Introduction
Moyamoya disease (MMD) was first defined by Suzuki
and Takaku in 1999(1) and presents
as the presence of an abnormal vascular network at the base of the
brain. MMD is characterized by progressive stenosis and even
occlusion of the terminal portion of the bilateral internal carotid
and intracerebral arteries (2). The
most common symptoms of MMD include headache, ischemia and
hemorrhage (3). The incidence of
MMD in Japan has increased from 0.35 per 100,000 individuals in
1995 to 0.94 per 100,000 individuals in 2006(4). MMD has mostly been reported to exhibit
a high incidence in various East Asian populations, including
Chinese, Japanese and Korean (5).
Although basic studies, including those using genomic and proteomic
approaches, have been performed, the precise etiology of MMD
remains unclear.
An increasing number of studies have demonstrated
that, although the occurrence and development of MMD are
multifactorial, genetic factors are closely associated with the
pathophysiology of MMD (6,7). More recently, a variety of loci were
found to be involved in MMD, including 17q25, 8q23, 6q25 and
3p24-p26 (8-11).
In particular, genome-wide and locus-specific association studies
have identified ring finger protein 213 (RNF213) in the 17q25-ter
region as a novel susceptibility gene for MMD (12). RNF213 has been shown to serve an
important role in angiogenesis and is closely associated with the
onset of MMD (13-15).
A previous study has revealed that the large trunk arteries have
irregular diameters and abnormal sprouting occurs in zebrafish
following RNF213 knockdown (16).
In addition, MMD is also frequently accompanied with hypertension
(17). A possible reason for this
is that changes in RNF213 expression can result in increased blood
pressure, which aggravates intracranial hemodynamic disorders
further and induce the formation of ‘smoky’ blood vessels (18,19). A
homozygous variant of RNF213 is considered to be the most
pathogenic and was found to be significantly associated with severe
manifestations of MMD (20). The
mutation rate of the rs112735431 locus of RNF213 was high among
Japanese patients with familial and sporadic Moyamoya (21), with similar findings observed in the
Han Chinese population (22,23).
RNF213 has also been recognized as an effective biomarker for
predicting MMD prognosis (24).
Alternatively, a variety of biomarkers and angiogenic factors,
including vascular endothelial growth factors (VEGF), cytokines,
such as matrix metallopeptidase 9, have been previously reportedly
implicated in MMD (25). Based on
this evidence, it was hypothesized that the RNF213 gene silencing
may promote the abnormal expression of MMD-associated factors
through a specific gene pathway, leading to the development of
MMD.
Mesenchymal stem cells (MSCs) possess high
self-renewal abilities and multidirectional differentiation
potentials (26). In certain
conditions, they can differentiate to form neuron-like and
microglia-like cells (27,28) MSCs has been reported not only to
increase endothelial cell growth, but also to promote skin wound
healing through vascular endothelial growth factor C-mediated
angiogenesis (29,30). In addition, MSCs have the advantage
of being easily obtained and reportedly exert neuroprotective
effects against ischemic brain damage (31), making their use increasingly popular
for basic research and clinical studies. Cerebral hemorrhage and
ischemia are common pathophysiological states in neurosurgical
diseases, including MMD and subarachnoid hemorrhage (32). After cerebrovascular events,
neurocognitive function is impaired (33). MSCs has been shown to confer
beneficial effects against cerebrovascular diseases (34). It has also been reported that there
were no significant complications within 5 years of MSC treatment
for ischemic stroke (35). However,
to the best of our knowledge, no studies have focused on the
effects of the susceptible gene RNF213 on MMD by using MSCs. The
rat bone marrow mesenchymal stem cells (rBMSCs) were therefore
selected for the present in vitro study, where the aim was
to determine the effects of RNF213 silencing on the expression of
VEGF and transforming growth factor β1 (TGF-β1) in rBMSCs and to
investigate the association between the RNF213 gene expression and
MMD occurrence.
Materials and methods
Subjects
A total of 40 patients with MMD were enrolled into
the Departments of Neurosurgery and Neurology of the Affiliated
Hospital of Jining Medical University (Jining, China) between
September 2014 and April 2015. The selection criteria were as
follows: i) Digital subtraction angiography (DSA) confirming MMD
and patients meeting the Guidelines for Diagnosis and Treatment of
Moyamoya Disease (Spontaneous Occlusion of the Circle of Willis)
(36); ii) CT or MRI examination
confirming cerebral infarction and cerebral hemorrhage or ischemia;
iii) patients with complete clinicopathological data. The exclusion
criteria were as follows: i) Patients with a history of autoimmune
disease or neuropsychiatric diseases; ii) patients with
intracranial tumors, severe brain injury and cardiovascular
disease; iii) use of any medications that could affect cognitive
function. An additional 40 healthy individuals were recruited as
the control group from the physical examination center of the
Affiliated Hospital of Jining Medical University during the same
period as aforementioned, and the individuals were confirmed to
exhibit no abnormality by laboratory and imaging examinations. The
study protocol and sample collection procedures were approved by
the Ethics Committee of the Affiliated Hospital of Jining Medical
University and written informed consent was obtained from each
participant.
Determination of serum VEGF, TGF-β1
and RNF213 levels
Morning fasting blood samples (5 ml each) were
collected from each subject by elbow venipuncture. Following
collection, the samples were left to stand at room temperature for
1 h and then centrifuged at 3,000 x g at 4˚C for 20 min. The
supernatant was removed and centrifuged for a second time at 3,000
x g at 4˚C for 10 min to obtain serum, which was stored at -80˚C
until analysis. The concentrations of VEGF (cat. no. DVE00; R&D
Systems, Inc.), TGF-β1 (cat. no. DB100B; R&D Systems, Inc.) and
RNF213 (cat. no. JL19472; Shanghai Jianglai Biological Technology
Co., Ltd.; http://www.jonln.com) were determined by
ELISA, according to the manufacturer's protocols.
Animals and rBMSC culture
A total of 25 male Sprague-Dawley rats (weight,
90-100 g; age, 4 weeks) were purchased from Jinan Pengyue
Experimental Animal Breeding, Co., Ltd. The rats were housed under
standard conditions of temperature (22±2˚C), light (12:12 h
light/dark cycle) and humidity (50±5%) with free access to food and
water. The rats were acclimatized for 7 days before the experiment.
All animal procedures were conducted in accordance with Guide for
the Care and Use of Laboratory Animals with the approval of
Affiliated Hospital of Jining Medical University. rBMSCs were
isolated according to the following method. Briefly, the rats were
anesthetized by 80% CO2 (45 sec), sacrificed by cervical
dislocation and then immersed in 75% alcohol for 10 min for
disinfection. Following removal of the double lower limb skin, the
femur and tibia were aseptically isolated and then soaked in PBS
with 1% penicillin-streptomycin for 1 min. The samples were
transferred to 15-ml centrifuge tubes. The ends of the bones were
removed and the bone marrow cavities exposed. The marrow cavity was
washed 5-10 times with complete medium, namely 89% Dulbecco's
modified Eagle's medium/Nutrient Mixture F-12 medium (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
and 1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific,
Inc.) until the medium was clear. All the flushing fluid was then
inoculated into a culture flask and incubated at 37˚C and 5%
CO2. The medium was replaced after 48 h. Based on
assessment of proliferation, the medium was changed every 2 days.
At ~80% confluence, the cells were harvested with 0.25%
trypsin-EDTA and sub-cultured at a ratio of 1:2.
Alizarin red and oil red O
staining
Cells were seeded into 6-well plates at a density of
2x104 cells/well and incubated overnight at 37˚C with 5%
CO2 in StemXVivo® Osteogenic/Adipogenic Base
Media (cat. no. CCM007; R&D Systems, Inc.). After reaching 100%
confluence, the medium was replaced with StemXVivo®
Adipogenic Differentiation Media (cat. no. CCM011; R&D Systems,
Inc.) to induce adipogenesis. After reaching 70% confluence, the
medium was replaced with StemXVivo® Osteogenic
Differentiation Media (cat. no. CCM009; R&D Systems, Inc.) to
induce osteogenesis. The differentiation medium was replaced every
3 days. After 14 days of adipogenic induction or 21 days of
osteogenic induction, MSCs were harvested and fixed with 4%
paraformaldehyde for 30 min at 4˚C. After washed twice in PBS,
cells were stained with Alizarin red (Sigma-Aldrich; Merck KGaA) or
oil red O (Sigma-Aldrich; Merck KGaA) at room temperature for 30
min and cells were observed under a light inverted microscope
(Olympus IX73; Olympus Corporation).
Flow cytometry analysis
Passage 3 rBMSCs were collected after 0.25% trypsin
(Gibco; Thermo Fisher Scientific, Inc.) treatment and washed with
PBS for three times. The cell concentration was adjusted to
1.0x106 cells/ml, and incubated for 25-30 min with
antibodies against the following cell surface antigens: CD29
(dilution, 1:50; cat. no. AF2405; R&D Systems), CD106
(dilution, 1:40; cat. no. ab134047; Abcam), CD34 (dilution, 1:50;
cat. no. ab81289; Abcam) and CD45 (dilution, 1:50; cat. no.
ab10558; Abcam). Cells were washed three times and incubated with
FITC-conjugated anti-goat (dilution, 1:200; cat. no. ab6881; Abcam)
or FITC-conjugated anti-rabbit (dilution, 1:200; cat. no. ab6717;
Abcam) secondary antibodies. Flow cytometry was performed using a
CytoFLEX flow cytometer (Beckman Coulter, Inc.), and the data were
analyzed with FlowJo software (version 10.4.2; FlowJo LLC).
Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 15 min
at room temperature and permeabilized with 0.2% Triton X-100 for 20
min at room temperature. After rinsing with PBS, cells were then
incubated with CD106 (dilution, 1:200; cat. no. ab134047; Abcam)
and CD34 (dilution, 1:200; cat. no. ab81289; Abcam) antibodies for
2 h at room temperature. After three washes with PBS, cells were
incubated with FITC-conjugated anti-rabbit (dilution, 1:200; cat.
no. ab6717; Abcam) secondary antibody. DAPI was used for nuclear
staining for 3 min at room temperature. The fluorescence staining
was captured with an inverted fluorescence microscope (Olympus
IX73; Olympus Corporation).
Lentiviral transfection
pSIH1-H1-copGFP was used to construct the lentiviral
vectors. The lentiviral vectors encoding the RNF213 short hairpin
RNA (RNF213-shRNA) were designed by Hanbio Biotechnology Co., Ltd.
A scramble sequence, named negative-shRNA, was used as the negative
control. The rBMSCs were divided into the following three groups:
i) RNF213-shRNA; ii) negative-shRNA; and iii) control. The control
group was not transfected with plasmids. For lentiviral infection,
rBMSCs in the logarithmic growth phase were seeded into 12-well
plates at a density of 5x104 cells/well. After 24 h,
lentivirus was added at a multiplicity of infection of 100. The
cells were maintained at 37˚C (5% CO2) for an additional
24 h, after which the medium was replaced with 1 ml fresh complete
medium (containing 10% fetal bovine serum and 1%
penicillin-streptomycin). The cells were incubated for another 24 h
and then used for further analysis. After 3 days, green fluorescent
protein (GFP) expression was assessed using an inverted
fluorescence microscope (magnification, x100).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from rBMSCs using an EZ-10
spin column total RNA isolation kit (cat. no. B610583; Sangon
Biotech Co., Ltd.), according to the manufacturer's protocol. cDNA
was generated using the Quant one step qRT-PCR Kit (cat. no. KR118;
Tiangen Biotech Co., Ltd.). The reverse transcription reaction was
carried out at 42˚C for 15 min, followed by 95˚C for 3 min. qPCR
was subsequently performed in a Bio-Rad CFX96 instrument (Bio-Rad
Laboratories, Inc.) using a SYBR SuperReal PreMix Plus (cat. no.
FP205; Tiangen Biotech Co., Ltd.) and gene-specific primers
(Table I). The thermocycling
conditions were as follows: Initial denaturation at 95˚C for 2 min,
followed by 40 cycles of amplification at 95˚C for 10 sec and 60˚C
for 32 sec. β-actin was used as the internal standard and each cDNA
was tested in triplicate. The 2-ΔΔCq method was used for
quantitative analysis (37).
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR analysis. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR analysis.
| Gene | Forward primer
sequence (5'-3') | Reverse primer
sequence (5'-3)' |
|---|
| RNF213 |
CAGCGTGTTAGGCAGATCAA |
TTGTACTGGCCCTGGTTAGC |
| VEGF |
CTGCTGTGGACTTGAGTTGG |
CAAACAGACTTCGGCCTCTC |
| TGF-1β |
CAATTCCTGGCGTTACCTTG |
AGCCCTGTATTCCGTCTCCT |
| β-actin |
TCAGGTCATCACTATCGGCAAT |
AAAGAAAGGGTGTAAAACGCA |
Western blotting (WB)
For WB, total protein was isolated from the rBMSCs
cells with the use of RIPA and PMSF buffer (Beijing Solarbio
Science & Technology Co., Ltd.) according to the manufacturer's
instructions. Protein concentration was determined using the
bicinchoninic acid protein assay kit (Beijing Solarbio Science
& Technology Co., Ltd.). The samples were separated by 12%
SDS-PAGE with ~50 µg protein in each lane and transferred onto PVDF
membranes. Non-specific binding was blocked by 5% skimmed milk in
TBS-0.1% Tween-20 (TBST) buffer and incubated overnight at 4˚C with
the following primary antibodies: TGF-β1 (dilution, 1:1,000; cat.
no. ab215715; Abcam), VEGF (dilution, 1:1,000; cat. no. ab231260;
Abcam) and β-actin (dilution, 1:1,000; cat. no. ab8227; Abcam).
Following washing with 0.1% TBST, the membranes were subsequently
probed with horseradish peroxidase-conjugated secondary antibody
for 1 h at room temperature (dilution, 1:2,000; cat. no. ab205718;
Abcam). Western blot data were detected using enhanced
chemiluminescence solution (Beijing Solarbio Science &
Technology Co., Ltd.) and quantified using ImageJ software (version
1.8.0; National Institutes of Health). β-actin was used as the
loading control.
Statistical analysis
The results are presented as the mean ± SD and were
analyzed using the SPSS statistics software (version 23.0; IBM
Corp.). Sex distribution was compared using the χ2 test.
Other groups were compared using an independent t-test for normally
distributed variables. Differences between groups were assessed by
one-way analysis of variance followed by Duncan's test. Pearson
correlation analysis was used to analyze the correlation between
various factors. P<0.05 was considered to indicate a
statistically significant difference.
Results
Demographic and clinical
characteristics of MMD
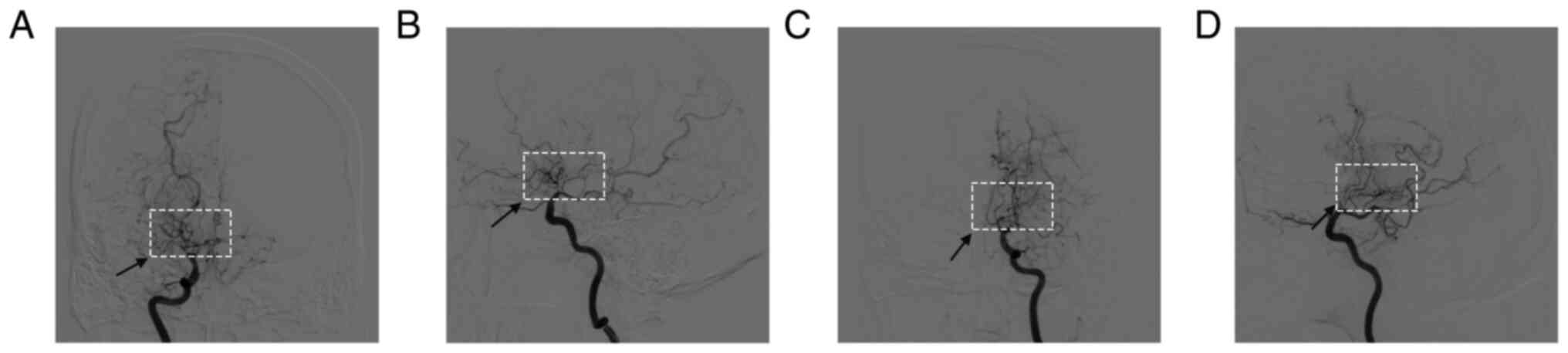
The typical morphology of MMD, as determined by DSA,
is shown in Fig. 1. The demographic
and clinical patient characteristics are presented in Table II. The age of MMD onset was 15-61
years, where the hemorrhage types involved included
intraventricular, cerebral lobe and subarachnoid; and the ischemic
types involved included cerebral infarction and insufficiency of
cerebral blood supply. No significant associations in age, sex and
body mass index were observed between the healthy control (aged
20-64 years) and MMD groups.
 | Table IIDemographic and clinicopathological
data of study subjects. |
Table II
Demographic and clinicopathological
data of study subjects.
