Introduction
Hepatocellular carcinoma (HCC) is a main type of
primary malignant tumor and is the third leading cause of
cancer-related death worldwide (1,2).
Despite advances in diagnosis and treatment, the long-term survival
rate of patients with HCC remains poor (~5 years) due to the early
metastasis and high recurrence rate (3). Therefore, identifying a novel
molecular mechanism and developing effective therapeutic strategies
for HCC is important.
Long non-coding RNAs (lncRNAs) are transcripts that
are >200 nucleotides in length and lack protein coding capacity
(4). Increasing evidence has
suggested that lncRNAs serve critical functions in diverse
biological and cellular processes, such as growth, invasion,
metastasis, differentiation and tumorigenesis (5-8).
The dysregulation of lncRNAs has been reported to be closely
associated with numerous tumors, including HCC (9). For example, Li et al (10) reported that lncRNA focally amplified
long non-coding RNA in epithelial cancer (FAL1) was upregulated in
HCC, and FAL1 overexpression promoted proliferation and metastasis
via regulating microRNA (miRNA/miR)-1236 expression. Ma et
al (11) demonstrated that
lncRNA CDKN2B antisense RNA 1 knockdown inhibited cell
proliferation, metastasis and invasion via competitively binding to
miR-122-5p in HCC. Double homeobox A pseudogene 8 (DUXAP8) is a
pseudogene-derived lncRNA that maps to chromosome 20q11, and has
been reported to be an oncogene and a potential biomarker in
various tumors (12,13). Moreover, previous reports have also
verified that DUXAP8 overexpression induced cell proliferation and
migration in renal cell carcinoma (14) and non-small-cell lung cancer
(15). However, the function and
molecular mechanism underlying DUXAP8 in HCC progression is not
completely understood.
miRNAs, a class of short non-coding RNAs, are
involved in the regulation of protein-coding genes by suppressing
mRNA translation (16). Previous
studies have demonstrated that miRNAs could exert a tumor
suppressor role in various types of cancer. For instance,
miR-574-3p has been indicated to inhibit the malignant behavior of
liver cancer cells via interacting with disintegrin and
metalloproteinase domain-containing protein 28(17). Moreover, miR-27b-5p has been
reported to repress the growth and metastasis of ovarian carcinoma
cells by targeting C-X-C motif chemokine 1(18). In addition, the suppressive effect
of miRNAs, such as miR-188-5p and miR-1271, on the progression of
HCC has also been demonstrated in previous studies (19,20).
Furthermore, a previous study indicated that miR-9-3p served as a
tumor suppressor and constrained cell proliferation by targeting
tafazzin expression in HCC cells (21). However, the mechanism underlying
miR-9-3p in HCC is not completely understood.
Insulin-like growth factor 1 receptor (IGF1R) has
tyrosine kinase activity and has been demonstrated to function as
an antiapoptotic agent by enhancing cell survival in a variety of
different types of cancer (22,23).
For example, knockdown of IGF1Rhas been indicated to decrease cell
proliferation, migration, and invasion in ovarian cancer cells
(24). Similarly, downregulation of
IGF1R has been revealed to suppress cell growth and improve
cisplatin sensitivity of head and neck squamous cell carcinoma
cells (25). Moreover, some studies
reported that IGF1R was upregulated, and promoted proliferation,
migration, invasion and EMT in HCC (26-30).
The results suggested that IGF1R served important roles in HCC
progression.
Therefore, the aim of the present study was to
investigate the function of DUXAP8 and to explore whether the
involvement of DUXAP8 in HCC was mediated via the miR-9-3p/IGF1R
axis.
Materials and methods
Clinical specimens and cell
culture
HCC pathologically confirmed tumor tissues and
adjacent healthy tissues (>5 cm from the tumor margin) were
obtained from 38 patients with HCC (31-85 years old) who had
undergone surgical treatment at People's Hospital of Dongying
between January 2011 and June 2014. The present study was approved
by People's Hospital of Dongying Ethics Committee (Dongying,
China). Written informed consent was obtained from all
patients.
Human normal liver cell line (THLE-2) and HCC cell
lines (SK-HEP-1 and Hep3B) were purchased from the American Type
Culture Collection. HCC cell lines (Huh-7 and Huh-1) were purchased
from the Japanese Collection of Research Bioresources Cell Bank.
All cells were maintained at 37˚C with 5% CO2 in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Invitrogen; Thermo Fisher Scientific, Inc.).
Cell transfection
DUXAP8 small interfering (si)RNA (si-DUXAP8#1,
5'-AGTTCAGTGTATTTGATAATA-3'; si-DUXAP8#2,
5'-GATTTGGTTTCAGAATCAAAG-3'; si-DUXAP8#3,
5'-GATGGTGTTGTACCACCTATA-3') and scramble siRNA control
[si-negative control (NC): 5'-TTCTCCGAACGTGTCACGTTT-3'] were
purchased from Shanghai GenePharma Co., Ltd. miR-9-3p mimic
(5'-ATAAAGCTAGATAACCGAAAGT-3'), scramble mimic control (miR-NC;
5'-TTCTCCGAACGTGTCACGTTT-3'), miR-9-3p inhibitor
(5'-ACTTTCGGTTATCTAGCTTTAT-3') and scramble inhibitor control
(anti-miR-NC; 5'-CAGTACTTTTGTGTAGTACAA-3') were purchased from
Applied Biosystems; Thermo Fisher Scientific, Inc. Furthermore, to
overexpress DUXAP8 or IGF1R, the full length sequence of DUXAP8 or
IGF1R was inserted into the pcDNA3.1 vector (Invitrogen; Thermo
Fisher Scientific, Inc.) to generate pcDNA3.1-DUXAP8 (DUXAP8) and
pcDNA3.1-IGF1R (IGF1R), respectively. pcDNA3.1 empty vector was
used as negative control. SK-HEP-1 and Huh-7 cells in 6-well plates
(2x105 cells/well) were transfected with
oligonucleotides (50 nM) and plasmids (2 µg) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Following incubation for 48 h, the transfected
cells were harvested and utilized for subsequent experiments.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from clinical tissues and
HCC cells using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). Subsequently, total RNA was reverse
transcribed into cDNA using M-MLV reverse transcriptase (Thermo
Fisher Scientific, Inc.) including 5x buffer, 10 mM dNTPs, and 50
µmol oligo dT primers (cat. no. 18418012; Thermo Fisher Scientific,
Inc.). The RT reaction was performed at 70˚C for 10 min followed by
42˚C for 1 h, according to the manufacturer's protocol. DUXAP8 and
IGF1R mRNA expression levels were measured via qPCR using SYBR
Premix Ex Taq (Takara Biotechnology Co., Ltd.). The thermocycling
conditions were as follows: Denaturation at 95˚C for 10 min,
followed by 40 cycles of denaturation at 95˚C for 30 sec, annealing
at 60˚C for 30 sec and extension at 72˚C for 1 min.
Has-miR-9-3p-specific TaqMan primer (5'-ATAAAGCTAGATAACCGAAAGT-3')
was purchased from Applied Biosystems (cat. no. 4427975; Thermo
Fisher Scientific, Inc.; Assay ID, 002231) for synthesis of cDNA.
miR-9-3p expression levels were measured via qPCR using a TaqMan
MicroRNA Assay Kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The thermocycling conditions were as follows: 16˚C for 30
min, 42˚C for 30 min, 85˚C for 5 min and 4˚C hold. The following
primers were used for qPCR: DUXAP8 forward, 5'-AGACGCCATGGAACAT-3'
and reverse, 5'-AAGCGGAGACCTGAGGAG-3'; GAPDH forward,
5'-AGAAGGCTGGGGCTCATTTG-3' and reverse, 5'-AGGGGCCATCCACAGTCTTC-3';
IGF1R forward, 5'-TTTCCCACAGCAGTCCACCTC-3' and reverse,
5'-AGCATCCTAGCCTTCTCACCC-3'; miR-9-3p forward,
5'-TCTTTGGTTATCTAGCTGTAT-3' and reverse, 5'-GAACATGTCTGCGTATCTC-3';
and U6 forward, 5'-GCTTCGGCAGCACATATACTAAAA-3' and reverse,
5'-CGCTTCACGAATTTGCGTGTCAT-3'. Moreover, miRNA and mRNA expression
levels were quantified using the 2-ΔΔCq method (31) and normalized to the internal
reference genes U6 and GAPDH, respectively.
Cell proliferation assay
Cell proliferation was detected by performing a Cell
Counting Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology)
according to the manufacturer's protocol. Briefly, transfected
SK-HEP-1 and Huh-7 cells (2.5-5x103 cells/well) were
inoculated into 96-well plates and incubated for 48 h at 37˚C.
Subsequently, CCK-8 (10 µl) solution was added to each well and
cultured at 37˚C for 0, 24, 48 or 72 h. The optical density value
of each well was measured at a wavelength of 450 nm using a
Varioskan Flash Microplate Reader (Thermo Fisher Scientific,
Inc.).
Cell migration and invasion
assays
Cell migration and invasion were detected using
24-well Transwell chambers (BD Biosciences) according to the
manufacturer's protocol. Transfected SK-HEP-1 and Huh-7 cells were
resuspended in serum-free DMEM overnight. Subsequently, cells
(5x104) were plated into the upper chamber, which was
pre-coated with Matrigel® at 4˚C overnight and
subsequently maintained at 37˚C for 4-5 h (BD Biosciences) for the
cell invasion assay. DMEM supplemented with 10% FBS (Invitrogen;
Thermo Fisher Scientific, Inc.) was plated in the lower chamber.
