Introduction
Cervical cancer is the fourth most common type of
cancer diagnosed in women worldwide, accounting for >500,000 new
cases and 311,365 deaths in 2018(1). Despite cervical cancer being one of
the more treatable malignant diseases, with multiple treatment
strategies available, including surgical resection, radiation and
chemotherapy, there are currently no effective targeted or
immunotherapeutic approaches for patients with advanced cervical
cancer (2,3). Therefore, it is important to identify
additional molecules that could provide novel treatment strategies
for cervical cancer.
B7 homolog 6 (B7-H6), which is a member of the B7
family, has been attracting increasing attention from researchers
due to its reported role in tumorigenesis (4,5).
B7-H6, as a ligand of NKp30, which is an activating receptor of
natural killer (NK) cells (6-8),
was found to play an important role in tumor immune surveillance
and escape (9-14).
Notably, B7-H6 expression is rarely reported in normal tissues and
is mainly expressed in numerous types of cancer, including
esophageal squamous cell carcinoma, oral squamous cell carcinoma
and breast cancer (4). In recent
years, B7-H6 expression has been reported to be upregulated in
certain cancers and to be associated with tumor differentiation,
stage and progression. For example, Zhou et al (15) retrospectively analyzed the clinical
data of 145 patients with esophageal squamous cell carcinoma and
found that B7-H6 expression levels were upregulated in esophageal
cancer tissues, and were associated with tumor size, stage and
lymph node metastasis. Sun et al (16) also used immunohistochemistry to
investigate the expression of B7-H6 in 305 patients with breast
cancer. The results revealed that the expression levels of B7-H6 in
breast cancer tissues were positively associated with tumor
progression. Furthermore, Wang et al (17) used immunohistochemistry to analyze
the expression of B7-H6 in 50 patients with oral squamous cell
carcinoma, and found that the expression levels of B7-H6 in oral
squamous cell carcinoma tissues were significantly upregulated
compared with those in the normal oral mucosa, and were
significantly associated with cancer differentiation. However, to
the best of our knowledge, the clinical significance and role of
B7-H6 in cervical cancer have not been investigated to date.
To explore whether B7-H6 may be a potential
therapeutic target for cervical cancer, the present study used
immunohistochemical analysis to determine the association between
B7-H6 expression and the physiology of cervical cancer.
Furthermore, short hairpin RNA (shRNA), cell function experiments,
apoptosis and cell cycle analysis were used to determine the
possible role of B7-H6 in the regulation of the biological behavior
of cervical cancer.
Materials and methods
Patients and samples
Archived cervical pathological tissue samples from
the Department of Pathology of Second Hospital of Tianjin Medical
University (Tianjin, China) obtained between November 2015 and
August 2019 were used in the present study. Each disease stage
comprised 30 samples. The age range of patients with cervical
intraepithelial neoplasia (CIN) grade I (CIN Ⅰ), CIN II, CIN III
and cervical cancer was 44-56, 42-57, 43-53 and 44-64 years,
respectively, with a median age of 50±8.11, 49±10.23, 47±6.02 and
54±9.42 years, respectively. Furthermore, 30 normal cervical
samples (median patient age, 53±2.53 years; range, 49-58 years)
were included in the present study. There were no statistically
significant differences in age among the groups (P=0.773). The
research protocol was approved (approval no. KY2019K023) by the
Ethics Committee of The Second Hospital of Tianjin Medical
University and the patients or their family members provided
written informed consent for their tissues to be used for research
purposes prior to participation.
The following inclusion criteria were used: i)
Normal cervical tissue samples were obtained from patients
undergoing hysterectomy with negative ThinPrep cytology test (TCT)
and human papillomavirus results; ii) CIN I and CIN II tissues were
obtained from patients who had undergone a cervical biopsy; iii)
CIN III (18) tissues were obtained
from patients who had undergone cervical conization; and iv)
cervical cancer tissues were obtained from patients who had
undergone a cervical diagnostic biopsy or therapeutic surgery. All
specimens were examined and the diagnosis was confirmed by
pathological analysis, and the TCTs revealed that <30% cervial
inflammatory cells were found in each sample. The following
exclusion criteria were used: i) Body mass index ≥25
kg/m2; ii) levels of biochemical markers (such as serum
lipid profile, creatinine and liver function tests) not within the
normal range; iii) smokers; and iv) presence of other systemic
diseases.
Immunohistochemistry
The cervical pathological tissue from the patients
recruited at our hospital were fixed with 10% formaldehyde for 24 h
at room temperature and embedded in paraffin. Every array block was
cut into 4-µm sections, deparaffinized with xylene and rehydrated
in a descending series of alcohol. Antigen retrieval was performed
by heating the tissue sections in sodium citrate buffer (0.01
mmol/l; pH 6.0) at 100˚C for 30 min. Cooled sections were
subsequently immersed in 3% hydrogen peroxide solution at 37˚C for
10 min to block the endogenous peroxidase activity and then
incubated with an anti-B7-H6 rabbit antibody (1:150; cat. no.
ab121794; Abcam) at 4˚C overnight. Following incubation with the
primary antibody, the sections were incubated with an
HRP-conjugated goat anti-rabbit secondary antibody (1:100; cat. no.
abs975; Absin Bioscience, Inc.) at room temperature for 1 h. The
sections were subsequently counterstained with hematoxylin for 30
sec at room temperature and DAB for 1 min was used for chromogen
detection at room temperature. Finally, the sections were
dehydrated using an ascending alcohol series, cleared and sealed
with neutral resin. Samples incubated with PBS instead of the
primary antibody served as negative controls (NC).
Evaluation of immunohistochemical
staining
Using the positive control cells as reference, the
immunostaining intensity of B7-H6, which stained the cytoplasm and
cell membrane, was assessed using the H-score. Notably, only 3%
interstitial cells were weakly stained with B7-H6, and these
interstitial cells were associated with the inflammatory reaction
that occurred around the lesion. In the process of counting, only
positive cells in the cervical epithelium were considered, while
the positive cells in the interstitium were not counted. The
H-score was calculated using the following equation: % of unstained
cells x (0 + % of weakly stained cells) x (1 + % of moderately
stained cells) x (2 + % of strongly stained cells) x3(10). A total of three high-density areas
were determined for each specimen under a low-power microscope
(magnification, x100), and each high-density area was subsequently
counted under a high-power light microscope (magnification, x200);
the mean value of the three areas was obtained.
