Introduction
Osteoporosis is a common systemic bone disease
particularly in the older population that is characterized by
diminished bone density and mass, coupled with destruction of the
bone microstructure (1,2). Human bone mesenchymal stem cells
(hBMSCs) are multipotent progenitor cells that have the potential
to differentiate into osteoblasts, which serve critical roles in
bone formation (3). It has been
previously reported that recovery of hBMSC osteogenic
differentiation ability may attenuate bone loss in osteoporosis
(4-6).
Therefore, directional differentiation of hBMSCs may be considered
to be a potential therapeutic strategy for osteoporosis.
Long non-coding RNAs (lncRNAs) are a class of
non-protein coding RNAs that are >200 nucleotides (nt) in length
(7,8). Increasing evidence has indicated that
lncRNAs are involved in the occurrence and development of a number
of diseases, including osteoporosis (9). Jiang et al (10) reported that the lncRNA small
nucleolar RNA host gene could inhibit the osteogenic
differentiation of BMSCs by negatively regulating the p38 MAPK
signaling pathway. In addition, Chen et al (11) demonstrated that bone marrow stem
cell-related lncRNA alleviated the progression of osteoporosis by
inhibiting osteoclast differentiation. A study reported that the
expression of long intergenic non-protein coding RNA 899
(LINC00899) was upregulated in the serum and bone marrow of
patients with acute myeloid leukemia, suggesting that LINC00899
expression may be a potential biomarker for the diagnosis and
prognosis of acute myeloid leukemia (12). However, the molecular mechanism
underlying the role of LINC00899 in the occurrence and development
of osteoporosis remain to be fully explored. Furthermore, lncRNAs
may act as competitive endogenous RNAs that sponge microRNAs
(miRNAs or miRs), thereby regulating osteogenic differentiation.
For example, depletion of the lncRNA KCNQ1 opposite
strand/antisense transcript 1 (KCNQ1OT1) was found to impede
osteogenic differentiation by regulating miR-214(13). Additionally, lncRNA MSC-antisense 1
could upregulate bone morphogenetic protein (BMP)2 expression to
modulate osteogenic differentiation by sponging miR-140-5p
(14). Therefore, the present study
aimed to investigate whether LINC00899 could affect the progression
of osteoporosis by regulating the expression of downstream
miRNAs.
miRNAs are short non-coding RNAs with a length of
~22 nt, which serve as vital regulators in a number of biological
processes, including cell proliferation and differentiation
(15). It has been reported that
some miRNAs are involved in osteoporosis by binding to the
3'-untranslated regions (3'-UTR) of their target mRNAs (16). For example, Wang et al
(17) showed that miR-765 exerted a
potential role in attenuating osteogenic differentiation by
targeting BMP6. Recently, Zhang et al (18) demonstrated that miR-664-5p could
promote the osteogenic differentiation by direct targeting the high
mobility group A2 protein. Additionally, Li et al (19) showed that miR-291a-3p improved cell
viability and promoted osteogenic differentiation by regulating
Dickkopf 1. In another study, Li et al (20) suggested that miR-374a was involved
in the progression of osteosarcoma (OS) and facilitated OS cell
migration by regulating Wnt/β-catenin signaling. However, the
biological role of miR-374a in osteoporosis remains unclear.
It has been suggested that runt-related
transcription factor 2 (RUNX2) is indispensable for bone formation
(21) and a potent inducer of
osteogenic differentiation (22).
In addition, RUNX2 was identified as a downstream target of several
miRNAs, such as miR-21(23),
miR-365a (24) and miR-217(25). Therefore, the present study
hypothesized that miR-374a may regulate osteoporosis progression by
regulating RUNX2. The present study was undertaken to determine the
biological role of LINC00899 in osteoporosis and investigated
whether LINC00899 could sponge miR-374a to enhance RUNX2
expression, with the aim of providing a novel therapeutic strategy
for alleviating osteoporosis.
