1. Introduction
The ongoing outbreak of the novel coronavirus
disease 2019 (COVID-19) caused by severe acute respiratory syndrome
coronavirus 2 (SARS-CoV-2) may be a potentially once-in-a-century
pandemic (1). The crude mortality
rate of COVID-19 is estimated at 3%; however, the mortality rate of
critical patients was at one point as high as 61.5% (2). No effective antiviral drugs specific
for treating SARS-CoV-2 infection are currently available. Early
identification of patients with severe COVID-19 and active organ
support remain the most efficient strategies for preventing its
progression and improving clinical outcomes (3). Additionally, strict preventative
measures to lower the risk of further disease transmission,
including social distancing and self-isolation, were adopted
quickly in a number of countries, which had a profoundly negative
impact on the physical and mental health and well-being of
individuals (4,5). Hence, there is strong concern
regarding the pathogenesis of COVID-19 amongst healthcare
professionals due to its high infectivity and lethality.
At present, the pathogenic mechanisms of human
COVID-19 remain to be fully elucidated. Recently, accumulating
evidence from clinical trials and experimental studies in
vitro and in vivo have increased our knowledge of the
potential molecular mechanisms of COVID-19 (6-18).
Additionally, previous work with other highly pathogenic
β-coronaviruses, such as SARS-CoV and Middle East respiratory
syndrome coronavirus (MERS-CoV) may provide insights that could
improve our understanding of the underlying mechanisms of COVID-19.
SARS, MERS and COVID-19 share various clinical, laboratory and
histopathological characteristics (11). Similar to SARS and MERS, there are
no significant distinguishing clinical characteristics of COVID-19
and symptoms overlap largely with other severe acute lower
respiratory infections (19).
SARS-CoV-2 has 75-80% genomic similarity to the SARS-CoV, and 50%
to the MERS-CoV (20,21). Moreover, SARS-CoV and SARS-CoV-2
attach to the same receptor, angiotensin-converting enzyme 2
(ACE2), suggesting a similar tissue tropism and route of entry
(8,22). A recent autopsy study revealed that
pathological changes in patients with COVID-19 are highly similar
to features observed in patients with SARS and MERS (23). Lymphopenia is a common event that
can predict pneumonia development and progression to respiratory
failure in patients with SARS, MERS and COVID-19 (6,24,25).
More importantly, although SARS-CoV-2 infection and host immune
patterns are incompletely characterized, elevated plasma levels of
TNF-α, IL-2, IL-7, IL-10, granulocyte colony stimulating factor
(G-CSF), interferon γ-induced protein 10 (IP10), monocyte
chemoattractant protein-1 (MCP-1), macrophage inflammatory protein
1 α (MIP-1A) and C-reactive protein (CRP) may be markers of severe
status in the early stages of infection (6,24),
suggesting that hypercytokinemia-related immunopathology may serve
a fundamental role in severe COVID-19. Although COVID-19, SARS and
MERS resemble each other clinically, in vitro studies have
highlighted notable differences between these viruses with respect
to their growth characteristics, receptor utilization and host
responses, suggesting that their pathogenesis may also
significantly differ. Additionally, dysregulation of the
cholinergic anti-inflammatory pathway may be involved in severe
COVID-19. Of note, it is speculated that as the SARS-CoV-2 virus
replicates, cell and viral debris or virions may interact with the
nicotinic acetylcholine receptors, thus blocking the action of the
cholinergic anti-inflammatory pathway (26-30).
It is difficult to elaborate the exact pathogenesis
of COVID-19. A growing body of studies have suggested the pivotal
role of a dysregulated or exacerbated immune response against
SARS-CoV-2, leading to an intense inflammatory response (6,18).
This dysregulated inflammatory response is systemic, but primarily
affects the lungs. The present review discusses and summarizes the
possible pathogenesis of SARS-CoV-2-mediated dysregulated immune
responses and the possible pathogenetic mechanisms of
SARS-CoV-2-mediated dysregulated immune inflammatory responses
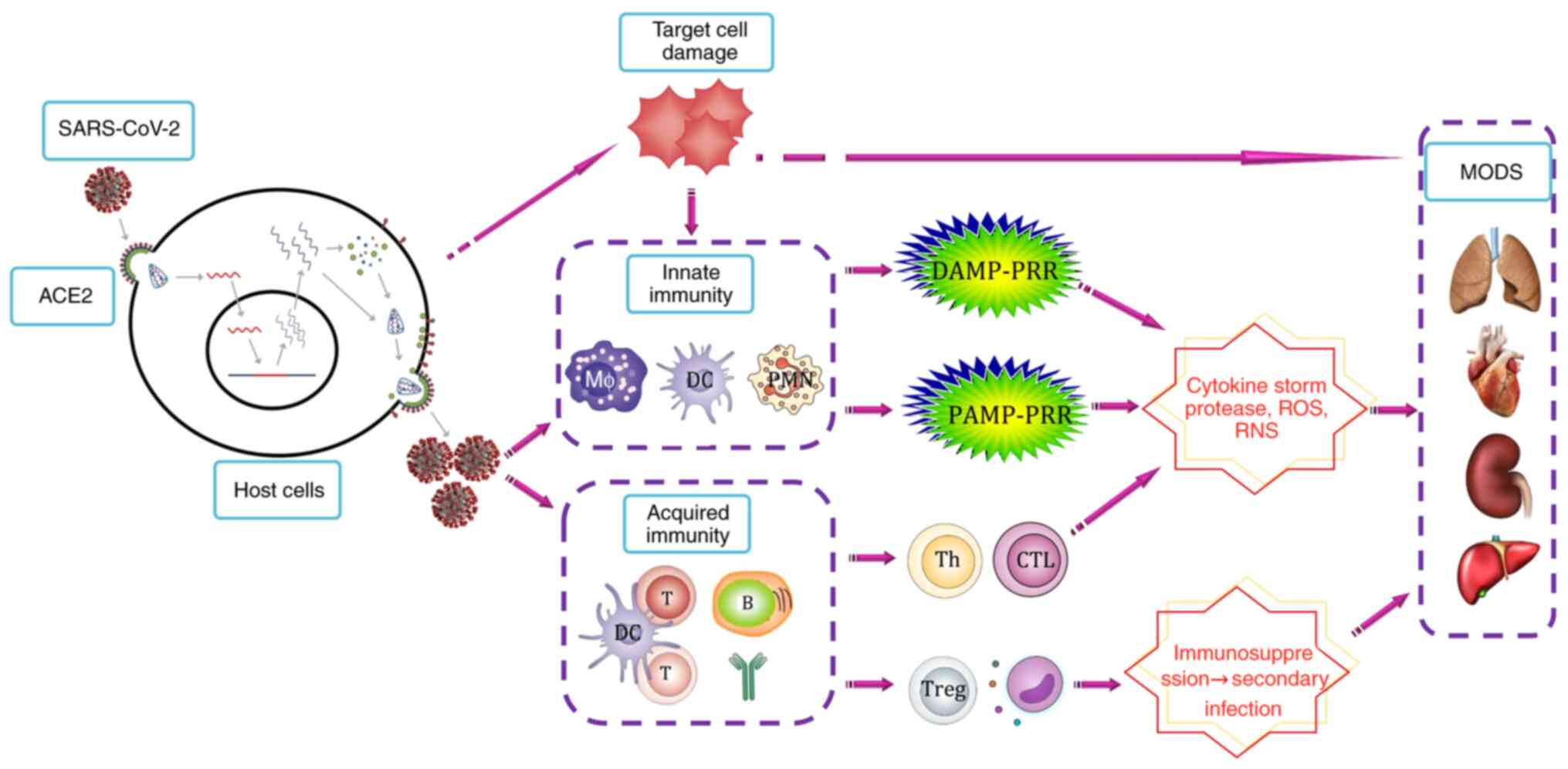
(Fig. 1). Further virus and
immune-related research is urgently required to improve our
understanding of the exact pathogenesis of COVID-19, and ultimately
lead to improvements in precise diagnosis, treatment and effective
vaccine design to manage COVID-19.
2. Overview of SARS-CoV-2
Coronaviruses (CoV) are a large family of enveloped
single positive-strand RNA viruses, which include α, β, γ and δ
genera with varying degrees of pathogenicity and immunogenicity
(31). Most CoVs only cause
self-limiting respiratory tract infections (32). By contrast, SARS-CoV, SARS-CoV-2 and
MERS-CoV, belong to the β-CoV genera, and may cause acute
respiratory distress syndrome (ARDS) and extrapulmonary
manifestations, such as diarrhea, shock, severe renal and liver
dysfunction, and multiple organ dysfunction syndrome (MODS)
(32).
The genomic structure of SARS-CoV-2 provides
important information regarding the pathogenicity and related
virulent factors. The entire genome of SARS-CoV-2 has been
sequenced, and has been demonstrated to contain 29,903 nucleotides
(21). The SARS-CoV-2 is
genetically similar to SARS-CoV and bat SARS-like coronaviruses.