Following incubation for 24 h at 37˚C, cells in the upper chamber
were removed using cotton swabs. Migratory or invading cells were
fixed using methanol for 30 min at 4˚C and stained with 0.1%
crystal violet solution for 20 min at 37˚C, followed by observation
using an inverted fluorescence microscope (magnification, x100;
Nikon Corporation).
Western blotting
Western blotting was performed to measure the
protein expression levels of IGF1R and EMT-related proteins
(E-cadherin, N-cadherin and vimentin) in SK-HEP-1 and Huh-7 cells.
Total protein was isolated from cells using pre-cold RIPA buffer
(Beyotime Institute of Biotechnology) including protease inhibitor.
Total protein was quantified using a BCA protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.). Proteins (50 µg) were separated
via 10% SDS-PAGE and electrotransferred to nitrocellulose membranes
(EMD Millipore). The membranes were blocked with 5% non-fat milk at
room temperature for 1 h. Subsequently, the membranes were
incubated overnight at 4˚C with primary antibodies targeted
against: IGF1R (1:1,000; cat. no. 3027), Snail (1:1,000; cat. no.
3879), Slug (1:1,000; cat. no. 9585), E-cadherin (1:1,000; cat. no.
14472), N-cadherin (1:500; cat. no. 14215), vimentin (1:1,000; cat.
no. 5741) and GAPDH (1:1,000; cat. no. 5174; all from Cell
Signaling Technology, Inc.). The membranes were incubated with a
horseradish peroxidase-labeled anti-rabbit IgG secondary antibody
(1:1,000; sc-2027; Santa Cruz Biotechnology, Inc.) at room
temperature for 1 h. Protein bands were visualized using an ECL
detection kit (Pierce; Thermo Fisher Scientific, Inc.) and analyzed
with Quantity One v4.6.2 software (Bio-Rad Laboratories, Inc.).
GAPDH was used as the loading control.
Dual-Luciferase reporter assay
The target gene of DUXAP8 or miR-9-3p was predicted
using StarBase v2.0 (http://starbase.sysu.edu.cn/starbase2/) and TargetScan
(www.targetscan.org) online prediction
software, respectively. Based on the bioinformatics prediction, a
partial DUXAP8 fragment containing the wild-type (WT) or mutant
(MUT) miR-9-3p binding site, or the IGF1R 3'untranslated region
(UTR) sequence containing the WT or MUT miR-9-3p binding site were
amplified from HCC cells via PCR with 1 unit AmpliTaq DNA
polymerase (Thermo Fisher Scientific, Inc.) using the following
thermo cycling conditions: Pre-denaturation at 94˚C for 10 min; 30
cycles at 94˚C for 30 sec, 53-57˚C for 30 sec and 72˚C for 45 sec;
and final extension at 72˚C for 10 min). The PCR product was
subsequently cloned into the psiCHECK-2 vector (Promega
Corporation) using the restriction endonuclease cleavage sites of
XhoI and NotI (Promega Corporation). The following
primers were used: DUXAP8-WT forward,
5'-CCGCTCGAGAACACTAATTGTAGACTATG-3' and reverse,
5'-ATAAGAATGCGGCCGCTATTGATGAGGATTTTCAAT-3'; DUXAP8-MUT forward,
5'-TCAAAACCCAGAAAACCATACAAGGACGATTTGTGA-3' and reverse,
5'-AACCCAGAAAACCATACAAGGACGATTTGTGAATT-3'; IGF1R-WT forward,
5'-CCGCTCGAGTAGTCAGTTGACGAAGATCT-3' and reverse,
5'-ATAAGAATGCGGCCGCTAGCTACACTCCAAAGGGAA-3'; and IGF1R-MUT forward,
5'-TTAGGACACCTGTTTACTAGAGGAACCGCAAATATGC-3' and reverse,
5'-GACACCTGTTTACTAGAGGAACCGCAAATATGCCAA-3'. SK-HEP-1 and Huh-7
cells in 24-well culture plates (2x105 cells/well) were
co-transfected using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) with 100 ng of DUXAP8-WT, DUXAP8-MUT, IGF1R-WT or
IGF1R-MUT and 100 nM miR-9-3p mimic or miR-NC. Following incubation
for 48 h, luciferase activities were assessed using a
Dual-Luciferase reporter assay kit (cat. no. E1910; Promega
Corporation), according to the manufacturer's protocol. Firefly
luciferase activity was normalized to that of Renilla
luciferase.
Bioinformatics analysis
The StarBase Pan-Cancer platform (http://starbase.sysu.edu.cn/panCancer.php) was used to
analyze the correlation between DUXAP8, miR-9-3p and IGF1R in liver
hepatocellular carcinoma (LIHC) (32,33).
RNA immunoprecipitation (RIP)
assay
To investigate the relationship between DUXAP8 and
miR-9-3p, RIP was performed using the Magna RIP RNA-Binding Protein
Immunoprecipitation kit (EMD Millipore) according to the
manufacturer's protocol. Briefly, at 80% confluence, SK-HEP-1 and
Huh-7 cells were harvested and lysed in RIP lysis buffer (EMD
Millipore). Subsequently, 100 µl cell extract was incubated with
RIP buffer including magnetic beads conjugated with anti-Argonaute2
(Ago2) antibody (1:1,000; cat. no. 03-248) or normal mouse IgG
antibody (1:1,000, cat. no. 12-371; both from EMD Millipore)
overnight at 4˚C. Proteinase K (Invitrogen; Thermo Fisher
Scientific, Inc.) was applied to digest the samples for 30 min at
55˚C, and the isolated RNAs were utilized for RT-qPCR analysis of
DUXAP8 and miR-9-3p expression levels.
Statistical analysis
All statistical analyses were performed using SPSS
software (version 18; SPSS, Inc.). Data are presented as the mean ±
SD. The χ2 test was used to analyze the association
between DUXAP8 expression levels and clinicopathological features
in HCC. Overall survival rates were evaluated using the
Kaplan-Meier method with the long rank test applied for
comparisons. The relationship among DUXAP8, miR-9-3p and IGF1R was
analyzed using Pearson's correlation coefficient. Comparisons
between two groups were analyzed using a paired Student's t-test.
Comparisons among multiple groups were analyzed using one-way ANOVA
followed by Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference. Experiments were
repeated at least three times.
Results
DUXAP8 expression is upregulated in
HCC tumor tissues and cells
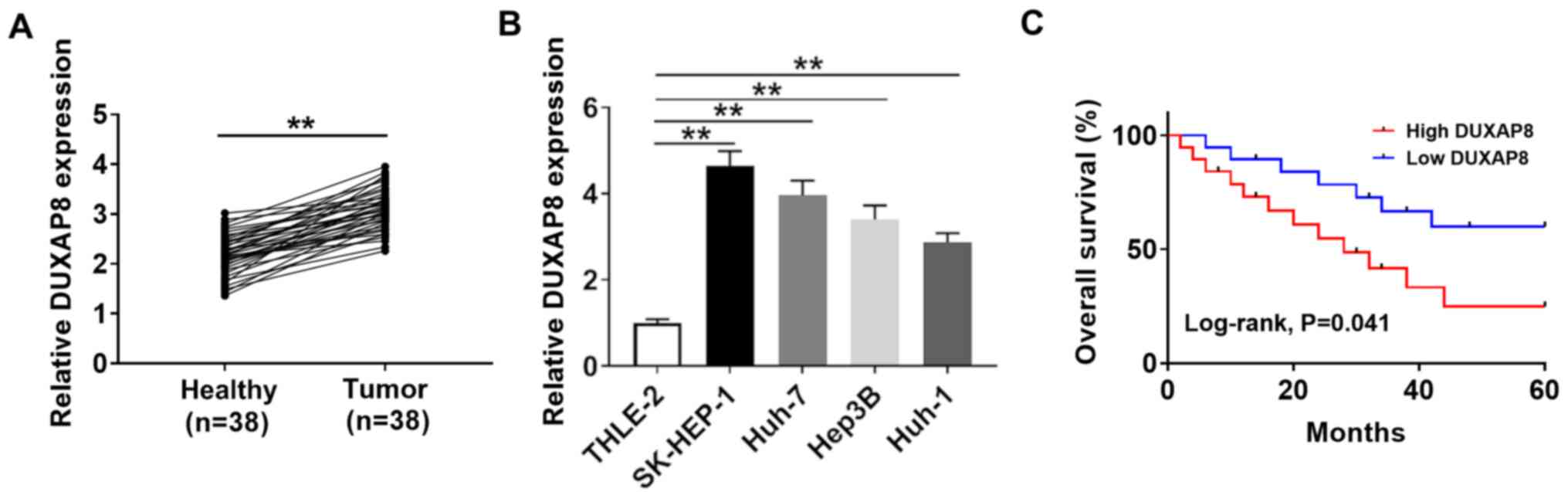
To investigate the function of DUXAP8 in HCC, its
expression was detected via RT-qPCR. Compared with adjacent healthy
tissues, the expression of DUXAP8 was significantly increased in 38
HCC tumor tissues (Fig. 1A). DUXAP8
expression levels were also significantly elevated in HCC cell
lines (SK-HEP-1, Huh-7, Hep38 and Huh-1) compared with the human
liver cell line (THLE-2; Fig. 1B),
with the highest expression levels observed in SK-HEP-1 and Huh-7
cells. Therefore, SK-HEP-1 and Huh-7 cells were selected for
subsequent experiments. The association between DUXAP8 expression
and clinicopathological factors in the 38 patients with HCC are
presented in Table I. DUXAP8
expression was significantly associated with tumor size, TNM stage
and metastasis (P<0.05), but was not significantly associated
with age, sex, HbsAg and cirrhosis. Moreover, the overall survival
curves suggested that DUXAP8 expression was inversely associated
with the prognosis of patients with HCC (Fig. 1C). The results indicated that DUXAP8
may serve as a novel biomarker for the prognosis of HCC.