| Variable | MMD, n=40 | Controls, n=40 | P-value |
|---|
| Age, years | 46.6±9.92 | 42.6±12.97 | >0.05 |
| Sex, male/female
(n) | 17/23 | 17/23 | >0.05 |
| BMI,
kg/m2 | 23.85±2.158 | 24.24±3.230 | >0.05 |
| Suzuki stage | | | |
|
I | 3 | | |
|
II | 7 | | |
|
III | 13 | | |
|
IV | 11 | | |
|
V | 6 | | |
| Clinical
classification | | | |
|
Ischemia | 29 | | |
|
Hemorrhage | 11 | | |
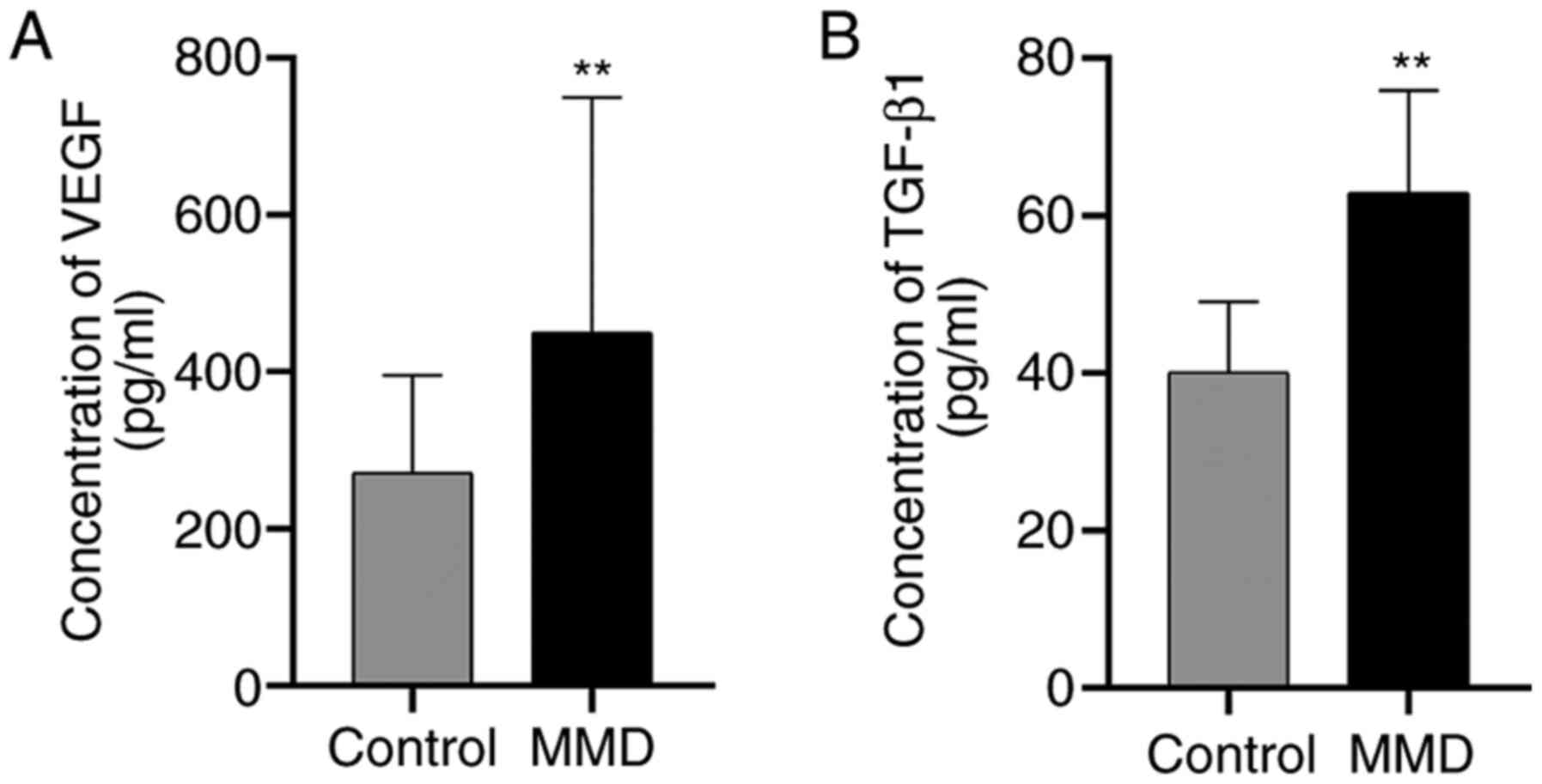
Serum VEGF and TGF-β1 levels
For patients with MMD, the VEGF level was
450.06±299.78 pg/ml, whilst that in the control group was
271.43±124.42 pg/ml. Similarly, increased levels of TGF-β1 were
observed in the MMD group, compared with those in the healthy
controls (Fig. 2A). The TGF-β1
level was 62.93±12.95 pg/ml in patients with MMD and 40.08±8.948.8
in the control group (Fig. 2B).
Accordingly, the levels of VEGF and TGF-β1 were significantly
higher in the patient group, as compared with those in the control
group (P<0.01; Fig. 2). The
expression levels of RNF213 in the MMD and healthy control groups
were also measured. However, there was no difference between the
two groups (Fig. S1A). In
addition, correlation between RNF213 and TGF-β1 in serum was
analyzed, where there was no significant correlation (Fig. S1B).
rBMSC morphological alterations and
identification
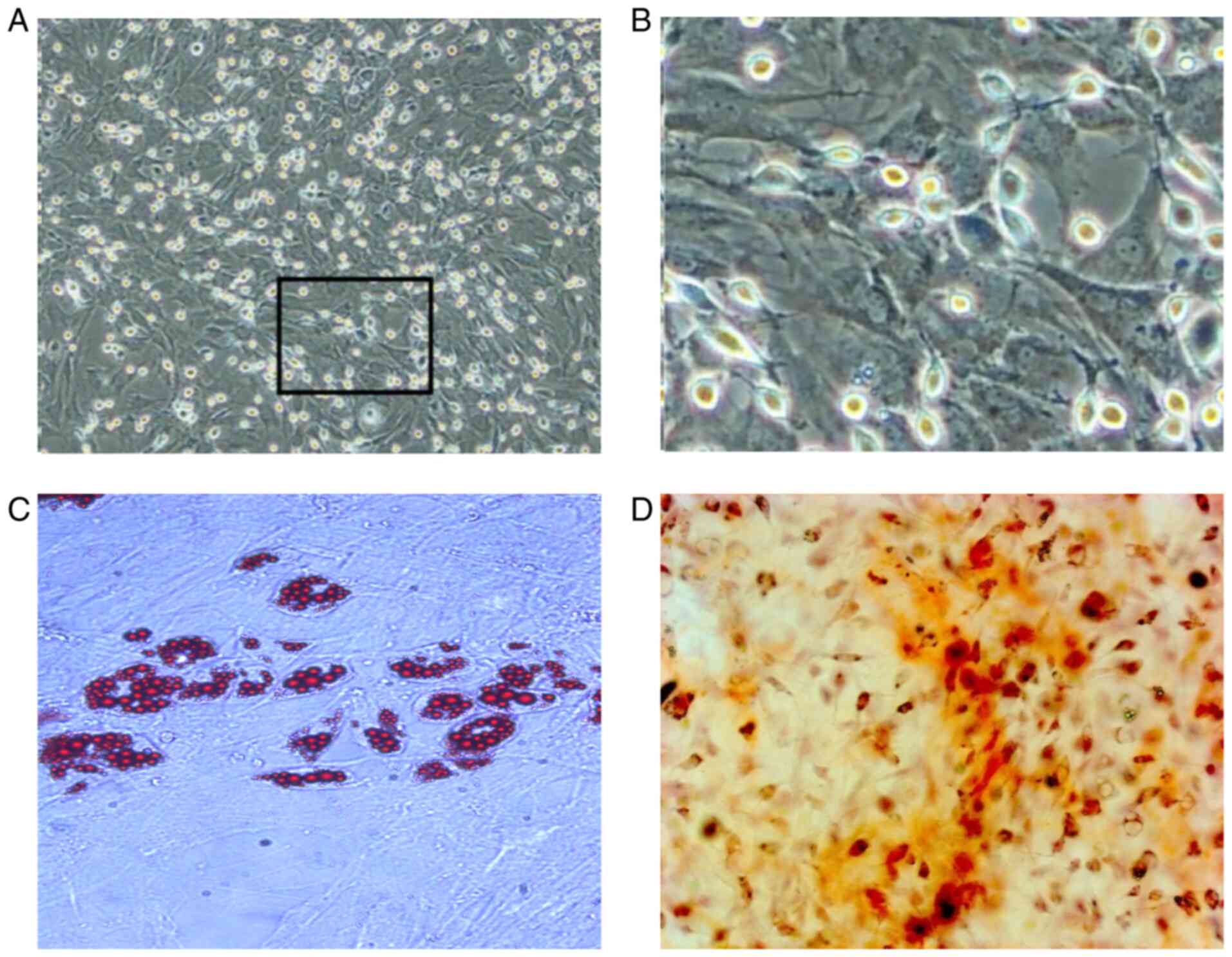
After 12 h of culture, the rBMSC began to adhere to
the culture flask, displayed acceptable proliferation and exhibited
a round or fusiform appearance with a clear outline (Fig. S2A). Following culture for 48 h, the
rBMSCs exhibited a polyhedral shape with evidence of cell
clustering, forming colonies after 3 days (Fig. S2B and C). The number of cell colonies at 4-7