Cell culture
The HeLa cervical cancer cell line was gifted by Dr
Xue Du (Department of Gynecology, The General Hospital of Tianjin
Medical University, Tianjin, China). HeLa cells were cultured in
RPMI-1640 medium (cat. no. 31870074; Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (cat. no. F8318;
Sigma-Aldrich; Merck KGaA), and maintained at 37˚C in a humidified
atmosphere containing 5% CO2.
Cell transfection
Two shRNAs of 28 µg/ml targeting B7-H6 (shRNA-1,
5'-CCCTGCTCTCCTAACAGTT-3'; and shRNA-2, 5'-GGTTCTACCCAGAGGCTAT-3')
and shRNA-NC (5'-TTC TCCGAACGTGTCACGT-3') were synthesized by
Suzhou GenePharma Co., Ltd. (cat. nos. 190331CZ, 190331DZ and
C03DZ, respectively). The pGLV3/H1/GFP (LV) lentiviral vectors
LV3-shRNA-1, LV3-shRNA-2 or LV3-shRNA-NC (MOI, 10) were transfected
into HeLa cells at room temperature, and screened with polybrene
(1.5 µg/ml, 1:1,000; Suzhou GenePharma Co., Ltd.). Viral infections
were performed serially. The fluorescence microscope was used to
observe whether GFP fluorescence transfection efficiency was
>80% after 48 h of transfection. Stable cell lines expressing
shRNAs were selected with 2 µg/ml puromycin 72 h after
transfection.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
RT-qPCR was used to detect the B7-H6 mRNA level in
HeLa cells transfected with the three different shRNAs. Total RNA
was extracted from cultured cells using TRIzol® reagent
(cat. no. 213407; Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Purified RNA was then
reversely transcribed to cDNA using RevertAid First Strand cDNA
Synthesis kit purchased from Thermo Fisher Scientific, Inc. Reverse
transcription was performed at 42˚C for 1 h and the reaction was
terminated by heating at 70˚C for 5 min. Next, qPCR was performed
with FastStart Universal SYBR-Green Master Mix (cat. no.
04913850001; Roche Diagnostics) following the manufacturer's
instructions. An initial amplification using B7-H6 specific primers
was performed with a denaturation step at 95˚C for 10 min, followed
by 35 cycles of denaturation at 95˚C for 60 sec, primer annealing
at 58˚C for 30 sec and primer extension at 72˚C for 30 sec. Data
were normalized to the geometric mean of the housekeeping gene
GAPDH to control the variability in expression levels and
calculated as 2-ΔΔCq method
(19). The primer sequences of
B7-H6 were as follows: Forward 5'-TTTTCCATTCATTGGTGGCCTA-3' and
reverse 5'-TTTTCCATTCATTGGTGGCCTA-3'. The primer sequences of GAPDH
were as follows: Forward 5'-CTGGAACGGTGA AGGTGACA-3' and reverse
5'-AAGGGACTTCCTGTAACA ATGCA-3'.
Western blotting
HeLa cells were lysed with RIPA lysis buffer (cat.
no. BL504A; Biosharp; http://www.biosharp.cn/index/product/details/language/en/product_id/1783.html),
and the protein concentration was determined using the Quick-Start
Bradford protein assay (Bio-Rad Laboratories, Inc.). Equal amounts
of protein (30 µg loaded per lane) were separated via 10% SDS-PAGE
and transferred onto PVDF membranes (cat. no. P2938-1ROL;
Sigma-Aldrich; Merck KGaA). The membranes were blocked at room
temperature for 1 h with TBS-0.05% Tween-20 containing 5% skimmed
milk powder. The membranes were subsequently incubated with an
anti-B7-H6 rabbit antibody (1:1,000; cat. no. ab121794; Abcam) or
an anti-GAPDH antibody (1:5,000; cat. no. G8795; Sigma-Aldrich;
Merck KGaA) at 4˚C overnight. Following primary antibody
incubation, the membranes were incubated with a HRP-conjugated goat
anti-rabbit (1:5,000; cat. no. abs20040; Absin Bioscience, Inc.) or
goat anti-mouse secondary antibody (1:4,000; cat. no. abs20039;
Absin Bioscience, Inc.) at room temperature for 1 h. Protein bands
were visualized using ECL reagent (cat. no. abs920; Absin
Bioscience, Inc.). The ImageJ software (version no. v1.8.0;
National Institutes of Health, Bethesda, MD) was used to quantify
the WB data. The experiment was repeated three times.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was determined using a CCK-8
assay (cat. no. 69092500; BioSharp Life Sciences). Briefly, the
cells were digested by 1 ml trypsin and seeded at a density of
2x103 cells/well into four 96-well plates. Following 1,
2, 3 and 4 days of incubation at room temperature, 10 µl CCK-8
reagent was added/well and incubated for 4 h at room temperature.
The optical density value of each well was measured at 450 nm using
plate reader to generate the cell proliferation curves. The
experiment was repeated three times.
Wound healing assay
The cells were digested, counted and seeded into a
6-well plate at a density of 5x105 cells/well. When the
cells reached 90% confluence, a linear scratch was created in the
cell monolayer using a 10-µl pipette tip. The non-adherent cells
were removed by rinsing with 1 ml PBS after creating the scratch.
The cells were subsequently incubated in serum-free RPMI-1640
medium at 37˚C for 0, 24 or 48 h. At the indicated time points, the
plate was imaged with a light microscope (Olympus Corporation) at a
magnification of x200 to observe the distance of cell migration,
which was calculated using the following formula: Migration
distance at n h = (scratch width at 0 h - scratch width at n h),
where n represents each time point. The measurements were made at
random intervals along the wound length. The experiment was
repeated three times.
Transwell invasion assay
In total, 60 µl Matrigel with 7 mg/ml (cat. no.
356237; Corning, Inc.) was evenly spread on the upper layer of the
chamber at 37˚C for 1 h. The cells were digested with 1 ml trypsin,
counted and 2x104 cells were subsequently centrifuged at
161 x g 3 min at room temperature, followed by removal of the
supernatant. The cells were resuspended in 100 µl serum-free
RPMI-1640 medium and plated into the upper chamber of 24-well
Transwell plate, while 500 µl RPMI-1640 medium supplemented with
10% FBS was plated into the lower chamber. Following incubation for
48 h at 37˚C, the cells were fixed with 600 µl paraformaldehyde
(4%) for 30 min at room temperature and stained at room temperature
with 600 µl crystal violet dye (0.1%; cat. no. G1061; Beijing
Solarbio Science & Technology Co., Ltd.) for 10 min. The
chambers were subsequently washed twice with PBS and dried. The
number of invasive cells was counted in five randomly selected
fields of view using an inverted fluorescent microscope
(magnification, x100; Olympus Corporation). The experiment was
repeated three times.