 | Table IAssociation between DUXAP8 expression
and clinicopathological factors in patients with hepatocellular
carcinoma. |
Table I
Association between DUXAP8 expression
and clinicopathological factors in patients with hepatocellular
carcinoma.
| | DUXAP8
expression | |
|---|
| Characteristic | n | High (n=19) | Low (n=19) | P-value |
|---|
| Age (years) | | | | 0.505 |
|
<50 | 17 | 10 | 7 | |
|
≥50 | 21 | 9 | 12 | |
| Sex | | | | 0.282 |
|
Male | 20 | 12 | 8 | |
|
Female | 18 | 7 | 11 | |
| HbsAg | | | | 0.304 |
|
Negative | 14 | 9 | 5 | |
|
Positive | 24 | 10 | 14 | |
| Cirrhosis | | | | 0.410 |
|
Absent | 16 | 9 | 7 | |
|
Present | 22 | 10 | 12 | |
| Tumor size
(cm) | | | | 0.031a |
|
<5 | 17 | 6 | 11 | |
|
≥5 | 21 | 13 | 8 | |
| TNM stage | | | | 0.012a |
|
I, II | 18 | 5 | 13 | |
|
III, IV | 20 | 14 | 6 | |
| Metastasis | | | | 0.026a |
|
Yes | 15 | 5 | 10 | |
|
No | 23 | 14 | 9 | |
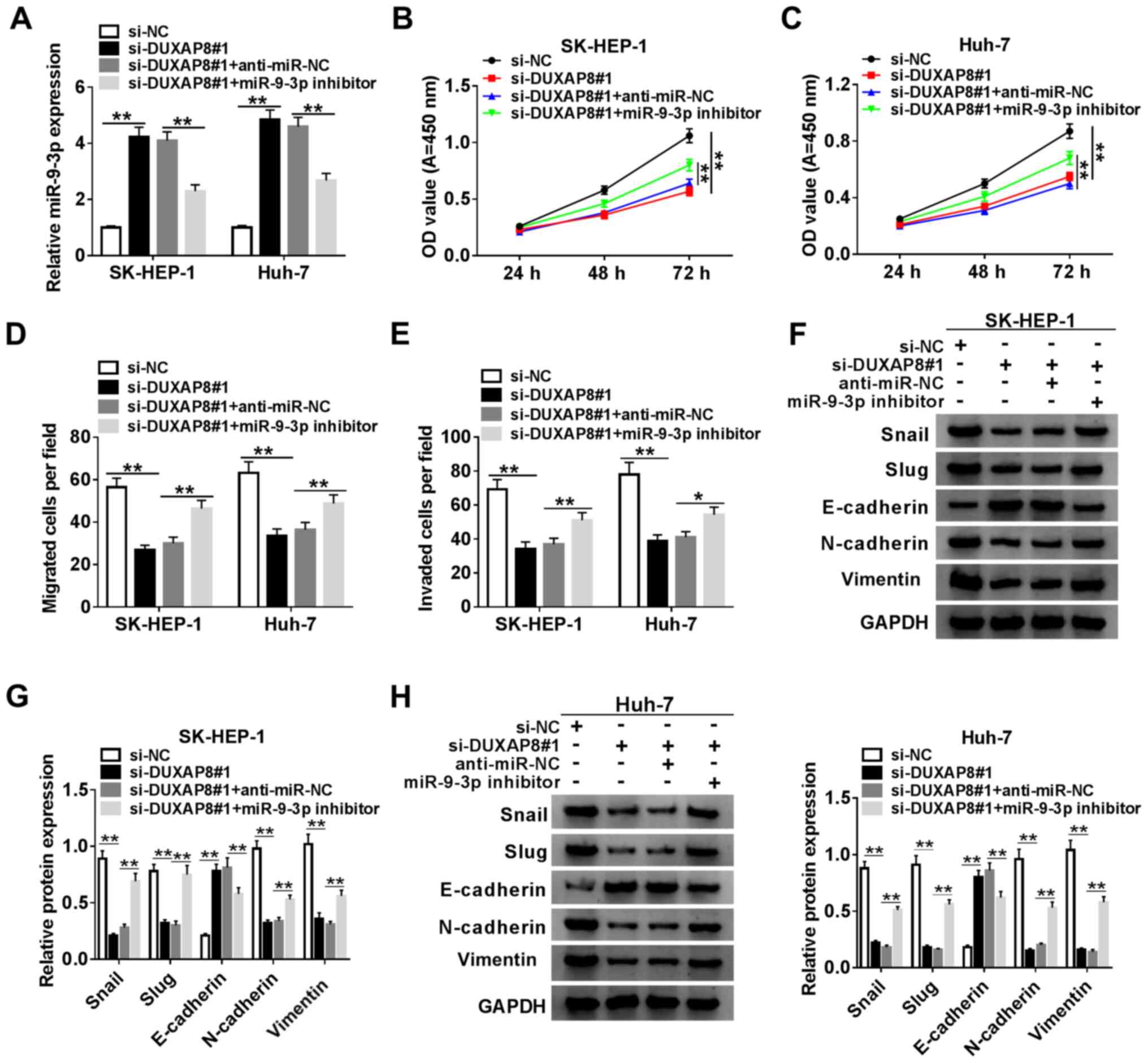
DUXAP8 induces HCC cell proliferation,
migration, invasion and EMT
To assess the function of DUXAP8 in HCC cells,
DUXAP8 knockdown was performed. The expression of DUXAP8 was
significantly decreased in si-DUXAP8-transfected SK-HEP-1 and Huh-7
cells compared with si-NC-transfected cells (Fig. 2A). Since si-DUXAP8#1 exhibited the
highest knockdown inefficiency, it was selected for subsequent
experiments. Therefore, the role of DUXAP8 in proliferation,
migration, invasion and EMT was investigated by knocking down
DUXAP8 expression in HCC cells. The CCK-8 and Transwell assay
results suggested that DUXAP8 knockdown significantly inhibited
SK-HEP-1 and Huh-7 cell proliferation (Fig. 2B and C), migration (Fig. 2D) and invasion (Fig. 2E) compared with si-NC. Subsequently,
the effect of DUXAP8 on HCC cell EMT was investigated. The western
blotting results indicated that DUXAP8 knockdown significantly
increased E-cadherin protein expression levels, but significantly
decreased Snail, Slug, N-cadherin and vimentin protein expression
levels compared with si-NC (Fig.
2F), which suggested that DUXAP8 promoted SK-HEP-1 and Huh-7
cell EMT. Collectively, the results suggested that DUXAP8 enhanced
HCC cell proliferation, migration, invasion and EMT.
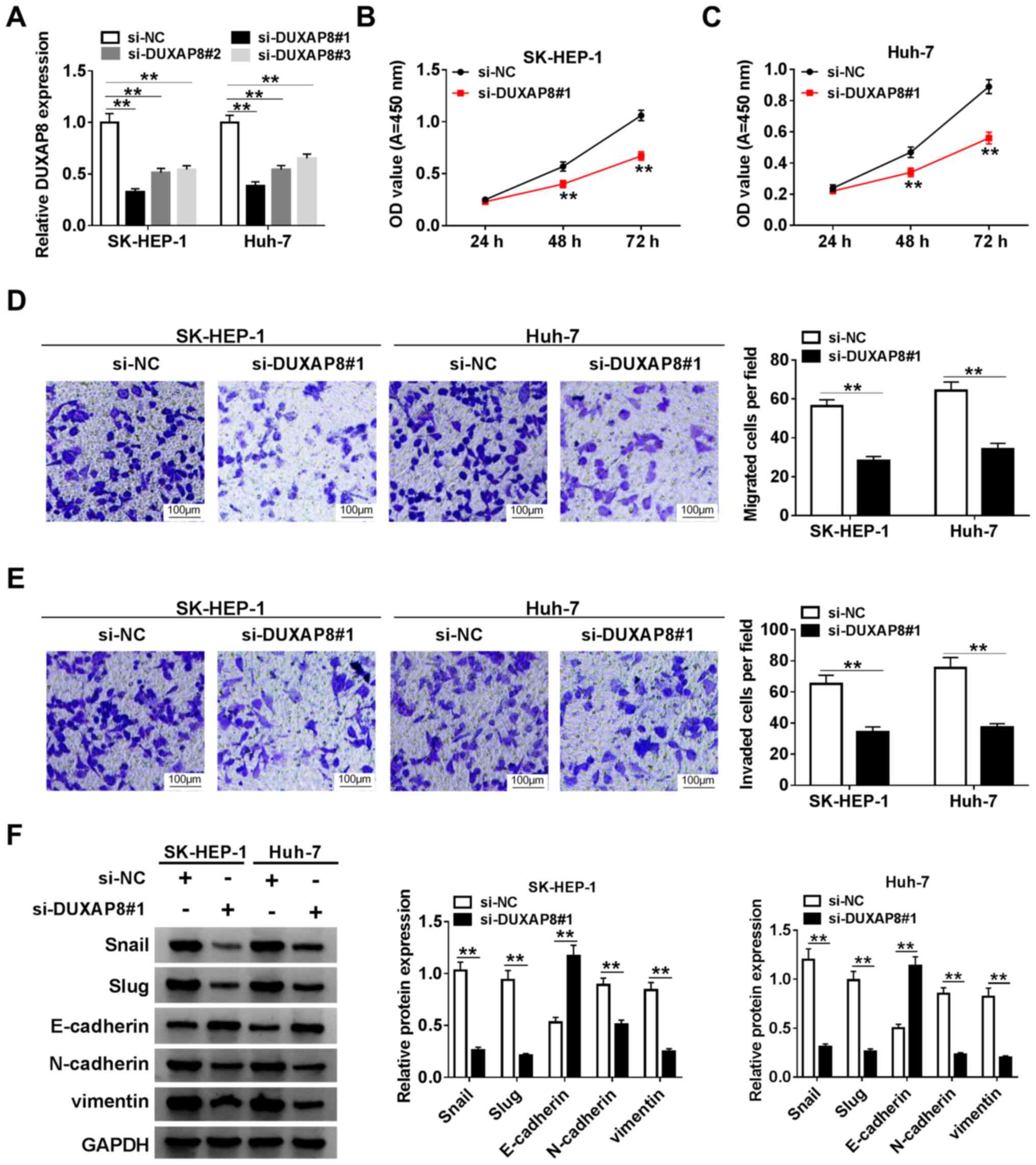 | Figure 2DUXAP8 induces HCC cell
proliferation, migration, invasion and EMT. (A) Knockdown
efficiency of si-DUXAP8 in SK-HEP-1 and Huh-7 cells. Cell
proliferation in si-DUXAP8-transfected (B) SK-HEP-1 and (C) Huh-7
cells. Cell (D) migration and (E) invasion in si-DUXAP8-transfected
SK-HEP-1 and Huh-7 cells. (F) Expression levels of EMT-related
proteins (Snail, Slug, E-cadherin, N-cadherin and vimentin) were
detected via western blotting in si-DUXAP8-transfected SK-HEP-1 and
Huh-7 cells. n=3. **P<0.01. DUXAP8, double homeobox A
pseudogene 8; HCC, hepatocellular carcinoma; EMT,
epithelial-mesenchymal transition; si, small interfering RNA; NC,
negative control; OD, optical density. |
miR-9-3p is a direct target of DUXAP8
in HCC cells
Increasing evidence has indicated that lncRNAs could
exert their role by interacting with miRNAs (34-36).