Materials and methods
Clinical samples
Bone tissues (100 mg) were collected from patients
with osteoporosis (n=15; female, 7; male, 8; age, 51-74 years) and
healthy controls (n=15; female, 7; male, 8; age, 50-73 years) at
the Affiliated Changzhou No. 2 People's Hospital of Nanjing Medical
University (Nanjing, China) between March 2016 and April 2018. The
inclusion criteria for patients were as follows: i) Patients were
diagnosed with osteoporosis, and ii) patients provided informed
consent. Patients with complications due to other diseases, such as
chronic inflammatory diseases, such as rheumatoid arthritis and
inflammatory bowel disease were excluded. The bone fragments
extracted from the transcervical region of the femoral neck were
dissected into smaller fragments, washed three times with PBS and
stored at -80˚C until further analysis. Written informed consent
was obtained from all patients prior to study enrollment. The
present study was approved by the Ethics Committee of the
Affiliated Changzhou No. 2 People's Hospital of the Nanjing Medical
University.
Cell culture and induction of
osteogenic differentiation
293T cells were purchased from The Cell Bank of Type
Culture Collection of The Chinese Academy of Sciences. Cells were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) and 1% penicillin-streptomycin (Thermo Fisher Scientific,
Inc.), and maintained in a humidified atmosphere containing 5%
CO2 at 37˚C. The hBMSCs were obtained from the BeNa
Culture Collection; Beijing Beina Chunglian Biotechnology Research
Institute (cat. no. BNCC100385; http://www.bnbio.com/pro/p1/6/p_242206.html) and
cultured in α-minimum essential medium (α-MEM; Gibco; Thermo Fisher
Scientific Inc.) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.), 100 mg/ml penicillin and
100 U/ml streptomycin at 37˚C in a humidified incubator containing
5% CO2. To induce osteogenic differentiation, cells were
treated with 10 µmol/l dexamethasone, 200 µM ascorbic acid and 10
mmol/l β-glycerophosphate (Sigma-Aldrich; Merck KGaA) at 37˚C in 5%
CO2 and the medium was changed every 3 days (26).
Cell transfection
The short hairpin RNA (shRNA) targeting LINC00899
(5'-GCACAUGAGAUCGACUGACUA-3') was cloned into the pGHP1/Neo vector
(Shanghai GenePharma Co., Ltd.) to generate shLINC00899. miR-374a
mimics (5'-CUUAUCAGAUUGUAUUGUAAUU-3'), miR-374a inhibitor
(5'-CACUUAUCAGGUUGUAUUAUAA-3'), and their corresponding negative
controls (shNC; NC mimics, 5'-UCACAACCUCCUAGAAAGAGUAGA-3'; NC
inhibitor, 5'-UAAGUACAAUAAUUGCGCCACU-3') were purchased from
Shanghai GenePharma Co., Ltd. To overexpress LINC00899, the
full-length LINC00899 sequence was sub-cloned into the pcDNA3.1
vector (Shanghai GenePharma Co., Ltd). Cell were transfected with
50 nM shLINC00899, 50 nM shNC, 50 nM miR-374a mimics, 50 nM
miR-374a inhibitor, 50 nM NC mimics or 50 nM NC inhibitor using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h prior to subsequent experiments.
Bioinformatic analysis and
dual-luciferase reporter assay
The StarBase 2.0 database (http://starbase.sysu.edu.cn/) was used to predict the
binding sites between miR-374a and LINC00899 or RUNX2. Mutants
within the miR-374a binding site were created using the QuikChange
II Site Directed Mutagenesis kit (Agilent Technologies, Inc.). The
wild-type (WT) or mutant (Mut) sequences of the miR-374a binding
site in the 3'-UTR of LINC00899 and RUNX2 were cloned into a
pmirGLO reporter vector (Shanghai GenePharma Co., Ltd.).
Subsequently, 293T cells were co-transfected with 100 nM miR-374a
mimics or NC mimics and 0.6 µg pmirGLO-LINC00899-WT/Mut or
pmirGLO-RUNX2-WT/Mut. Following incubation for 48 h, the relative
luciferase activity was detected using a dual-luciferase reporter
assay system (Promega Corporation). Firefly luciferase activity was
normalized to Renilla luciferase activity.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted from tissues and hBMSCs
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols.
