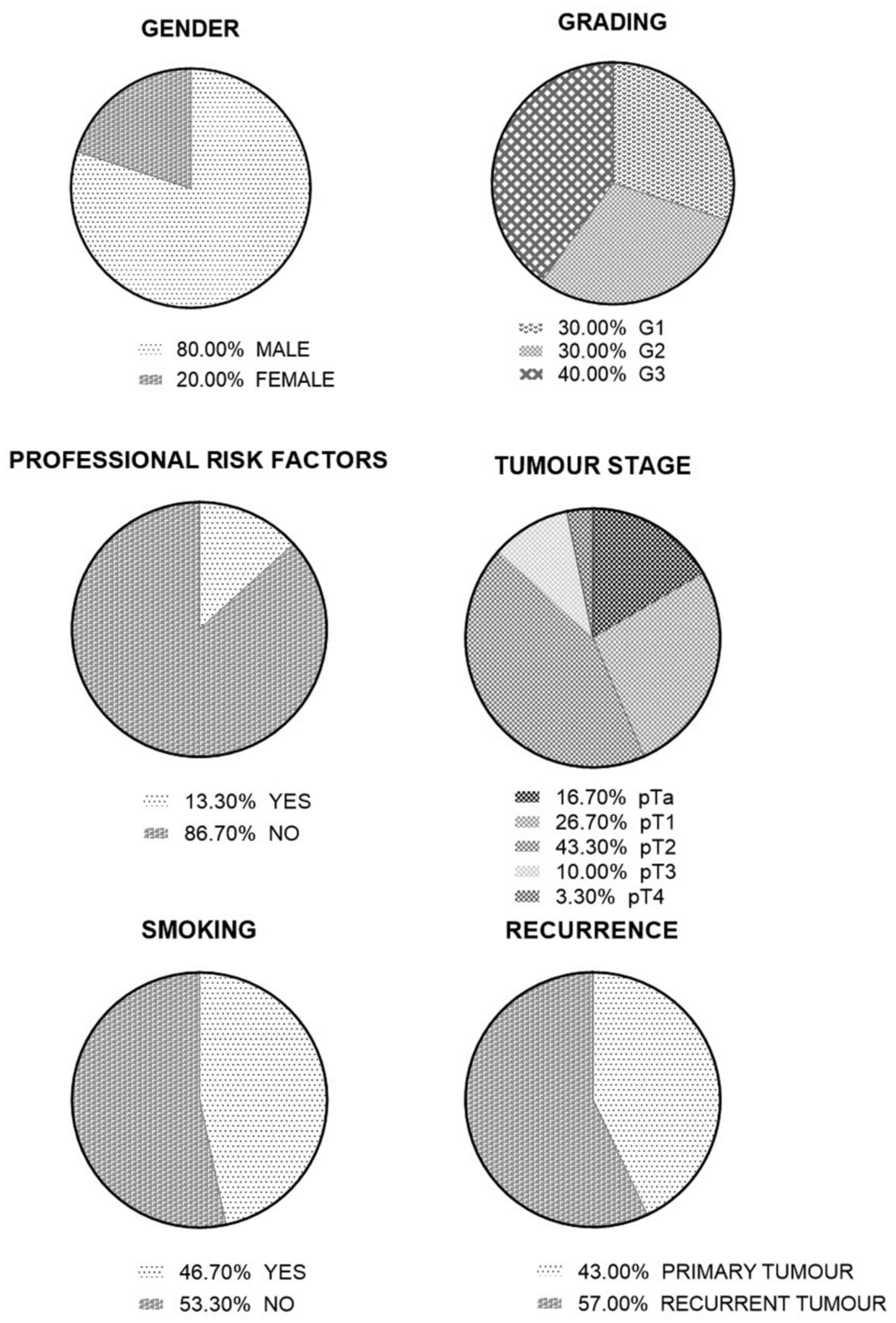
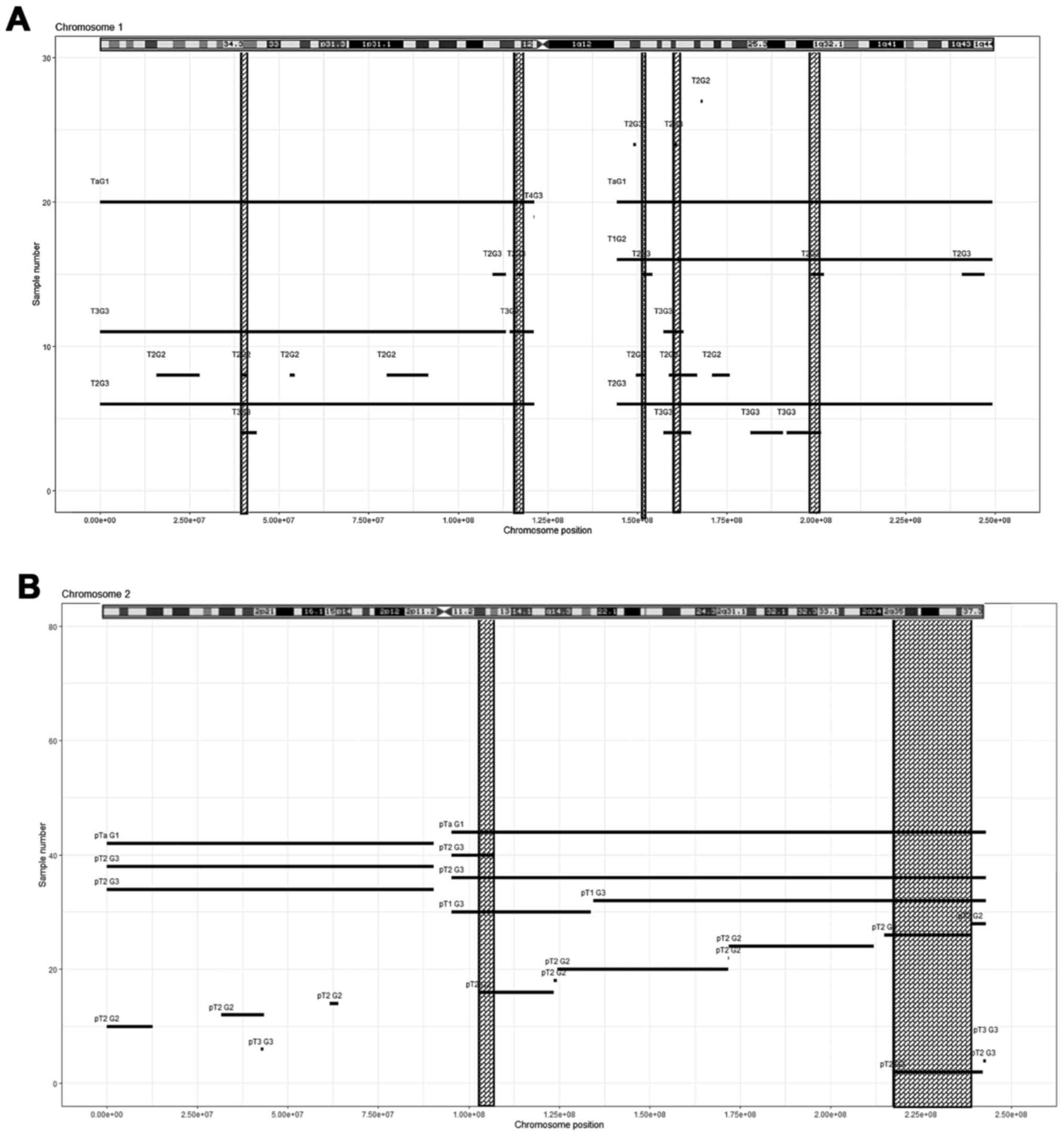
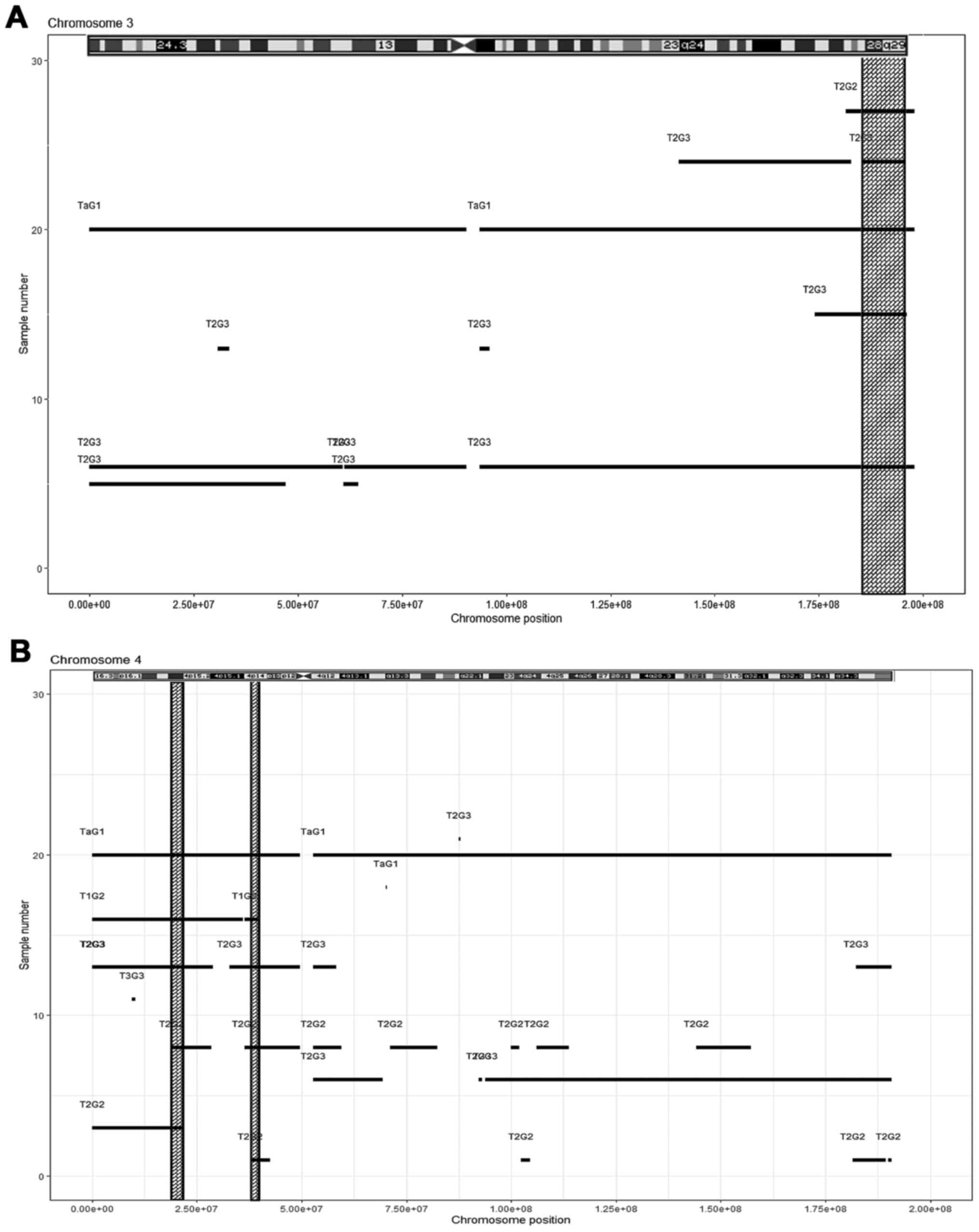