Therefore, bioinformatics-based target prediction analysis using
StarBase was performed to identify the target miRNAs of DUXAP8.
miR-9-3p was found to bind with DUXAP8 (Fig. 3D). To further assess the interaction
between DUXAP8 and miR-9-3p, SK-HEP-1 and Huh-7 cells were
co-transfected with DUXAP8-WT or DUXAP8-MUT reporter vector and
miR-NC or miR-9-3p mimics, and the subsequent luciferase activities
were measured. The results suggested that miR-9-3p overexpression
significantly decreased the luciferase activity of the DUXAP8-WT
reporter compared with miR-NC, but miR-9-3p overexpression had no
significant effect on the luciferase activity of the DUXAP8-MUT
reporter compared with miR-NC (Fig.
3E and F). Moreover, in
accordance with the bioinformatics analysis and luciferase assay,
the RIP assay results suggested that DUXAP8 and miR-9-3p were
significantly enriched in Ago2 pellets of SK-HEP-1 and Huh-7 cell
extracts compared with the IgG control group (Fig. 3G and H). Meanwhile, the transfection
efficiencies of miR-9-3p mimics and DUXAP8 overexpression were
detected (Fig. S1). Moreover,
miR-9-3p expression was significantly decreased in HCC tumor
tissues and cell lines compared with adjacent healthy tissues and a
human liver cell line, respectively (Fig. 3A and B). miR-9-3p expression levels were
inversely correlated with DUXAP8 expression levels in HCC tumor
tissues (Fig. 3C). Subsequently,
miR-9-3p expression levels were detected in DUXAP8-transfected and
si-DUXAP8-transfected SK-HEP-1 and Huh-7 cells. The RT-qPCR results
suggested that DUXAP8 overexpression significantly downregulated
miR-9-3p expression compared with pcDNA3.1, whereas DUXAP8
knockdown significantly upregulated miR-9-3p expression levels
compared with si-NC (Fig. 3I and
J), suggesting that manipulation of
DUXAP8 expression altered the expression of miR-9-3p. The results
suggested that DUXAP8 interacted with miR-9-3p to repress its
expression.
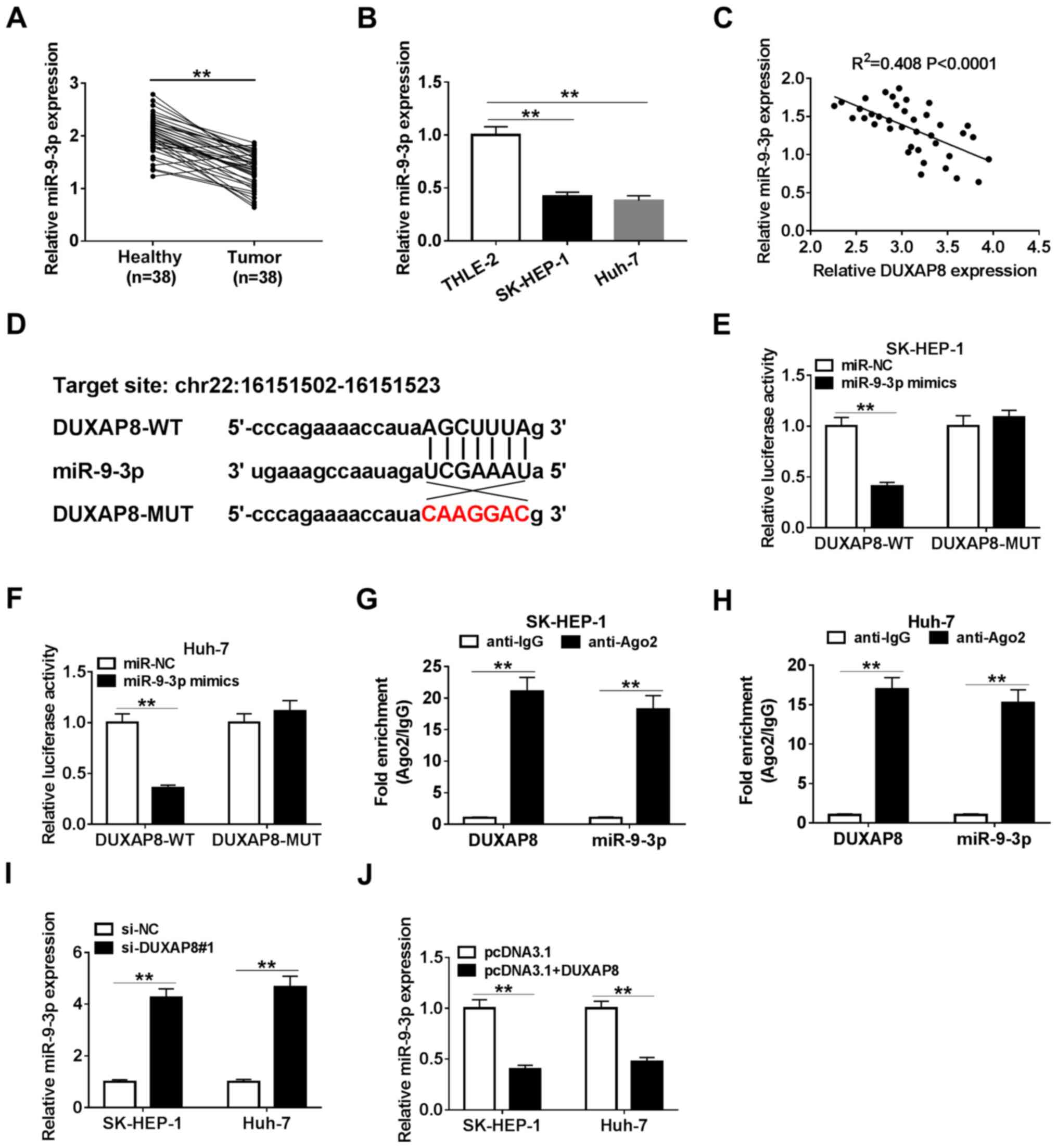 | Figure 3miR-9-3p is a direct target of DUXAP8
in HCC cells. (A) Expression of miR-9-3p in 38 paired HCC tumor and
adjacent healthy tissues. (B) Expression of miR-9-3p in HCC cell
lines (SK-HEP-1, Huh-7, Hep38 and Huh-1) and a normal human liver
cell line (THLE-2). (C) Correlation between DUXAP8 and miR-9-3p in
HCC tissues. (D) Binding sites between DUXAP8 and miR-9-3p. The
relative luciferase activity in (E) SK-HEP-1 and (F) Huh-7 cells
co-transfected with DUXAP8-WT or DUXAP8-MUT vectors and miR-NC or
miR-9-3p mimics. An RNA immunoprecipitation assay was conducted in
(G) SK-HEP-1 and (H) Huh-7 cell extracts to examine whether
miR-9-3p endogenously associated with DUXAP8. The effects of DUXAP8
(I) knockdown and (J) overexpression on miR-9-3p expression were
detected via reverse transcription-quantitative PCR in SK-HEP-1 and
Huh-7 cells. n=3. **P<0.01. miR, microRNA; DUXAP8,
double homeobox A pseudogene 8; HCC, hepatocellular carcinoma; WT,
wild-type; MUT, mutant; NC, negative control; si, small interfering
RNA; Ago2, Argonaute2. |
DUXAP8 facilitates HCC progression by
targeting miR-9-3p
To further investigate the mechanism underlying
DUXAP8 in HCC progression, rescue experiments were performed by
transfecting SK-HEP-1 and Huh-7 cells with si-NC, si-DUXAP8,
si-DUXAP8 + anti-miR-NC or si-DUXAP8 + miR-9-3p inhibitor. The
RT-qPCR results indicated that DUXAP8 knockdown significantly
increased miR-9-3p expression levels compared with si-NC, which
were significantly reversed by co-transfection with miR-9-3p
inhibitor in SK-HEP-1 and Huh-7 cells (Fig. 4A). The transfection efficiency of
miR-9-3p inhibitor was examined (Fig.