Colony formation assay
Cells at the logarithmic growth phase were collected
when they reached a confluence of 90%. After staining with 0.4%
trypan blue dye for 3 min at room temperature, the number of living
cells that were released into 1x103/ml suspension were
counted. Complete culture medium (1 ml) and cell suspension (1 ml)
was added into the 6-well plate (3,000 cells per well)
successively, and the cells were dispersed by rotating slightly.
The 6-well plate was placed in the incubator for further culture
for 1 week at 37˚C in a humidified atmosphere containing 5%
CO2. After removing the medium in the 6-well plate, the
cells were washed twice with PBS and then fixed with 4%
paraformaldehyde for 20 min at room temperature. After removing the
paraformaldehyde and washing twice with PBS, 0.1% crystal violet
dye was added to the 6-well plate for 10 min at room temperature,
then the excess dye was washed with PBS and the plate was dried
with air. The transparent plate with a gridwas put on the bottom of
the plate. The cell clone clusters containing >50 cells number
in each hole was counted under a light microscope (Olympus
Corporation) at a magnification of x100. Finally, the clone
formation rate was calculated as follows: Clone formation rate =
(clone number/inoculated cell number) x 100%. After analyzing the
data of three independent repeat experiments, statistical analysis
was carried out and a histogram was drawn.
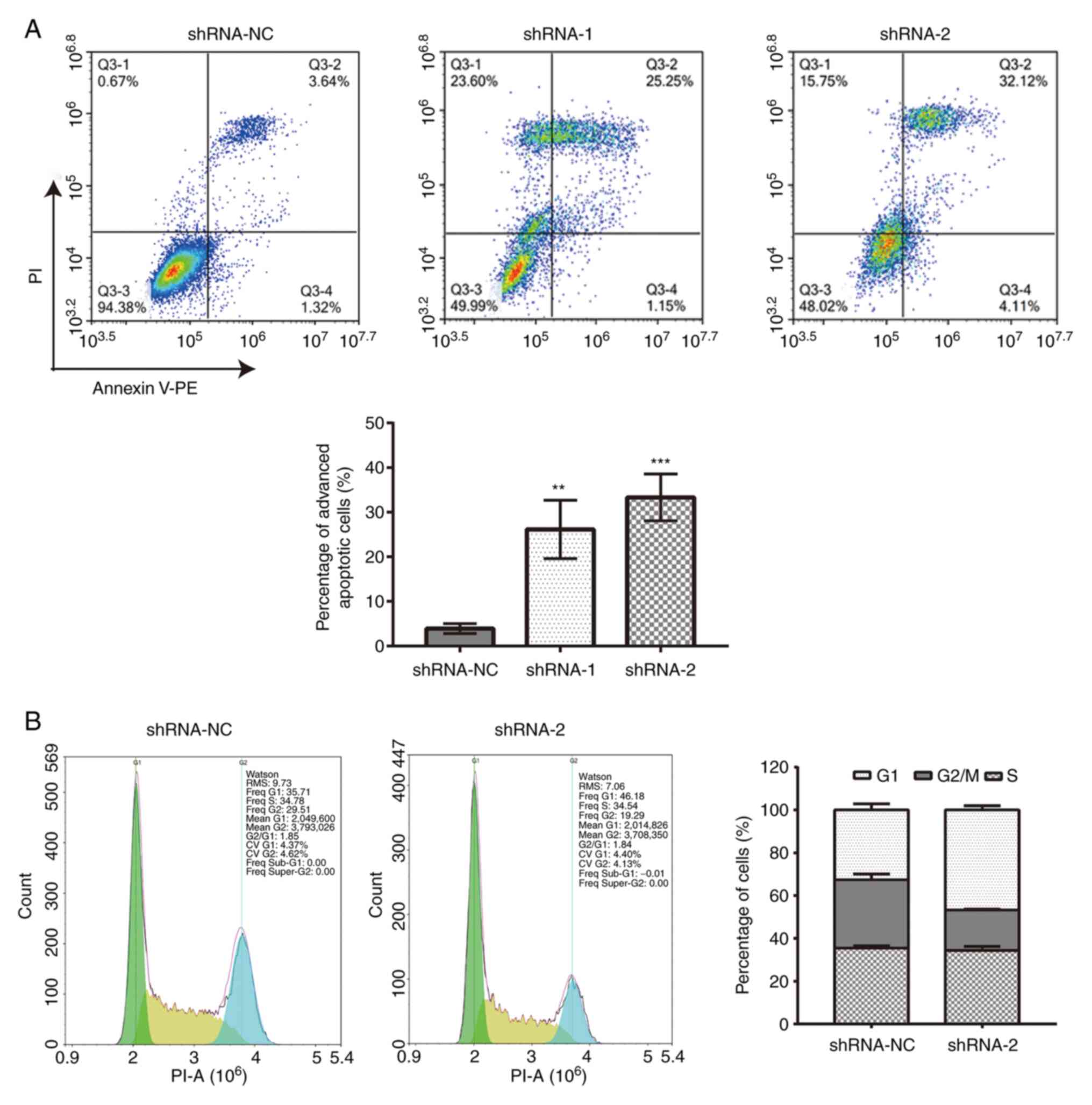
Apoptosis and cell cycle analyses
Briefly, 1x106 cells from the shRNA-NC
and shRNA-2 groups were washed with PBS. Apoptosis was detected
using a PE-Annexin V apoptosis detection kit (cat. no. 559763; BD
Biosciences). In total, 1x106 cells in a 100 µl cell
suspension was incubated with 5 µl PE-Annexin V solution and 5 µl
propidium iodide (PI) solution (concentration, 10 µg/ml) for 15 min
in the dark at 4˚C. The experiment was repeated three times.
For cell cycle analysis, 1x106 cells in a
100 µl cell suspension were stained with 0.5 ml PI/RNase staining
buffer (cat. no. 550825; BD Biosciences) for 20 min in the dark at
4˚C. Finally, the stained cells of 1x106 in 100 µl cell
suspension were detected by the NovoCyte D3000 flow cytometer (cat.
no. CA92121; ACEA Biosciences, Inc.) and the data were analyzed by
the NovoExpress software (version no. 1.5; ACEA Biosciences, Inc.).