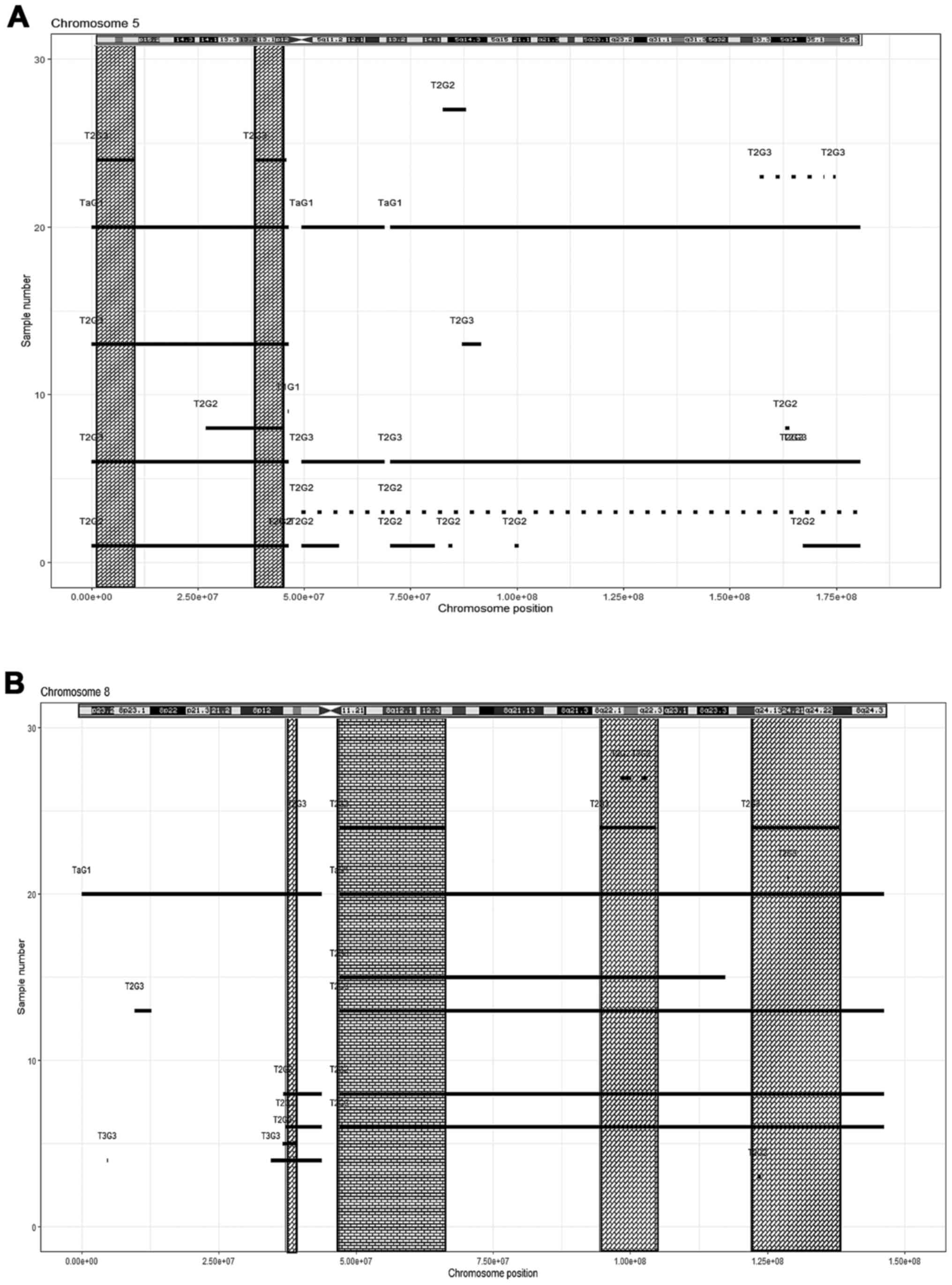
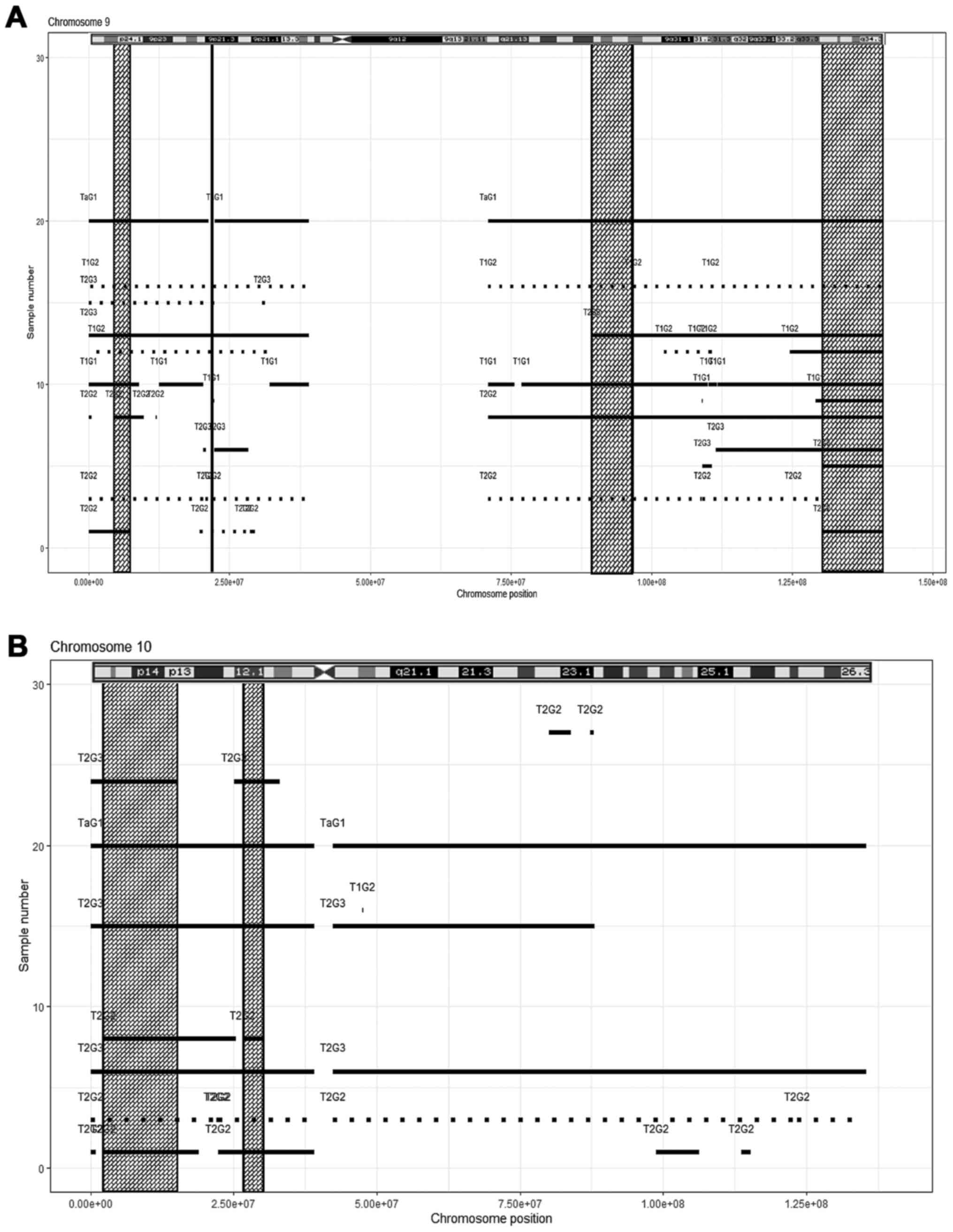
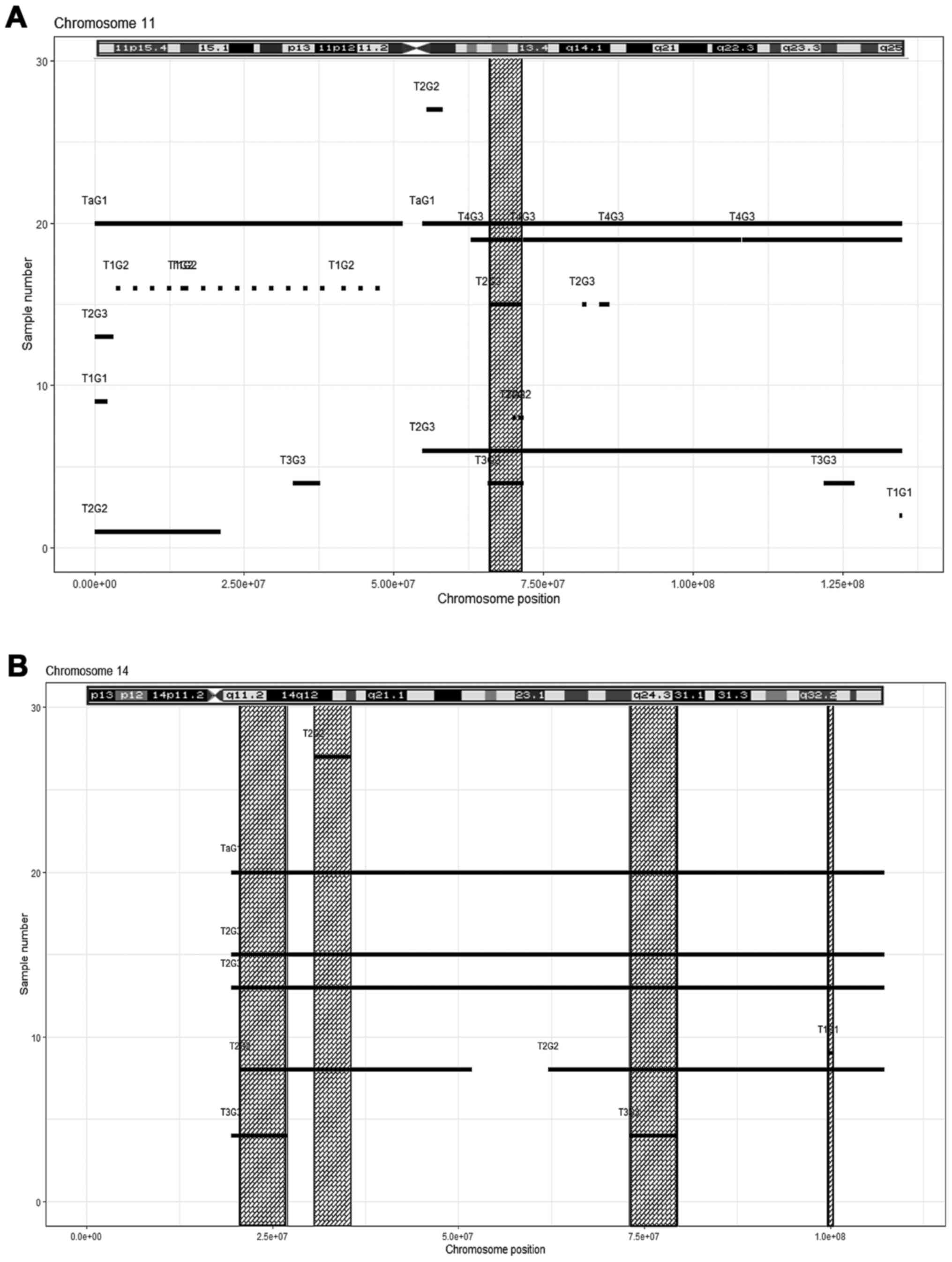
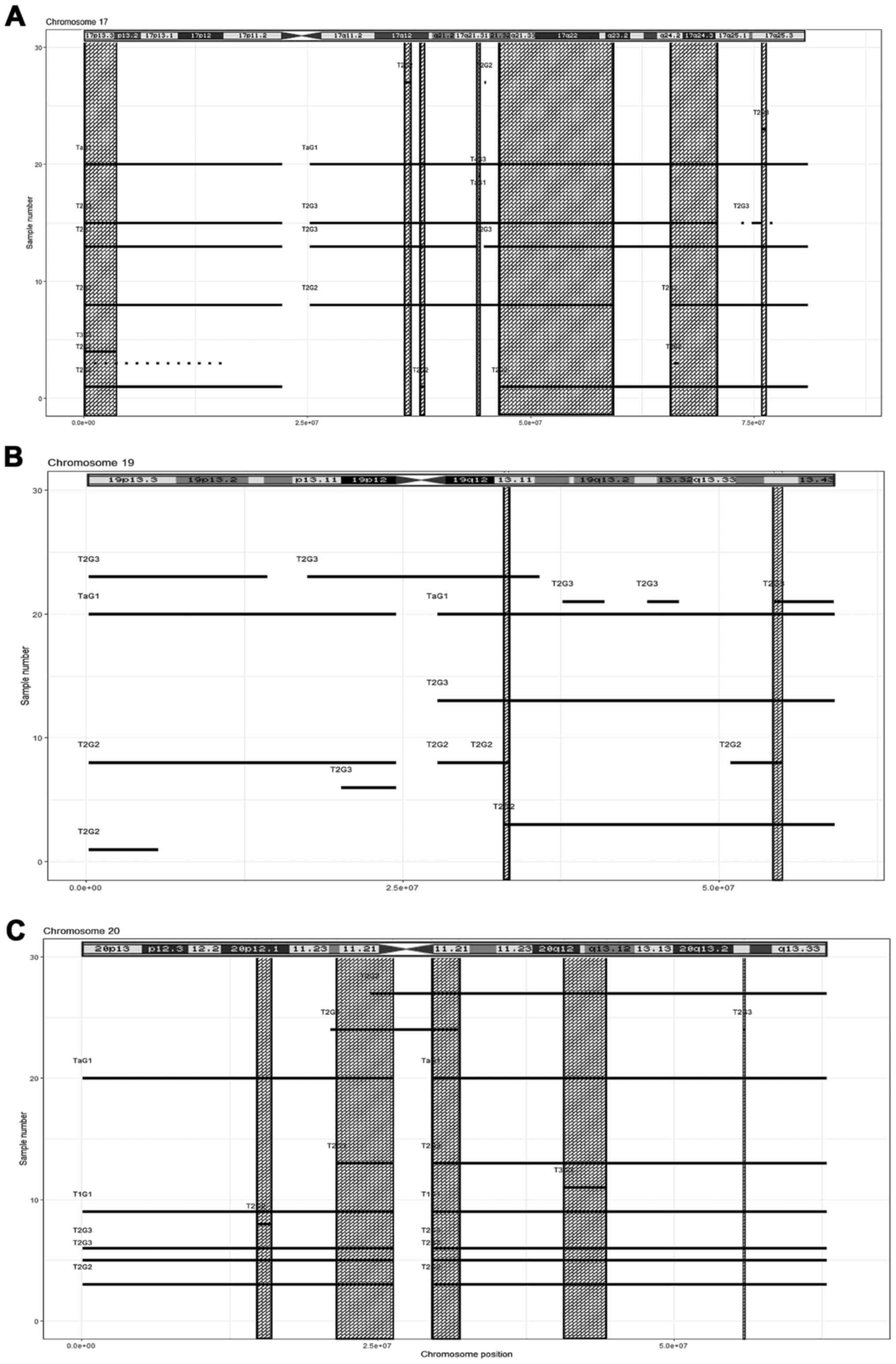
|
1
|
Conconi D, Panzeri E, Redaelli S, Bovo G,
Viganò P, Strada G, Dalprà L and Bentivegna A: Chromosomal
imbalances in human bladder urothelial carcinoma: Similarities and
differences between biopsy samples and cancer stem-like cells. BMC
Cancer. 14(646)2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kawanishi H, Takahashi T, Ito M, Matsui Y,
Watanabe J, Ito N, Kamoto T, Kadowaki T, Tsujimoto G, Imoto I, et
al: Genetic analysis of multifocal superficial urothelial cancers
by array-based comparative genomic hybridisation. Br J Cancer.