S1). Furthermore, compared with si-NC, DUXAP8 knockdown
significantly inhibited SK-HEP-1 and Huh-7 cell proliferation,
migration and invasion, and miR-9-3p knockdown reversed DUXAP8
knockdown-mediated effects (Figs.
4B-E, S2A and S2B). The western blotting results
suggested that miR-9-3p inhibition significantly decreased
si-DUXAP8-induced increases in E-cadherin protein expression
levels, and decreases in Snail, Slug, N-cadherin and vimentin
protein expression levels in SK-HEP-1 and Huh-7 cells (Fig. 4F-I). Collectively, the results
suggested that DUXAP8 may exert its carcinogenic effects partially
by regulating miR-9-3p.
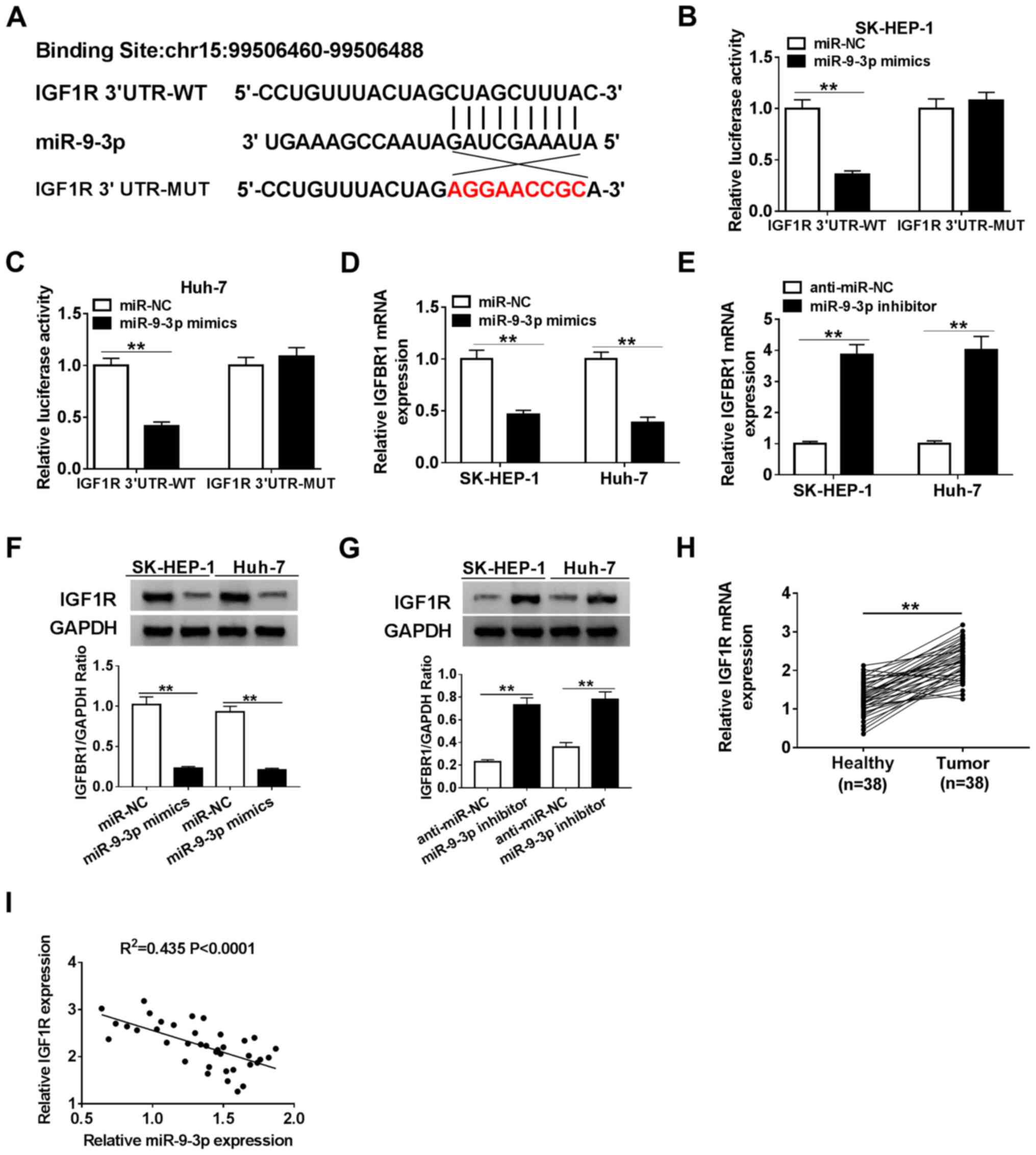
IGF1R is a target of miR-9-3p in HCC
cells
Previous reports have demonstrated that miRNAs may
exert their function by interacting with mRNAs (37-39).
Therefore, TargetScan was used to analyze the potential downstream
target of miR-9-3p. IGF1R was predicted to possess binding sites
with miR-9-3p (Fig. 5A). To
validate the bioinformatics prediction, SK-HEP-1 and Huh-7 cells
were co-transfected with IGF1R-WT or IGF1R-MUT reporter plasmids
and miR-NC or miR-9-3p mimics. The luciferase assay suggested that
compared with miR-NC, miR-9-3p overexpression significantly
inhibited the luciferase activity of the IGF1R-WT reporter, but had
no significant effect on the luciferase activity of the IGF1R-MUT
reporter (Fig. 5B and C). Subsequently, IGF1R expression in
miR-9-3p mimics- or miR-9-3p inhibitor-transfected SK-HEP-1 and
Huh-7 cells was detected. The RT-qPCR and western blotting results
suggested that miR-9-3p expression altered the expression of IGF1R,
as evidenced by miR-9-3p overexpression significantly decreasing
IGF1R expression levels compared with miR-NC, and miR-9-3p
knockdown significantly increasing IGF1R expression levels compared
with anti-miR-NC in SK-HEP-1 and Huh-7 cells (Fig. 5D-G). Moreover, IGF1R expression was
significantly upregulated and negatively correlated with miR-9-3p
expression in HCC tumor tissues (Fig.
5H and I). Collectively, the
results suggested that IGF1R was a target of miR-9-3p in HCC
cells.
miR-9-3p suppresses HCC progression by
targeting IGF1R
To further investigate whether miR-9-3p could exert
its function via targeting in HCC cells, SK-HEP-1 and Huh-7 cells
were transfected with miR-NC, miR-9-3p mimics, miR-9-3p mimics +
pcDNA3.1 or miR-9-3p mimics + pcDNA3.1-IGF1R. The RT-qPCR and
western blotting results indicated that miR-9-3p overexpression
significantly decreased the expression of IGF1R in SK-HEP-1 and
Huh-7 cells compared with miR-NC, which was reversed by
co-transfection with pcDNA3.1-IGF1R (Fig. 6A-C). In addition, according to the
analysis performed using the Pan-Cancer platform of StarBase, the
expression correlation of DUXAP8, IGF1R and miR-9-3p in LIHC
samples was assessed. The expression of miR-9-3p was positively
correlated with that of DUXAP8 or IGF1R, and DUXAP8 expression was
positively correlated with that of IGF1R (Fig. S3). Simultaneously, the transfection
efficiency of IGF1R overexpression was measured (Fig. S1). Compared with miR-NC, miR-9-3p
overexpression significantly inhibited SK-HEP-1 and Huh-7 cell
proliferation, migration and invasion, which was reversed by IGF1R
overexpression (Figs. 6D-G,
S2C and S2D). In addition, compared with miR-NC,
miR-9-3p overexpression significantly increased E-cadherin protein
expression levels, and decreased Snail, Slug, N-cadherin and
vimentin protein expression levels, which were reversed by
co-transfection with pcDNA3.1-IGF1R (Fig. 6H and I), indicating that IGF1R abolished
miR-9-3p-mediated inhibition of SK-HEP-1 and Huh-7 cell EMT. The
results indicated that miR-9-3p repressed HCC development by
modulating IGF1R.