days were gradually increased, where the colony arrangement was
vortex- or chrysanthemum-shaped, with mutual integration with the
neighboring cell colonies observed. At ~8 days of primary culture,
the cell fusion rate had reached 80% (Fig. 3A and B), where sub-cultured cells proliferated
at a higher rate compared with those in the primary culture. After
3-4 days, the cell fusion rate had reached 80-90%. Passage three
rBMSCs underwent adipogenic induction for 14 days, which appeared
as red lipid droplets with oil red O staining (Fig. 3C). Osteogenesis was also induced in
third-passage rBMSCs for 21 days, presenting as red, clear,
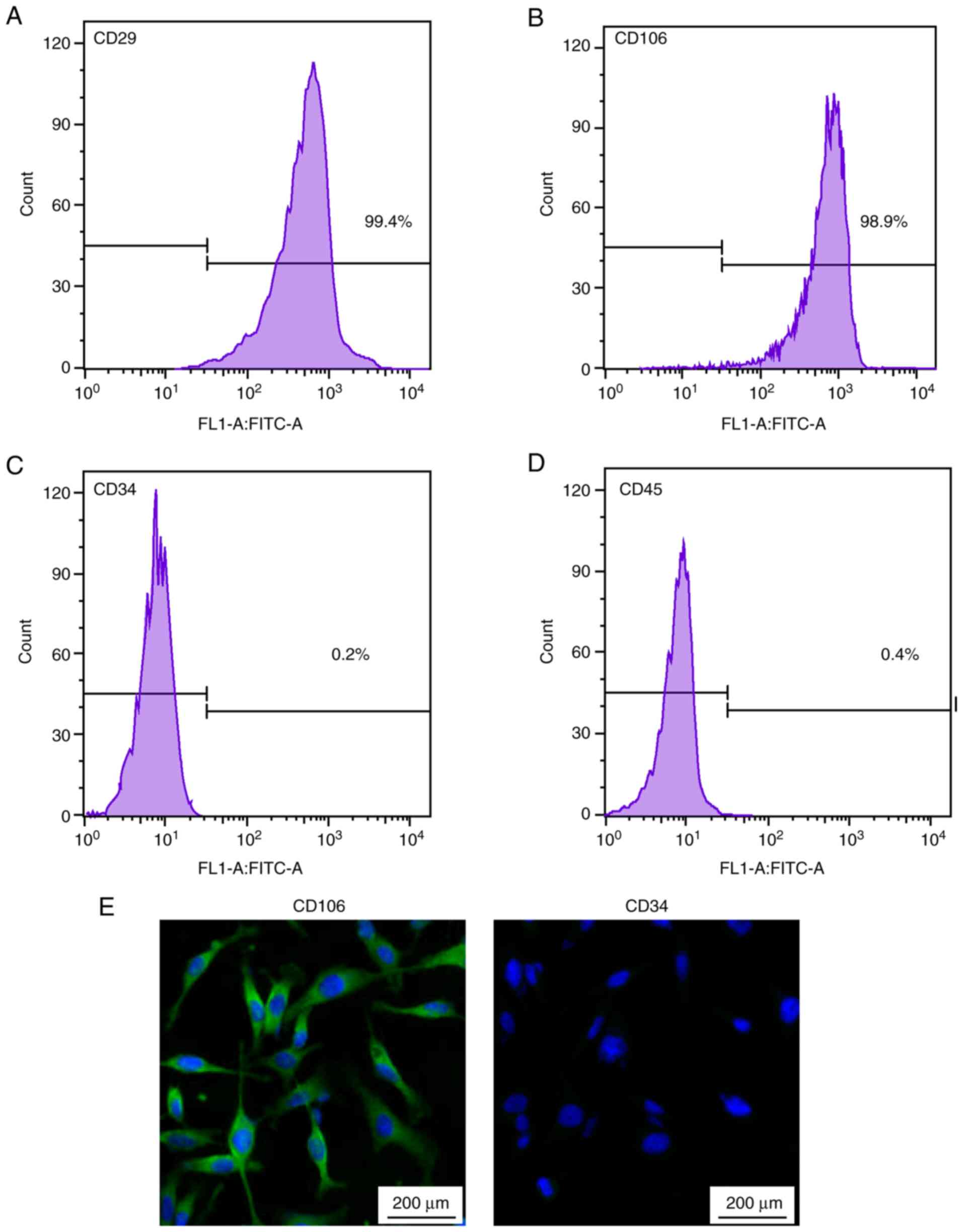
densely-calcified nodules following Alizarin red staining (Fig. 3D). Subsequently, flow cytometry
confirmed that CD29 (Fig. 4A) and
CD106 (Fig. 4B) were positively
expressed, with no expression of CD34 (Fig. 4C) and CD45 (Fig. 4D). The immunofluorescence analysis
indicated that CD106 were highly expressed, but not CD34 (Fig. 4E). All the results indicated that
rBMSCs were successfully isolated in high purity.
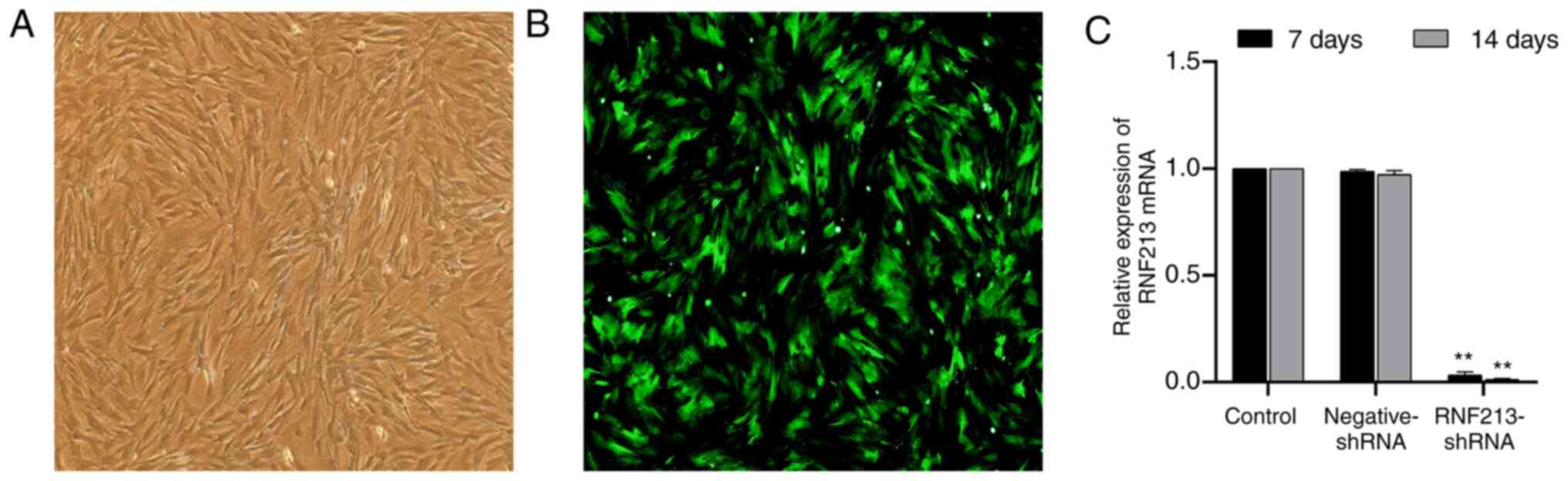
RNF213 silencing in rBMSCs
At 2 days post-transfection, the rBMSCs exhibited
normal morphology under an optical microscope (Fig. 5A), where the expression of GFP was
detected by fluorescence microscopy (Fig. 5B). RNF213 mRNA expression analysis
was subsequently conducted. Compared with control group, rBMSCs
transfected with RNF213-shRNA displayed a significantly lower
RNF213 expression on days 7 and 14, and there was no significant
difference between the negative-shRNA and control groups (Fig. 5C). These data suggested that the
successful silencing of the RNF213 gene was achieved in rBMSCs.
