Revealing protein-lncRNA interaction. Brief Bioinform.
17:106–116. 2016.PubMed/NCBI View Article : Google Scholar
|