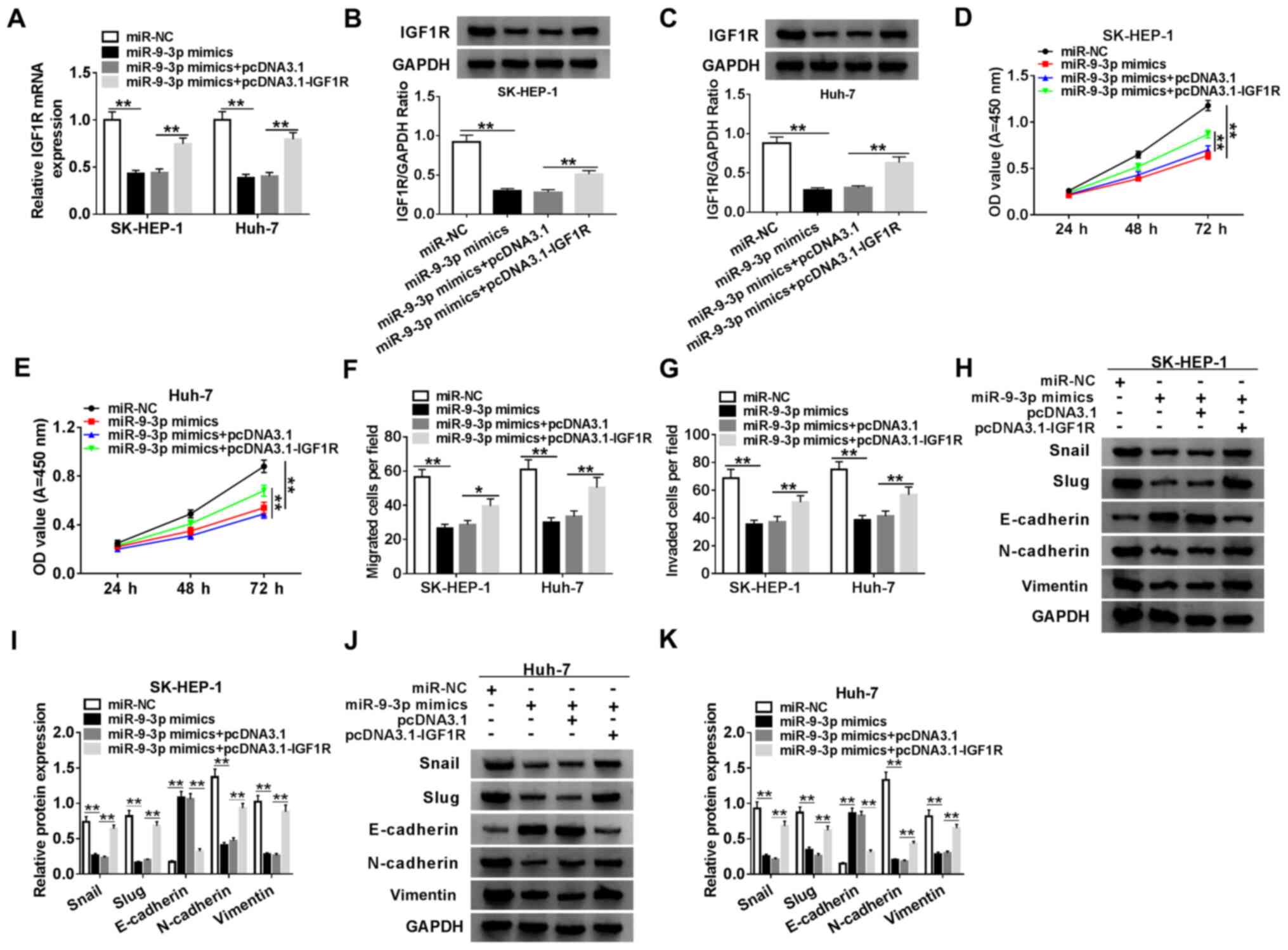 | Figure 6miR-9-3p suppresses HCC progression
by targeting IGF1R. (A) IGF1R expression in SK-HEP-1 and Huh-7
cells. IGF1R protein expression levels in (B) SK-HEP-1 and (C)
Huh-7 cells. Cell proliferation in (D) SK-HEP-1 and (E) Huh-7 cells
transfected with miR-NC, miR-9-3p mimics, miR-9-3p mimics +
pcDNA3.1 or miR-9-3p mimics + IGF1R. Cell (F) migration and (G)
invasion in SK-HEP-1 and Huh-7 cells transfected with miR-NC,
miR-9-3p mimics, miR-9-3p mimics + pcDNA3.1 or miR-9-3p mimics +
IGF1R. (H and I) EMT-related protein expression levels were (H)
determined by western blotting and (I) semi-quantified in SK-HEP-1
cells. EMT-related protein expression levels were (J) determined by
western blotting and (K) semi-quantified in Huh-7 cells. n=3.
*P<0.05; **P<0.01. miR, microRNA; HCC,
hepatocellular carcinoma; IGF1R, insulin-like growth factor 1
receptor; NC, negative control; EMT, epithelial-mesenchymal
transition; OD, optical density. |
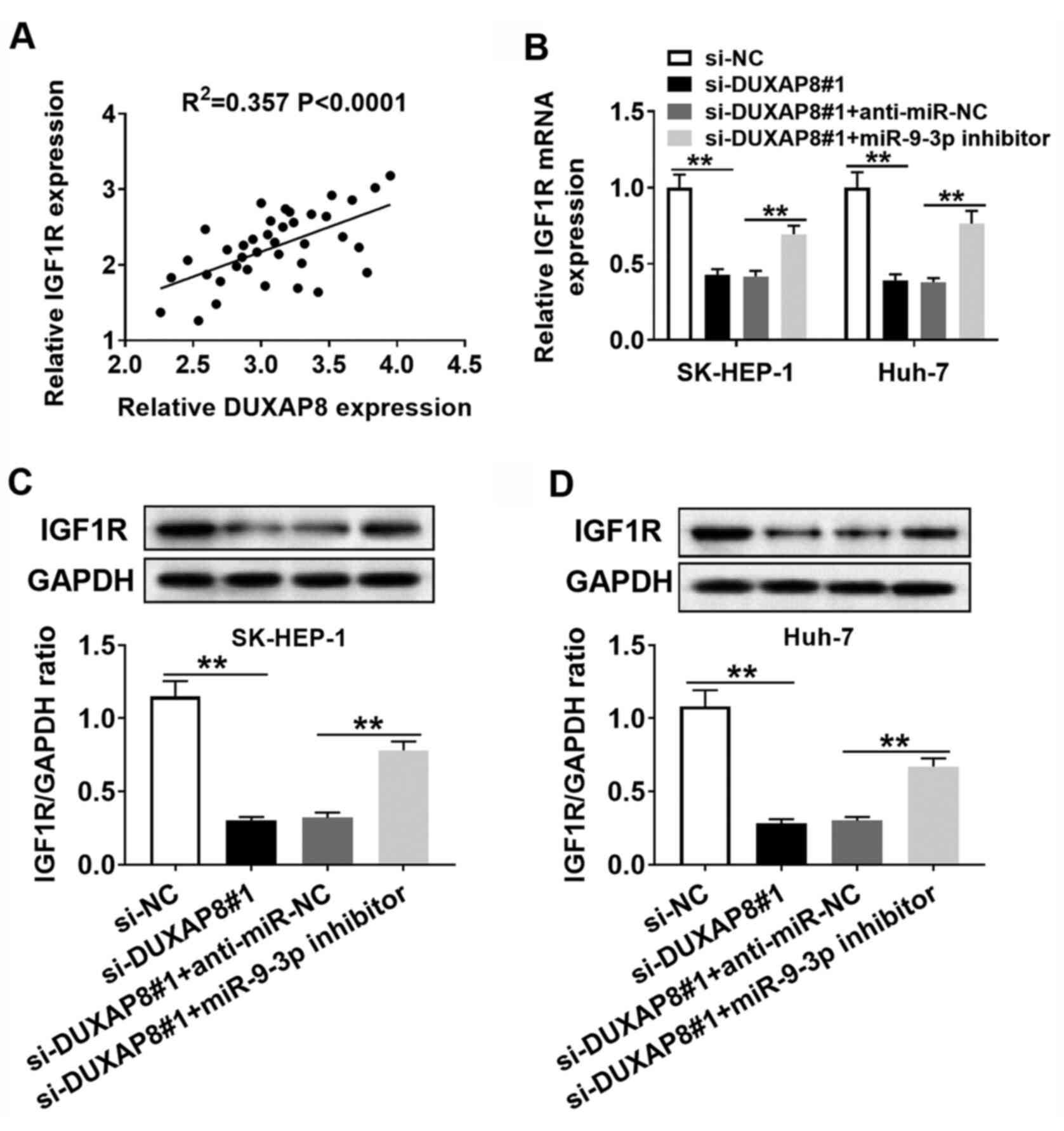
Verification of the
DUXAP8/miR-9-3p/IGF1R regulatory axis in HCC cells
According to the aforementioned results, it was
hypothesized that DUXAP8 could exert its carcinogenic effects via
the DUXAP8/miR-9-3p/IGF1R regulatory signaling pathway.
Furthermore, DUXAP8 expression was positively correlated with IGF1R
expression in HCC tumor tissues (Fig.
7A). Subsequently, whether DUXAP8 could regulate the expression
of IGF1R via miR-9-3p was investigated. The RT-qPCR and western
blotting results suggested that DUXAP8 knockdown reduced the
expression of IGF1R compared with si-NC, and co-transfection with
miR-9-3p inhibitor reversed si-DUXAP8-mediated inhibitory effects
on IGF1R expression in SK-HEP-1 and Huh-7 cells (Fig. 7B-D). Collectively, the results
suggested that DUXAP8 may serve as a molecular sponge to sequester
miR-9-3p from IGF1R in HCC cells.
Discussion
Numerous studies have indicated that lncRNAs are
aberrantly expressed and serve as diagnostic and prognostic
biomarkers in HCC (40-42).
LncRNA DUXAP8, as an oncogene, is dysregulated in multiple tumors,
such as bladder cancer (13),
non-small-cell lung cancer (43)
and ovarian cancer (44). For
instance, Xu et al (45)
demonstrated that DUXAP8 was overexpressed in esophageal squamous
cell cancer tissues, and DUXAP8 knockdown blocked proliferation,
invasion and colony formation in vitro. Lin et al
(46) reported that DUXAP8
downregulation inhibited proliferation by binding to phosphatase
and tensin homolog in bladder cancer. Nevertheless, the exact
function and mechanisms underlying DUXAP8 in HCC progression are
not completely understood.
miR-9-3p has been confirmed as a tumor suppressor in
a variety of different types of cancer, such as glioma (47), nasopharyngeal carcinoma (48) and bladder cancer (49). Of interest, in HCC cells, Yang et
al (50) demonstrated that
miR-9-3p overexpression reduced the migration and invasion rates of
HCC in vitro. Tang et al (51) demonstrated that miR-9-3p, a
functional biomarker, inhibited cell proliferation by suppressing
human fibroblast growth factor-5 expression in HCC.