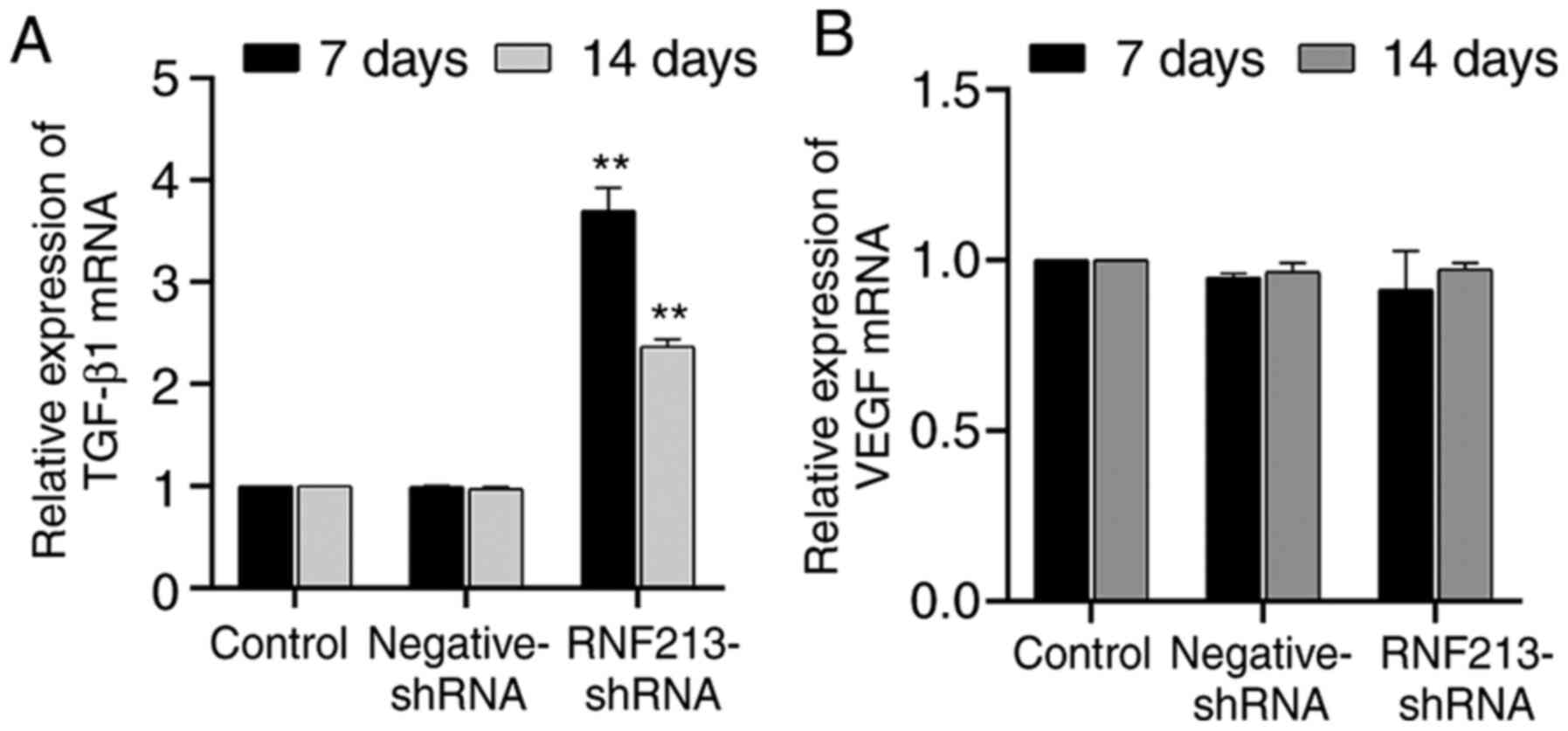
Effects of RNF213 silencing on TGF-β1
and VEGF mRNA expression
As shown in Fig. 6,
the TGF-β1 expression levels in the RNF213-shRNA group were
significantly higher compared with those in the control groups, and
there was no significant difference between the negative-shRNA and
control groups. The expression level of TGF-β1 was > three times
greater compared with that in the control group on day 7, and >
two times greater compared with that in the control group on day 14
(Fig. 6A). However, no significant
differences in the VEGF expression were observed between the
RNF213-shRNA and the control groups on both days (Fig. 6B).
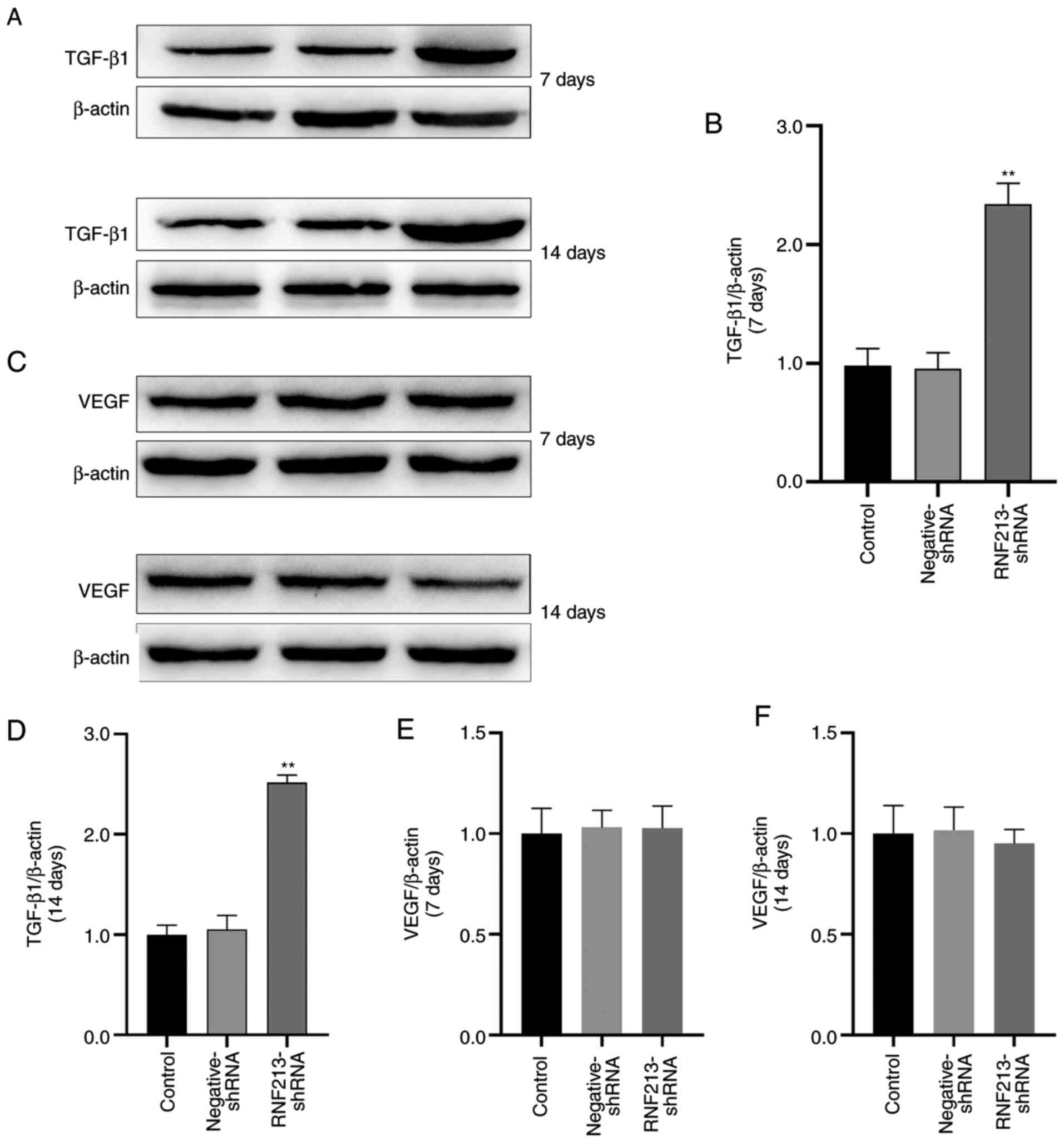
Effects of RNF213 knockdown on TGF-β1
and VEGF protein expression
RNF213 silencing increased TGF-β1 expression, as
compared with that in the control group on days 7 and 14 after
transfection, respectively, and there was no significant difference
between the negative-shRNA and control groups (Fig. 7A, B
and D). The VEGF protein expression
level in the negative-shRNA group and RNF213-shRNA group did not
significantly differ from the control group. (Fig. 7C, E
and F).
Discussion
MMD is a chronic and progressive cerebrovascular
occlusion disorder, where the etiology and pathogenic mechanism of
which remain poorly understood. The basic pathological feature of
MMD is an abnormal vascular network formed at the base of the skull
(38). The age of onset
distribution for MMD has been suggested to have two peaks, one at 5
and another at ~40 years of age (39). Regarding gender, two previous
studies mentioned the female-to-male ratio. One survey conducted in
hospitals throughout Japan reported that the ratio was 1.8(40), whilst another survey conducted in
Hokkaido showed that the ratio was 2.18(41). Epidemiological analyses have
reported distinctive features of familial Moyamoya disease, with
the male to female ratio at 5.0 in familial cases (42). Surgical treatment constitutes the
main available treatment for MMD with no pharmacological options
currently available (43). In a
previous study that included 10 years of follow-ups, antiplatelet
medications were not found to affect the incidence of cerebral
infarction in patients with Moyamoya disease (44). Surgical revascularization is the
most successful therapy used to improve cerebral hemodynamics and
reduce the risk of subsequent strokes (45). Typical revascularization surgeries
for MMD include direct, indirect and combined procedures (46). However, due to the heterogeneity of
MMD, the optimal surgical procedures for both ischemic and
hemorrhagic MMD remain unclear.
Previous studies have successively determined that
VEGF and TGF-β1 are abnormally expressed in the plasma of patients
with MMD (47-49).
To the best of our knowledge, VEGF is the most prominent promoting
factor of angiogenesis, with the ability to induce endothelial cell
proliferation and increase vascular permeability (50). It has also been reported to
stimulate the proliferation of neuronal precursors both in
vitro and in vivo (51).
Vascular endothelial growth factor is a key positive regulator of
both physiological and pathological angiogenesis (52,53).
Therefore, VEGF serves an important role in promoting the formation
of new ‘smoke-like’ blood vessels and the establishment of
collateral circulation in the brain. Perl et al (54) found that the expression of VEGF and
its receptor were significantly increased in the brains of patients
with ischemic cerebrovascular disease following long-term ischemia
and hypoxia stimulation. In the present study, VEGF was found to be
more highly expressed in the serum of patients with MMD compared
with that in the control group. This suggests that serum VEGF
elevation may be involved in the development of abnormal vascular
networks at the base of the brain.
