Introduction
Breast cancer is the most common malignant tumor in
women worldwide; the incidence rate increases by 0.3% per year and
it has been estimated that there will be 2.3 million new cases of
breast cancer in 2020 and breast cancer has surpassed lung cancer
in the number of new cases (1-3).
Recurrence and metastasis following systematic therapy are the most
common causes of mortality (4).
Thus, it is important to identify novel therapeutic targets for
breast cancer.
Inhibitor of growth 3 (ING3) is a member of the ING
family (5), which consists of five
members with different subtypes, according to alternative splicing
(6). Their encoded proteins
comprise a highly conserved plant homeodomain, a Cys4-His-Cys3 form
of zinc finger that directly interacts with histone H3, and a
nuclear localization sequence (6,7). ING
proteins play significant roles in several biological processes,
including apoptosis, DNA repair, cell cycle regulation and histone
methylation (8,9). Recent studies have reported that the
ING family are closely associated with cancer (10,11).
ING family members are tumor suppressors that decrease invasion,
migration and proliferation of different types of cancer (12-14).
ING3 is in 7q31 of chromosome seven, and is
considered a suppressor in different types of cancer (10,15).
Li et al (16) suggested
that downregulation of ING3 expression promotes the
proliferation of head and neck squamous cell carcinoma cells.
Furthermore, Lu et al (17)
demonstrated that downregulation of ING3 expression promotes
tumorigenesis in hepatocellular carcinoma. However, ING3 has
been reported to act as an oncogene in prostate cancer, which
promotes cell proliferation (18,19).
In melanoma, ING3 nuclear expression is downregulated and
associated with low disease-specific 5-year survival rates
(20), and the nuclear localization
sequence of ING3 is critical to its function as a tumor suppressor
(21). However, the role of ING3 in
breast cancer remains unknown. Previous studies have demonstrated
that ING3 is frequently expressed in breast cancer and
gynecological cancers; however, ING3 expression has not been
detected in the nucleus of breast cancer tissues (7,22).
In most cases, ING3 is considered a tumor suppressor
(16,17,23),
thus it was hypothesized that ING3 does not play an inhibitory role
in breast cancer due to loss of nuclear localization capacity.
To further investigate the effect of ING3 on the
biological behavior of breast cancer, ING3 expression was
analyzed in breast cancer tissues and normal tissues to determine
its influence on the prognosis of patients with breast cancer. The
role of ING3 on the migration and invasion of breast cancer cells
was also investigated.
Materials and methods
Patients, tissue samples and
follow-up
The present study was approved by the Ethics
Committees of the Third Affiliated Hospital of Kunming Medical
University, Yunnan Cancer Hospital (Kunming, China; approval no.
QT202002), and written informed consent was provided by all
patients prior to the study start. The UALCAN database (http://ualcan.path.uab.edu) was used to analyze
ING3 expression in cancer tissues and normal tissues.
Follow-up and survival analyses were performed using the UALCAN and
KM-plot (http://kmplot.com/analysis)
databases. Patient data and ING3 expression data (FPKM) were
downloaded from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov) (TCGA-BRCA) and UCSC
Xena (http://xena.ucsc.edu/public) databases
(TCGA.BRCA.sampleMap/BRCA_clinicalMatrix). R (4.0.2) (24) software and Perl (5.28.1; https://www.activestate.com/products/perl/downloads)
software were to determine whether the long-term survival of
patients with different clinical tumor-node-metastasis (TNM) stages
(25) and subtypes (PAM50) were
associated with ING3 expression levels. Median ING3
expression levels between each subgroup was used to distinguish
between the high and low expression groups, as follows: Stage I,
2.02357250; stage II, 1.9492; stage III, 1.894254; stage IV,
1.794536; luminal A, 1.803995; luminal B, 1.824826, human epidermal
growth factor receptor 2 (HER2)-enriched, 1.497432; basal-like,
2.346218.
Primary cell separation
Normal breast epithelial cells (NBECs) were
separated from tissue samples following surgical resection. Tissues
were transported on ice in RPMI-1640 medium (Corning, Inc.)
supplemented with 1% penicillin/streptomycin (Biological
Industries), and used to isolate primary cells within 2 h. The
tissues were washed three times with DPBS (Beijing Solarbio Science
& Technology Co., Ltd.) and trimmed of excess fat, prior to
cutting into 1-2 mm thick sections on ice. Type I collagenase (1.5
mg/ml, Sigma-Aldrich; Merck KGaA) was dissolved in DPBS containing
5% fetal bovine serum (FBS; Corning, Inc.) to digest tissues into
cells. Tissues were dissociated by manual agitation for 20-40 min
at 37˚C, and digestion was observed under a light microscope
(magnification, x100). Cells were washed three times with DPBS
containing 0.04% FBS to stop digestion and collected via
centrifugation at 4˚C 3,000 x g for 5 min. Red Blood Cell lysis
buffer (Invitrogen; Thermo Fisher Scientific, Inc.) was used to
lyse erythrocytes on ice. Cells were re-washed three times with
DPBS and cultured in RPMI-1640 medium supplemented with 10% FBS
(Corning, Inc.), at 37˚C with 95% air and 5% CO2.
Cell lines and culture
Human breast cancer cell lines, HCC1937 and MCF7,
were purchased from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. Cells were maintained in RPMI-1640
medium supplemented with 10% FBS (both purchased from Corning,
Inc.), at 37˚C with 95% air and 5% CO2.
Lentiviral transfection
The overexpressing ING3 lentivirus (LV5-ING3) and
the negative control lentivirus (LV5-NC) were synthesized by
Shanghai GenePharma Co., Ltd. The breast cancer cells were
transduced with lentivirus (LV5-ING3 or LV5-NC), MCF7 and HCC1937
cells were inoculated into 6-well plates at a density of
5x105/3 ml 24 h prior to transfection. On the day of
transfection, 25x105 lentivirus was added to MCF7 cells
and 50x105 lentivirus was added to HCC1937 cells, and
polybrene (8 µg/ml; Shanghai GenePharma Co., Ltd.) was added to the
culture medium. After 72 h of screening with puromycin (1 µg/ml;
Beijing Solarbio Science & Technology Co., Ltd.), transfection
efficiency was determined via eGFP expression by fluorescence
microscopy. Cell viability was determined using the cell counting
device (JIMBIO-FIL). Cell suspension (5 µl) was stained with 0.4%
trypan blue dye (5 µl) at room temperature and immediately used for
cell viability determination; overexpression of ING3 was
detected via reverse transcription-quantitative (RT-q)PCR and
western blot analyses.
RT-qPCR
Total RNA was extracted from breast cancer cells and
NBECs using the RNAprep Pure cell kit (Tiangen Biotech Co., Ltd.),
and reverse transcribed into cDNA using the FastQuant RT kit with
gDNase (Tiangen Biotech Co., Ltd.), the RNA was mixed with the
genomic DNA removal system and incubated at 42˚C for 3 min,
following which, the reverse transcription reaction solution was
added and incubated at 42˚C for 15 min and 95˚C for 3 min to
synthesize cDNA. qPCR was subsequently performed using SuperReal
PreMix Plus (SYBR Green, Tiangen Biotech Co., Ltd.). The following
primer sequences were used for qPCR: ING3 forward,
5'-GCTGGATCAGGAACTGGCTAA-3' and reverse,
5'-TCTGTTGTCGTATGGTGAGAAGT-3'; and GAPDH forward,
5'-CAGGAGCGAGATCCCTCCAAAAT-3' and reverse,
5'-AGATGATGACCCTTTTGGCTCCC-3'. The following thermocycling
conditions were used for qPCR: 95˚C for 15 min for 1 cycle and 95˚C
for 10 sec following 62˚C for 32 sec for 40 cycles. Relative
expression levels were calculated using the 2-∆∆Cq
method (18) and normalized to the
internal reference gene GAPDH.
Western blotting
Total protein was extracted from the cultured cells
using RIPA buffer (Beyotime Institute of Biotechnology) and PMSF
(Biosharp Life Sciences) mixed at a 100:1 ratio. Total protein was
quantified using BCA protein quantification reagent (Beijing
Dingguo Changsheng Biotechnology Co., Ltd.). Proteins (30 µg) were
separated via electrophoresis using Spacer on a 12% SDS-PAGE gel.
The separated proteins were subsequently transferred onto PVDF
membranes (EMD Millipore), washed the PVDF membrane with TBST
containing 0.1% Tween-20 and blocked in western blocking fluid
(Beyotime Institute of Biotechnology) for 1.5 h at room
temperature. The membranes were incubated with primary antibodies
against ING3 (1:1,000 dilution; cat. no. GTX102480; GeneTex, Inc.)
and GAPDH (1:5,000 dilution; cat. no. GTX100118; GeneTex, Inc.) at
overnight 4˚C. Following the primary incubation, membranes were
washed three times with TBST (10 min each), and subsequently
incubated with secondary antibody (1:5,000 dilution; cat. no.