In the present study, the expression level of DUXAP8
in HCC tumor tissues and cells was measured. The results indicated
that DUXAP8 expression was significantly upregulated in HCC tumor
tissues and cells compared with adjacent healthy tissues and a
normal liver cell line, respectively. Moreover, the
clinicopathologic factors and the prognostic value of HCC were
assessed. The results indicated that DUXAP8 expression was
significantly associated with tumor size, TNM stage and metastasis,
and patients with high DUXAP8 expression displayed a worse overall
survival rate compared with patients with low DUXAP8 expression,
suggesting that DUXAP8 served a vital role in the development and
progression of HCC.
The effect of DUXAP8 on HCC cell proliferation,
migration, invasion and EMT was evaluated. Functional analyses
indicated that DUXAP8 enhanced HCC cell proliferation, migration,
invasion and EMT, indicating that DUXAP8 performed as an oncogene
in HCC progression. Subsequently, the potential molecular
mechanisms underlying DUXAP8 in HCC development were investigated.
In recent decades, several studies have identified that lncRNA
could serve as a miRNA sponge to interact with miRNAs (52,53).
Therefore, StarBase was used to predict the underlying target
miRNAs of DUXAP8. As a result, miR-9-3p was found to possess
binding sites of DUXAP8, which was further indicated by the
luciferase reporter assay. Moreover, in the present study, miR-9-3p
expression was downregulated and inversely correlated with DUXAP8
expression in HCC tumor tissues and cells. Consequently, whether
the effect of DUXAP8 on HCC progression was mediated via the
regulation of miR-9-3p was investigated. In the present study, the
results verified that silencing DUXAP8 inhibited HCC cell
proliferation, migration, invasion and EMT, whereas miR-9-3p
knockdown attenuated the effects of DUXAP8 on HCC cells. Therefore,
to the best of our knowledge, the present study indicated for the
first time that DUXAP8 served as an oncogenic factor in HCC
development potentially via targeting the expression of
miR-9-3p.
Increasing studies have focused on lncRNA-miRNA-mRNA
regulatory axes, suggesting that lncRNAs serve as competing
endogenous RNAs (ceRNAs) to sequester miRNAs away from target
mRNAs, leading to the upregulation of mRNA expression (54-56).
Therefore, IGF1R as the target gene of miR-9-3p in HCC cells was
identified using TargetScan software and verified by performing
luciferase reporter assays. As a subunit of the transmembrane
receptor family, IGF1R participates in various biological
processes, including embryogenesis, tissue repair and metastatic
diffusion of tumor cells (57). A
previous study also indicated that IGF1R triggered HCC cell
metastasis and proliferation (58).
In the present study, compared with adjacent healthy tissues, IGF1R
expression was upregulated and negatively correlated with miR-9-3p
expression in HCC tumor tissues. Therefore, whether IGF1R was
involved in mediating miR-9-3p expression in HCC development was
investigated. The results indicated that, compared with miR-NC,
miR-9-3p overexpression inhibited proliferation, migration,
invasion and EMT and IGF1R overexpression partly reversed
miR-9-3p-induced suppressive effects in HCC cells, indicating that
miR-9-3p may inhibit tumor progression partially by targeting
IGF1R.
In addition, to further assess whether DUXAP8 served
as a ceRNA of miR-9-3p by interacting with IGF1R, the effect of
DUXAP8 on IGF1R expression was investigated. In the present study,
IGF1R expression was positively correlated with DUXAP8 expression
in HCC tumor tissues. Compared with si-NC, DUXAP8 knockdown
decreased IGF1R expression, which was reversed by co-transfection
with miR-9-3p inhibitor in HCC cells. In addition, the association
among DUXAP8, IGF1R and miR-9-3p was also analyzed using the
Pan-Cancer platform of StarBase, and indicated that miR-9-3p was
positively correlated with DUXAP8 or IGF1R, and DUXAP8 expression
was positively correlated with that of IGF1R. The aforementioned
results differed from the other results of the present study, which
might be due to the difference between high-throughput predicted
data and actual gene expression. In the present study, RT-qPCR was
performed to detect the expression of each gene in patient samples,
and the obtained data should be more accurate.
In conclusion, the present study suggested that
DUXAP8 served as a miR-9-3p sponge to upregulate IGF1R expression,
thereby enhancing the development of HCC. The present study also
indicated that targeting the DUXAP8/miR-9-3p/IGF1R regulatory axis
may serve as a promising therapeutic approach for HCC.
Supplementary Material
Transfection efficiencies of miR-9-3p
mimic, miR-9-3p inhibitor, IGF1R overexpression and DUXAP8
overexpression in hepatocellular carcinoma cells. Transfection
efficiency of (A) miR-9-3p mimic, (B) miR-9-3p inhibitor, (C)
pcDNA3-1-IGF1R and (D) pcDNA3-1-DUXAP8 in SK-HEP-1 and Huh-7 cells.
n=3. **P<0.01. miR, microRNA; IGF1R, insulin-like
growth factor 1 receptor; DUXAP8, double homeobox A pseudogene 8;
NC, negative control.
Pan-cancer analysis was performed to
detect the correlation among DUXAP8, IGF1R and miR-9-3p in LIHC
samples. DUXAP8, double homeobox A pseudogene 8; IGF1R,
insulin-like growth factor 1 receptor; miR, microRNA; LIHC, liver
hepatocellular carcinoma.
Representative images of Transwell
migration and invasion assays. Cell (A) migration and (B) invasion
in si-DUXAP8. and miR-9-3p inhibitor-transfected SK-HEP-1 and Huh-7
cells. Cell (C) migration and (D) invasion in miR-9-3p mimics. and
pcDNA3-1-IGF1R-transfected SK-HEP-1 and Huh-7 cells. si, small
interfering RNA; DUXAP8, double homeobox A pseudogene 8; miR,
microRNA; IGF1R, insulin-like growth factor 1 receptor; NC,
negative control.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QG and BY designed the study. XZ and TY performed
the experiments. JL and WX analyzed and interpreted the data. WX
drafted the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by People's Hospital
of Dongying Ethics Committee (Dongying, China). Written informed
consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Erstad DJ and Tanabe KK: Prognostic and
therapeutic implications of microvascular invasion in
hepatocellular carcinoma. Ann Surg Oncol. 26:1474–1493.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mercer TR, Dinger ME and Mattick JS: Long
Non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Marchese FP, Raimondi I and Huarte M: The
multidimensional mechanisms of long noncoding RNA function. Genome
Biol. 18(206)2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kong L and Zhang C: LncRNA DLX6-AS1
aggravates the development of ovarian cancer via modulating FHL2 by
sponging miR-195-5p. Cancer Cell Int. 20(370)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yang Q, Wan Q, Zhang L, Li Y, Zhang P, Li
D, Feng C, Yi F, Zhang L, Ding X, et al: Analysis of LncRNA
expression in cell differentiation. RNA Biol. 15:413–422.
2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wu Y and Qian Z: Long non-coding RNAs
(lncRNAs) and microRNAs regulatory pathways in the tumorigenesis
and pathogenesis of glioma. Discov Med. 28:129–138. 2019.PubMed/NCBI
|
|
9
|
Yang Y, Chen L, Gu J, Zhang H, Yuan J,
Lian Q, Lv G, Wang S, Wu Y, Yang YT, et al: Recurrently deregulated
lncRNAs in hepatocellular carcinoma. Nat Commun.
8(14421)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li B, Mao R, Liu C, Zhang W, Tang Y and
Guo Z: LncRNA FAL1 promotes cell proliferation and migration by
acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells.
Life Sci. 197:122–129. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ma J, Li T and Han X: Knockdown of LncRNA
ANRIL suppresses cell proliferation, metastasis, and invasion via
regulating miR-122-5p expression in hepatocellular carcinoma. J
Cancer Res Clin Oncol. 144:205–214. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhao X, Hao S, Wang M, Xing D and Wang C:
Knockdown of pseudogene DUXAP8 expression in glioma suppresses
tumor cell proliferation. Oncol Lett. 17:3511–3516. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Jiang B, Hailong S, Yuan J, Zhao H, Xia W,
Zha Z, Bin W and Liu Z: Identification of oncogenic long noncoding
RNA SNHG12 and DUXAP8 in human bladder cancer through a
comprehensive profiling analysis. Biomed Pharmacother. 108:500–507.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Huang T, Wang X, Yang X, Ji J, Wang Q, Yue
X and Dong Z: Long non-coding RNA DUXAP8 enhances renal cell
carcinoma progression via downregulating miR-126. Med Sci Monit.
24:7340–7347. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sun M, Nie FQ, Zang C, Wang Y, Hou J, Wei
C, Li W, He X and Lu KH: The pseudogene DUXAP8 promotes
non-small-cell lung cancer cell proliferation and invasion by
epigenetically silencing EGR1 and RHOB. Mol Ther. 25:739–751.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Afonso-Grunz F and Muller S: Principles of
miRNA-mRNA interactions: Beyond sequence complementarity. Cell Mol
Life Sci. 72:3127–3141. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zha Z, Jia F, Hu P, Mai E and Lei T:
MicroRNA-574-3p inhibits the malignant behavior of liver cancer
cells by targeting ADAM28. Oncol Lett. 20:3015–3023.
2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Liu CH, Jing XN, Liu XL, Qin SY, Liu MW
and Hou CH: Tumor-suppressor miRNA-27b-5p regulates the growth and
metastatic behaviors of ovarian carcinoma cells by targeting CXCL1.