TGF-β1 serves an important role in promoting
angiogenesis and neuroprotection, in addition to promoting
anti-inflammatory and chemotactic processes (55,56). A
previous study reported that the delivery of a TGF-β1 expression
plasmid into the arteries resulted in intimal and medial
hyperplasia (57). This phenomenon
is similar to the pathological changes observed on the arteries of
patients with MMD. The expression of TGF-β1 was found to be
significantly higher in the superficial temporal artery smooth
muscle cells of patients with MMD compared with those in patients
with cerebral arteriosclerosis and healthy controls, but, there was
no difference in TGF-β1 expression in the superficial temporal
artery smooth muscle cells between patients with cerebral
arteriosclerosis and healthy individuals (58). These aforementioned studies
suggested that TGF-β1 promotes angiogenesis and intimal hyperplasia
in MMD. The results of the present study revealed that TGF-β1
expression was significantly higher in patients with MMD compared
with that in healthy individuals, suggesting an influence on the
formation of smoke-like blood vessels. The RNF213 gene is a
susceptibility gene for MMD, with rs112735431 particularly relevant
to the occurrence and development of MMD (59). In addition to rs112735431, numerous
variant sites have been identified, including rs148731719,
rs371441113 and rs138130613 (60,61).
One possible explanation for the lack of a significant correlation
between RNF213 and TGF-β1 is that RNF213 polymorphism in patients
with MMD is a gain-of-function mutation (62).
In scientific research, the use of stably
transfected cell lines remains to be the most reliable routine
method for delivering genes of interest. Although several types of
stem cells, such as embryonic stem cells have also been used, their
use comes with several limitations, including the length of time
required to obtain a sufficient number of stem cells, as well as
legal and ethical restrictions surrounding transplantation and gene
therapy (63). Therefore, stem
cells do not fully meet current experimental requirements. In this
regard, BMSCs were a suitable alternative for the present study,
with advantages including a lower rejection rate, convenience and
no moral or ethical restrictions (64). It has also been reported that BMSCs
can promote central nervous system repair (65). However, there is no standard
protocol for the isolation, culture and identification of BMSCs.
Currently, four primary methods are used to isolate and purify
BMSCs, namely the whole bone marrow adherent method, density
gradient centrifugation, flow cytometric separation and
immunomagnetic beads (66). Due to
the indeterminate phenotype and susceptibly of BMSCs to damage, the
latter of the two methods are rarely used for practical application
(67). Although higher-purity BMSCs
can be obtained by density gradient centrifugation, this method is
complex in practice and delivers the lowest yield of all four
methods (68). Whole bone marrow
adhesion is easy to conduct and most accurately simulates the
natural BMSC environment (69).
Therefore, in the present study, the whole bone marrow adherent
method was used to isolate rBMSCs. Currently, there are three
methods used to identify MSC (70).
In the present study, changes in rBMSC morphology were observed
under a microscope, whilst surface antigen expression (CD29, CD106,
CD34 and CD45) was assessed by flow cytometry. Furthermore,
adipogenic and osteogenic differentiation of rBMSCs was assessed by
oil red O and Alizarin red staining, respectively. The results from
the present study indicated that the whole bone marrow adherent
method was successfully used to isolate and culture BMSCs in
rats.
In the present study, the RNF213 gene was
silenced in rBMSCs using lentiviral vectors, which was confirmed by
RT-qPCR. The RNF213 gene encodes a 5,256-amino acid protein
containing a zinc finger and an ATPase domain, which exhibit E3
ubiquitin ligase and energy-dependent chaperone activities,
respectively (71). Previous
studies have shown that RNF213 is associated with the onset and
severity of MMD and may therefore be a potential marker for the
evaluation of MMD prognosis (72-74).
Animal experiments have highlighted that the intima-media is more
fragile following the ligation of the common carotid arteries in
RNF213-knockdown mice compared with that in normal mice, with
altered hemodynamics (75). The
pathological changes to the lesion vessels in MMD primarily exhibit
an irregular smooth muscle cell shape, vacuoles, in addition to
fragile vessels with a reduced wall thickness (3). Smooth muscle cell proliferation is the
major cause of lesion vessel thickening and medial thinning in
patients with MMD (76). Recent
data also suggested that various proinflammatory molecules (such as
C-reactive protein, interleukin-6) and angiogenic factors are
involved in the pathogenesis of MMD (77,78).
Combined with suggestions from the present study on MMD etiology,
it was hypothesized that the aberrant expression of VEGF and TGF-β1
following genetic and/or environmental alterations promotes
abnormal smooth muscle cell proliferation, resulting in arterial
stenosis and occlusion. This ultimately leads to the clinical
symptoms of hemorrhage or ischemia. To verify this speculation, the
mRNA expression levels of VEGF and TGF-β1 in rBMSCs were determined
following RNF213 silencing. The expression of TGF-β1 was
significantly elevated in RNF213-silenced rBMSCs, supporting the
aforementioned hypothesis. RNF213 regulates TGF-β1 expression
through ≥ one pathways, which are also implicated in the occurrence
and development of MMD. However, the VEGF expression did not
experience significant changes in rBMSCs, rendering association
with RNF213 unlikely. It can therefore be concluded that RNF213 may
not be the only susceptibility gene for MMD and that VEGF may be
involved in the occurrence and development of MMD under the
influence of other such genes. Previous studies have identified
that MMD patients with the CC genotype of VEGF-634 exhibited an
increased collateral vessel formation following surgery (79). An alternative explanation is
therefore that VEGF is not involved in the occurrence of MMD but
participates in neovascularization following disease onset.
In conclusion, the present study provided evidence
that RNF213 silencing influenced TGF-β1 expression. The association
between RNF213 and the mechanisms of MMD development should be
investigated further and applied to MMD screening and gene therapy
in the early stages of this disease.
Supplementary Material
Expression of RNF213 and its
correlation with TGF-β1. (A) Comparison of serum RNF213 levels in
MMD and healthy control using ELISA. Data are expressed as the mean
± SD and were analyzed by one-way ANOVA. (B) Correlation between
serum RNF213 and serum TGF-β1 expression. RNF213, ring finger
protein 213; MMD, Moyamoya disease; TGF-β1, transforming growth
factor β1.
rBMSC morphological alterations after
culture. (A) 12 h, (B) 48 h and (C) 3 days after culture
(magnification, x200).
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by the Key Research and
Development Program of Jining Science and Technology (grant no.
2018SMNS005) and the National Natural Science Foundation of China
(grant no. 81901954).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CW wrote the initial draft of the manuscript. CW,
CS, YZ and ZL collected the samples and performed the experiments.
HS and ZL supervised the methods of all the experiments and
analyzed the data. ZL revised the manuscript. FJ and CC contributed
to the conception and design of the study. FJ and CC confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
All participants provided written informed consent
for the collection and use of their serum samples. All studies
involving animals were performed according to protocols approved by
the Ethics Committee of the Affiliated Hospital of Jining Medical
University (Jining, China). The ethics reference number was
2020C053.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Suzuki J and Takaku A: Cerebrovascular
‘moyamoya’ disease. Disease showing abnormal net-like vessels in
base of brain. Arch Neurol. 20:288–299. 1969.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Han W, Jin F, Zhang H, Yang M, Cui C, Wang
C and Jiang P: Association of Brain-Gut Peptides with Inflammatory
Cytokines in Moyamoya Disease. Mediators Inflamm.
2020(5847478)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Geng C, Cui C, Guo Y, Wang C, Zhang J, Han
W, Jin F, Chen D and Jiang P: Metabolomic Profiling Revealed
Potential Biomarkers in Patients With Moyamoya Disease. Front
Neurosci. 14(308)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Huang S, Guo Z, Shi M, Yang Y and Rao M:
Etiology and pathogenesis of Moyamoya Disease: An update on disease
prevalence. Int J Stroke. 12:246–253. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Fang YC, Wei LF, Hu CJ and Tu YK:
Pathological Circulating Factors in Moyamoya Disease. Int J Mol
Sci. 22(22)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kapoor S: The genetics of moyamoya
disease: Recent insights into the pathogenesis of the disease.
Neurosurgery. 72:E320–E321. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Roder C, Nayak NR, Khan N, Tatagiba M,
Inoue I and Krischek B: Genetics of Moyamoya disease. J Hum Genet.
55:711–716. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yamauchi T, Tada M, Houkin K, Tanaka T,
Nakamura Y, Kuroda S, Abe H, Inoue T, Ikezaki K, Matsushima T, et
al: Linkage of familial moyamoya disease (spontaneous occlusion of
the circle of Willis) to chromosome 17q25. Stroke. 31:930–935.
2000.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sakurai K, Horiuchi Y, Ikeda H, Ikezaki K,
Yoshimoto T, Fukui M and Arinami T: A novel susceptibility locus
for moyamoya disease on chromosome 8q23. J Hum Genet. 49:278–281.
2004.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Inoue TK, Ikezaki K, Sasazuki T,
Matsushima T and Fukui M: Linkage analysis of moyamoya disease on
chromosome 6. J Child Neurol. 15:179–182. 2000.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ikeda H, Sasaki T, Yoshimoto T, Fukui M
and Arinami T: Mapping of a familial moyamoya disease gene to
chromosome 3p24.2-p26. Am J Hum Genet. 64:533–537. 1999.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Kamada F, Aoki Y, Narisawa A, Abe Y,
Komatsuzaki S, Kikuchi A, Kanno J, Niihori T, Ono M, Ishii N, et
al: A genome-wide association study identifies RNF213 as the first
Moyamoya disease gene. J Hum Genet. 56:34–40. 2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang X, Zhang Z, Liu W, Xiong Y, Sun W,
Huang X, Jiang Y, Ni G, Sun W, Zhou L, et al: Impacts and
interactions of PDGFRB, MMP-3, TIMP-2, and RNF213 polymorphisms on
the risk of Moyamoya disease in Han Chinese human subjects. Gene.