Chan et al (32) found that
the genome of SARS-CoV-2 has 82% nucleotide similarity with that of
human SARS-CoV. Further genetic analysis confirmed that SARS-CoV-2
was ~79% homologous to SARS-CoV and ~50% homologous to MERS-CoV
(20).
Structural proteins, including spike (S), envelope
(E), membrane (M) and nucleocapsid (N) proteins, serve a crucial
role in the pathogenesis of viruses, as well as virion assembly and
structure (33). The S glycoprotein
has a very potent influence on viral tropism and pathogenic
phenotype. It has been confirmed that the S protein is the primary
protein that mediates the binding of SARS-CoV-2 to the receptor
ACE2 of the host cells and causes membrane fusion, which serves a
key role in viral entry into cells (7,8). The S
protein is the primary target of neutralizing antibodies (Abs) and
the focus of treatment and vaccine development. In SARS-CoV, the
nucleocapsid (N) protein binds to viral RNA and participates in
viral replication, M protein serves an important role in
stabilizing the viral structure, envelope formation, as well as
viral budding and release. The E protein has been demonstrated to
be a virulence domain that activates immunopathology in SARS-CoV
infection (34). However, it is
currently unclear whether these structural proteins undergo similar
functions in COVID-19.
It appears that SARS-CoV-2 may be less pathogenic
than MERS-CoV and is closer to that of SARS-CoV. The basic
reproductive number ‘R0’ is defined as the number of additional
individuals one case infects during the course of their illness.
The estimated average R0 for COVID-19 ranges between 2 and 6.47
(35-40).
In comparison, the estimated average R0 for SARS was 2, and 1.3 for
MERS (36). The mean serial
interval, in the epidemiology of infectious diseases, refers to the
duration between symptom onset of a secondary case and that of its
primary case (37). A recent study
reported that the mean serial interval (the duration between
symptom onset of a secondary case and that of its primary case) of
COVID-19 was 3.96 days, considerably shorter than that for SARS
(8.4 days) or MERS (14.6 days), suggesting that SARS-CoV-2 spreads
far more rapidly than SARS-CoV and MERS-CoV. SARS-CoV-2 appears to
have higher transmissibility (a higher R0) and a similar case
fatality rate to that of SARS-CoV (40,41).
There are some differences in the viral load
kinetics between SARS-CoV, MERS-CoV and SARS-CoV-2 infections
(42). For the majority of patients
with COVID-19, the peak viral load of SARS-CoV-2 is very high at
presentation, and declines steadily. By contrast, the viral load of
SARS-CoV peaks at ~10 days, and that of MERS-CoVin the second week
after symptom onset (43). Of note,
the peak viral load of SARS-CoV-2 is positively correlated with age
(43). High viral loads in the
upper respiratory tract samples in patients with COVID-19 are
suggestive of a significant shedding of SARS-CoV-2 and a
potentially high risk of transmissibility during the first few days
of clinical symptoms (44).
3. Pathogenesis of COVID-19
The pathogenic phases of COVID-19 remain
incompletely understood. Previous studies have proposed SARS may
consist of three phases: Viral replication, immune hyperactivity
and pulmonary destruction (45).
The clinical phases of COVID-19 have been recently proposed:
Viremia phase, acute phase and recovery phase (14). It is generally hypothesized that the
course of infection goes through the following stages (33,45-48):
Viral invasion and replication, dysregulated immune response,
multiple organ damage and recovery. Firstly, the virus enters the
host cells, where it replicates, assembles and is released
extracellularly to target cells, and this directly causes the
damage and destruction of parenchymal cells such as alveolar
epithelial cells. At the same time, a large number of pathogen
associated molecular pattern (PAMP) and damage associated molecular
pattern (DAMP) molecules are released to stimulate the innate
immune response, induce inflammatory cell infiltration, release
large quantities of cytokines, chemokines, proteases and free
radicals, causing ARDS, sepsis and MODS. It has been observed that
the pathological findings of COVID-19-induced pneumonia appear to
resemble those seen in SARS-CoV and MERS-CoV infection including
bilateral acute changes with diffuse alveolar damage and vascular
congestion, patchy inflammatory cellular infiltration,
intra-alveolar edema, hemorrhage, proteinaceous exudate, denudation
and reactive hyperplasia of pneumocytes, as well as the presence of
multinucleated giant cells, but hyaline membrane formation was is
not prominent observed (49,50).
After the initial critical stage, the inflammatory response is
gradually resolved, the damaged organ gradually recovers, and some
of the damaged organs enter fibrosis and chronic stage, such as
chronic critical illness, persistent inflammation,
immunosuppression and catabolism syndrome.
It is speculated that the major pathological
alterations that take place in the vital organs during COVID-19 may
be caused directly by the cytopathic effect mediated by SARS-CoV-2,
and indirectly as a result of the harmful immune responses induced
by SARS-CoV-2, but the relative importance of each of these
requires further study. There is some evidence supporting the more
important role of an abnormal immune response (rather than a direct
viral cytopathic effect) in the effects of COVID-19. It has been
observed that patients with COVID-19 had the highest viral load
during the early stage (43). The
timeline of COVID-19 infection showed that the median time from
onset of symptoms to first hospital admission was 7 days, 9 days
till ARDS, and 10.5 days till ICU (24). The association of worsening clinical
progression with declining viral loads (42) and the onset of an immunological
response, plus the presence of significantly elevated cytokines
levels suggested that severe lung damage was largely
immunopathological in nature (6,24,42,44).
SARS-CoV-2 invades host cells
It is widely accepted that human CoV
transmissibility and pathogenesis primarily depends on the
interactions between the virus and specific host cells (46,51).
Receptor recognition and entry is the first step of viral infection
and is the key determinant of tissue tropism. Enhanced binding
affinity between SARS-CoV-2 and ACE2 has been proposed to correlate
with elevated virus transmissibility and disease severity in humans
(7,52). CoV entry into host cells is a
multi-step process involving several distinct domains in the S
protein that mediates viral attachment to the target cell surface,
receptor engagement, protease processing and membrane fusion.
Subsequently, the viral genome is released into the cytoplasm, and
the virus replicates within the host cells (53). Notably, three CoV (human CoV-NL63,
SARS-CoV and SARS-CoV-2) that bind to the same receptor (ACE2)
cause diseases of varying severity, indicating that there may be
other pathogenic factors underlying the differences between these
three coronaviruses (54). It has
been demonstrated that the overall ACE2-binding mode of the
SARS-CoV-2 S receptor-binding domain (RBD) is nearly identical to
that of the SARS-CoV RBD, but SARS-CoV-2 RBD takes a more compact
conformation, which enhances its ACE2-binding affinity (8,9). Walls
et al (7) showed that the
RBD of SARS-CoV-2 S protein and SARS-CoV S protein bind with
similar affinities to human ACE2 to enter cells. However, another
study observed that SARS-CoV-2 and ACE2 have an affinity that is
10-20 times that of SARS-CoV, which may be related to the higher
transmissibility seen in SARS-CoV-2(55).
The characteristic distribution of SARS-CoV-2 and
ACE2 may contribute to revealing the pathogenic mechanisms of
COVID-19. SARS-CoV-2 viral RNA can be detected in respiratory
secretions, peripheral blood, urine and stool specimens of some
patients with COVID-19, which coincides with various transmission
pathways in SARS-CoV-2 infection (56). Virions in the blood that are
released from the primary target (for example the lung) may
circulate and infect host cells in the remote secondary organs and
tissues.
On the other hand, ACE2 is expressed in the lungs,
heart, renal system and gastrointestinal tract, of which it is
abundantly present in the epithelia of the human lungs and small
intestines (57-59).
These observations may indicate that ACE2 serves an important role
in extrapulmonary manifestations of COVID-19, such as
gastrointestinal symptoms (57,60,61).
It is noteworthy that gut-lung crosstalk may be involved in the
pathogenesis of COVID-19; however, the potential efficacy of
probiotics as one of the novel therapeutic approaches of COVID-19
requires further exploration (62).
In addition, ACE2 is widely expressed in the vascular endothelial
cells and smooth muscle cells in all organs, which may cause
extensive vascular endothelial cell injury and this may be the
molecular basis by which multiple organ lesions are formed in
COVID-19-infected patients (59,63).
Cardiac injury has been reported in 7-23% of patients with
COVID-19, which is associated with a higher mortality (64). A more recent study showed that
patients with basic heart failure disease showed increased ACE2
expression, suggesting that cardiac cells with high expression of
ACE2 may act as the target cells of SARS-CoV-2(65).
Direct cytopathic effect of
SARS-CoV-2
After entering the host cells, the virus can
replicate and survive within the target cells. It is speculated
that the life cycle of SARS-CoV-2 may be similar to other single
positive-strand RNA coronaviruses to a certain extent (33,66,67).