J Ovarian Res. 13(92)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Fang F, Chang RM, Yu L, Lei X, Xiao S,
Yang H and Yang LY: MicroRNA-188-5p suppresses tumor cell
proliferation and metastasis by directly targeting FGF5 in
hepatocellular carcinoma. J Hepatol. 63:874–885. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chen Y, Zhao ZX, Huang F, Yuan XW, Deng L
and Tang D: MicroRNA-1271 functions as a potential tumor suppressor
in hepatitis B virus-associated hepatocellular carcinoma through
the AMPK signaling pathway by binding to CCNA1. J Cell Physiol.
234:3555–3569. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Higashi T, Hayashi H, Ishimoto T, Takeyama
H, Kaida T, Arima K, Taki K, Sakamoto K, Kuroki H, Okabe H, et al:
MiR-9-3p plays a tumour-suppressor role by targeting TAZ (WWTR1) in
hepatocellular carcinoma cells. Br J Cancer. 113:252–258.
2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Erickson KE and Rukhlenko OS: Modeling
cell line-specific recruitment of signaling proteins to the
insulin-like growth factor 1 receptor. PLoS Comput Biol.
15(e1006706)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lin JF, Tsai TF, Lin YC, Chen HE, Chou KY
and Hwang TI: Benzyl isothiocyanate suppresses IGF1R, FGFR3 and
mTOR expression by upregulation of miR-99a-5p in human bladder
cancer cells. Int J Oncol. 54:2106–2116. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang Y, Huang S, Guo Y and Li L: MiR-1294
confers cisplatin resistance in ovarian Cancer cells by targeting
IGF1R. Biomed Pharmacother. 106:1357–1363. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Khalil A and Jameson MJ: Downregulation of
igf1r expression inhibits growth and enhances cisplatin sensitivity
of head and neck squamous cell carcinoma cells in vitro. Horm
Cancer. 10:11–23. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liu W, Kang L, Han J, Wang Y, Shen C, Yan
Z, Tai Y and Zhao C: MiR-342-3p suppresses hepatocellular carcinoma
proliferation through inhibition of IGF-1R-mediated Warburg effect.
Onco Targets Ther. 11:1643–1653. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ren L, Yao Y, Wang Y and Wang S: MiR-505
suppressed the growth of hepatocellular carcinoma cells via
targeting IGF-1R. Biosci Rep. 39(BSR20182442)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ye Y, Zhuang J, Wang G, He S, Zhang S,
Wang G, Ni J, Wang J and Xia W: MicroRNA-495 suppresses cell
proliferation and invasion of hepatocellular carcinoma by directly
targeting insulin-like growth factor receptor-1. Exp Ther Med.
15:1150–1158. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Liu MD, Wu H, Wang S, Pang P, Jin S, Sun
CF and Liu FY: MiR-1275 promotes cell migration, invasion and
proliferation in squamous cell carcinoma of head and neck via
up-regulating IGF-1R and CCR7. Gene. 646:1–7. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhou Y, Zhang Z, Wang N, Chen J, Zhang X,
Guo M, John Zhong L and Wang Q: Suppressor of cytokine signalling-2
limits IGF1R-mediated regulation of epithelial-mesenchymal
transition in lung adenocarcinoma. Cell Death Dis.
9(429)2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42 (Database Issue):D92–D97. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yang JH, Li JH, Shao P, Zhou H, Chen YQ
and Qu LH: StarBase: A database for exploring microRNA-mRNA
interaction maps from Argonaute CLIP-Seq and Degradome-Seq data.
Nucleic Acids Res. 39 (Database Issue):D202–D209. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li Y: MIR31HG exhibits oncogenic property
and acts as a sponge for miR-361-3p in cervical carcinoma. Biochem
Biophys Res Commun. 529:890–897. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Sun F and Wu K: Long Noncoding RNA PVT1
promotes prostate cancer metastasis by increasing NOP2 expression
via targeting tumor suppressor MicroRNAs. Onco Targets Ther.
13:6755–6765. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Shi X, Huo J, Gao X, Cai H and Zhu W: A
newly identified lncRNA H1FX-AS1 targets DACT1 to inhibit cervical
cancer via sponging miR-324-3p. Cancer Cell Int.
20(358)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wu R, Zhao B, Ren X, Wu S, Liu M, Wang Z
and Liu W: MiR-27a-3p targeting GSK3β promotes triple-negative
breast cancer proliferation and migration through Wnt/β-catenin
pathway. Cancer Manag Res. 12:6241–6249. 2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li G, Qi HW, Dong HG, Bai P, Sun M and Liu
HY: Targeting deubiquitinating enzyme USP26 by microRNA-203
regulates Snail1's pro-metastatic functions in esophageal cancer.
Cancer Cell Int. 20(355)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ma HF, Lv GX and Zhang DH: miR-381
mediates the development of head and neck squamous cell carcinoma
via targeting STC2. Onco Targets Ther. 13:4485–4493.
2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Huo X, Han S, Wu G, Latchoumanin O, Zhou
G, Hebbard L, George J and Qiao L: Dysregulated long noncoding RNAs
(lncRNAs) in hepatocellular carcinoma: Implications for
tumorigenesis, disease progression, and liver cancer stem cells.
Mol Cancer. 16(165)2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhang H, Xu HB, Kurban E and Luo HW:
LncRNA SNHG14 promotes hepatocellular carcinoma progression via
H3K27 acetylation activated PABPC1 by PTEN signaling. Cell Death
Dis. 11(646)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
He H, Wang Y, Ye P, Yi D, Cheng Y, Tang H,
Zhu Z, Wang X and Jin S: Long noncoding RNA ZFPM2-AS1 acts as a
miRNA sponge and promotes cell invasion through regulation of
miR-139/GDF10 in hepatocellular carcinoma. J Exp Clin Cancer Res.
39(159)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yin D, Hua L, Wang J and Liu Y: Long
non-coding RNA DUXAP8 facilitates cell viability, migration, and
glycolysis in non-small-cell lung cancer via regulating HK2 and
LDHA by inhibition of miR-409-3p. Onco Targets Ther. 13:7111–7123.
2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Meng Q, Li Z, Pan J and Sun X: Long
noncoding RNA DUXAP8 regulates proliferation and apoptosis of
ovarian cancer cells via targeting miR-590-5p. Hum Cell.
33:1240–1251. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Xu LJ, Yu XJ, Wei B, Hui HX, Sun Y, Dai J
and Chen XF: Long non-coding RNA DUXAP8 regulates proliferation and
invasion of esophageal squamous cell cancer. Eur Rev Med Pharmacol
Sci. 22:2646–2652. 2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Lin MG, Hong YK, Zhang Y, Lin BB and He
XJ: Mechanism of lncRNA DUXAP8 in promoting proliferation of
bladder cancer cells by regulating PTEN. Eur Rev Med Pharmacol Sci.
22:3370–3377. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Yang L, Mu Y, Cui H, Liang Y and Su X:
MiR-9-3p augments apoptosis induced by H2O2 through down regulation
of Herpud1 in glioma. PLoS One. 12(e0174839)2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Ding Y, Pan Y, Liu S, Jiang F and Jiao J:
Elevation of MiR-9-3p suppresses the epithelial-mesenchymal
transition of nasopharyngeal carcinoma cells via down-regulating
FN1, ITGB1 and ITGAV. Cancer Biol Ther. 18:414–424. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Cai H, Yang X, Gao Y, Xu Z, Yu B, Xu T, Li
X, Xu W, Wang X and Hua L: Exosomal MicroRNA-9-3p secreted from
BMSCs downregulates ESM1 to suppress the development of bladder
cancer. Mol Ther Nucleic Acids. 18:787–800. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yang S, Cai H, Hu B and Tu J: LncRNA
SAMMSON negatively regulates miR-9-3p in hepatocellular carcinoma
cells and has prognostic values. Biosci Rep.
39(BSR20190615)2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Tang J, Li Y, Liu K, Zhu Q, Yang WH, Xiong
LK and Guo DL: Exosomal miR-9-3p suppresses HBGF-5 expression and
is a functional biomarker in hepatocellular carcinoma. Minerva Med.
109:15–23. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Militello G, Weirick T, John D, Döring C,
Dimmeler S and Uchida S: Screening and validation of lncRNAs and
circRNAs as miRNA sponges. Brief Bioinform. 18:780–788.
2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Olgun G, Sahin O and Tastan O: Discovering
lncRNA mediated sponge interactions in breast cancer molecular
subtypes. BMC Genomics. 19(650)2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Yoon JH, Abdelmohsen K and Gorospe M:
Functional interactions among microRNAs and long noncoding RNAs.
Semin Cell Dev Biol. 34:9–14. 2014.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z
and Zhang LW: Identification of key long non-coding RNAs as
competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma.
Eur Rev Med Pharmacol Sci. 20:2285–2295. 2016.PubMed/NCBI
|
|
56
|
Lin P, Wen DY, Li Q, He Y, Yang H and Chen
G: Genome-wide analysis of prognostic lncRNAs, miRNAs, and mRNAs
forming a competing endogenous RNA Network in hepatocellular
carcinoma. Cell Physiol Biochem. 48:1953–1967. 2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Schwartz MA and Ginsberg MH: Networks and
crosstalk: Integrin signalling spreads. Nat Cell Biol. 4:E65–E68.
2002.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Han X, Wang X, Zhao B, Chen G, Sheng Y,
Wang W and Teng M: MicroRNA-187 inhibits tumor growth and
metastasis via targeting of IGF-1R in hepatocellular carcinoma. Mol
Med Rep. 16:2241–2246. 2017.PubMed/NCBI View Article : Google Scholar
|