97:260–266. 2007.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kallioniemi A, Kallioniemi OP, Citro G,
Sauter G, DeVries S, Kerschmann R, Caroll P and Waldman F:
Identification of gains and losses of DNA sequences in primary
bladder cancer by comparative genomic hybridization. Genes
Chromosomes Cancer. 12:213–219. 1995.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sauter G: Rudolf Virchow Prize 1997.
Molecular cytogenetic analysis of superficial urothelial cancer of
the bladder. Verh Dtsch Ges Pathol. 81:18–27. 1997.PubMed/NCBI(In German).
|
|
5
|
Voorter C, Joos S, Bringuier PP, Vallinga
M, Poddighe P, Schalken J, du Manoir S, Ramaekers F, Lichter P and
Hopman A: Detection of chromosomal imbalances in transitional cell
carcinoma of the bladder by comparative genomic hybridization. Am J
Pathol. 146:1341–1354. 1995.PubMed/NCBI
|
|
6
|
Zaharieva BM, Simon R, Diener PA,
Ackermann D, Maurer R, Alund G, Knönagel H, Rist M, Wilber K,
Hering F, et al: High-throughput tissue microarray analysis of
11q13 gene amplification (CCND1, FGF3, FGF4, EMS1) in urinary
bladder cancer. J Pathol. 201:603–608. 2003.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Huang WC, Taylor S, Nguyen TB, Tomaszewski
JE, Libertino JA, Malkowicz SB and McGarvey TW: KIAA1096, a gene on
chromosome 1q, is amplified and overexpressed in bladder cancer.
DNA Cell Biol. 21:707–715. 2002.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Veltman JA, Fridlyand J, Pejavar S, Olshen
AB, Korkola JE, DeVries S, Carroll P, Kuo WL, Pinkel D, Albertson
D, et al: Array-based comparative genomic hybridization for
genome-wide screening of DNA copy number in bladder tumors. Cancer
Res. 63:2872–2880. 2003.PubMed/NCBI
|
|
9
|
Bellmunt J, Selvarajah S, Rodig S, Salido
M, de Muga S, Costa I, Bellosillo B, Werner L, Mullane S, Fay AP,
et al: Identification of ALK gene alterations in urothelial
carcinoma. PLoS One. 9(e103325)2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Scaravilli M, Asero P, Tammela TL,
Visakorpi T and Saramäki OR: Mapping of the chromosomal
amplification 1p21-22 in bladder cancer. BMC Res Notes.
7(547)2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Weilandt M, Koch A, Rieder H, Deenen R,
Schwender H, Niegisch G and Schulz WA: Target genes of recurrent
chromosomal amplification and deletion in urothelial carcinoma.
Cancer Genomics Proteomics. 11:141–153. 2014.PubMed/NCBI
|
|
12
|
Ewald JA, Downs TM, Cetnar JP and Ricke
WA: Expression microarray meta-analysis identifies genes associated
with Ras/MAPK and related pathways in progression of
muscle-invasive bladder transition cell carcinoma. PLoS One.
8(e55414)2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hussain SA, Palmer DH, Syn WK, Sacco JJ,
Greensmith RM, Elmetwali T, Aachi V, Lloyd BH, Jithesh PV, Arrand
J, et al: Gene expression profiling in bladder cancer identifies
potential therapeutic targets. Int J Oncol. 50:1147–1159.