After replication is complete, new virus particles are assembled in
the endoplasmic reticulum, after which they are released outside of
the cell. At the same time, target cells lyse or form syncytia and
other lesions occur. SARS-CoV-2 may induce a substantial cytopathic
effect on host cells, thus early effective antiviral treatment may
reduce the risk of progression, and thereby mortality (68). It is unclear whether SARS-CoV-2
interferes with target cells in other ways to cause host cell
damage or apoptosis, including mitochondrial damage, endoplasmic
reticulum stress, intracellular environment alterations (such as pH
changes) or enzyme dysfunction.
In view of the expression of ACE2 in immune cells,
including monocytes/macrophages and lymphocytes (59), it is unclear whether SARS-CoV-2 can
directly infect certain immune cells to cause immune cell damage.
More importantly, immune cells may migrate within the body.
Therefore, the SARS-CoV-2-infected immune cells may allow the virus
to disseminate systemically. Pathological studies using COVID-19
models have shown that the common type of damage caused by
SARS-CoV-2 infection also occurs in the immune system, and spleen
and lymphoid atrophy have been shown to be associated with marked
cytokine activation, suggesting that SARS-CoV-2 might directly
damage immune cells (6,24,25,69).
Initiation of the innate immune
response
The innate immune response, which uses various
pattern recognition receptors (PRRs) to recognize and respond to
viruses, is an important barrier to viral infection (70). The intensity of the host immune and
inflammatory responses are closely related to the type of invading
virus, the viral load, and the age and immune status of the host
(71). In general, host innate
immune cells are stimulated to produce antiviral and
proinflammatory cytokines and chemokines to eliminate the invading
viruses (71,72).
PAMP-PRR pathway
The viral RNA that is present within the infected
cells is detected by various PRRs in the immune cells, which leads
to the secretion of type I interferons (IFNs), proinflammatory
cytokines and chemokines (70,73).
Previous studies have demonstrated that key components of the
innate immune signaling pathways serve important roles as
protective factors against SARS-CoV disease, including STAT1 and
myeloid differentiation primary response protein MyD88(74). Gralinski et al (75) identified an adaptor protein (TIR
domain-containing adapter molecule 2) in the toll-like receptor
signaling pathway that may be involved in the development of SARS.
The IFN response, a key component of antiviral innate immunity, is
initiated by retinoic acid-inducible gene-I-like receptor-mediated
recognition of viral replicative intermediates in the cytosol
(73). However, Channappanavar
et al (76) showed that
robust SARS-CoV replication and delayed IFN-I signaling promotes
severe SARS, as IFN-I could promote the accumulation of pathogenic
macrophages, thus causing lung immunopathology and vascular
leakage. In this regard, the specific pathogenic PAMPs of
SARS-CoV-2 and the corresponding PRRs and signaling pathways remain
to be systemically identified.
Macrophages are crucial components of innate
immunity and potential mediators of immunopathology (77). Moreover, macrophages are the main
target cells for SARS-CoV replication (78). MERS-CoV and SARS-CoV can easily
infect and robustly replicate in human macrophages and dendritic
cells, inducing the aberrant production of proinflammatory
cytokines and chemokines (77,79,80).
In SARS-CoV infection, viroporin 3a has also been shown to induce
the activation of nucleotide oligomerization domain-like receptor
protein 3 inflammasome and the secretion of IL-1β in macrophages,
suggesting that PAMP-PRR signaling in macrophages may result in the
release of proinflammatory cytokines in COVID-19(15).
DAMP-PRR pathway
Following cellular injury and necrosis, endogenous
DAMPs can be released, such as DNA, RNA, ATP, heat shock proteins,
high mobility group protein B1 and the extracellular matrix, which
could be recognized and activated by corresponding PRRs, and
promote the release of cytokines and chemokines, and this may
further aggravate the inflammatory response and tissue damage,
forming a vicious cycle (81). It
is speculated that both DAMPs and PAMPs may also contribute to the
systemic dysregulation of the innate immune response and may be
involved in the development of MODS in COVID-19. After SARS-CoV-2
activates PRRs, it may induce the antiviral innate immune response,
and also lead to cell damage and organ dysfunction.
Adaptive immune response
Antigen-presenting cells present antigen peptides to
T and B cells for recognition, thereby inducing cellular and
humoral immunity. Ni et al (82) characterized SARS-CoV-2-specific
humoral and cellular immunity in recovered patients with Covid-19.
Both T cells and B cells were detected in newly discharged patients
(82). In addition, Spearmen's
correlation showed that the neutralizing antibody titers were
significantly positively correlated with the numbers of NP-specific
T cells (82). These findings
suggested both B and T cells participate in immune-mediated
protection to viral infection.
Cellular immune response
The role of T cells and its subsets in resisting
COVID-19 remains unclear. Previous studies have confirmed that the
S protein of SARS-CoV is the primary antigen protein that induces
the host immune response, and serves an important role in
activating cytotoxic T cell responses and causing humoral immune
responses. Xu et al (23)
found that the proportions of circulating CD4+ and
CD8+ T cells were substantially decreased in patients
infected with COVID-19, but their status was hyperactivated. In
addition, there is an increased percentage of highly proinflammatory
T helper 17 (Th17) cells and high numbers of cytotoxic
CD8+ T cells, indicating that the overactivation of T
cells may partly account for the severe inflammatory response
(23). However, the disease is more
severe when lymphocytopenia is present in COVID-19, suggesting that
the T cell response may be necessary for SARS-CoV-2 clearance. Diao
et al (83) observed that in
addition to a reduction in the number of T cells, surviving T cells
are functionally exhausted in COVID-19. In addition, T cell
subpopulation differentiation and functional imbalance are key
factors in the development of some inflammatory diseases.
Therefore, an imbalance in the ratio of Th1/Th2 and Th17/regulatory
T cells in COVID-19 may be a research topic that requires further
study.
Humoral immune response
The host humoral response against SARS-CoV-2
comprises specific IgA, IgM and IgG responses. Most patients with
COVID-19 have a specific Ab response ≥10 days following the onset
of symptoms (41). In a recent
study of 82 confirmed and 58 probable COVID-19 cases, the specific
IgM and IgA Abs were detected on day 5 (IQR 3-6), while IgG was
detected on day 14 (IQR 10-18) after symptom onset (84). However, the persistence of
neutralizing Abs for SARS-CoV-2 requires further study.
Antiviral neutralizing Abs play a pivotal role in
viral clearance. The S protein RBD is specific for SARS-CoV-2 and
may be the direct target for neutralizing Abs (43). Tian et al (17) assessed the cross-reactivity of
anti-SARS-CoV Abs with SARS-CoV-2 S protein. This previous study
revealed that the epitope of CR3022, a SARS-CoV-specific human
monoclonal Ab, which does not overlap with the ACE2 binding site,
could bind potently with SARS-CoV-2 RBD. Most recently, the
neutralizing Ab from three convalescent SARS patients was reported
to reduce SARS-CoV-2-driven cell entry, although with lower
efficiency compared with SARS-CoV, suggesting that Ab responses
raised against SARS-CoV S protein during infection or vaccination
could at least partially protect against SARS-CoV-2 infection
(22). It has also been suggested
that convalescent plasma in patients with COVID-19 might be useful
as a potential therapy (85). On
the other hand, Ab-dependent cell-mediated cytotoxicity may also be
involved in cellular damage and organ injury (15). The Fc receptor-mediated Ab-dependent
enhancement of SARS-CoV-2 infection may additionally lead to
inflammatory responses (15).
Hypercytokinemia and organ damage
COVID-19 can cause both pulmonary and systemic
inflammation, leading to MODS in high risk patients (86). Organ dysfunction is the key
diagnostic criterion for severe or critical SARS-CoV-2 pneumonia
(87,88). The most frequent organ dysfunction
in patients with severe and critical COVID-19 includes ARDS, shock,
acute myocardial injury, liver injury, kidney injury and MODS
(2,25,86,88-90).
The most frequent type of organ dysfunction in patients with severe
and critical COVID-19 admitted to the ICU includes ARDS (61.1%),
arrhythmia (44.4%), shock (30.6%), myocardial injury (22.2%) and
acute kidney injury (8.3%) (82).
Another clinical trial indicated that the majority of critically
ill patients with COVID-19 had organ function injury, including
ARDS (67%), acute kidney injury (29%), liver dysfunction (29%) and
cardiac injury (23%), and 71% of these patients required mechanical
ventilation (2). It is generally
assumed that the fundamental pathophysiology of critical COVID-19
is severe ARDS (2).