The results in second (PI+ AV+; late apoptotic cells) and fourth
(PI- AV+; early apoptotic cells) quadrants was counted for this
analysis. The experiment was repeated three times.
Statistical analysis
The experimental data were processed by SPSS 19.0
statistical software (SPSS, Inc.). GraphPad Prism 8 (GraphPad
Software, Inc.) was used to plot figures and verify the results.
The data are shown as mean ± standard deviation. One way ANOVA and
Bonferonni's test were used for the comparison among multiple
groups. Independent unpaired samples t test was used for comparison
between two groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
B7-H6 expression is associated with
cervical cancer grade
B7-H6 expression levels were analyzed in normal
cervical, CIN I, CIN II, CIN III and cervical cancer tissues using
immunohistochemistry. The results revealed positive B7-H6
expression to differing intensities in CIN lesions and cervical
cancer tissues. In further detail, no cells were found to express
B7-H6 in normal cervical tissues (Fig.
1A), whereas in CIN I, CIN II, CIN III and cervical cancer
tissues, the H-score of B7-H6 expression increased alongside the
advancement in disease stage (Fig.
1B and C). Notably, the H-score
of B7-H6 expression in CIN II tissues (0.53±4.33) was significantly
upregulated compared with that in CIN I tissues (0.086±6.19); the
H-score of B7-H6 expression in CIN III tissues (1.85±5.21) was
significantly upregulated compared with in CIN II tissues; and the
H-score of B7-H6 expression in cervical cancer tissues (2.34±2.93)
was significantly upregulated compared with in CIN III tissues.
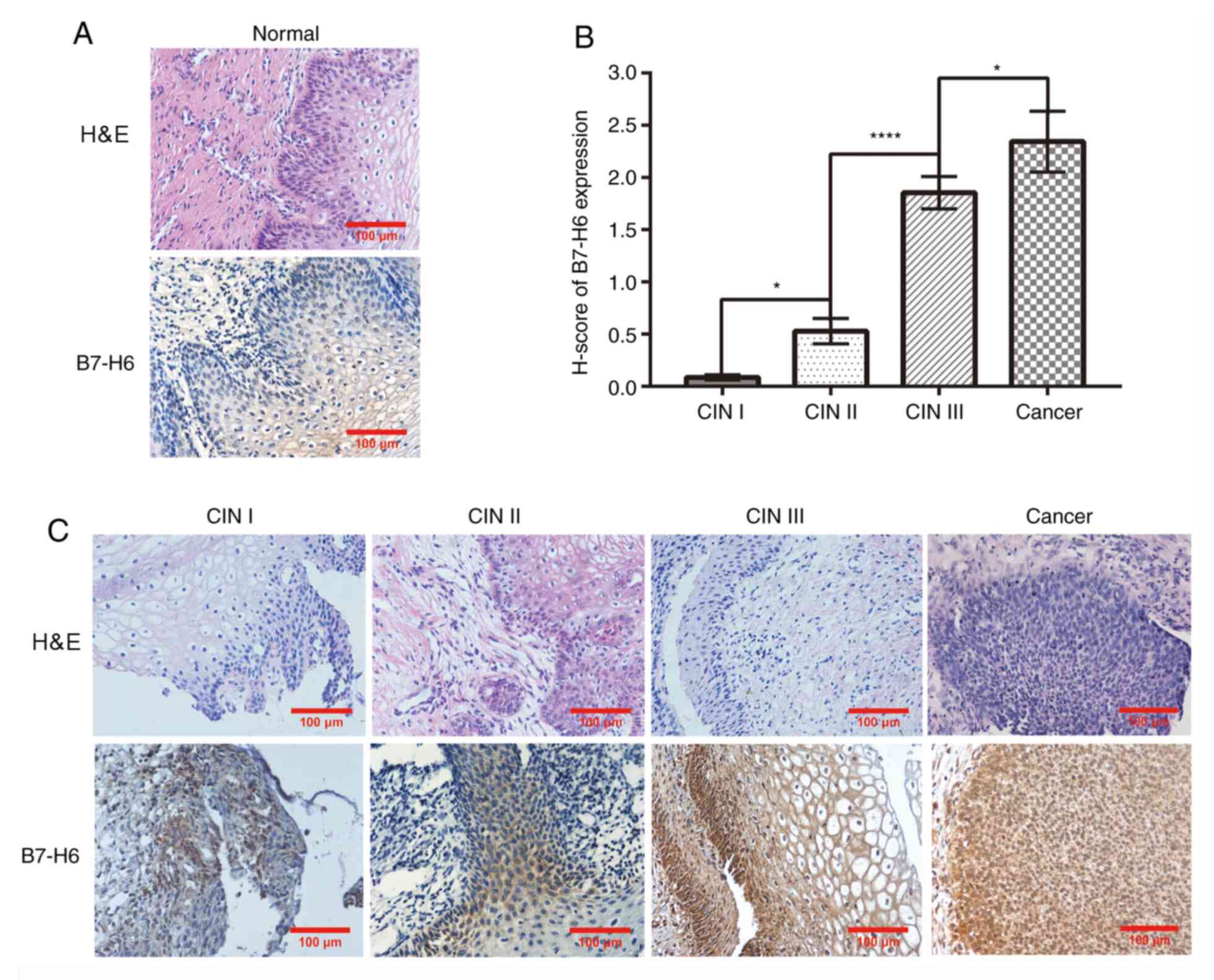 | Figure 1B7-H6 expression is associated with
cervical cancer grade. (A) H&E and immunohistochemical staining
of B7-H6 in normal cervical tissue (magnification, x200; scale bar,
100 µm). (B) B7-H6 differential expression was analyzed using
H-score (CIN II compared with CINⅠ, *P<0.05; CIN III
compared with CINII, ****P<0.0001; cervical cancer
compared with CIN III, *P<0.05). (C) H&E and
immunohistochemical staining of B7-H6 in CIN I, CIN II, CIN III and
cervical cancer lesion tissues (magnification, x200; scale bar, 100
µm). B7-H6, B7 homolog 6; CIN, cervical intraepithelial
neoplasia. |
Knockdown of B7-H6 expression in HeLa
cells
The efficiency of infection of the HeLa cells with
three groups of recombinant lentiviral vectors harboring B7-H6
shRNA was confirmed by analysis of GFP expression using
fluorescence microscopy (Fig. 2A).