PrimeScript™ RT reagent kit (Takara Bio, Inc.) was used
for cDNA generation of LINC00899 and mRNA, using the following
reaction conditions: 42˚C for 15 min followed by 3 cycles at 85˚C
for 5 sec. For miR-374a, a TaqMan™ MicroRNA Reverse
Transcription kit (Thermo Fisher Scientific, Inc.) was used for
cDNA generation using the following reaction conditions: 50˚C for 5
min and 80˚C for 2 min. qPCR was performed on the ABI 7900
Detection System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using the SYBR-Green PCR Master Mix kit (Thermo Fisher
Scientific, Inc.). The thermocycling conditions were as follows:
Pre-denaturation at 95˚C for 1 min, followed by 40 cycles of 95˚C
for 15 sec, 60˚C for 30 sec and 72˚C for 30 sec. The data were
analyzed using the 2-ΔΔCq method (27). The primer sequences were as follows:
LINC00899 forward, 5'-CAGTCAGCCTCAGTTTCCAA-3' and reverse,
5'-AGGCAGGGCTGTGCTGAT-3'; miR-374a forward,
5'-GGTCACAGTGAACCGGTC-3' and reverse, 5'-GTGCAGGGTCCGAGGT-3'; RUNX2
forward, 5'-CTTATACAATGTCAACAGCC-3' and reverse,
5'-TCCTTATGCTCTTTCTTCC-3'; OPN forward, 5'-CAAATACCCAGATGCTGTGGC-3'
and reverse, 5'-TCCTGGCTGTCCACATGGTC-3'; OCN forward,
5'-GCCGAGAAATGTTGGAGAAA-3' and reverse, 5'-CTCCTTAATCTGGCCAACCA-3';
GAPDH forward, 5'-CCACTCCTCCACCTTTGAC-3' and reverse,
5'-ACCCTGTTGCTGTAGCCA-3' and U6 forward, 5'-CTTCGGCAGCACATATACT-3'
and reverse, 5'-AAAATATGGAACGCTTCACG-3'. The expression of
LINC00899, RUNX2, OCN and OPN expression was normalized to the
internal reference gene GAPDH, whereas that of miR-374a expression
was normalized to U6.
RNA immunoprecipitation (RIP)
assay
RIP assay was performed using the Magna RNA-binding
protein immunoprecipitation kit (EMD Millipore). Briefly, hBMSCs
were lysed with 100 µl Lysis Buffer (EMD Millipore) for 5 min at
4˚C. Argonaute 2 (Ago2; 5 µg; cat. no. ab32381; Abcam.) and IgG (5
µg; cat. no. ab172730; Abcam) were incubated with 50 µl A/G
magnetic beads for 1 h at 4˚C. Then, hBMSCs cell lysates was mixed
with the beads to incubate for 4 h at 4˚C. RNA was extracted from
the immunoprecipitation complex using 50 µg/ml Proteinase K (cat.
no. P2308; Sigma-Aldrich; Merck KGaA) at 55˚C for 30 min to digest
the protein. Subsequently, the enrichment of LINC00899 and miR-374a
was measured by RT-qPCR.
Alkaline phosphatase (ALP)
activity
The ALP activity was measured at 0, 7 and 14 days
following cell culture, using the ALP assay kit (cat. no. P0321;
Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. Briefly, hBMSCs (1x105) were
collected, rinsed with PBS, incubated and lysed with RIPA buffer
(Beyotime Institute of Biotechnology). The cell lysate was
centrifuged at 15,000 x g at 4˚C for 8 min before ALP activity was
determined in the supernatant. The optical absorbance was finally
measured at 450 nm using a microplate reader (Thermo Fisher
Scientific, Inc.).
Alizarin Red S (ARS) staining
Following osteogenic differentiation, treated hBMSCs
(1x105) were washed twice with PBS and then fixed with
4% paraformaldehyde for 20 min at room temperature. Subsequently,
the cells were washed twice with PBS and stained with 1 ml ARS
(Sigma-Aldrich; Merck KGaA) at 37˚C for 10 min. After washing with
distilled water, images of the cells were captured under a light
microscope (magnification, x200; Olympus Corporation).
Western blot analysis
Total proteins were extracted from hBMSCs using a
RIPA buffer (Beyotime Institute of Biotechnology) and quantified
using the (BCA) Protein Assay kit (Pierce; Thermo Fisher
Scientific, Inc.). The protein samples (20 µg g/lane) were
separated by 10% SDS-PAGE and transferred onto PVDF membranes.
Following blocking for 2 h with 5% non-fat dry milk at room
temperature, the membranes were incubated with antibodies against
RUNX2 (1:1,000; cat. no. 12556; Cell Signaling Technology, Inc.),
osteopontin (OPN; 1:1,000; cat. no. ab214050; Abcam), osteocalcin
(OCN; 1:1,000; cat. no. ab93876; Abcam) and GAPDH (1:1,000; cat.
no. ab8245; Abcam) at 4˚C overnight. The next day, the membranes
were incubated with a horseradish peroxidase-conjugated secondary
antibody (1:1,000; cat. no. ab205718; Abcam) for 1 h at 37˚C.