2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mengual L, Burset M, Ars E, Lozano JJ,
Villavicencio H, Ribal MJ and Alcaraz A: DNA microarray expression
profiling of bladder cancer allows identification of noninvasive
diagnostic markers. J Urol. 182:741–748. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sanchez-Carbayo M: Use of high-throughput
DNA microarrays to identify biomarkers for bladder cancer. Clin
Chem. 49:23–31. 2003.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
de Ravel TJ, Devriendt K, Fryns JP and
Vermeesch JR: What's new in karyotyping? The move towards array
comparative genomic hybridisation (CGH). Eur J Pediatr.
166:637–643. 2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Letasiova S, Medve'ova A, Sovcikova A,
Dušinská M, Volkovová K, Mosoiu C and Bartonová A: Bladder cancer,
a review of the environmental risk factors. Environ Health. 11
Suppl 1(Suppl 1)(S11)2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Minner S, Kilgue A, Stahl P, Weikert S,
Rink M, Dahlem R, Fisch M, Höppner W, Wagner W, Bokemeyer C, et al:
Y chromosome loss is a frequent early event in urothelial bladder
cancer. Pathology. 42:356–359. 2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Panani AD and Roussos C: Sex chromosome
abnormalities in bladder cancer: Y polysomies are linked to
PT1-grade III transitional cell carcinoma. Anticancer Res.
26(1A):319–323. 2006.PubMed/NCBI
|
|
20
|
Forsberg LA: Loss of chromosome Y (LOY) in
blood cells is associated with increased risk for disease and
mortality in aging men. Hum Genet. 136:657–663. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lopez V, Gonzalez-Peramato P, Suela J,
Serrano A, Algaba F, Cigudosa JC, Vidal A, Bellmunt J, Heredero O
and Sánchez-Carbayo M: Identification of prefoldin amplification
(1q23.3-q24.1) in bladder cancer using comparative genomic
hybridization (CGH) arrays of urinary DNA. J Transl Med.
11(182)2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Huang Y, Li G, Wang K, Mu Z, Xie Q, Qu H,
Lv H and Hu B: Collagen Type VI Alpha 3 Chain promotes
epithelial-mesenchymal transition in bladder cancer cells via
transforming growth factor β (TGF-β)/Smad pathway. Med Sci Monit.
24:5346–5354. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liang Y, Luo H, Zhang H, Dong Y and Bao Y:
Oncogene Delta/Notch-Like EGF-Related receptor promotes cell
proliferation, invasion, and migration in hepatocellular carcinoma
and predicts a poor prognosis. Cancer Biother Radiopharm.
33:380–386. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Deng S, He SY, Zhao P and Zhang P: The
role of oncostatin M receptor gene polymorphisms in bladder cancer.
World J Surg Oncol. 17(30)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sun J, Zhang H, Tao D, Xie F, Liu F, Gu C,
Wang M, Wang L, Jiang G, Wang Z and Xiao X: CircCDYL inhibits the
expression of C-MYC to suppress cell growth and migration in
bladder cancer. Artif Cells Nanomed Biotechnol. 47:1349–1356.
2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chen Y, Peng Y, Xu Z, Ge B, Xiang X, Zhang
T, Gao L, Shi H, Wang C and Huang J: Knockdown of lncRNA SNHG7
inhibited cell proliferation and migration in bladder cancer
through activating Wnt/β-catenin pathway. Pathol Res Pract.
215:302–307. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Daizumoto K, Yoshimaru T, Matsushita Y,
Fukawa T, Uehara H, Ono M, Komatsu M, Kanayama HO and Katagiri T: A
DDX31/Mutant-p53/EGFR Axis promotes multistep progression of
Muscle-Invasive bladder cancer. Cancer Res. 78:2233–2247.
2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wu L, Chaffee KG, Parker AS, Sicotte H and
Petersen GM: Zinc transporter genes and urological cancers:
Integrated analysis suggests a role for ZIP11 in bladder cancer.
Tumour Biol. 36:7431–7437. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kim YW, Yoon HY, Seo SP, Lee SK, Kang HW,
Kim WT, Bang HJ, Ryu DH, Yun SJ, Lee SC, et al: Clinical
implications and prognostic values of prostate cancer
susceptibility candidate methylation in primary nonmuscle invasive
bladder cancer. Dis Markers. 2015(402963)2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chen Z, Zhou L, Wang L, Kazobinka G, Zhang
X, Han X, Li B and Hou T: HBO1 promotes cell proliferation in
bladder cancer via activation of Wnt/β-catenin signaling. Mol
Carcinog. 57:12–21. 2018.PubMed/NCBI View Article : Google Scholar
|