Using RT-qPCR and western blotting analyses, the transfection of
shRNA-1 (RT-qPCR, 0.22±0.02; western blotting, 28,538±5,872) or
shRNA-2 (RT-qPCR, 0.11±0.01; western blotting, 15,917±3,750) into
HeLa cells significantly downregulated the expression levels of
B7-H6 compared with HeLa cells transfected with shRNA-NC (RT-qPCR,
1.00±0.09; western blotting, 83,808±5,282; Fig. 2B and C).
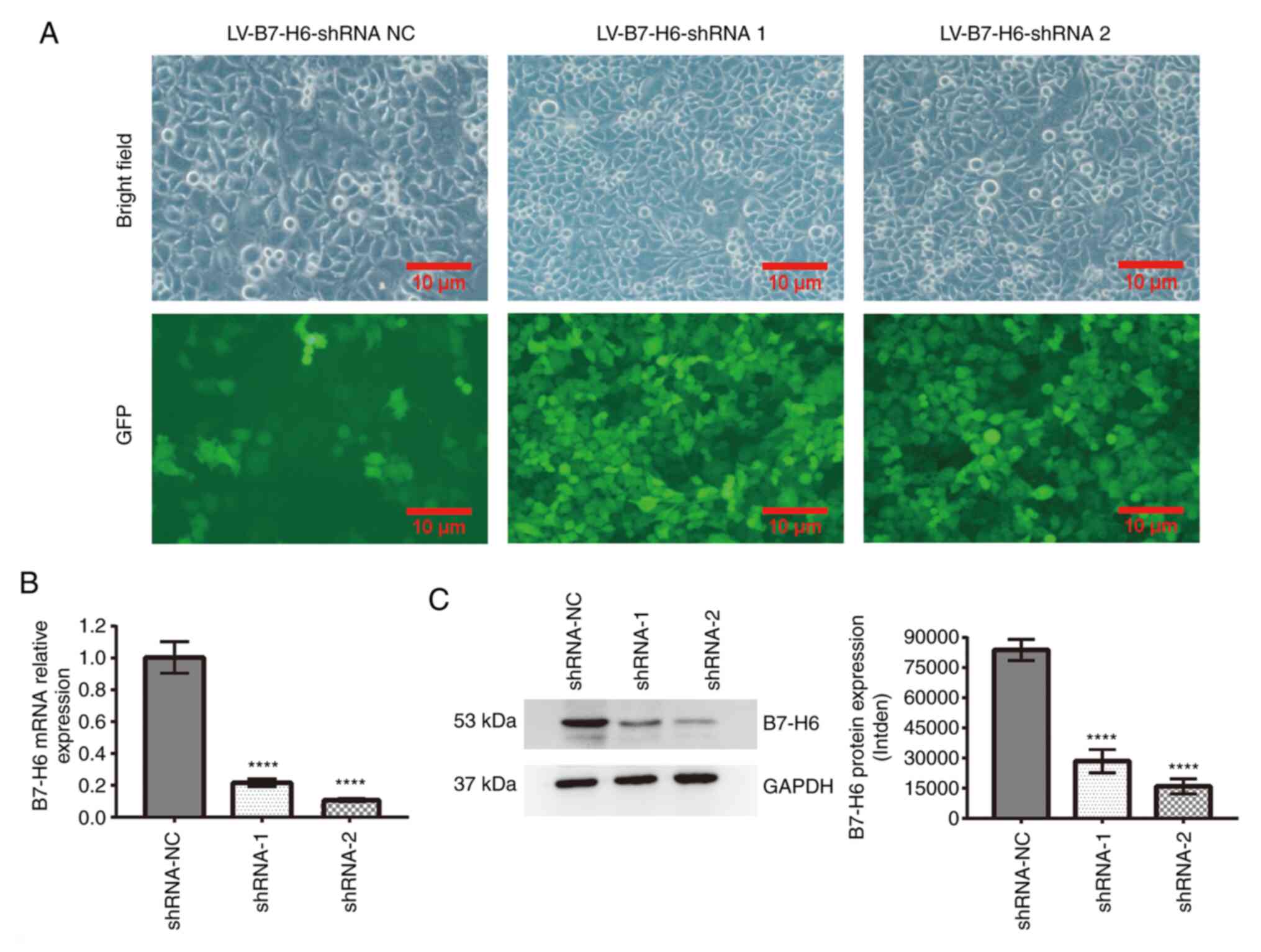 | Figure 2Knockdown of B7-H6 expression in HeLa
cells. (A) Confirmation of the efficiency of infection of the HeLa
cells with three groups of recombinant LV vector harboring B7-H6
shRNA by analysis of GFP expression using fluorescence microscopy
(scale bar, 100 µm). (B) Compared with shRNA-NC, the B7-H6 mRNA
level decreased in the shRNA-1 and shRNA-2 groups, as shown by
reverse transcription-quantitative PCR analysis
(****P<0.0001). (C) Compared with shRNA-NC, B7-H6
expression decreased in the shRNA-1 and shRNA-2 groups, as shown
using western blotting (****P<0.0001). B7-H6, B7
homolog 6; shRNA, short hairpin RNA; NC, negative control; LV,
lentivirus; GFP, green fluorescent protein; intden, integrated
density. |
Effect of B7-H6 on HeLa cell
biological functions
The proliferation rate of HeLa cells following
knockdown of B7-H6 expression was analyzed using CCK-8 and colony
formation assays. The results of the CCK-8 assay revealed that the
viability of cells transfected with shRNA-1 (3 days, 1.40±0.08; 4
days, 2.48±0.14) or shRNA-2 (3 days, 0.96±0.11; 4 days, 1.55±0.24)
was significantly decreased compared with the cells transfected
with shRNA-NC (3 days, 1.70±0.28; 4 days, 3.36±0.16; Fig. 3A). The results of the colony
formation assay also demonstrated that, compared with the cells
transfected with shRNA-NC (89±0.91%), the colony formation rate of
cells transfected with shRNA-1 (37±1.25%) or shRNA-2 (15±1.69%) was
decreased following 1 week of culture (Fig. 3B).