Finally, the protein bands were visualized using the Pierce™ ECL
Western Blotting substrate (Thermo Fisher Scientific, Inc.).
Statistical analysis
All experiments were repeated ≥ three times. The
data were analyzed using the GraphPad Prism 6.0 software (GraphPad
Software, Inc.) and presented as the mean ± standard deviation. The
differences between two groups were analyzed using unpaired
Student's t-test, whilst those among multiple groups were compared
using one-way ANOVA followed by Tukey's post hoc test. P<0.05
was considered to indicate a statistically different
difference.
Results
Expression levels of LINC00899 and
miR-374a during osteogenic differentiation
The dynamic expression pattern of LINC00899 and
miR-374a was determined on days 0, 7 and 14 after osteogenic
differentiation. As shown in Fig.
1A and B, the expression levels
of LINC00899 were increased in a time dependent manner, whilst
those of miR-374a were decreased as the osteogenic induction
process progressed. Furthermore, RT-qPCR and western blot analysis
revealed that the mRNA and protein expression levels of RUNX2, OPN
and OCN were also gradually increased during the process of
osteogenic differentiation in a time dependent manner (Fig. 1C-F). In addition, ALP activity and
the area of mineralized nodules were markedly increased during
osteogenic induction, as shown by ALP assay and ARS, respectively
(Fig. 1G and H). These aforementioned results suggest
that LINC00899 expression was upregulated whereas miR-374a
expression was downregulated during osteogenic differentiation.
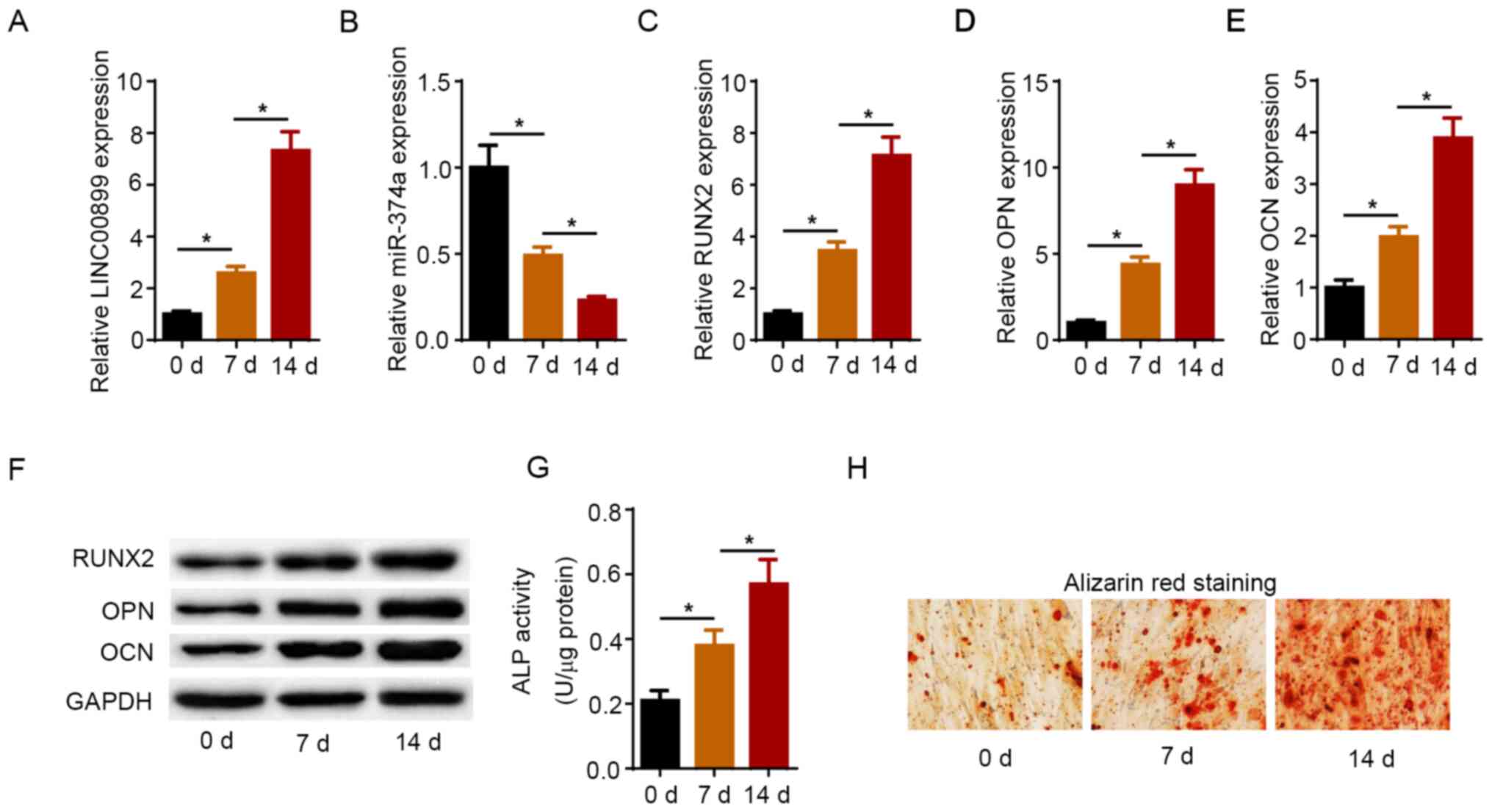 | Figure 1Expression levels of LINC00899 and
miR-374a during osteogenic differentiation. Expression levels of
(A) LINC00899 and (B) miR-374a in hBMSCs treated with osteogenic
differentiation medium for 0, 7 and 14 days as detected by RT-qPCR.