GTX2131110-01; GeneTex, Inc.) at room temperature for 2 h. Protein
bands were visualized using the ECL kit (Suzhou Xinsaimei
Biotechnology Co., Ltd.) and analyzed using ImageJ software (1.42q;
National Institutes of Health).
Migration and invasion assays
Cells were collected and resuspended in culture
medium without serum. For the migration assay, 8x104
MCF7 cells and HCC1937 cells transfected with LV5-ING3 and LV5-NC
were plated in the upper chambers of Transwell plates without
Matrigel, cell culture medium containing 20% FBS (Corning, Inc.)
was added to the lower chamber. For the invasion assay, Matrigel
was diluted on ice with RPMI-1640 medium (Corning, Inc.) at a ratio
of 1:8, and solidified at 37℃ for 2 h. Following coating,
8x104 MCF7 cells and HCC1937 cells transfected with
LV5-ING3 and LV5-NC were plated in the upper chambers of 8-µm pore
size plates coated with 60 µl Matrigel, cell culture medium
containing 20% FBS (Corning, Inc.) was added to the lower chamber.
Following incubation for 24 h, at 37˚C with 95% air and 5%
CO2, the non-invasive and non-migratory cells were
removed. The invasive and migratory cells were fixed with 4%
polysorbate 30 min and stained with Giemsa stain 30 min at room
temperature. Stained cells were counted for quantification and
images captured using a light microscope (magnification, x100)
(26).
Wound healing assay
Cells transfected with LV5-ING3 and LV5-NC were
suspended and seeded into 12-well plates. Following cell adhesion,
sterile 200 µl pipette tips were used to scratch the cell
monolayers of each well. Plates were washed with PBS to remove
detached cells. Cells were cultured in serum-free medium. Wound
closure was observed using a light microscope (magnification, x100)
and measured at 0, 12 and 24 h. Image-pro Plus 6.0 software (Media
Cybernetics, Inc.) was used to measure the distance of the wound,
using the following formula: Percent wound closure = wound closure
distance of 12 or 24 h/wound closure distance of 0 h.
GeneMANIA protein interaction and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
analyses
Homo ING3 protein and other core protein
interactions were analyzed using the GeneMANIA database (http://genemania.org). Pathway analysis was performed
using the KEGG database (https://www.kegg.jp). All analyses were completed on
March 12, 2020, using the default parameters of the databases.
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corp.) and GraphPad Prism 6.0 software (GraphPad
Software, Inc.). Unpaired Student's t-test was used to compare
differences between two group, while one-way ANOVA and Bonferroni
post hoc test were used to compare differences between multiple
groups. Survival analysis was performed using the Kaplan-Meier
method and log-rank test. P<0.05 was considered to indicate a
statistically significant difference.
Results
ING3 expression in tissues and
prognosis