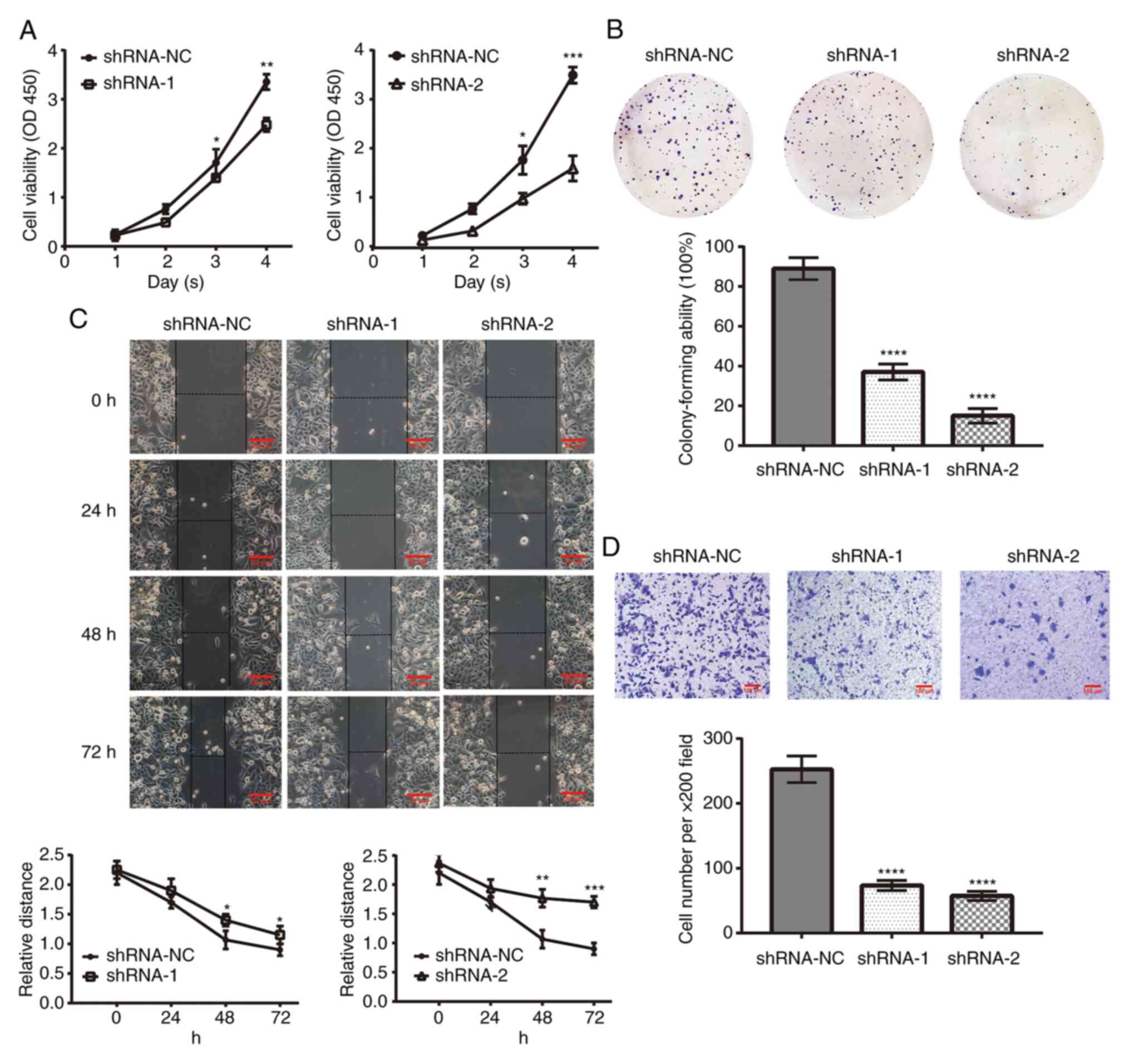 | Figure 3Effect of B7-H6 on HeLa cell
biological functions. (A) B7-H6 knockdown by shRNA-1 and shRNA-2
inhibited cell viability, as shown by the Cell Counting Kit-8 assay
(shRNA-1 vs. shRNA-NC: 3 days, *P<0.05; 4 days,
**P<0.01; and shRNA-2 vs. shRNA-NC: 3 days,
*P<0.05; 4 days, ***P<0.001). (B)
Colony formation assay demonstrated that B7-H6 knockdown inhibited
the colony-forming ability of shRNA-1 and shRNA-2 cells (compared
with shRNA-NC, ****P<0.0001). (C) Wound healing assay
demonstrated that the migration ability of HeLa cell was
significantly weakened following B7-H6 knockdown (shRNA-1 vs.
shRNA-NC at 48 and 72 h, *P<0.05; shRNA-2 vs.
shRNA-NC at 48 h, **P<0.01; at 72 h,
***P<0.001) (scale bar, 20 µm). (D) Transwell
invasion assay demonstrated that the number of crystal
violet-stained HeLa cells was decreased following B7-H6 knockdown
(compared with shRNA-NC, ****P<0.0001) (scale bar,
100 µm). B7-H6, B7 homolog 6; shRNA, short hairpin RNA; NC,
negative control. |
Subsequently, wound healing and Transwell invasion
assays were performed to determine the role of B7-H6 in HeLa cell
migration and invasion. The wound healing assay results
demonstrated that the cell-free area in plates containing cells
transfected with shRNA-1 (48 h, 1.40±0.10; 72 h, 1.15±0.15) or
shRNA-2 (48 h, 1.77±0.15; 72 h, 1.70±0.10) was significantly wider
compared with cells transfected with shRNA-NC (48 h, 1.07±0.15; 72
h, 0.90±0.10) (Fig. 3C). The
results of the Transwell invasion assay revealed that the invasive
ability of cells transfected with shRNA-1 (73.33±7.37) or shRNA-2
(57.33±3.48) was decreased compared with cells transfected with
shRNA-NC (252.7±4.62) at 48 h (Fig.
3D).
Effect of B7-H6 knockdown on HeLa cell
apoptosis and cell cycle distribution
Flow cytometric analysis demonstrated that, compared
with cells transfected with shRNA-NC (3.91±4.22%), an increased
proportion of cells transfected with shRNA-1 (26.15±3.99%) or
shRNA-2 (33.32±7.24%) underwent late cell apoptosis (Fig. 4A). The results also demonstrated
that the transfection with shRNA-2 increased the percentage of G1
cells (46.78±1.99%) and decreased the percentage of G2/M cells
(18.83±0.40%) compared with cells transfected with shRNA-NC (G1,
32.63±2.84%; G2/M, 31.80±2.69%) (Fig.
4B).