RT-qPCR was used to detect the mRNA levels of (C) RUNX2, (D) OPN
and (E) OCN during osteoblast differentiation of hBMSCs. (F)
Western blotting was used to measure the protein levels of RUNX2,
OPN and OCN during osteoblast differentiation of hBMSCs. (G) ALP
activity and (H) Alizarin red S staining were detected on 0, 7 and
14 days after osteogenic induction (magnification, x200). The data
were presented as mean ± SD. *P<0.05. LINC00899, long
intergenic non-protein coding RNA 899; miR, microRNA; RUNX2,
runt-related transcription factor 2; OPN, osteopontin; OCN,
osteocalcin; RT-qPCR, reverse transcription-quantitative PCR;
hBMSCs, human bone mesenchymal stem cells; ALP, alkaline
phosphatase. |
Knockdown of LINC00899 promotes the
progression of osteoporosis
RT-qPCR assay results demonstrated that the
LINC00899 expression levels were significantly higher in
non-osteoporotic tissues compared with those in tissues from
patients with osteoporosis (Fig.
2A). Subsequently, to investigate the effect of LINC00899 on
osteogenic differentiation, LINC00899 expression was knocked down
in hBMSCs on day 14 following osteogenic induction using shRNA, the
transfection efficiency of which was confirmed by RT-qPCR (Fig. 2B). Subsequently, LINC00899 knockdown
was found to have downregulated the expression of
osteogenesis-related genes, RUNX2, OPN and OCN, at both mRNA and
protein levels (Fig. 2C-F). In
addition, results from ALP assay demonstrated that LINC00899
knockdown significantly decreased ALP activity compared with that
in cells transfected with shNC (Fig.
2G). ARS staining revealed that the number of mineralized
nodules was reduced following transfection of hBMSCs with
shLINC00899 (Fig. 2H). Taken
together, these findings suggest that LINC00899 knockdown reduced
the expression of osteogenesis-related genes, ALP activity and
osteogenic capacity of hBMSCs.