ING3 expression in breast cancer tissues and
normal tissues was determined using TCGA and UALCAN databases. As
presented in Fig. 1A, ING3
expression was significantly downregulated in cancer tissues
compared with normal tissue (P<0.001). ING3 was expressed
across different races, TNM stages, subclasses, menopause statuses,
ages, sex and histological subtypes, as presented in Fig. 1B-H. No difference in ING3
mRNA expression among different stages, menopausal states and sex
of patient with breast cancer was observed, however, ING3
mRNA expression was significantly higher in triple negative breast
cancer than in luminal and HER2 positive breast cancer
(P<0.001), ING3 mRNA expression was also higher in
Caucasians than in African Americans (P<0.01), the expression of
ING3 was also higher in ILC breast cancer than mixed breast
cancer (P<0.05).
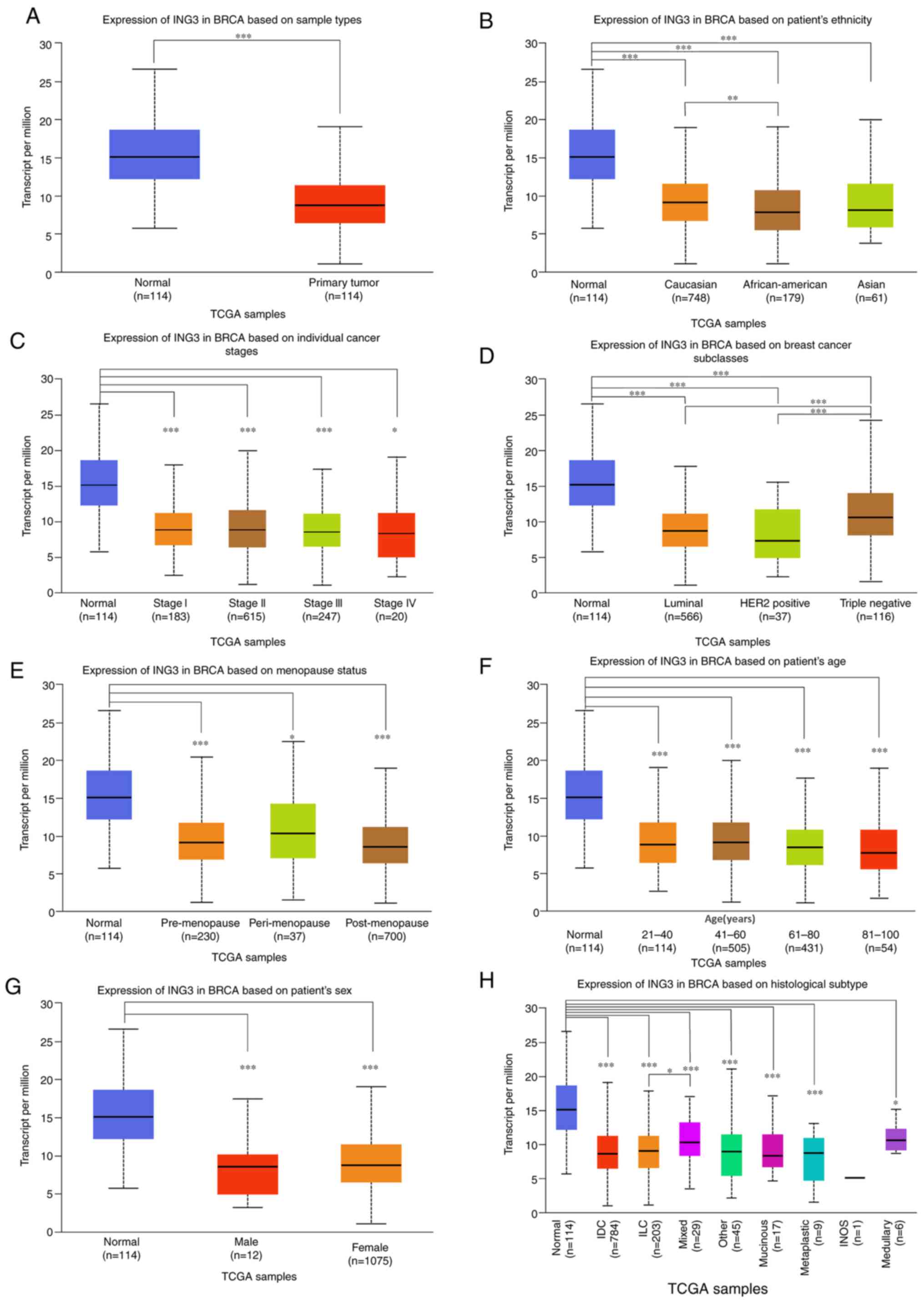 | Figure 1ING3 expression in breast
cancer tissues and normal tissues. (A) ING3 expression in
breast cancer tissues compared with normal tissues. ING3
expression in normal tissues compared with breast cancer tissues
among (B) different races, (C) different tumor-node-metastasis
stages, (D) different subclasses, (E) different menopause status,
(F) different ages, (G) different sex and (H) different
histological subtypes. *P<0.05;
**P<0.01; ***P<0.001. ING3, inhibitor
of growth 3; TCGA, The Cancer Genome Atlas; HER2, human epidermal
growth factor receptor 2; IDC (Infiltrating Ductal Carcinoma), ILC
(Infiltrating Lobular Carcinoma), INOS (Infiltrating Carcinoma
NOS). |
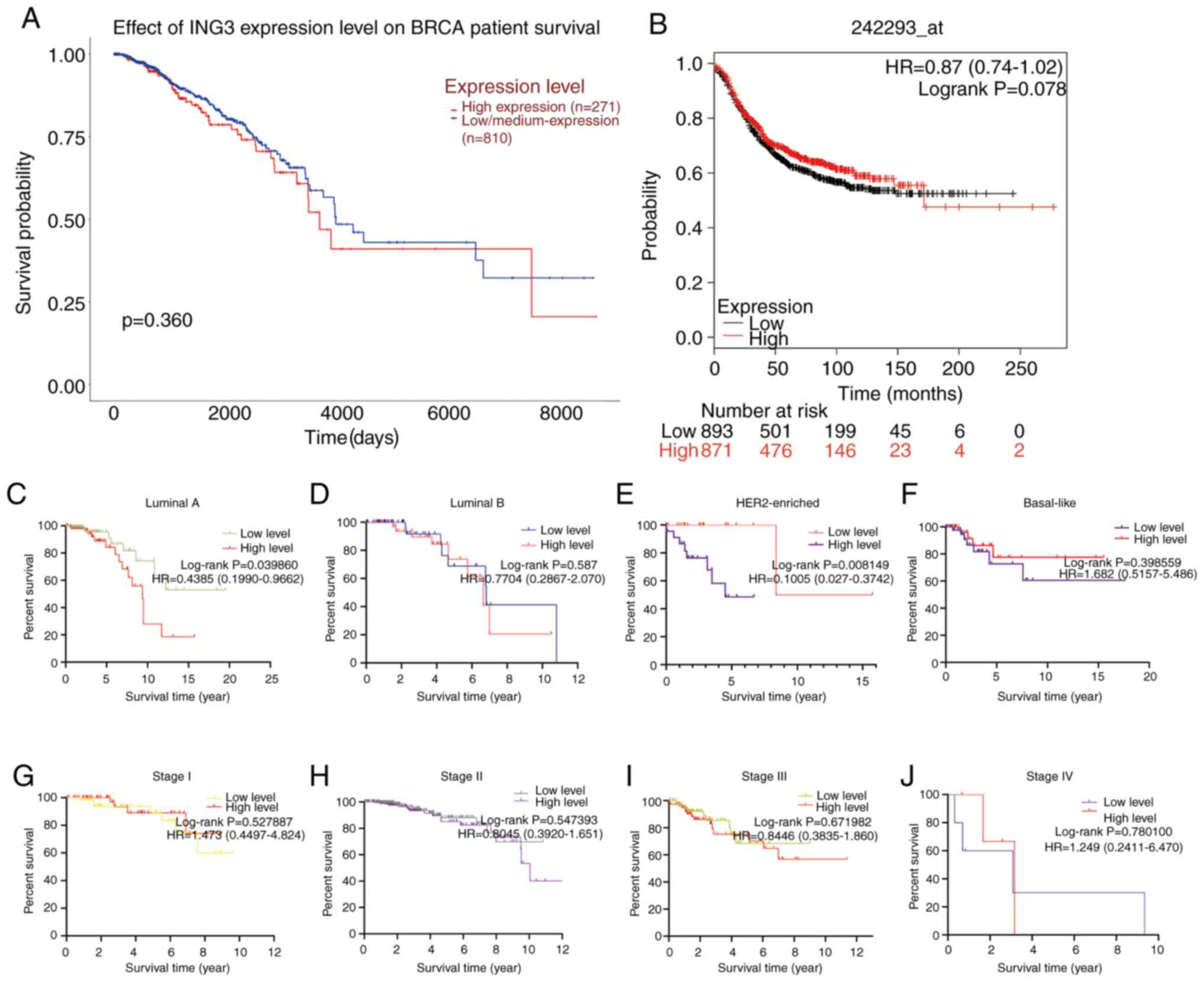
The present study investigated whether ING3
expression influences the prognosis of patients with breast cancer.
Survival analysis was performed using the UALCAN, TCGA, UCSC Xena
and KM-plot databases. Notably, no significant differences in
long-term survival were observed between patients with low and high
ING3 expression, respectively (P=0.360 by UALCAN; Fig. 2A and P=0.078 by KM-plot; Fig. 2B). However, ING3 expression
was significantly associated with prognosis in the luminal A
(P=0.0039860; Fig. 2C) and
HER2-enriched (P=0.008149; Fig. 2E)
subtypes. Conversely, ING3 expression was not associated with
prognosis in patients with different clinical stages of breast
cancer (Fig. 2G-J). In addition,
there was no significant difference between luminal B and
basal-like breast cancer (Fig. 2D
and F).
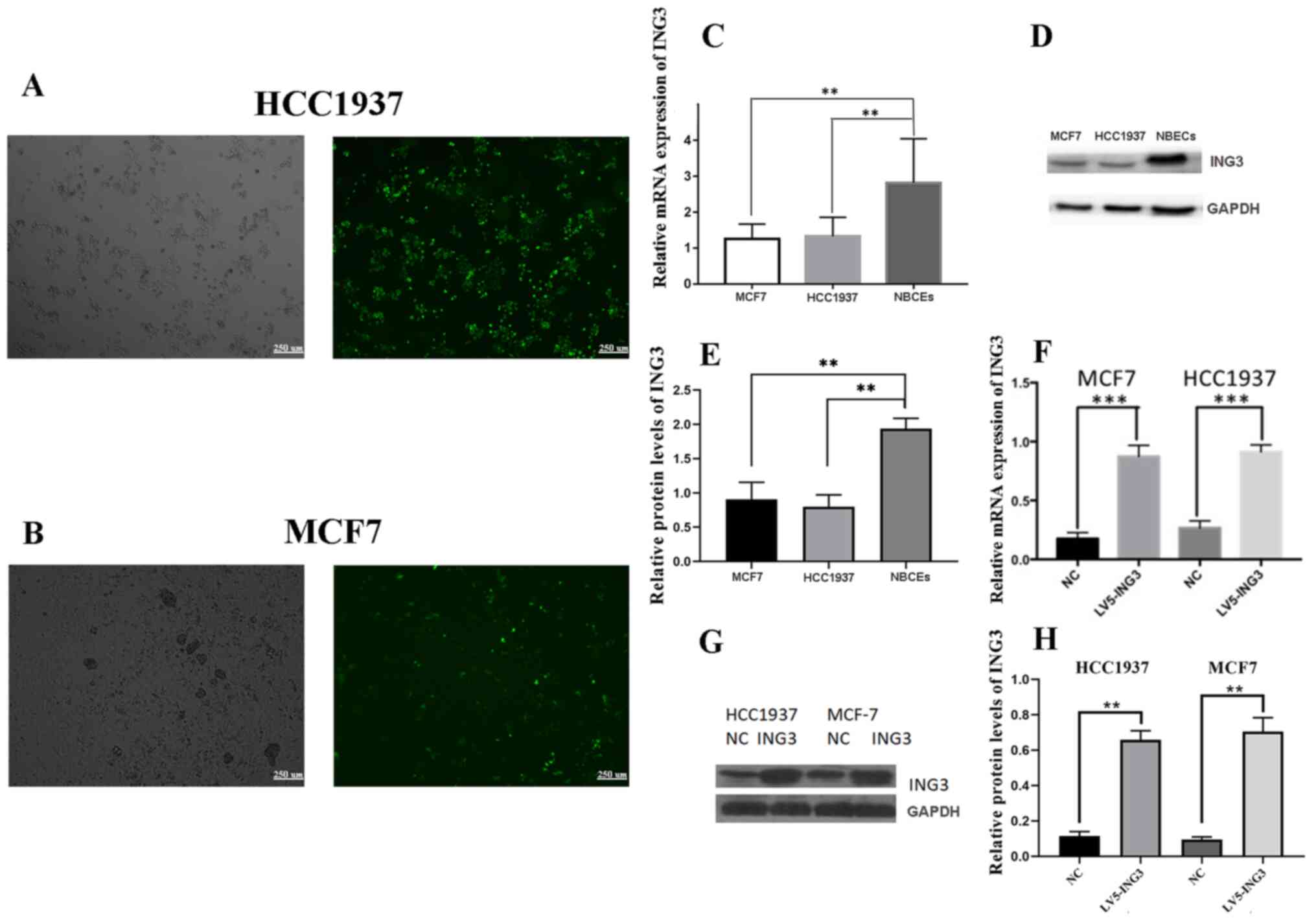
ING3 expression in cell lines and
lentiviral transduction
The results of the present study demonstrated that
both ING3 mRNA and protein expression levels were higher in
NBECs compared with MCF7 and HCC1937 cells (Fig. 3C-E). Lentiviral vectors
overexpressing ING3 (LV5-ING3) and LV5-NC were transfected
into MCF7 and HCC1937 cells. No significant differences were
observed in cell viability following transfection with the
corresponding lentivirus vectors (Fig.