526:437–442. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ma J, Liu Y, Ma L, Huang S, Li H and You
C: RNF213 polymorphism and Moyamoya disease: A systematic review
and meta-analysis. Neurol India. 61:35–39. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wu Z, Jiang H, Zhang L, Xu X, Zhang X,
Kang Z, Song D, Zhang J, Guan M and Gu Y: Molecular analysis of
RNF213 gene for moyamoya disease in the Chinese Han population.
PLoS One. 7(e48179)2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu W, Morito D, Takashima S, Mineharu Y,
Kobayashi H, Hitomi T, Hashikata H, Matsuura N, Yamazaki S, Toyoda
A, et al: Identification of RNF213 as a susceptibility gene for
moyamoya disease and its possible role in vascular development.
PLoS One. 6(e22542)2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Togao O, Mihara F, Yoshiura T, Tanaka A,
Kuwabara Y, Morioka T, Matsushima T, Sasaki T and Honda H:
Prevalence of stenoocclusive lesions in the renal and abdominal
arteries in moyamoya disease. AJR Am J Roentgenol. 183:119–122.
2004.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Koizumi A, Kobayashi H, Liu W, Fujii Y,
Senevirathna ST, Nanayakkara S, Okuda H, Hitomi T, Harada KH,
Takenaka K, et al: P.R4810K, a polymorphism of RNF213, the
susceptibility gene for moyamoya disease, is associated with blood
pressure. Environ Health Prev Med. 18:121–129. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu S, Liu M, Li Q, Liu X, Wang Y, Mambiya
M, Zhang K, Yang L, Zhang Q, Shang M, et al: Association of single
nucleotide polymorphisms of MTHFR, TCN2, RNF213 with susceptibility
to hypertension and blood pressure. Biosci Rep.
39(39)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Nomura S, Aihara Y, Akagawa H, Chiba K,
Yamaguchi K, Kawashima A, Okada Y and Kawamata T: Can Moyamoya
Disease Susceptibility Gene Affect Extracranial Systemic Artery
Stenosis? J Stroke Cerebrovasc Dis. 29(104532)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang X, Wang Y, Nie F, Li Q, Zhang K, Liu
M, Yang L, Zhang Q, Liu S, Zeng F, et al: Association of Genetic
Variants With Moyamoya Disease in 13 000 Individuals: A
Meta-Analysis. Stroke. 51:1647–1655. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sun X, Luo M, Li J, Lai R, Lin J, Wang Y,
Xu X, Wu S and Sheng W: Prevalence of RNF213 variants in
symptomatic intracranial arterial stenosis/occlusion in China. Mol
Genet Genomics. 295:635–643. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang Y, Zhang Z, Wei L, Zhang Q, Zou Z,
Yang L, Li D, Shang M, Han C, Mambiya M, et al: Predictive role of
heterozygous p.R4810K of RNF213in the phenotype of Chinese moyamoya
disease. Neurology. 94:e678–e686. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Smith ER: Moyamoya Biomarkers. J Korean
Neurosurg Soc. 57:415–421. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Hu JT, Luo J and Chen QX: The
Susceptibility Pathogenesis of Moyamoya Disease. World Neurosurg.
101:731–741. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang P, Xing C, Rhodes SD, He Y, Deng K,
Li Z, He F, Zhu C, Nguyen L, Zhou Y, et al: Loss of Asxl1 Alters
Self-Renewal and Cell Fate of Bone Marrow Stromal Cell, Leading to
Bohring-Opitz-like Syndrome in Mice. Stem Cell Reports. 6:914–925.
2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hu Y, Li X, Huang G, Wang J and Lu W:
Fasudil may induce the differentiation of bone marrow mesenchymal
stem cells into neuron like cells via the Wnt/β catenin pathway.
Mol Med Rep. 19:3095–3104. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kawanishi S, Takata K, Itezono S, Nagayama
H, Konoya S, Chisaki Y, Toda Y, Nakata S, Yano Y, Kitamura Y, et
al: Bone-Marrow-Derived Microglia-Like Cells Ameliorate Brain
Amyloid Pathology and Cognitive Impairment in a Mouse Model of
Alzheimer's Disease. J Alzheimers Dis. 64:563–585. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kinnaird T, Stabile E, Burnett MS, Lee CW,
Barr S, Fuchs S and Epstein SE: Marrow-derived stromal cells
express genes encoding a broad spectrum of arteriogenic cytokines
and promote in vitro and in vivo arteriogenesis through paracrine
mechanisms. Circ Res. 94:678–685. 2004.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhu M, Chu Y, Shang Q, Zheng Z, Li Y, Cao
L, Chen Y, Cao J, Lee OK, Wang Y, et al: Mesenchymal stromal cells
pretreated with pro-inflammatory cytokines promote skin wound
healing through VEGFC-mediated angiogenesis. Stem Cells Transl Med.
9:1218–1232. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zheng Z, Zhang L, Qu Y, Xiao G, Li S, Bao
S, Lu QR and Mu D: Mesenchymal Stem Cells Protect Against
Hypoxia-Ischemia Brain Damage by Enhancing Autophagy Through Brain
Derived Neurotrophic Factor/Mammalin Target of Rapamycin Signaling
Pathway. Stem Cells. 36:1109–1121. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Griessenauer CJ, Farrell S, Sarkar A, Zand
R, Abedi V, Holland N, Michael A, Cummings CL, Metpally R, Carey
DJ, et al: Genetic susceptibility to cerebrovascular disease: A
systematic review. J Cereb Blood Flow Metab. 38:1853–1871.
2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Stienen MN, Smoll NR, Weisshaupt R,
Fandino J, Hildebrandt G, Studerus-Germann A and Schatlo B: Delayed
cerebral ischemia predicts neurocognitive impairment following
aneurysmal subarachnoid hemorrhage. World Neurosurg. 82:e599–e605.
2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Baek HJ, Chung SY, Park MS, Kim SM, Park
KS and Son HU: Preliminary study of neurocognitive dysfunction in
adult moyamoya disease and improvement after superficial temporal
artery-middle cerebral artery bypass. J Korean Neurosurg Soc.
56:188–193. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li F, Zhang J, Liao R, Duan Y, Tao L, Xu Y
and Chen A: Mesenchymal stem cell derived extracellular vesicles
prevent neural stem cell hypoxia injury via promoting miR-210-3p
expression. Mol Med Rep. 22:3813–3821. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Research Committee on the Pathology and
Treatment of Spontaneous Occlusion of the Circle of Willis; Health
Labour Sciences Research Grant for Research on Measures for
Infractable Diseases. Guidelines for diagnosis and treatment of
moyamoya disease (spontaneous occlusion of the circle of Willis).
Neurol Med Chir (Tokyo). 52:245–266. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Fujimura M and Tominaga T: Significance of
Cerebral Blood Flow Analysis in the Acute Stage after
Revascularization Surgery for Moyamoya Disease. Neurol Med Chir
(Tokyo). 55:775–781. 2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Lee JS, Hong JM, Moon GJ, Lee PH, Ahn YH
and Bang OY: STARTING collaborators. A long-term follow-up study of
intravenous autologous mesenchymal stem cell transplantation in
patients with ischemic stroke. Stem Cells. 28:1099–1106.
2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wakai K, Tamakoshi A, Ikezaki K, Fukui M,
Kawamura T, Aoki R, Kojima M, Lin Y and Ohno Y: Epidemiological
features of moyamoya disease in Japan: Findings from a nationwide
survey. Clin Neurol Neurosurg. 99 (Suppl 2):S1–S5. 1997.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Baba T, Houkin K and Kuroda S: Novel
epidemiological features of moyamoya disease. J Neurol Neurosurg
Psychiatry. 79:900–904. 2008.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kuroda S and Houkin K: Moyamoya disease:
Current concepts and future perspectives. Lancet Neurol.
7:1056–1066. 2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Mayeku J and Lopez-Gonzalez MA: Current
Surgical Options for Moyamoya Disease. Cureus.
12(e11332)2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Yamada S, Oki K, Itoh Y, Kuroda S and
Suzuki N: Effects of Surgery and Antiplatelet Therapy in Ten-Year
Follow-Up from the Registry Study of Research Committee on Moyamoya
Disease in Japan. J Stroke Cerebrovasc Dis. 25:340–349.
2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Arias EJ, Derdeyn CP, Dacey RG and Zipfel
GJ: Advances and surgical considerations in the treatment of
moyamoya disease. Neurosurgery. 74 (Suppl 1):S116–125.
2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Tokairin K, Kazumata K, Gotoh S, Sugiyama
T and Kobayashi H: Neuroendoscope-Assisted Aneurysm Trapping for
Ruptured Intraventricular Aneurysms in Moyamoya Disease Patients.