The involvement of multiple organs may be related to
the direct damage of target cells by SARS-CoV-2 and improper host
responses, such as the immune-inflammatory response (Fig. 1). The effects of the host immune
response are a double-edged sword, both protecting the host
(immunity) by clearing the infection, and harming the host by
inducing tissue and cell damage, resulting in immunopathology and
worse clinical outcomes (91). In
other words, cytokines and chemokines released from activated
immune cells not only participate in the antiviral immune response,
but can also cause cell damage and organ dysfunction. The optimal
objective is to achieve a careful balance in the immune response,
which could eliminate the virus, whilst avoiding
inflammatory-mediated organ injury.
Hypercytokinemiais an uncontrolled host inflammatory
state that is characterized by fulminant MOD and elevated
proinflammatory cytokine responses (92). Hypercytokinemia serves a key role in
pathogenic inflammation both in severe SARS and COVID-19 (11,92-96).
The cytokines and chemokines found in MERS-CoV-infected cells share
a similar expression profile to SARS-CoV-infected cells (56). Several studies from humans who
succumbed to highly pathogenic human CoV infections, such as SARS
and MERS, have also suggested that a dysregulated immune response
and immunopathology occurred, resulting in excessive inflammation
and lethal consequences during human CoV infections (92,95).
Macrophages in the lung tissue are proposed to be the primary
inducer of hypercytokinemiaand underlie the pathogenesis of MERS
and SARS (54). In serum from
patients with COVID-19 with a poor outcome, there was a significant
increase in CRP, IL-2, IL-7, IL-10, G-CSF, IP10, MCP-1, MIP-1A and
TNF-α, characterized as hypercytokinemia (24). Chen et al (6) also demonstrated elevated cytokine
levels (IL-6, IL-10 and TNF-α) in severe COVID-19. A recent study
reported that COVID-19 is associated with an elevated cytokine
profile that is similar to that observed in secondary
hemophagocytic lymphohistiocytosis (97). Findings from autopsies and serum of
patients with COVID-19 suggest a crucial immune-inflammatory
implication in the progression to ARDS and MODS (23). ARDS caused by SARS-CoV-2 infection
seems to primarily result from exaggerated and uncontrollable
inflammation initiated by viral replication. High levels of
proinflammatory cytokines may lead to tissue damage in the heart,
liver, kidney and the central nervous system, causing sepsis, shock
or multiple organ failure (92).
The detailed expression profile of the cytokine and chemokine
responses in COVID-19 requires further investigation and comparison
with that in MERS and SARS.
Acquired immune-induced proinflammatory reactions
(including Th17 and cytotoxic T lymphocyte accumulation) may also
serve an important role in tissue damage caused by hypercytokinemia
(23). This exacerbated detrimental
inflammatory response towards invading viruses is termed sepsis
(98). It is suggested that
appropriate immunomodulatory treatments according to the changes of
patients' immune status may be the key breakthrough in treatment.
Most recently, preliminary data have shown that dexamethasone
resulted in lower 28-day mortality amongst patients hospitalized
with COVID-19 who were receiving respiratory support (99). In addition, proteolytic enzymes
(such as elastase, collagenase, cathepsin and matrix
metalloproteinase) released at the site of inflammation may also
mediate tissue and organ damage (100). Oxidative stress (such as increased
reactive oxygen species and reactive nitrogen species) is an
important pathway that contributes to numerous inflammatory
pathological processes, including in patients infected with
COVID-19. The oxidative damage imposed on host tissues via
polymorphonuclear cells and macrophage activation may lead to
tissue damage and organ dysfunction (101-103).
Considering the harmful effects of oxidative stress in COVID-19,
antioxidant therapies using bioactive compounds, as well as
encouraging healthy lifestyles as a potential treatment is an
attractive and practical strategy that warrants further study in
the treatment of COVID-19 (103-106).
Immunosuppression
It has been observed that lymphopenia (defective
acquired immunity) is a common feature in patients with COVID-19,
and it is related to disease severity and mortality (10,87,88,107).
Immunosuppression may lead to difficulty in removing the virus or
secondary infections. Hospital-acquired secondary infection is
frequent in patients with severe COVID-19 (5-15.5%) (2,24,108).
A recent meta-analysis (109),
including 3,448 patients from 28 studies, showed that secondary
bacterial infection was identified in 14.3% of patients with
COVID-19. Moreover, it has been suggested that immunocompromised
patients may have a higher viral load of SARS-CoV-2, prolonged
viral shedding and impaired Ab responses (10,43).
Liang et al (110) found
that patients with cancer may be more susceptible to infection with
SARS-CoV-2 than healthy individuals, and had a worse prognosis, as
their immune systems were suppressed by the effects of the tumors
and anticancer treatment.
The reason for significant lymphopenia in patients
with severe COVID-19 remains unclear. It is speculated that the
underlying mechanisms of lymphopenia may include hemopoietic tissue
depression, as well as direct invasion by viral particles, which
damages the lymphocytes and results in its destruction (2). It has been postulated that SARS-CoV-2
may directly infect T cells and lead to T cell depletion (79). Pathological studies on biopsy
tissues from patients with COVID-19 have revealed that the cell
damage caused by SARS-CoV-2 infection often occurs in the immune
system (50). Furthermore, it is
hypothesized that the underlying mechanism includes increased
apoptosis or necrosis of immune cells (2), and lymphocyte recruitment and
sequestration in the infection sites or lymphoid tissues
(lymphocyte redistribution). However, these speculations require
experimental confirmation. In addition, several other factors may
also contribute to the development of immune suppression, such as a
reduction in the number or function of antigen presenting cells,
increased anti-inflammatory cytokines (such as IL-10 and TGF-β),
neuroendocrine responses (such as glucocorticoids), elevated
regulatory T cells and myeloid-derived suppressor cells (111). Of note, lymphopenia and
hypercytokinemia were observed in patients with critical SARS-CoV
in 2003, Swine flu in 2009, and COVID-19 in 2019, which may
indicate that there is a particular dysregulated immunological
phenotype associated with significantly elevated severity (25).
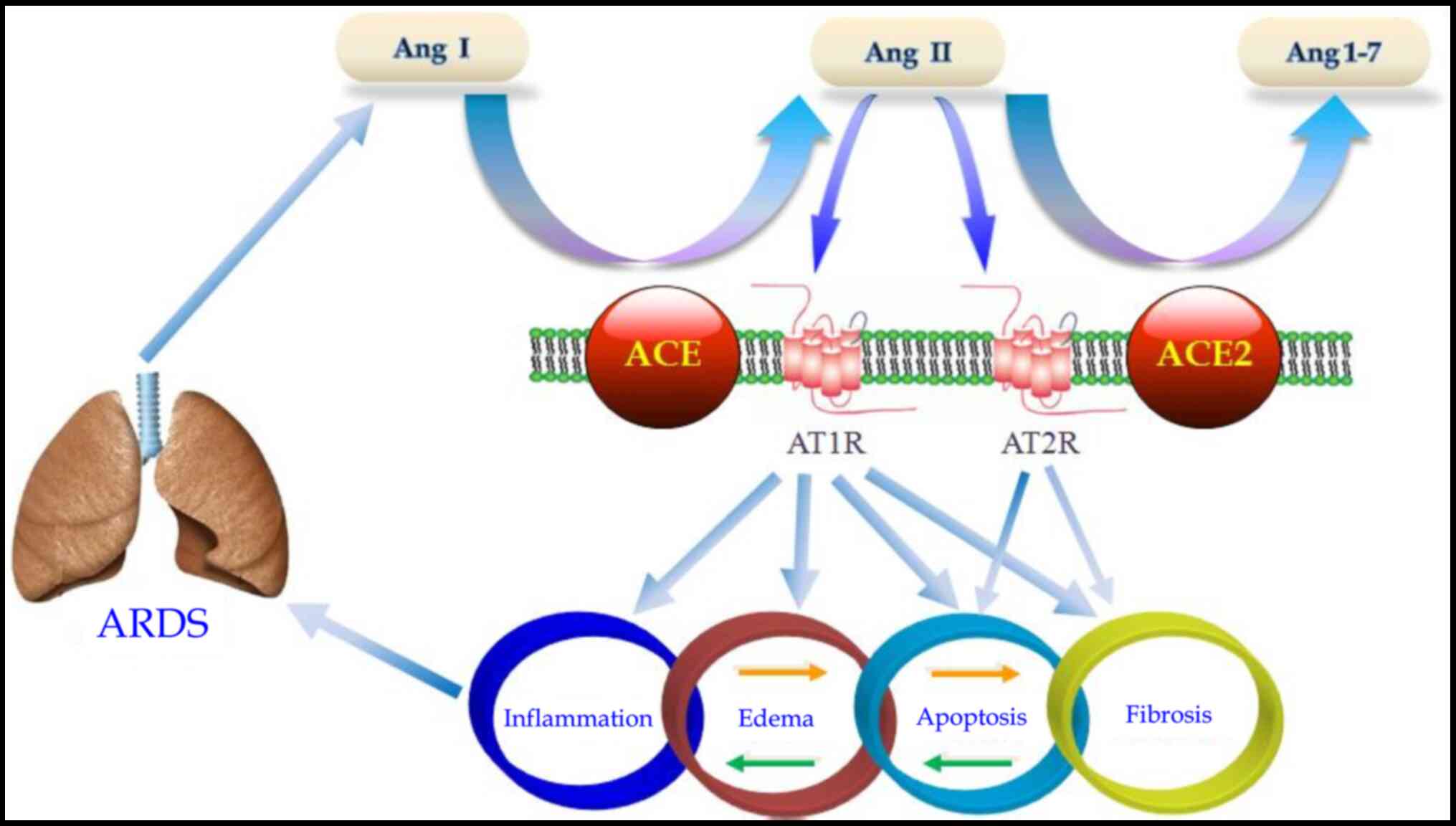
Renin-angiotensin system in
COVID-19
ACE2 is an important component of the
renin-angiotensin-aldosterone system, which converts angiotensin II
into angiotensin 1-7 and angiotensin I into angiotensin
1-9(112). Notably, in addition to
mediating viral entry, the SARS-CoV S protein also has effects on
the downregulated expression of ACE2, leading to aggravated lung
injury (33). These results have
led to the hypothesis that the binding of SARS-CoV-2 S protein is a
virulence factor for COVID-19 outside of its role in viral
attachment and entry.