S1). The transfection efficiency was detected via eGFP
expression by fluorescence microscopy (Fig. 3A and B). The mRNA and protein expression levels
of ING3 increased in MCF7 and HCC1937 cells following
transfection with LV5-ING3 compared with the LV5-NC group (Fig. 3F-H).
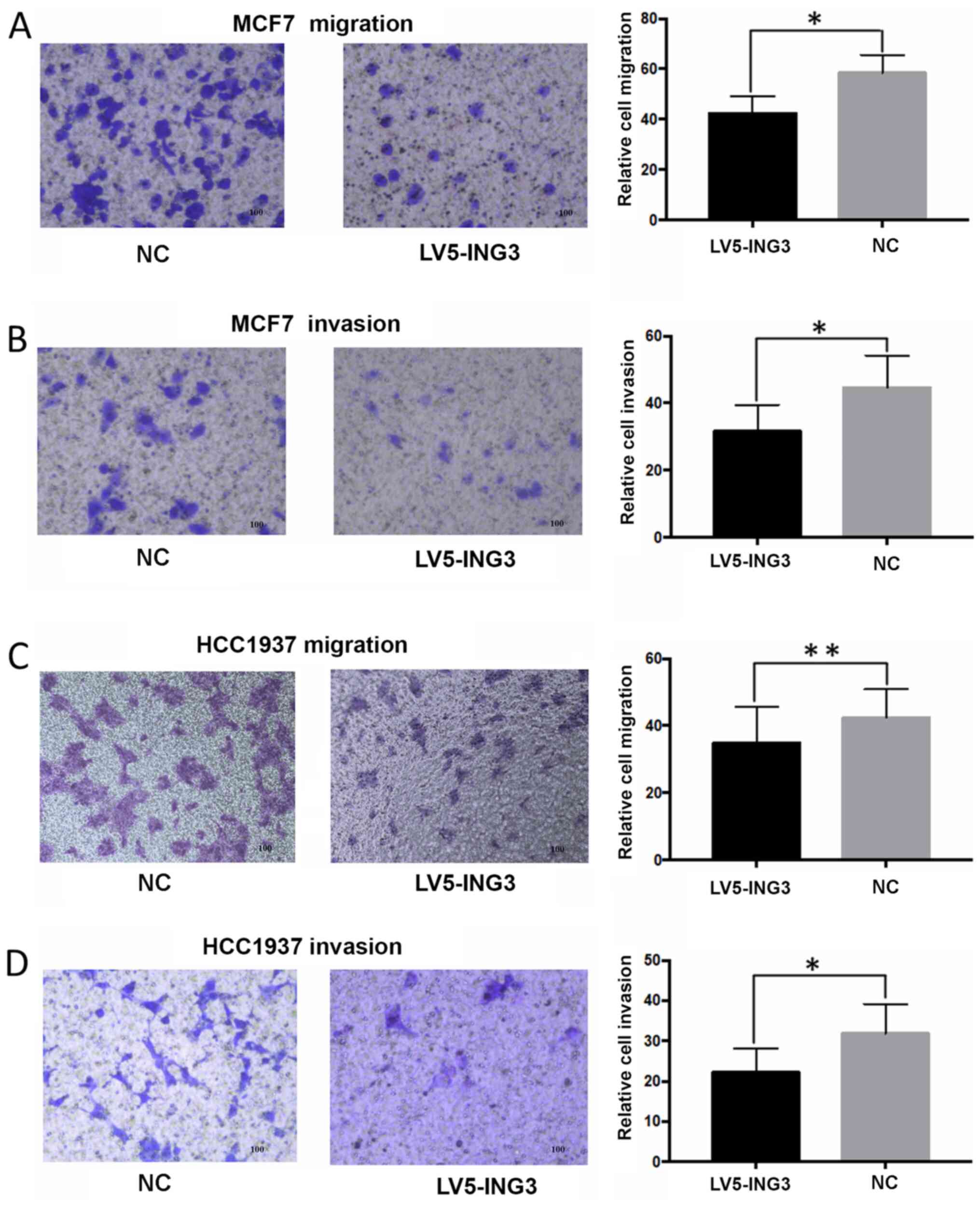
Overexpression of ING3 inhibits
migration and invasion
Overexpression of ING3 inhibited the
migratory and invasive abilities of MCF7 cells. The average
migration cell counts were 41±8 for the LV5-ING3 group compared
with 58±6 for the NC group (P<0.05; Fig. 4A). The average invasion cell counts
were 33±7 for the LV5-ING3 group compared with 42±8 for the NC
group (P<0.05; Fig. 4B).
Furthermore, overexpression of ING3 inhibited the migratory
and invasive abilities of HCC1937 cells. The average migration cell
counts were 37±7 for the LV5-ING3 group compared with 41±6 for the
NC group (P<0.01; Fig. 4C). The
average invasion cell counts were 23±6 for the LV5-ING3 group
compared with 32±7 for the NC group (P<0.05; Fig. 4D).
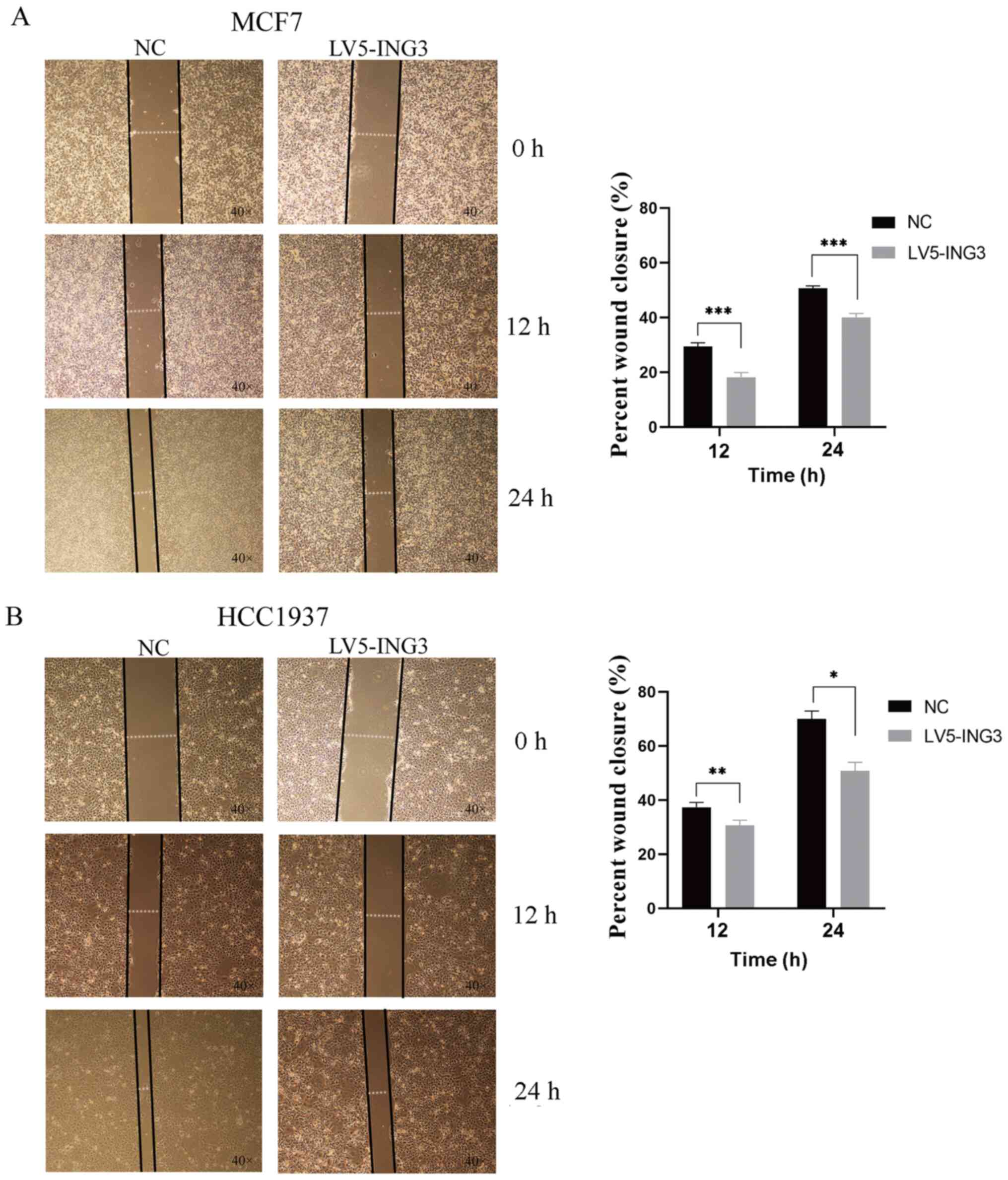
The results of the wound healing assay suggested a
similar phenomenon. In MCF7 cells, the percentage wound closure was
18.16±1.76 in the LV5-ING3 group vs. 29.40±1.37 in the NC group in
12 h (P<0.001; Fig. 5A).