Discussion
B7-H6, also known as NK cell toxicity receptor 3
ligand 1, belongs to the B7 immunoglobulin superfamily and, in
recent years, has been discovered to act as a co-stimulator through
bioinformatics analysis and mass spectrometry. The specific
expression of B7-H6 in tumor cells has been shown to promote
tumorigenesis (16) and to be
associated with clinicopathological parameters (19,20).
The B7-H6 gene encodes a 454-amino acid type I transmembrane
protein, with a predicted molecular mass of 51 kDa. Similar to all
known B7 family members, B7-H6 comprises two immunoglobulin domains
with adjacent phase 1 introns in the extracellular region (21). Although B7-H6 is expressed at very
low levels, if at all, in normal tissues, its expression was found
to be upregulated in several types of cancer, including lymphoma,
leukemia, hepatocellular carcinoma, gastric cancer, brain cancer,
ovarian cancer and breast cancer (19,22-27).
However, to the best of our knowledge, the clinical significance
and role of B7-H6 in cervical cancer remain unclear.
The present study used immunohistochemistry analysis
to demonstrate that the expression levels of B7-H6 were gradually
upregulated as precancerous lesions developed into cervical cancer.
These findings are consistent with previous findings in esophageal
squamous cell carcinoma (16),
breast cancer (17) and oral
squamous cell carcinoma (19).
B7-H6 was not only reported to be associated with the progression
of these malignant tumors, but it was also closely associated with
the clinicopathological parameters of the tumors and patient
prognosis. Li et al (20)
reported that the high expression of B7-H6 in gastric cancer
tissues was significantly associated with overall survival. These
aforementioned findings combined with the results of the present
study suggested that the transcription of B7-H6 may be activated in
cervical precancerous lesions and thereby serve an important role
in the development of cervical cancer. B7-H6 expression may also be
associated with clinicopathological parameters and prognosis in
cervical cancer.
In the present study, subsequent cell experiments
demonstrated that the expression levels of B7-H6 were successfully
knocked down in HeLa cells using shRNA. CCK-8 and colony formation
assays demonstrated that the proliferative ability of HeLa cells
following B7-H6 knockdown was reduced. In addition, the migratory
and invasive abilities of HeLa cells following B7-H6 knockdown were
also significantly decreased, as determined using wound healing and
Transwell invasion assays, respectively. Flow cytometry analysis
also demonstrated that B7-H6 knockdown in HeLa cells promoted late
apoptosis and cell cycle arrest at the G1 phase. These findings
were consistent with the results of Chen et al (22), which reported that the gene
silencing of B7-H6 in the HepG2 and SMMC-7721 cell lines using
shRNA significantly inhibited cell proliferation, migration and
invasion, and induced cell apoptosis and cell cycle arrest by
downregulating the expression levels of c-Myc, c-Fos and cyclin D1.
Similarly, Che et al (27)
reported that the proliferative and invasive abilities of glioma
cell lines, including CRT, U251, SHG-44, SF-295, TG-905 and U373,
were decreased following B7-H6 knockdown. In addition, the study
also found that the expression levels of proteins associated with
apoptosis, migration and invasion, including E-cadherin, Bcl-2,
vimentin, N-cadherin, matrix metalloproteinase (MMP)-2, MMP-9 and
survivin, were altered. Li et al (28) also suggested that B7-H6 may exert
anti-apoptotic effects by upregulating the expression levels of
MMP-9 and activating the STAT3 signaling pathway. In addition, in B
cell lymphoma, B7-H6 gene silencing inhibited tumor cell colony
formation, growth, invasion and migration, and induced cell
apoptosis (29). These
aforementioned studies and the results of the present study
indicated that B7-H6 may promote cervical cancer tumorigenesis by
regulating cell proliferation, cell cycle progression and
apoptosis.
In conclusion, the results of the present study
suggested that B7-H6 may be an oncogene and represent a novel tumor
biomarker and potential target for cervical cancer treatment.
Therefore, further investigations into the role of B7-H6 may
provide novel insights into the treatment of cervical cancer.
However, there were certain limitations to the present study.
First, the possible mechanisms promoting B7-H6 transcription and
the signaling pathway through which B7-H6 may regulate the
proliferation, migration and apoptosis of cervical cancer cells
remain unclear. Second, the mechanism of action of B7-H6 should be
investigated in additional cervical cancer cell lines, and in
vivo animal models should be used in future studies to verify
the effect of B7-H6 in cervical cancer. Finally, further studies
are required to determine the association between B7-H6 and the
clinicopathological parameters and prognosis of cervical
cancer.
Acknowledgements
The authors would like to thank Dr Xue Du,
Department of Gynecology, The General Hospital of Tianjin Medical
University, Tianjin, China for providing the HeLa cell line.
Funding
Funding: The present study was supported by the Youth Fund of
the Second Hospital of Tianjin Medical University, the Second
Hospital of Tianjin Medical University (grant no. 2018ydey16) and
The Natural Science Foundation of Tianjin, Tianjin Municipal
Science and Technology Beaureau (grant no. 18JCYBJC28000).
Availability of data and materials
The datasets used and analysed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
RG: Project development, data collection, data
analysis, manuscript writing. GL: Project development. CL:
Experimental technical support. XL: Data collection, conduction of
experiments, data analysis. YX, WY and FW: Data collection. All the
authors have read and approved the final manuscript. Authentication
of the raw data: CL and XL.
Ethics approval and consent to
participate
The present study was approved by the the Ethics
Committee of the Second Hospital of Tianjin Medical University, and
it is registered under the approval no. KY2019K023. All patients or
their family members provided written informed consent for their
tissues to be used for research purposes prior to
participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Stelzle D, Tanaka LF, Lee KK, Ibrahim
Khalil A, Baussano I, Shah ASV, McAllister DA, Gottlieb SL, Klug
SJ, et al: Estimates of the global burden of cervical cancer
associated with HIV. Lancet Glob Health. 2:e161–e169.
2020.PubMed/NCBI View Article : Google Scholar : Erratum in: Lancet
Glob Health 2, e119, 2021.
|
|
2
|
Abu Rustum NR, Yashar CM, Bradley K,
Campos SM, Chon HS, Chu C, Clinton L, Cohn D, Crispens MA, et al:
NCCN Guidelines Version 1.2021 Cervical Cancer. J Natl Compr Canc
Netw. 6:660–666. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chitsike L and Duerksen-Hughes P: The
potential of immune checkpoint blockade in cervical cancer: Can
combinatorial regimens maximize response? A review of the
literature. Curr Treat Options Oncol. 21(95)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ni L and Dong C: New B7 family checkpoints
in human cancers. Mol Cancer Ther. 16:1203–1211. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hu Y, Zeng T, Xiao Z, Hu Q, Li Y, Tan X,
Yue H, Wang W, Tan H and Zou J: Immunological role and underlying
mechanisms of B7-H6 in tumorigenesis. Clin Chim Acta. 502:191–198.