Our previous data and other studies have
demonstrated that angiotensin II is involved in the
pathophysiological processes of pulmonary inflammation, pulmonary
edema, pulmonary fibrosis and parenchymal cell apoptosis in a
lipopolysaccharide-induced ARDS animal model (Fig. 2) (113-117).
Blocking the angiotensin II receptor may inhibit the function of
mature lung dendritic cells, reducing lipopolysaccharide-induced
ARDS (118), and thus guide the
development of potentially beneficial drugs.
4. Recovery of immune homeostasis and repair
of organ damage
There are distinct long-term outcomes observed in
patients with COVID-19, including recovery, organ fibrosis and
dysfunction, chronic critical illness or persistent inflammation,
immunosuppression and catabolism syndrome, and possibly even death.
A retrospective study of 1,591 consecutive patients with COVID-19
referred to ICU for admission in Italy revealed that 58% of
patients were still in the ICU, 16% patients were discharged, and
26% succumbed to the disease whilst in ICU (119). The long-term prognosis of patients
with COVID-19 depends on a variety of factors, including whether
the virus is cleared in time, and whether the inflammatory response
subsides and inflammatory cells and cytokines are cleared. During
the recovery of COVID-19, the number of CD4+ T cells,
CD8+ T cells, B cells and NK cells, and the markers of
CD8+ T cell exhaustion may gradually normalize.
Additionally, SARS-CoV-2-specific Abs can be identified. Long-term
prognosis also depends on the regeneration and repair of
parenchymal cells in damaged organ tissues. Pulmonary fibrosis
appears frequently in COVID-19, including in patients who survived
the infection (23,120,121). However, at present it is unknown
whether patients with COVID-19 will develop chronic critical
illnesses or persistent inflammation-immunosuppression and
catabolism syndrome. There are a number of problems that require
solving even after the patient clears the acute phase. For example,
how can chronic critical illness, persistent inflammation,
immunosuppression and catabolism syndrome be avoided? What are the
roles and mechanisms of specialized pro-resolving mediators in
COVID-19? These gaps in our knowledge urgently require further
investigation in order to contribute to an improved understanding
of the pathogenesis of COVID-19.
Of note, patients with COVID-19 can relapse or
become reinfected. Relapse in patients with COVID-19 refers to the
reappearance of symptoms in survivors due to the persistence of the
SARS-CoV-2 at immunologically segregated body sites. Reinfection
refers to survivors being susceptible to acquiring new infections
after recovery. Patients reinfected with a strain determined to be
of a different genotype or subtype than the previous strain they
were originally infected with can easily be identified using
genotyping assays. Elsayed et al (122) reported that there were 11 cases of
relapse for COVID-19 at the time of study. The reason for this is
currently unknown, but it may involve factors such as age and
immune status of the host, the presence of underlying lung disease,
and the severity of SARS-CoV2 infection, all of which could affect
the elimination of the virus (122). It is noteworthy to speculate that
an inflammatory rebound triggered by an inappropriate immune
response could constitute a probable explanation of the recurrence
of clinical symptoms (123).
5. Conclusions
In summary, the pathogenic mechanisms of COVID-19 as
a novel severe respiratory infectious disease are not yet fully
determined, which is largely due to the novelty this disease.
Although a number of crucial questions remain unanswered at
present, it is obvious that we are only beginning to understand the
pathogenic mechanisms of COVID-19. The present review discussed the
pathogenesis of COVID-19. It is assumed that SARS-CoV-2
dysregulates the immune inflammatory response in a manner similar
to SARS-CoV and MERS-CoV infections. Severe COVID-19 is
characterized by organ dysfunction, hypercytokinemia and
lymphopenia. Immune dysfunction in patients with COVID-19,
including lymphopenia, decreased numbers of CD4+ T cells
and abnormal cytokine levels, is a common feature and may be a
crucial factor associated with disease severity and worse outcomes
(6,117). The direct damage and lysis of host
target cells by the virus and the inappropriate innate and acquired
immune responses of the host may be the key pathogenic mechanisms
underlying the severity of SARS-CoV-2. The molecular determinants
that may account for the important differences in pathogenesis
between the highly pathogenic human coronaviruses (SARS-CoV,
MERS-CoV and SARS-CoV-2) are currently unknown. Further in-depth
studies on the pathogenesis of COVID-19 will be crucial for
devising novel treatment strategies and designing effective
vaccines for this highly fatal emerging infectious disease. As our
knowledge of the pathogenesis improves, a more reasonable approach
to therapeutic treatments and vaccine development can be designed
in order to combat this novel and fatal illness.
Acknowledgements
Not applicable.
Funding
Funding: The Fifth Program of the ‘333’ Project of Jiangsu
Province (grant no. BRA2016070), Scientific and Technological
Development Project of Suzhou (grant no. SS201874).
Availability of data and materials
Not applicable.
Authors' contributions
JL conceived the subject of the review. CL and QH
drafted the manuscript, and JL and HQ revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gates B: Responding to Covid-19 - A
Once-in-a-century pandemic. N Engl J Med. 382:1677–1679.
2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H,
Wu Y, Zhang L, Yu Z, Fang M, et al: Clinical course and outcomes of
critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China:
A single-centered, retrospective, observational study. Lancet
Respir Med. 8:475–481. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Poston JT, Patel BK and Davis AM:
Management of critically ill adults with COVID-19. JAMA.
323:1839–1841. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Maugeri G, Castrogiovanni P, Battaglia G,
Pippi R, D'Agata V, Palma A, Di Rosa M and Musumeci G: The impact
of physical activity on psychological health during Covid-19
pandemic in Italy. Heliyon. 6(e04315)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ravalli S and Musumeci G: Coronavirus
outbreak in Italy: Physiological benefits of home-based exercise
during pandemic. J Funct Morphol Kinesiol. 5(31)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chen G, Wu D, Guo W, Cao Y, Huang D, Wang
H, Wang T, Zhang X, Chen H, Yu H, et al: Clinical and immunological
features of severe and moderate coronavirus disease 2019. J Clin
Invest. 130:2620–2629. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Walls AC, Park YJ, Tortorici MA, Wall A,
McGuire AT and Veesler D: Structure, function, and antigenicity of
the SARS-CoV-2 spike glycoprotein. Cell. 181:281–292.e6.
2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S,
Zhang Q, Shi X, Wang Q, Zhang L and Wang X: Structure of the
SARS-CoV-2 spike receptor-binding domain bound to the ACE2
receptor. Nature. 581:215–220. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara
H, Geng Q, Auerbach A and Li F: Structural basis of receptor
recognition by SARS-CoV-2. Nature. 581:221–224. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen Y and Li L: SARS-CoV-2: Virus
dynamics and host response. Lancet Infect Dis. 20:515–516.
2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Pedersen SF and Ho YC: SARS-CoV-2: A storm
is raging. J Clin Invest. 130:2202–2205. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yan R, Zhang Y, Li Y, Xia L, Guo Y and
Zhou Q: . Structural basis for the recognition of SARS-CoV-2 by
full-length human ACE2. Science. 367:1444–1448. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vaduganathan M, Vardeny O, Michel T,
McMurray J, Pfeffer MA and Solomon SD:
Renin-angiotensin-aldosterone system inhibitors in patients with
Covid-19. N Engl J Med. 382:1653–1659. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lin L, Lu L, Cao W and Li T: Hypothesis
for potential pathogenesis of SARS-CoV-2 infection-a review of
immune changes in patients with viral pneumonia. Emerg Microbes
Infect. 9:727–732. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Fu Y, Cheng Y and Wu Y: Understanding
SARS-CoV-2-mediated inflammatory responses: From mechanisms to
potential therapeutic tools. Virol Sin. 35:266–271. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zheng M and Song L: Novel antibody
epitopes dominate the antigenicity of spike glycoprotein in
SARS-CoV-2 compared to SARS-CoV. Cell Mol Immunol. 17:536–538.