Furthermore, the percentage wound closure was 40.03±1.48 in the
LV5-ING3 group vs. 50.63±0.95 in the NC group in 24 h (P<0.001;
Fig. 5A). In HCC1937 cells, the
percentage wound closure was 30.69±1.85 in the LV5-ING3 group vs.
37.35±1.78 in the NC group at 12 h (P<0.01; Fig. 5B). Furthermore, the percentage wound
closure was 50.76±3.22 in the LV5-ING3 group vs. 70.03±2.89 in the
NC group at 24 h (P<0.05; Fig.
5B).
Potential pathways regulated by
ING3
The present study aimed to investigate the potential
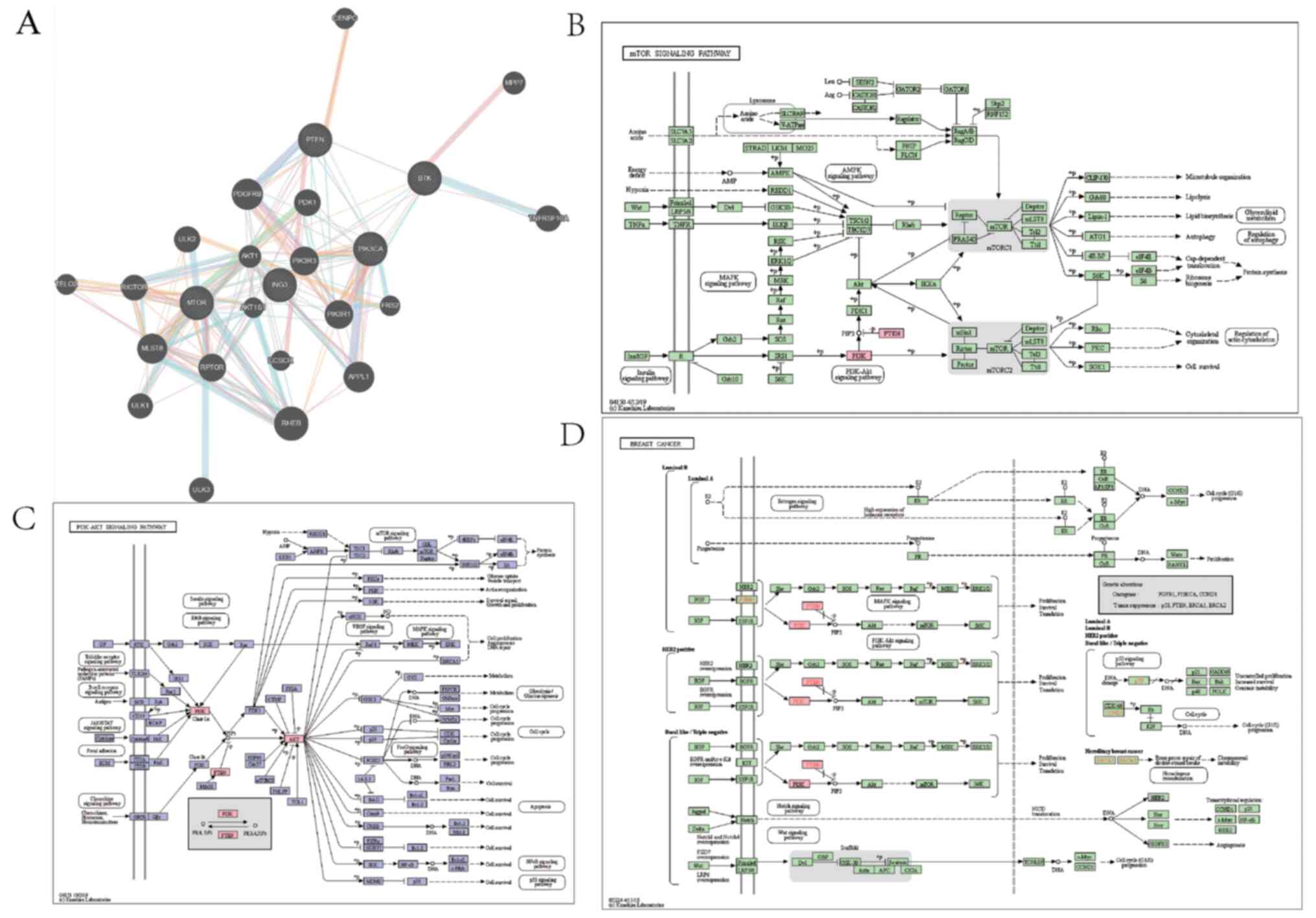
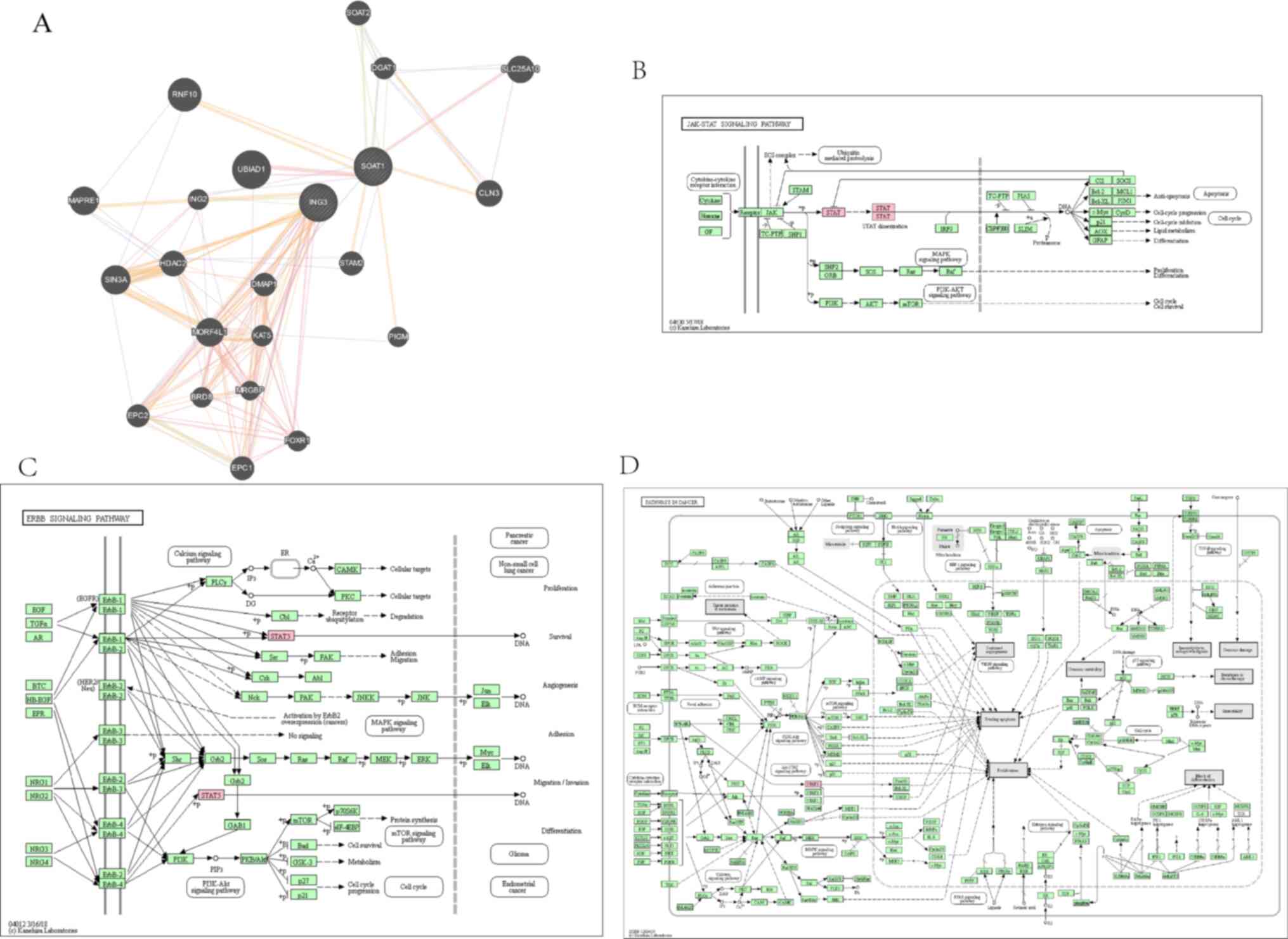
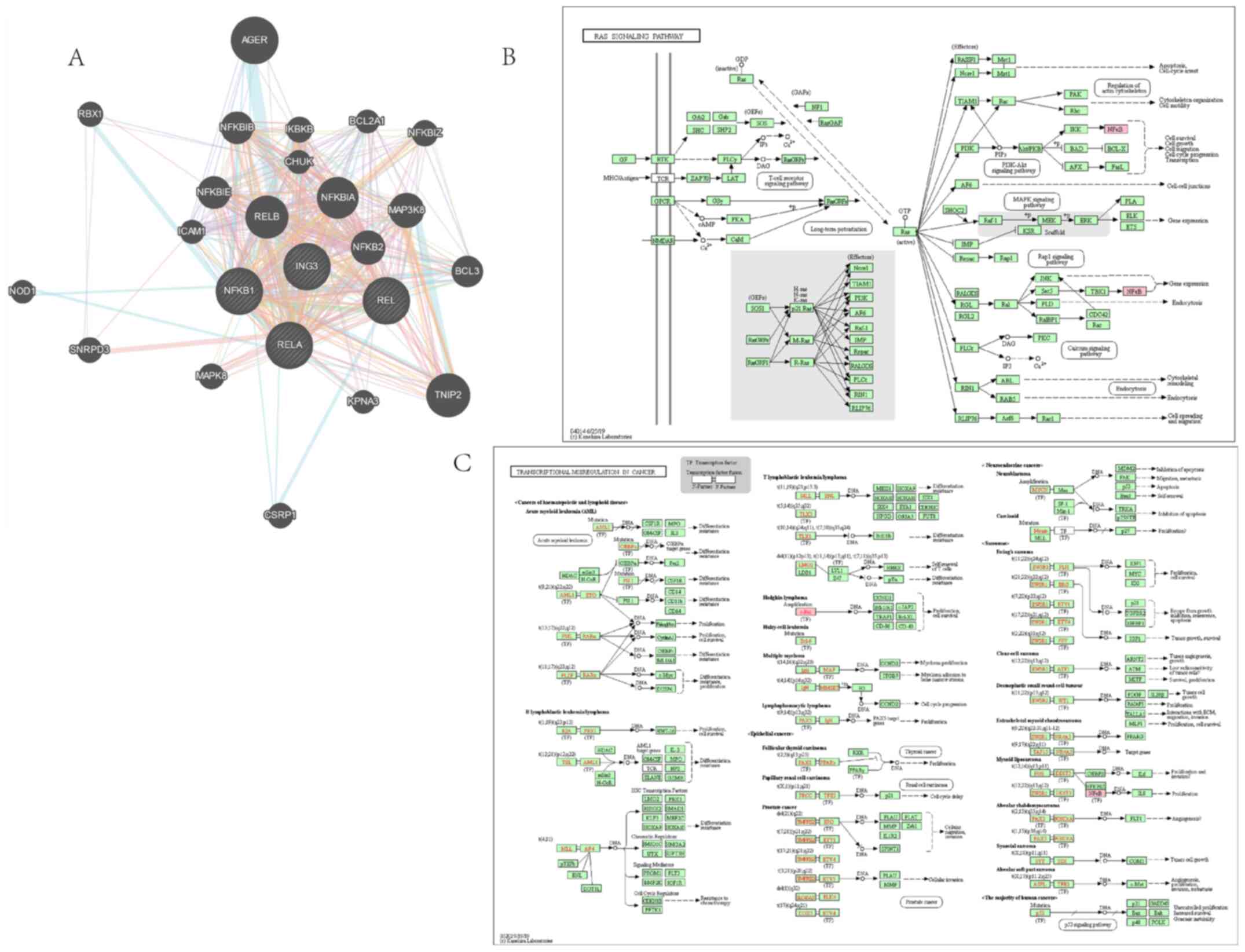
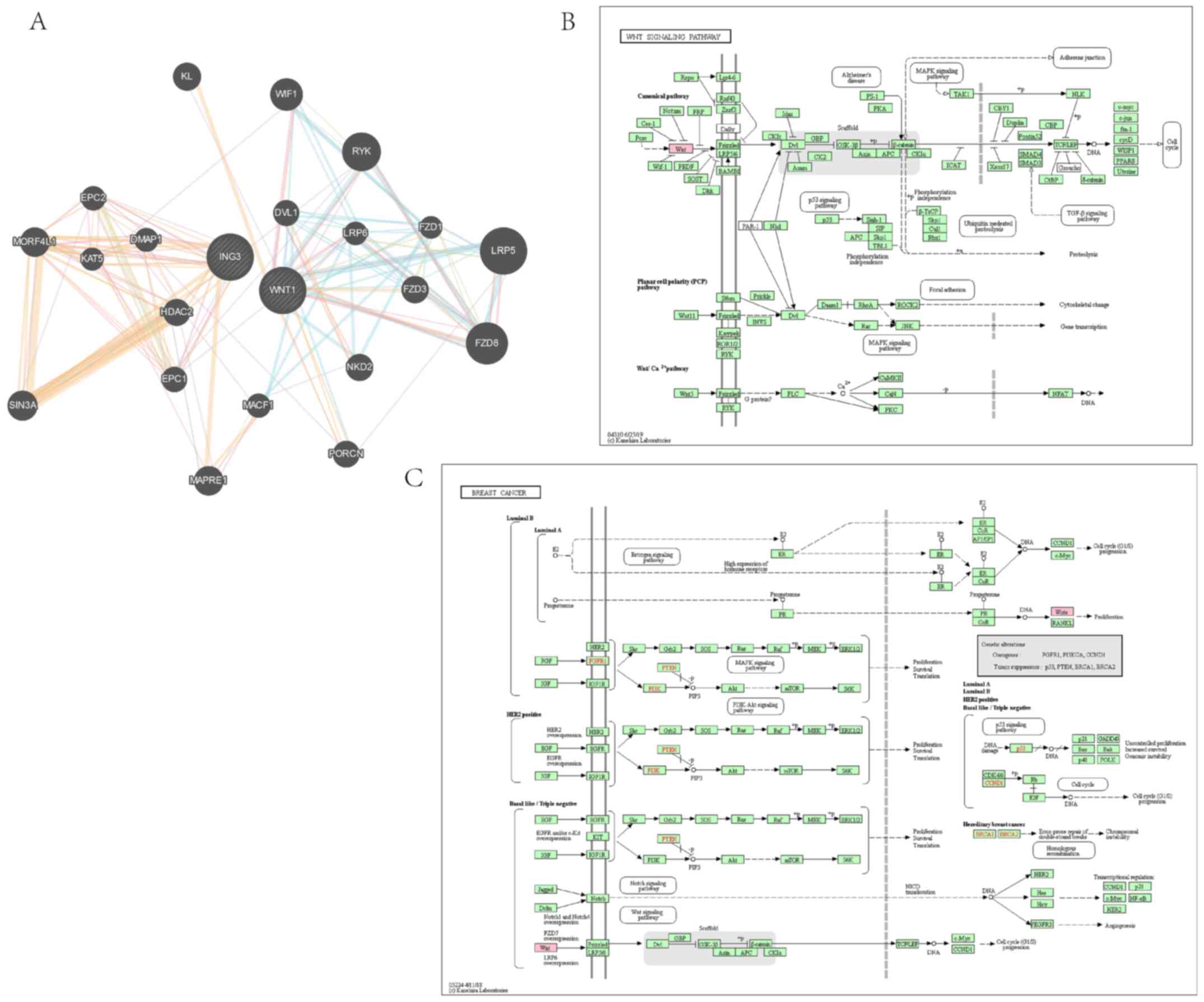
regulating mechanism of ING3. The PI3K/AKT, JAK/STAT, NF-κB and
Wnt/β-catenin pathways are closely associated with different types
of cancer, including breast cancer (27-33).
In the present study, GeneMANIA analysis exhibited interaction
between ING3 and the core protein of the PI3K/AKT pathway, while
KEGG pathway analysis demonstrated that ING3 may regulate the
PI3K/AKT pathway (Fig. 6). Similar
results were observed for the JAK/STAT (Fig. 7), NF-κB (Fig. 8) and Wnt/β-catenin (Fig. 9) pathways.
Discussion
Data from TCGA and UALCAN databases confirmed that
ING3 expression is downregulated in breast cancer tissues compared
with normal breast tissues, and similar results were observed in
breast cancer cell lines and NBECs. In the present study, ING3 had
prognostic significance in certain types of breast cancer, such as
luminal A and HER2-enriched breast cancer. To the best of the
authors' knowledge, the present study was the first to demonstrate
that overexpression of ING3 inhibits the migratory and invasive
abilities of breast cancer cells. The results demonstrated that the
PI3K/AKT, JAK/STAT, NF-κB and Wnt/β-catenin pathways are potential
pathways regulated by ING3.