2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li Y, Wang Q and Mariuzza RA: Structure of
the human activating natural cytotoxicity receptor NKp30 bound to
its tumor cell ligand B7-H6. J Exp Med. 208:703–714.
2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Barrow AD, Martin CJ and Colonna M: The
Natural cytotoxicity receptors in health and disease. Front
Immunol. 10:909–913. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Joyce MG, Tran P, Zhuravleva MA, Jaw J,
Colonna M and Sun PD: Crystal structure of human natural
cytotoxicity receptor NKp30 and identification of its ligand
binding site. Proc Natl Acad Sci USA. 108:6223–6228.
2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Fiegler N, Textor S, Arnold A, Rölle A,
Oehme I, Breuhahn K, Moldenhauer G, Witzens-Harig M and Cerwenka A:
Downregulation of the activating NKp30 ligand B7-H6 by HDAC
inhibitors impairs tumor cell recognition by NK cells. Blood.
122:684–693. 2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pesce S, Tabellini G, Cantoni C, Patrizi
O, Coltrini D, Rampinelli F, Matta J, Vivier E, Moretta A, Parolini
S, et al: B7-H6-mediated downregulation of NKp30 in NK cells
contributes to ovarian carcinoma immune escape. OncoImmunology.
4(e1001224)2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Schlecker E, Fiegler N, Arnold A, Altevogt
P, Rose-John S, Moldenhauer G, Sucker A, Paschen A, von Strandmann
EP, Textor S, et al: Metalloprotease-mediated tumor cell shedding
of B7-H6, the ligand of the natural killer cell-activating receptor
NKp30. Cancer Res. 74:3429–3440. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Phillips M, Romeo F, Bitsaktsis C and
Sabatino D: B7H6-derived peptides trigger TNF-α-dependent
immunostimulatory activity of lymphocytic NK92-MI cells.
Biopolymers. 106:658–672. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cao G, Wang J, Zheng X, Wei H, Tian Z and
Sun R: Tumor therapeutics work as stress inducers to enhance tumor
sensitivity to natural killer (NK) cell cytolysis by up-regulating
NKp30 ligand B7-H6. J Biol Chem. 290:29964–29973. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Textor S, Bossler F, Henrich KO,
Gartlgruber M, Pollmann J, Fiegler N, Arnold A, Westermann F,
Waldburger N, Breuhahn K, et al: The proto-oncogene Myc drives
expression of the NK cell-activating NKp30 ligand B7-H6 in tumor
cells. Oncoimmunology. 5(e1116674)2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhou H, Dong J, Guo L, Wang X, Wang K, Cai
X and Yang S: The prognostic value of B7-H6 in esophageal squamous
cell carcinoma. Sci Rep. 9:18122–18130. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sun J, Tao H, Li X, Wang L, Yang J, Wu P,
Zhang Y and Guo Y: Clinical significance of novel costimulatory
molecule B7-H6 in human breast cancer. Oncol Lett. 14:2405–2409.
2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang J, Jin X, Liu J, Zhao K, Xu H, Wen J,
Jiang L, Zeng X, Li J and Chen Q: The prognostic value of B7-H6
protein expression in human oral squamous cell carcinoma. J Oral
Pathol Med. 46:766–772. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Waxman AG, Chelmow D, Darragh TM, Lawson H
and Moscicki AB: Revised terminology for cervical histopathology
and its implications for management of high-grade squamous
intraepithelial lesions of the cervix. Obstet Gynecol.
120:1465–1471. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li D, Xiang S, Shen J, Xiao M, Zhao Y, Wu
X, Du F, Ji H, Li M, Zhao Q, et al: Comprehensive understanding of
B7 family in gastric cancer: Expression profile, association with
clinicopathological parameters and downstream targets. Int J Biol
Sci. 16:568–582. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chen Y, Mo J, Jia X and He Y: The B7
Family Member B7-H6: A new bane of tumor. Pathol Oncol Res.
24:717–721. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chen L, Feng J, Xu B, Zhou Y, Zheng X, Wu
C and Jiang J: B7-H6 expression in human hepatocellular carcinoma
and its clinical significance [corrected]. Cancer Cell Int.
18:126–136. 2018.PubMed/NCBI View Article : Google Scholar : Erratum in: Cancer
Cell Int 18, 134 2018.
|
|
23
|
Jiang T, Wu W, Zhang H, Zhang X, Zhang D,
Wang Q, Huang L, Wang Y and Hang C: High expression of B7-H6 in
human glioma tissues promotes tumor progression. Oncotarget.
8:37435–37447. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu MR, Zhang T, Gacerez AT, Coupet TA,
DeMars LR and Sentman CL: B7H6-specific bispecific T cell engagers
lead to tumor elimination and host antitumor immunity. J Immunol.
194:5305–5311. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chen XJ, Shen J, Zhang GB and Chen WC:
B7-H6 protein expression has no prognostic significance in human
gastric carcinoma. Pathol Oncol Res. 20:203–207. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xu Z, Shen J, Wang MH, Yi T, Yu Y, Zhu Y,
Chen B, Chen J, Li L, Li M, et al: Comprehensive molecular
profiling of the B7 family of immune-regulatory ligands in breast
cancer. OncoImmunology. 5(e1207841)2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Che F, Xie X, Wang L, Su Q, Jia F, Ye Y,
Zang L, Wang J, Li H, Quan Y, et al: B7-H6 expression is induced by
lipopolysaccharide and facilitates cancer invasion and metastasis
in human gliomas. Int Immunopharmacol. 59:318–327. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li YM, Liu ZY, Li ZC, Wang JC, Yu JM, Yang
HJ, Chen ZN and Tang J: Alterations of the immunologic
co-stimulator B7 and TNFR families correlate with hepatocellular
carcinoma prognosis and metastasis by inactivating STAT3. Int J Mol
Sci. 20:156–174. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wu F, Wang J and Ke X: Knockdown of B7-H6
inhibits tumor progression and enhances chemosensitivity in B-cell
non-Hodgkin lymphoma. Int J Oncol. 48:1561–1570. 2016.PubMed/NCBI View Article : Google Scholar
|