2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tian X, Li C, Huang A, Xia S, Lu S, Shi Z,
Lu L, Jiang S, Yang Z, Wu Y and Ying T: Potent binding of 2019
novel coronavirus spike protein by a SARS coronavirus-specific
human monoclonal antibody. Emerg Microbes Infect. 9:382–385.
2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cao X: COVID-19: Immunopathology and its
implications for therapy. Nat Rev Immunol. 20:269–270.
2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Arabi YM, Murthy S and Webb S: COVID-19: A
novel coronavirus and a novel challenge for critical care.
Intensive Care Med. 46:833–836. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H,
Wang W, Song H, Huang B, Zhu N, et al: Genomic characterisation and
epidemiology of 2019 novel coronavirus: Implications for virus
origins and receptor binding. Lancet. 395:565–574. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song
ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al: A new coronavirus
associated with human respiratory disease in China. Nature.
579:265–269. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hoffmann M, Kleine-Weber H, Schroeder S,
Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH,
Nitsche A, et al: SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2
and is blocked by a clinically proven protease inhibitor. Cell.
181:271–280.e8. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Xu Z, Shi L, Wang Y, Zhang J, Huang L,
Zhang C, Liu S, Zhao P, Liu H, Zhu L, et al: Pathological findings
of COVID-19 associated with acute respiratory distress syndrome.
Lancet Respir Med. 8:420–422. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu
Y, Zhang L, Fan G, Xu J, Gu X, et al: Clinical features of patients
infected with 2019 novel coronavirus in Wuhan, China. Lancet.
395:497–506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bermejo-Martin JF, Almansa R, Menéndez R,
Mendez R, Kelvin DJ and Torres A: Lymphopenic community acquired
pneumonia as signature of severe COVID-19 infection. J Infect.
80:e23–e24. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tsatsakis A, Petrakis D, Nikolouzakis TK,
Docea AO, Calina D, Vinceti M, Goumenou M, Kostoff RN, Mamoulakis
C, Aschner M and Hernández AF: COVID-19, an opportunity to
reevaluate the correlation between long-term effects of
anthropogenic pollutants on viral epidemic/pandemic events and
prevalence. Food Chem Toxicol. 141(111418)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sarkar C, Mondal M, Torequl Islam M,
Martorell M, Docea AO, Maroyi A, Sharifi-Rad J and Calina D:
Potential therapeutic options for COVID-19: Current status,
challenges, and future perspectives. Front Pharmacol.
11(572870)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kwong KK and Chan ST: The role of carbon
monoxide and heme oxygenase-1 in COVID-19. Toxicol Rep.
7:1170–1171. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Farsalinos K, Niaura R, Le Houezec J,
Barbouni A, Tsatsakis A, Kouretas D, Vantarakis A and Poulas K:
Editorial: Nicotine and SARS-CoV-2: COVID-19 may be a disease of
the nicotinic cholinergic system. Toxicol Rep. 7:658–663.
2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Nasi A, McArdle S, Gaudernack G, Westman
G, Melief C, Rockberg J, Arens R, Kouretas D, Sjölin J and Mangsbo
S: Reactive oxygen species as an initiator of toxic innate immune
responses in retort to SARS-CoV-2 in an ageing population, consider
N-acetylcysteine as early therapeutic intervention. Toxicol Rep.
7:768–771. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Peeri NC, Shrestha N, Rahman MS, Zaki R,
Tan Z, Bibi S, Baghbanzadeh M, Aghamohammadi N, Zhang W and Haque
U: The SARS, MERS and novel coronavirus (COVID-19) epidemics, the
newest and biggest global health threats: What lessons have we
learned. Int J Epidemiol. 49:717–726. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan
S and Yuen KY: Genomic characterization of the 2019 novel
human-pathogenic coronavirus isolated from a patient with atypical
pneumonia after visiting Wuhan. Emerg Microbes Infect. 9:221–236.
2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Weiss SR and Leibowitz JL: Coronavirus
pathogenesis. Adv Virus Res. 81:85–164. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Jimenez-Guardeño JM, Nieto-Torres JL,
DeDiego ML, Regla-Nava JA, Fernandez-Delgado R, Castaño-Rodriguez C
and Enjuanes L: The PDZ-binding motif of severe acute respiratory
syndrome coronavirus envelope protein is a determinant of viral
pathogenesis. PLoS Pathog. 10(e1004320)2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Riou J and Althaus CL: Pattern of early
human-to-human transmission of Wuhan 2019 novel coronavirus
(2019-nCoV), December 2019 to January 2020. Euro Surveill.
25(2000058)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wu JT, Leung K and Leung GM: Nowcasting
and forecasting the potential domestic and international spread of
the 2019-nCoV outbreak originating in Wuhan, China: A modelling
study. Lancet. 395:689–697. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Vink MA, Bootsma MC and Wallinga J: Serial
intervals of respiratory infectious diseases: A systematic review
and analysis. Am J Epidemiol. 180:865–875. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li Q, Guan X, Wu P, Wang X, Zhou L, Tong
Y, Ren R, Leung K, Lau E, Wong JY, et al: Early transmission
Ddynamics in Wuhan, China, of novel coronavirus-infected pneumonia.
N Engl J Med. 382:1199–1207. 2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhao S, Lin Q, Ran J, Musa SS, Yang G,
Wang W, Lou Y, Gao D, Yang L, He D, et al: Preliminary estimation
of the basic reproduction number of novel coronavirus (2019-nCoV)
in China, from 2019 to 2020: A data-driven analysis in the early
phase of the outbreak. Int J Infect Dis. 92:214–217.
2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Munster VJ, Koopmans M, van Doremalen N,
van Riel D and de Wit E: A novel coronavirus emerging in China-key
questions for impact assessment. N Engl J Med. 382:692–694.
2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Swerdlow DL and Finelli L: Preparation for
possible sustained transmission of 2019 novel coronavirus: Lessons
from pevious epidemics. JAMA. 323:1129–1130. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zou L, Ruan F, Huang M, Liang L, Huang H,
Hong Z, Yu J, Kang M, Song Y, Xia J, et al: SARS-CoV-2 viral load
in upper respiratory specimens of infected patients. N Engl J Med.
382:1177–1179. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
To KK, Tsang OT, Leung WS, Tam AR, Wu TC,
Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, et al: Temporal profiles
of viral load in posterior oropharyngeal saliva samples and serum
antibody responses during infection by SARS-CoV-2: An observational
cohort study. Lancet Infect Dis. 20:565–574. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Lescure FX, Bouadma L, Nguyen D, Parisey
M, Wicky PH, Behillil S, Gaymard A, Bouscambert-Duchamp M, Donati
F, Le Hingrat Q, et al: Clinical and virological data of the first
cases of COVID-19 in Europe: A case series. Lancet Infect Dis.
20:697–706. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Navas-Martín SR and Weiss S: Coronavirus
replication and pathogenesis: Implications for the recent outbreak
of severe acute respiratory syndrome (SARS), and the challenge for
vaccine development. J Neurovirol. 10:75–85. 2004.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Skariyachan S, Challapilli SB, Packirisamy
S, Kumargowda ST and Sridhar VS: Recent aspects on the pathogenesis
mechanism, animal models and novel therapeutic interventions for
Middle East Respiratory Syndrome Coronavirus infections. Front
Microbiol. 10(569)2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
van den Brand JM, Smits SL and Haagmans
BL: Pathogenesis of Middle East respiratory syndrome coronavirus. J
Pathol. 235:175–184. 2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Millet JK and Whittaker GR: Host cell
proteases: Critical determinants of coronavirus tropism and
pathogenesis. Virus Res. 202:120–134. 2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
von der Thüsen J and van der Eerden M:
Histopathology and genetic susceptibility in COVID-19 pneumonia.
Eur J Clin Invest. 50(e13259)2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Hanley B, Lucas SB, Youd E, Swift B and
Osborn M: Autopsy in suspected COVID-19 cases. J Clin Pathol.
73:239–242. 2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Weiss SR and Navas-Martin S: Coronavirus
pathogenesis and the emerging pathogen severe acute respiratory
syndrome coronavirus. Microbiol Mol Biol Rev. 69:635–664.
2005.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Wan Y, Shang J, Sun S, Tai W, Chen J, Geng
Q, He L, Chen Y, Wu J, Shi Z, et al: Molecular Mechanism for
antibody-dependent enhancement of coronavirus entry. J Virol.
94:e02015–19. 2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Letko M, Marzi A and Munster V: Functional
assessment of cell entry and receptor usage for SARS-CoV-2 and
other lineage B betacoronaviruses. Nat Microbiol. 5:562–569.