World Neurosurg. 141:278–283. 2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Kang HS, Kim JH, Phi JH, Kim YY, Kim JE,
Wang KC, Cho BK and Kim SK: Plasma matrix metalloproteinases,
cytokines and angiogenic factors in moyamoya disease. J Neurol
Neurosurg Psychiatry. 81:673–678. 2010.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Sakamoto S, Kiura Y, Yamasaki F, Shibukawa
M, Ohba S, Shrestha P, Sugiyama K and Kurisu K: Expression of
vascular endothelial growth factor in dura mater of patients with
moyamoya disease. Neurosurg Rev. 31:77–81; discussion 81.
2008.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Yamamoto M, Aoyagi M, Tajima S, Wachi H,
Fukai N, Matsushima Y and Yamamoto K: Increase in elastin gene
expression and protein synthesis in arterial smooth muscle cells
derived from patients with Moyamoya disease. Stroke. 28:1733–1738.
1997.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Ng YS, Krilleke D and Shima DT: VEGF
function in vascular pathogenesis. Exp Cell Res. 312:527–537.
2006.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Jin K, Zhu Y, Sun Y, Mao XO, Xie L and
Greenberg DA: Vascular endothelial growth factor (VEGF) stimulates
neurogenesis in vitro and in vivo. Proc Natl Acad Sci USA.
99:11946–11950. 2002.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Fujii T, Yonemitsu Y, Onimaru M, Inoue M,
Hasegawa M, Kuwano H and Sueishi K: VEGF function for upregulation
of endogenous PlGF expression during FGF-2-mediated therapeutic
angiogenesis. Atherosclerosis. 200:51–57. 2008.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Chen CY, Rao SS, Ren L, Hu XK, Tan YJ, Hu
Y, Luo J, Liu YW, Yin H, Huang J, et al: Exosomal DMBT1 from human
urine-derived stem cells facilitates diabetic wound repair by
promoting angiogenesis. Theranostics. 8:1607–1623. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Perl M, Chung CS and Ayala A: Apoptosis.
Crit Care Med. 33 (Suppl):S526–S529. 2005.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Kim DW, Jo YY, Garagiola U, Choi JY, Kang
YJ, Oh JH and Kim SG: Increased Level of Vascular Endothelial
Growth Factors by 4-hexylresorcinol is Mediated by Transforming
Growth Factor-β1 and Accelerates Capillary Regeneration in the
Burns in Diabetic Animals. Int J Mol Sci. 21(21)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
König HG, Kögel D, Rami A and Prehn JH:
TGF-{beta}1 activates two distinct type I receptors in neurons:
Implications for neuronal NF-{kappa}B signaling. J Cell Biol.
168:1077–1086. 2005.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Nabel EG, Shum L, Pompili VJ, Yang ZY, San
H, Shu HB, Liptay S, Gold L, Gordon D and Derynck R: Direct
transfer of transforming growth factor beta 1 gene into arteries
stimulates fibrocellular hyperplasia. Proc Natl Acad Sci USA.
90:10759–10763. 1993.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Hojo M, Hoshimaru M, Miyamoto S, Taki W,
Nagata I, Asahi M, Matsuura N, Ishizaki R, Kikuchi H and Hashimoto
N: Role of transforming growth factor-β1 in the pathogenesis of
moyamoya disease. J Neurosurg. 89:623–629. 1998.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Huang Y, Cheng D, Zhang J and Zhao W:
Association between the rs112735431 polymorphism of the RNF213 gene
and moyamoya disease: A case-control study and meta-analysis. J
Clin Neurosci. 32:14–18. 2016.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Park MG, Shin JH, Lee SW, Park HR and Park
KP: RNF213 rs112735431 polymorphism in intracranial artery
steno-occlusive disease and moyamoya disease in Koreans. J Neurol
Sci. 375:331–334. 2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Zhang Q, Liu Y, Zhang D, Wang R, Zhang Y,
Wang S, Yu L, Lu C, Liu F, Zhou J, et al: RNF213 as the major
susceptibility gene for Chinese patients with moyamoya disease and
its clinical relevance. J Neurosurg. 126:1106–1113. 2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Hitomi T, Habu T, Kobayashi H, Okuda H,
Harada KH, Osafune K, Taura D, Sone M, Asaka I, Ameku T, et al: The
moyamoya disease susceptibility variant RNF213 R4810K (rs112735431)
induces genomic instability by mitotic abnormality. Biochem Biophys
Res Commun. 439:419–426. 2013.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Liu G, Beggs H, Jürgensen C, Park HT, Tang
H, Gorski J, Jones KR, Reichardt LF, Wu J and Rao Y: Netrin
requires focal adhesion kinase and Src family kinases for axon
outgrowth and attraction. Nat Neurosci. 7:1222–1232.
2004.PubMed/NCBI View
Article : Google Scholar
|
|
64
|
Abe K, Yamashita T, Takizawa S, Kuroda S,
Kinouchi H and Kawahara N: Stem cell therapy for cerebral ischemia:
From basic science to clinical applications. J Cereb Blood Flow
Metab. 32:1317–1331. 2012.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Harris VK, Yan QJ, Vyshkina T, Sahabi S,
Liu X and Sadiq SA: Clinical and pathological effects of
intrathecal injection of mesenchymal stem cell-derived neural
progenitors in an experimental model of multiple sclerosis. J
Neurol Sci. 313:167–177. 2012.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Gamie Z, Tran GT, Vyzas G, Korres N,
Heliotis M, Mantalaris A and Tsiridis E: Stem cells combined with
bone graft substitutes in skeletal tissue engineering. Expert Opin
Biol Ther. 12:713–729. 2012.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Deryugina EI and Müller-Sieburg CE:
Stromal cells in long-term cultures: Keys to the elucidation of
hematopoietic development? Crit Rev Immunol. 13:115–150.
1993.PubMed/NCBI
|
|
68
|
Lisignoli G, Remiddi G, Cattini L,
Cocchini B, Zini N, Fini M, Grassi F, Piacentini A and Facchini A:
An elevated number of differentiated osteoblast colonies can be
obtained from rat bone marrow stromal cells using a gradient
isolation procedure. Connect Tissue Res. 42:49–58. 2001.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Li YH, Wang ZD, Wang W, Ding CW, Zhang HX
and Li JM: The biocompatibility of calcium phosphate cements
containing alendronate-loaded PLGA microparticles in vitro. Exp
Biol Med (Maywood). 240:1465–1471. 2015.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Dominici M, Le Blanc K, Mueller I,
Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A,
Prockop Dj and Horwitz E: Minimal criteria for defining multipotent
mesenchymal stromal cells. The International Society for Cellular
Therapy position statement. Cytotherapy. 8:315–317. 2006.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Moteki Y, Onda H, Kasuya H, Yoneyama T,
Okada Y, Hirota K, Mukawa M, Nariai T, Mitani S and Akagawa H:
Systematic Validation of RNF213 Coding Variants in Japanese
Patients With Moyamoya Disease. J Am Heart Assoc.
4(e001862)2015.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Bang OY, Ryoo S, Kim SJ, Yoon CH, Cha J,
Yeon JY, Kim KH, Kim GM, Chung CS, Lee KH, et al: Adult Moyamoya
Disease: A Burden of Intracranial Stenosis in East Asians? PLoS
One. 10(e0130663)2015.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Nomura S, Yamaguchi K, Akagawa H,
Kawashima A, Moteki Y, Ishikawa T, Aihara Y, Saito T, Okada Y and
Kawamata T: Genotype-Phenotype Correlation in Long-Term Cohort of
Japanese Patients with Moyamoya Disease. Cerebrovasc Dis.
47:105–111. 2019.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Kim EH, Yum MS, Ra YS, Park JB, Ahn JS,
Kim GH, Goo HW, Ko TS and Yoo HW: Importance of RNF213 polymorphism
on clinical features and long-term outcome in moyamoya disease. J
Neurosurg. 124:1221–1227. 2016.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Fujimura M, Sonobe S, Nishijima Y, Niizuma
K, Sakata H, Kure S and Tominaga T: Genetics and Biomarkers of
Moyamoya Disease: Significance of RNF213 as a Susceptibility Gene.
J Stroke. 16:65–72. 2014.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Reid AJ, Bhattacharjee MB, Regalado ES,
Milewicz AL, El-Hakam LM, Dauser RC and Milewicz DM: Diffuse and
uncontrolled vascular smooth muscle cell proliferation in rapidly
progressing pediatric moyamoya disease. J Neurosurg Pediatr.
6:244–249. 2010.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Han W, Qiao Y, Zhang H, Geng C, Zhu X,
Liao D, Guo Y, Yang M, Chen D and Jiang P: Circulating sortilin
levels are associated with inflammation in patients with moyamoya
disease. Metab Brain Dis. 36:103–109. 2021.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Corey S and Luo Y: Circular RNAs and
neutrophils: Key factors in tackling asymptomatic moyamoya disease.
Brain Circ. 5:150–155. 2019.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Park YS, Jeon YJ, Kim HS, Chae KY, Oh SH,
Han IB, Kim HS, Kim WC, Kim OJ, Kim TG, et al: The role of VEGF and
KDR polymorphisms in moyamoya disease and collateral
revascularization. PLoS One. 7(e47158)2012.PubMed/NCBI View Article : Google Scholar
|