Studies investigating the association between ING3
and cancer are gaining significant interest (18,34-36).
Increasing evidence suggest that ING3 is a key protein in cell
apoptosis (37), cell proliferation
and renewal (36), and tumor
biological behaviors (18). Yang
et al (35) reported that
ING3 expression is downregulated in gastric cancer. However, Nabbi
et al (19) demonstrated
that ING3 expression is upregulated in prostate cancer. The role of
ING3 in breast cancer remains largely unknown. The present study
compared ING3 expression across different races, TNM stages,
subclasses, menopause status, ages, sex and histological subtypes.
Notably, no significant differences were observed in the respective
comparisons. To the best of our knowledge, the present study is the
first to reveal that ING3 may act as a tumor suppressor gene in
breast cancer. The results of the present study differ from
previous findings on prostate cancer (18,19),
but are similar to studies on head and neck cancer (16,23).
The results of the present study suggest that
ING3 may be a prognostic biomarker for patients with breast
cancer. In the present study, high ING3 expression predicted poor
prognosis in patients with luminal A and HER2-enriched breast
cancer. However, ING3 expression was not associated with prognosis
in patients with breast cancer without classification, and no
significant association was observed between ING3 expression and
prognosis in patients with different clinical stages.
Metastasis is a key feature of malignancies
(38,39). Migration and invasion initiate
metastasis in vitro (40-42).
ING3 is considered a tumor suppressor in hepatocellular carcinoma,
which attenuates proliferation, migration and invasion (17). However, it is considered a
tumorigenesis promoter in prostate cancer (18,19).
The results of the present study demonstrated that ING3 mRNA and
protein expression levels were higher in NBECs compared with MCF7
and HCC1937 cells. Similar results were observed between breast
cancer tissues and normal tissues. The results of the Transwell
migration and invasion, and wound healing assays demonstrated that
overexpression of ING3 inhibited the migratory and invasive
abilities of MCF7 cells. Collectively, these results suggest that
ING3 acts as a tumor suppressor in breast cancer, influencing
biological behaviors, particularly attenuating migration and
invasion.
In the present study, high ING3 mRNA expression was
associated with poor prognosis, while overexpression of ING3
inhibited the metastasis of breast cancer cells. However, gene
expression is a complex biological process, and further studies are
required to validate gene function at the mRNA level, as only a
weak association was observed between mRNA and protein expression
(43,44). Previous studies have demonstrated
that ING3 protein can be rapidly degraded by the
SCFskp2-mediated ubiquitin-protease system (45,46).
Thus, prospective studies will focus on investigating the
association between ING3 protein expression and prognosis.
Studying the mechanisms of metastasis is important
in identifying novel anti-cancer drugs and developing cancer
therapy (47-49).
The activation of cancer-related pathways as a promotor of
tumorigenesis, proliferation, migration and invasion in several
types of cancer is generally accepted (50,51).
ING3 regulates cell proliferation, apoptosis and cell cycle in
gastric cancer via the PI3K/AKT pathway (23). However, the mechanisms by which ING3
regulates pathways in breast cancer remain unclear. The results of
the present study revealed interactions between ING3 and core
proteins of the PI3K/AKT, JAK/STAT, NF-κB and Wnt/β-catenin
pathways. In addition, KEGG pathway analysis indicated that ING3
potentially regulates the PI3K/AKT, JAK/STAT, NF-κB and
Wnt/β-catenin pathways. Although these assumptions were not proven
in the present study, they remain valid hypotheses and will be the
focus of prospective studies.
In conclusion, the results of the present study
suggest that ING3 plays a key role in breast cancer. ING3
expression was downregulated in breast cancer tissues compared with
normal tissues. In addition, ING3 expression influenced the
prognosis of patients with different molecular subtypes. Notably,
overexpression of ING3 inhibited migration and invasion in
vitro. Thus, ING3 may be used to regulate the biological
behavior of breast cancer via tumor-related pathways.
Supplementary Material
Cell viability following transfection.
No significant differences were observed in cell viability
following transfection of MCF7 and HCC1937 cells with LV5-NC or
LV5-ING3 lentivirus vectors. ING3, inhibitor of growth 3; NC,
negative control.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant nos. 81960542 and 81960517), the
Science and Technology Project of Yunnan Provincial Science and
Technology Department (grant nos. 202001AU070053 and
202001AU070093), the Scientific Research Foundation of Yunnan
Education Department (grant nos. 2019J1288 and 2020J0198), the
Research Institution Project of the Health Science and Technology
Plan of Yunnan Province (grant no. 2018NS0055) and the Yunnan
Health Training Project of High Level Talents (grant no.
H-2019075).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
The study was designed by ST, RL, DL and HZ. DL, MW,
YT, KZ and WC carried out the funding obtain and manuscript review.
HL (first author), RG, DL, RL downloaded the gene expression and
clinical data from the TCGA database.ST, HL (first author), XT in
charge of bioinformatics analysis. HZ, YT and WC is responsible for
cell transfection, molecular biology experiments and cell function
experiments. The data were statistically analyzed by MW, KZ, HL, MT
and KW. HL(first author) and ST write the manuscript, and all
authors participate in the revision. ST and HZ confirm the
authenticity of all the raw data. All authors reviewed and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committees of the Third Affiliated Hospital of Kunming Medical
University, Yunnan Cancer Hospital, Kunming, China; approval no.
QT202002, and written informed consent was provided by all patients
prior to the beginning of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin: Feb 4, 2021 (Epub ahead
of print). doi: 10.3322/caac.21660.
|
|
4
|
Schwartz RS and Erban JK: Timing of
metastasis in breast cancer. N Engl J Med. 376:2486–2488.
2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mouche A, Archambeau J, Ricordel C,
Chaillot L, Bigot N, Guillaudeux T, Grenon M and Pedeux R: ING3 is
required for ATM signaling and DNA repair in response to DNA double
strand breaks. Cell Death Differ. 26:2344–2357. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ludwig S, Klitzsch A and Baniahmad A: The
ING tumor suppressors in cellular senescence and chromatin. Cell
Biosci. 1(25)2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gou WF, Yang XF, Shen DF, Zhao S, Sun HZ,
Luo JS and Zheng HC: Immunohistochemical profile of ING3 protein in
normal and cancerous tissues. Oncol Lett. 13:1631–1636.
2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Shiseki M, Nagashima M, Pedeux RM,
Kitahama-Shiseki M, Miura K, Okamura S, Onogi H, Higashimoto Y,
Appella E, Yokota J and Harris CC: p29ING4 and p28ING5 bind to p53
and p300, and enhance p53 activity. Cancer Res. 63:2373–2378.
2003.PubMed/NCBI
|
|
9
|
Bose P, Thakur SS, Brockton NT, Klimowicz
AC, Kornaga E, Nakoneshny SC, Riabowol KT and Dort JC: Tumor cell
apoptosis mediated by cytoplasmic ING1 is associated with improved
survival in oral squamous cell carcinoma patients. Oncotarget.
5:3210–3219. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang R, Jin J, Shi J and Hou Y: INGs are
potential drug targets for cancer. J Cancer Res Clin Oncol.
143:189–197. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Dantas A, Al Shueili B, Yang Y, Nabbi A,
Fink D and Riabowol K: Biological functions of the ING proteins.
Cancers (Basel). 11(1817)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liu XL, Meng J, Zhang XT, Liang XH, Zhang
F, Zhao GR and Zhang T: ING5 inhibits lung cancer invasion and
epithelial-mesenchymal transition by inhibiting the WNT/β-catenin
pathway. Thorac Cancer. 10:848–855. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu XL, Zhang XT, Meng J, Zhang HF, Zhao
Y, Li C, Sun Y, Mei QB, Zhang F and Zhang T: ING5 knockdown
enhances migration and invasion of lung cancer cells by inducing
EMT via EGFR/PI3K/Akt and IL-6/STAT3 signaling pathways.
Oncotarget. 8:54265–54276. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cai L, Li H, Chen C, Cheng X, Wang Y, Liu
J, Wang Y and Hao L: Role of inhibitor of growth 4 in the
suppression of human melanoma cells through the Fas/FasL-mediated
apoptosis pathway. Int J Mol Med. 41:1055–1061. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Suzuki S, Nozawa Y, Tsukamoto S, Kaneko T,
Imai H and Minami N: ING3 is essential for asymmetric cell division
during mouse oocyte maturation. PLoS One. 8(e74749)2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li X, Zhang Q, Zhang M, Luo Y and Fu Y:
Downregulation of nuclear ING3 expression and translocalization to
cytoplasm promotes tumorigenesis and progression in head and neck
squamous cell carcinoma (HNSCC). Histol Histopathol. 35:681–690.