2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Davidson AM, Wysocki J and Batlle D:
Interaction of SARS-CoV-2 and other coronavirus with ACE
(Angiotensin-converting enzyme)-2 as their main receptor:
Therapeutic implications. Hypertension. 76:1339–1349.
2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Wrapp D, Wang N, Corbett KS, Goldsmith JA,
Hsieh CL, Abiona O, Graham BS and McLellan JS: Cryo-EM structure of
the 2019-nCoV spike in the prefusion conformation. Science.
367:1260–1263. 2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Zhou J, Chu H, Li C, Wong BH, Cheng ZS,
Poon VK, Sun T, Lau CC, Wong KK, Chan JY, et al: Active replication
of Middle East respiratory syndrome coronavirus and aberrant
induction of inflammatory cytokines and chemokines in human
macrophages: Implications for pathogenesis. J Infect Dis.
209:1331–1342. 2014.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Hamming I, Timens W, Bulthuis ML, Lely AT,
Navis G and van Goor H: Tissue distribution of ACE2 protein, the
functional receptor for SARS coronavirus. A first step in
understanding SARS pathogenesis. J Pathol. 203:631–637.
2004.PubMed/NCBI View Article : Google Scholar
|
|
58
|
To KF and Lo AW: Exploring the
pathogenesis of severe acute respiratory syndrome (SARS): The
tissue distribution of the coronavirus (SARS-CoV) and its putative
receptor, angiotensin-converting enzyme 2 (ACE2). J Pathol.
203:740–743. 2004.PubMed/NCBI View Article : Google Scholar
|
|
59
|
He L, Ding Y, Zhang Q, Che X, He Y, Shen
H, Wang H, Li Z, Zhao L, Geng J, et al: Expression of elevated
levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+
cells in SARS patients: Relation to the acute lung injury and
pathogenesis of SARS. J Pathol. 210:288–297. 2006.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Gu J, Han B and Wang J: COVID-19:
Gastrointestinal manifestations and potential fecal-oral
transmission. Gastroenterology. 158:1518–1519. 2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Xiao F, Tang M, Zheng X, Liu Y, Li X and
Shan H: Evidence for gastrointestinal infection of SARS-CoV-2.
Gastroenterology. 158:1831–1833.e3. 2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Mak J, Chan F and Ng SC: Probiotics and
COVID-19: One size does not fit all. Lancet Gastroenterol Hepatol.
5:644–645. 2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Liang W, Feng Z, Rao S, Xiao C, Xue X, Lin
Z, Zhang Q and Qi W: Diarrhoea may be underestimated: A missing
link in 2019 novel coronavirus. Gut. 69:1141–1143. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Shi S, Qin M, Shen B, Cai Y, Liu T, Yang
F, Gong W, Liu X, Liang J, Zhao Q, et al: Association of cardiac
injury with mortality in hospitalized patients with COVID-19 in
Wuhan, China. JAMA Cardiol. 5:802–810. 2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Chen L, Li X, Chen M, Feng Y and Xiong C:
The ACE2 expression in human heart indicates new potential
mechanism of heart injury among patients infected with SARS-CoV-2.
Cardiovasc Res. 116:1097–1100. 2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Graham RL, Sparks JS, Eckerle LD, Sims AC
and Denison MR: SARS coronavirus replicase proteins in
pathogenesis. Virus Res. 133:88–100. 2008.PubMed/NCBI View Article : Google Scholar
|
|
67
|
McBride R and Fielding BC: The role of
severe acute respiratory syndrome (SARS)-coronavirus accessory
proteins in virus pathogenesis. Viruses. 4:2902–2923.
2012.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Yu X, Sun S, Shi Y, Wang H, Zhao R and
Sheng J: SARS-CoV-2 viral load in sputum correlates with risk of
COVID-19 progression. Crit Care. 24(170)2020.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Chan JF, Zhang AJ, Yuan S, Poon VK, Chan
CC, Lee AC, Chan WM, Fan Z, Tsoi HW, Wen L, et al: Simulation of
the clinical and pathological manifestations of Coronavirus Disease
2019 (COVID-19) in golden Syrian hamster model: Implications for
disease pathogenesis and transmissibility. Clin Infect Dis.
71:2428–2446. 2020.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Iwasaki A and Pillai PS: Innate immunity
to influenza virus infection. Nat Rev Immunol. 14:315–328.
2014.PubMed/NCBI View Article : Google Scholar
|
|
71
|
van der Poll T and Opal SM: Host-pathogen
interactions in sepsis. Lancet Infect Dis. 8:32–43. 2008.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Asehnoune K, Villadangos J and Hotchkiss
RS: Understanding host-pathogen interaction. Intensive Care Med.
42:2084–2086. 2016.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Streicher F and Jouvenet N: Stimulation of
innate immunity by host and viral RNAs. Trends Immunol.
40:1134–1148. 2019.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Totura AL and Baric RS: SARS coronavirus
pathogenesis: Host innate immune responses and viral antagonism of
interferon. Curr Opin Virol. 2:264–275. 2012.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Gralinski LE, Menachery VD, Morgan AP,
Totura AL, Beall A, Kocher J, Plante J, Harrison-Shostak DC,
Schäfer A, Pardo-Manuel de Villena F, et al: Allelic variation in
the toll-like receptor adaptor protein Ticam2 contributes to
SARS-coronavirus pathogenesis in mice. G3 (Bethesda). 7:1653–1663.
2017.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Channappanavar R, Fehr AR, Vijay R, Mack
M, Zhao J, Meyerholz DK and Perlman S: Dysregulated type I
interferon and inflammatory monocyte-macrophage responses cause
lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe.
19:181–193. 2016.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Page C, Goicochea L, Matthews K, Zhang Y,
Klover P, Holtzman MJ, Hennighausen L and Frieman M: Induction of
alternatively activated macrophages enhances pathogenesis during
severe acute respiratory syndrome coronavirus infection. J Virol.
86:13334–13349. 2012.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Peiris JS and Cheung CY: The macrophage in
the pathogenesis of severe acute respiratory syndrome coronavirus
infection. Hong Kong Med J. 15 (Suppl 6):S21–S25. 2009.PubMed/NCBI
|
|
79
|
Zhou J, Chu H, Chan JF and Yuen KY: Middle
East respiratory syndrome coronavirus infection: Virus-host cell
interactions and implications on pathogenesis. Virol J.
12(218)2015.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Yoshikawa T, Hill T, Li K, Peters CJ and
Tseng CT: Severe acute respiratory syndrome (SARS)
coronavirus-induced lung epithelial cytokines exacerbate SARS
pathogenesis by modulating intrinsic functions of monocyte-derived
macrophages and dendritic cells. J Virol. 83:3039–3048.
2009.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Eppensteiner J, Kwun J, Scheuermann U,
Barbas A, Limkakeng AT, Kuchibhatla M, Elster EA, Kirk AD and Lee
J: Damage- and pathogen-associated molecular patterns play
differential roles in late mortality after critical illness. JCI
Insight. 4(e127925)2019.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Ni L, Ye F, Cheng ML, Feng Y, Deng YQ,
Zhao H, Wei P, Ge J, Gou M, Li X, et al: Detection of
SARS-CoV-2-Specific humoral and cellular immunity in COVID-19
convalescent individuals. Immunity. 52:971–977.e3. 2020.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning
L, Chen L, Li M, Liu Y, Wang G, et al: Reduction and functional
exhaustion of T cells in patients with Coronavirus disease 2019
(COVID-19). Front Immunol. 11(827)2020.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Guo L, Ren L, Yang S, Xiao M, Chang Yang
F, Dela Cruz CS, Wang Y, Wu C, Xiao Y, et al: Profiling early
humoral response to diagnose novel coronavirus disease (COVID-19).
Clin Infect Dis. 71:778–785. 2020.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Chen L, Xiong J, Bao L and Shi Y:
Convalescent plasma as a potential therapy for COVID-19. Lancet
Infect Dis. 20:398–400. 2020.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Chen T, Wu D, Chen H, Yan W, Yang D, Chen
G, Ma K, Xu D, Yu H, Wang H, et al: Clinical characteristics of 113
deceased patients with coronavirus disease 2019: Retrospective
study. BMJ. 368(m1091)2020.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Xie J, Tong Z, Guan X, Du B, Qiu H and
Slutsky AS: Critical care crisis and some recommendations during
the COVID-19 epidemic in China. Intensive Care Med. 46:837–840.