2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lu M, Chen F, Wang Q, Wang K, Pan Q and
Zhang X: Downregulation of inhibitor of growth 3 is correlated with
tumorigenesis and progression of hepatocellular carcinoma. Oncol
Lett. 4:47–52. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
McClurg UL, Nabbi A, Ricordel C, Korolchuk
S, McCracken S, Heer R, Wilson L, Butler LM, Irving-Hooper BK,
Pedeux R, et al: Human ex vivo prostate tissue model system
identifies ING3 as an oncoprotein. Br J Cancer. 118:713–726.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Nabbi A, McClurg UL, Thalappilly S, Almami
A, Mobahat M, Bismar TA, Binda O and Riabowol KT: ING3 promotes
prostate cancer growth by activating the androgen receptor. BMC
Med. 15(103)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang Y, Dai DL, Martinka M and Li G:
Prognostic significance of nuclear ING3 expression in human
cutaneous melanoma. Clin Cancer Res. 13:4111–4116. 2007.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhou R, Rotte A, Li G, Chen X, Chen G and
Bhandaru M: Nuclear localization of ING3 is required to suppress
melanoma cell migration, invasion and angiogenesis. Biochem Biophys
Res Commun. 527:418–424. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wu X, Chen C, Luo B, Yan D, Yan H, Chen F,
Guan F, Wu H and Yuan J: Nuclear ING3 expression is correlated with
a good prognosis of breast cancer. Front Oncol.
10(589009)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhao S, Wang L, Zhang C, Deng Y, Zhao B,
Ren Y, Fu Y and Meng X: Inhibitor of growth 3 induces cell death by
regulating cell proliferation, apoptosis and cell cycle arrest by
blocking the PI3K/AKT pathway. Cancer Gene Ther. 25:240–247.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Team C. Team RDC.R: A Language and
environment for statistical computing. R Foundation for Statistical
Computing, Vienna, Austria, 2012.
|
|
25
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474.
2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tang S, Pan H, Wei W, Yang H, Liu J and
Yang R: GOLPH3: A novel biomarker that correlates with poor
survival and resistance to chemotherapy in breast cancer.
Oncotarget. 8:105155–105169. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Mayer IA and Arteaga CL: The PI3K/AKT
pathway as a target for cancer treatment. Annu Rev Med. 67:11–28.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Torres-Arzayus MI, Font de Mora J, Yuan J,
Vazquez F, Bronson R, Rue M, Sellers WR and Brown M: High tumor
incidence and activation of the PI3K/AKT pathway in transgenic mice
define AIB1 as an oncogene. Cancer Cell. 6:263–274. 2004.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Laurent C, Nicolae A, Laurent C, Le Bras
F, Haioun C, Fataccioli V, Amara N, Adélaïde J, Guille A, Schiano
JM, et al: Gene alterations in epigenetic modifiers and JAK-STAT
signaling are frequent in breast implant-associated ALCL. Blood.
135:360–370. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jiang L, Zhao XH, Mao YL, Wang JF, Zheng
HJ and You QS: Long non-coding RNA RP11-468E2.5 curtails colorectal
cancer cell proliferation and stimulates apoptosis via the JAK/STAT
signaling pathway by targeting STAT5 and STAT6. J Exp Clin Cancer
Res. 38(465)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Davis RT, Blake K, Ma D, Gabra M,
Hernandez GA, Phung AT, Yang Y, Maurer D, Lefebvre A, Alshetaiwi H,
et al: Transcriptional diversity and bioenergetic shift in human
breast cancer metastasis revealed by single-cell RNA sequencing.
Nat Cell Biol. 22:310–320. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Matsumoto S, Yamamichi T, Shinzawa K,
Kasahara Y, Nojima S, Kodama T, Obika S, Takehara T, Morii E,
Okuyama H, et al: GREB1 induced by Wnt signaling promotes
development of hepatoblastoma by suppressing TGFβ signaling. Nat
Commun. 10(3882)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wellenstein MD, Coffelt SB, Duits D, van
Miltenburg MH, Slagter M, de Rink I, Henneman L, Kas SM, Prekovic
S, Hau CS, et al: Loss of p53 triggers WNT-dependent systemic
inflammation to drive breast cancer metastasis. Nature.
572:538–542. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Fink D, Yau T, Nabbi A, Wagner B, Wagner
C, Hu SM, Lang V, Handschuh S, Riabowol K and Rülicke T: Loss of
Ing3 expression results in growth retardation and embryonic death.
Cancers (Basel). 12(80)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Yang C, Gao J, Yan N, Wu B, Ren Y, Li H
and Liang J: Propofol inhibits the growth and survival of gastric
cancer cells in vitro through the upregulation of ING3. Oncol Rep.
37:587–593. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Nabbi A, Almami A, Thakur S, Suzuki K,
Boland D, Bismar TA and Riabowol K: ING3 protein expression
profiling in normal human tissues suggest its role in cellular
growth and self-renewal. Eur J Cell Biol. 94:214–222.
2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Luo J, Shah S, Riabowol K and Mains PE:
The Caenorhabditis elegans ing-3 gene regulates ionizing
radiation-induced germ-cell apoptosis in a p53-associated pathway.
Genetics. 181:473–482. 2009.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Padmanaban V, Krol I, Suhail Y, Szczerba
BM, Aceto N, Bader JS and Ewald AJ: E-cadherin is required for
metastasis in multiple models of breast cancer. Nature.
573:439–444. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zeng Q, Michael IP, Zhang P, Saghafinia S,
Knott G, Jiao W, McCabe BD, Galván JA, Robinson H, Zlobec I, et al:
Synaptic proximity enables NMDAR signalling to promote brain
metastasis. Nature. 573:526–531. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chen L, Yang S, Jakoncic J, Zhang JJ and
Huang XY: Migrastatin analogues target fascin to block tumour
metastasis. Nature. 464:1062–1066. 2010.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Goetz JG, Minguet S, Navarro-Lérida I,
Lazcano JJ, Samaniego R, Calvo E, Tello M, Osteso-Ibáñez T,
Pellinen T, Echarri A, et al: Biomechanical remodeling of the
microenvironment by stromal caveolin-1 favors tumor invasion and
metastasis. Cell. 146:148–163. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
de Sousa Abreu R, Penalva LO, Marcotte EM
and Vogel C: Global signatures of protein and mRNA expression
levels. Mol Biosyst. 5:1512–1526. 2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Maier T, Güell M and Serrano L:
Correlation of mRNA and protein in complex biological samples. FEBS
Lett. 583:3966–3973. 2009.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chen G, Wang Y, Garate M, Zhou J and Li G:
The tumor suppressor ING3 is degraded by SCF(Skp2)-mediated
ubiquitin-proteasome system. Oncogene. 29:1498–1508.
2010.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Tallen G and Riabowol K: Keep-ING balance:
Tumor suppression by epigenetic regulation. FEBS Lett.
588:2728–2742. 2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Roe JS, Hwang CI, Somerville TDD, Milazzo
JP, Lee EJ, Da Silva B, Maiorino L, Tiriac H, Young CM, Miyabayashi
K, et al: Enhancer reprogramming promotes pancreatic cancer
metastasis. Cell. 170:875–888.e20. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Weissmueller S, Manchado E, Saborowski M,
Morris JP VI, Wagenblast E, Davis CA, Moon SH, Pfister NT,
Tschaharganeh DF, Kitzing T, et al: Mutant p53 drives pancreatic
cancer metastasis through cell-autonomous PDGF receptor β
signaling. Cell. 157:382–394. 2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Santen RJ, Veldhuis J, Samojlik E, Lipton
A, Harvey H and Wells SA: Mechanism of action of aminoglutethimide
in breast cancer. Lancet. 1:44–45. 1979.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li J, Cheng D, Zhu M, Yu H, Pan Z, Liu L,
Geng Q, Pan H, Yan M and Yao M: OTUB2 stabilizes U2AF2 to promote
the Warburg effect and tumorigenesis via the AKT/mTOR signaling
pathway in non-small cell lung cancer. Theranostics. 9:179–195.
2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wu X, Zhou Z, Xu S, Liao C, Chen X, Li B,
Peng J, Li D and Yang L: Extracellular vesicle packaged
LMP1-activated fibroblasts promote tumor progression via autophagy
and stroma-tumor metabolism coupling. Cancer Lett. 478:93–106.
2020.PubMed/NCBI View Article : Google Scholar
|