2020.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J,
Wang B, Xiang H, Cheng Z, Xiong Y, et al: Clinical characteristics
of 138 hospitalized patients with 2019 novel coronavirus-infected
pneumonia in Wuhan, China. JAMA. 323:1061–1069. 2020.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Yang F, Shi S, Zhu J, Shi J, Dai K and
Chen X: Analysis of 92 deceased patients with COVID-19. J Med
Virol. 92:2511–2515. 2020.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Giannitsi S, Tsinivizov P, Poulimenos LE,
Kallistratos MS, Varvarousis D, Manolis AJ, Tsamakis K, Rizos E,
Spandidos DA, Tsiptsios D and Triantafyllis AS: [Case Report]
Stress induced (Takotsubo) cardiomyopathy triggered by the COVID-19
pandemic. Exp Ther Med. 20:2812–2814. 2020.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Germain RN: Maintaining system
homeostasis: The third law of Newtonian immunology. Nat Immunol.
13:902–906. 2012.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Channappanavar R and Perlman S: .
Pathogenic human coronavirus infections: Causes and consequences of
cytokine storm and immunopathology. Semin Immunopathol. 39:529–539.
2017.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Nieto-Torres JL, DeDiego ML,
Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA,
Fernandez-Delgado R, Castaño-Rodriguez C, Alcaraz A, Torres J,
Aguilella VM and Enjuanes L: Severe acute respiratory syndrome
coronavirus envelope protein ion channel activity promotes virus
fitness and pathogenesis. PLoS Pathog. 10(e1004077)2014.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Bermejo JF and Muñoz-Fernandez MA: Severe
acute respiratory syndrome, a pathological immune response to the
new coronavirus-implications for understanding of pathogenesis,
therapy, design of vaccines, and epidemiology. Viral Immunol.
17:535–544. 2004.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Chousterman BG, Swirski FK and Weber GF:
Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol.
39:517–528. 2017.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Yuen KS, Ye ZW, Fung SY, Chan CP and Jin
DY: SARS-CoV-2 and COVID-19: The most important research questions.
Cell Biosci. 10(40)2020.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Misra DP, Agarwal V, Gasparyan AY and
Zimba O: Rheumatologists' perspective on coronavirus disease 19
(COVID-19) and potential therapeutic targets. Clin Rheumatol.
39:2055–2062. 2020.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Cecconi M, Evans L, Levy M and Rhodes A:
Sepsis and septic shock. Lancet. 392:75–87. 2018.PubMed/NCBI View Article : Google Scholar
|
|
99
|
RECOVERY Collaborative Group. Horby P, Lim
WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N,
Brightling C, Ustianowski A, et al: Dexamethasone in hospitalized
patients with Covid-19-preliminary report. N Engl J Med, 2020.
|
|
100
|
Gourd NM and Nikitas N: Multiple organ
dysfunction syndrome. J Intensive Care Med. 35:1564–1575.
2019.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Padureanu R, Albu CV, Mititelu RR,
Bacanoiu MV, Docea AO, Calina D, Padureanu V, Olaru G, Sandu RE,
Malin RD and Buga AM: Oxidative stress and inflammation
interdependence in multiple sclerosis. J Clin Med.
8(1815)2019.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Laforge M, Elbim C, Frère C, Hémadi M,
Massaad C, Nuss P, Benoliel JJ and Becker C: Tissue damage from
neutrophil-induced oxidative stress in COVID-19. Nat Rev Immunol.
20:515–516. 2020.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Sharifi-Rad M, Anil Kumar NV, Zucca P,
Varoni EM, Dini L, Panzarini E, Rajkovic J, TsouhFokou PV, Azzini
E, Peluso I, et al: Lifestyle, oxidative stress, and antioxidants:
Back and forth in the pathophysiology of chronic diseases. Front
Physiol. 11(694)2020.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Sharifi-Rad J, Rodrigues CF, Sharopov F,
Docea AO, Can Karaca A, Sharifi-Rad M, KahveciKarıncaoglu D,
Gülseren G, Şenol E, Demircan E, et al: Diet, Lifestyle and
cardiovascular diseases: Linking pathophysiology to
cardioprotective effects of natural bioactive compounds. Int J
Environ Res Public Health. 17(2326)2020.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Salehi B, Rescigno A, Dettori T, Calina D,
Docea AO, Singh L, Cebeci F, Özçelik B, Bhia M, Dowlati Beirami A,
et al: Avocado-soybean unsaponifiables: A panoply of potentialities
to be exploited. Biomolecules. 10(130)2020.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Salehi B, Capanoglu E, Adrar N, Catalkaya
G, Shaheen S, Jaffer M, Giri L, Suyal R, Jugran AK, Calina D, et
al: Cucurbits Plants: A key emphasis to its pharmacological
potential. Molecules. 24(1854)2019.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He
JX, Liu L, Shan H, Lei CL, Hui D, et al: Clinical characteristics
of coronavirus disease 2019 in China. N Engl J Med. 382:1708–1720.
2020.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Chen N, Zhou M, Dong X, Qu J, Gong F, Han
Y, Qiu Y, Wang J, Liu Y, Wei Y, et al: Epidemiological and clinical
characteristics of 99 cases of 2019 novel coronavirus pneumonia in
Wuhan, China: A descriptive study. Lancet. 395:507–513.
2020.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Langford BJ, So M, Raybardhan S, Leung V,
Westwood D, MacFadden DR, Soucy JR and Daneman N: Bacterial
co-infection and secondary infection in patients with COVID-19: A
living rapid review and meta-analysis. Clin Microbiol Infect.
26:1622–1629. 2020.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Liang W, Guan W, Chen R, Wang W, Li J, Xu
K, Li C, Ai Q, Lu W, Liang H, et al: Cancer patients in SARS-CoV-2
infection: A nationwide analysis in China. Lancet Oncol.
21:335–337. 2020.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Fattahi F and Ward PA: Understanding
immunosuppression after sepsis. Immunity. 47:3–5. 2017.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Shenoy V, Ferreira AJ, Qi Y, Fraga-Silva
RA, Díez-Freire C, Dooies A, Jun JY, Sriramula S, Mariappan N,
Pourang D, et al: The angiotensin-converting enzyme
2/angiogenesis-(1-7)/Mas axis confers cardiopulmonary protection
against lung fibrosis and pulmonary hypertension. Am J Respir Crit
Care Med. 182:1065–1072. 2010.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Liu L, Qiu HB, Yang Y, Wang L, Ding HM and
Li HP: Losartan, an antagonist of AT1 receptor for angiotensin II,
attenuates lipopolysaccharide-induced acute lung injury in rat.
Arch Biochem Biophys. 481:131–136. 2009.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Zhu Y, Qiu HB, Yang Y, Liu L, Zhao MM,
Chen QH and Guo T: Angiotensin II type 2 receptor expression and
its modulation in angiotensin II induced acute lung injury in rat.
Zhongguo Wei Zhong Bing Ji Jiu Yi Xue. 20:585–587. 2008.PubMed/NCBI(In Chinese).
|
|
115
|
Imai Y, Kuba K and Penninger JM:
Angiotensin-converting enzyme 2 in acute respiratory distress
syndrome. Cell Mol Life Sci. 64:2006–2012. 2007.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Kuba K, Imai Y and Penninger JM:
Angiotensin-converting enzyme 2 in lung diseases. Curr Opin
Pharmacol. 6:271–276. 2006.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Busse LW, Chow JH, McCurdy MT and Khanna
AK: COVID-19 and the RAAS-a potential role for angiotensin II. Crit
Care. 24(136)2020.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Liu J, Zhang PS, Yu Q, Liu L, Yang Y, Guo
FM and Qiu HB: Losartan inhibits conventional dendritic cell
maturation and Th1 andTh17 polarization responses: Νovel mechanisms
of preventive effects on lipopolysaccharide-inducedacute lung
injury. Int J Mol Med. 29:269–276. 2012.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Grasselli G, Zangrillo A, Zanella A,
Antonelli M, Cabrini L, Castelli A, Cereda D, Coluccello A, Foti G,
Fumagalli R, et al: Baseline characteristics and outcomes of 1591
patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy
Region, Italy. JAMA. 323:1574–1581. 2020.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Tian S, Hu W, Niu L, Liu H, Xu H and Xiao
SY: Pulmonary pathology of early-phase 2019 novel coronavirus
(COVID-19) pneumonia in two patients with lung cancer. J Thorac
Oncol. 15:700–704. 2020.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Vasarmidi E, Tsitoura E, Spandidos DA,
Tzanakis N and Antoniou KM: Pulmonary fibrosis in the aftermath of
the COVID-19 era. Exp Ther Med. 20:2557–2560. 2020.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Elsayed SM, Reddy MK, Murthy PM, Gupta I,
Valiuskyte M, Sánchez DF and Diaz MA: The possibility and cause of
relapse after previously recovering from COVID-19: A systematic
review. Cureus. 12(e10264)2020.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Gousseff M, Penot P, Gallay L, Batisse D,
Benech N, Bouiller K, Collarino R, Conrad A, Slama D, Joseph C, et
al: Clinical recurrences of COVID-19 symptoms after recovery: Viral
relapse, reinfection or inflammatory rebound. J Infect. 81:816–846.
2020.PubMed/NCBI View Article : Google Scholar
|