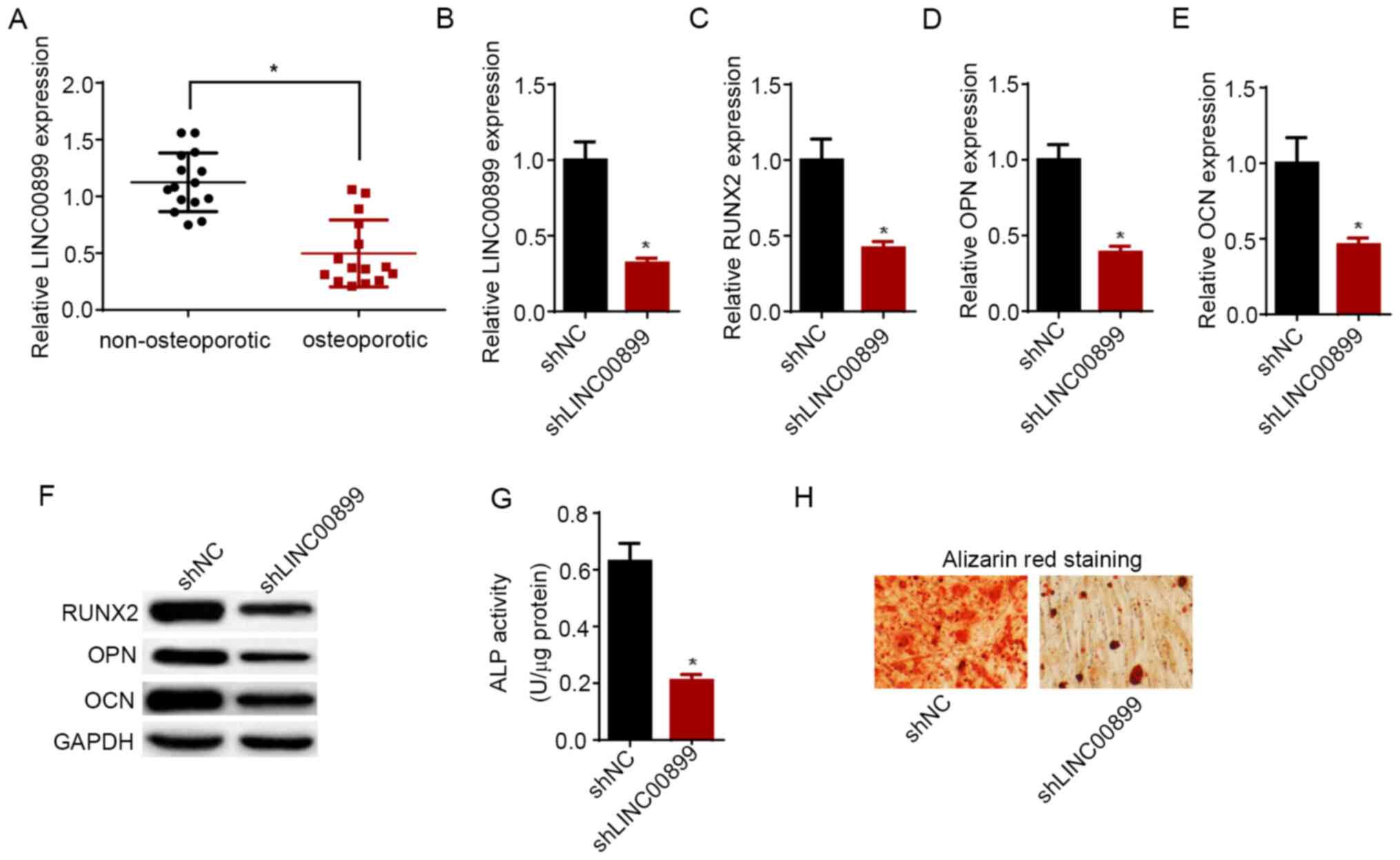 | Figure 2Knockdown of LINC00899 expression
promotes the progression of osteoporosis. (A) RT-qPCR was used to
detect the expression of LINC00899 in femoral bone tissues from
patients with osteoporosis and health individuals.
*P<0.05. (B) Expression of LINC00899 in hBMSCs
transfected with shNC and shLINC00899 was detected by RT-qPCR. mRNA
expression of osteogenesis-related genes (C) RUNX2, (D) OPN and (E)
OCN in hBMSCs transfected with shNC and shLINC00899 was detected by
RT-qPCR. (F) Western blotting was utilized to detect the protein
levels of RUNX2, OPN and OCN in hBMSCs transfected with shNC and
shLINC00899. (G) ALP activity was detected in hBMSCs transfected
with shNC and shLINC00899. (H) Alizarin red S staining was measured
in hBMSCs transfected with shNC and shLINC00899 (magnification,
x200). The data were presented as mean ± SD. *P<0.05
vs. shNC. RT-qPCR, reverse transcription-quantitative PCR;
LINC00899, long intergenic non-protein coding RNA 899; hBMSCs,
human bone mesenchymal stem cells; ALP, alkaline phosphatase; sh,
short hairpin; NC, negative control; RUNX2, runt-related
transcription factor 2; OPN, osteopontin; OCN, osteocalcin. |
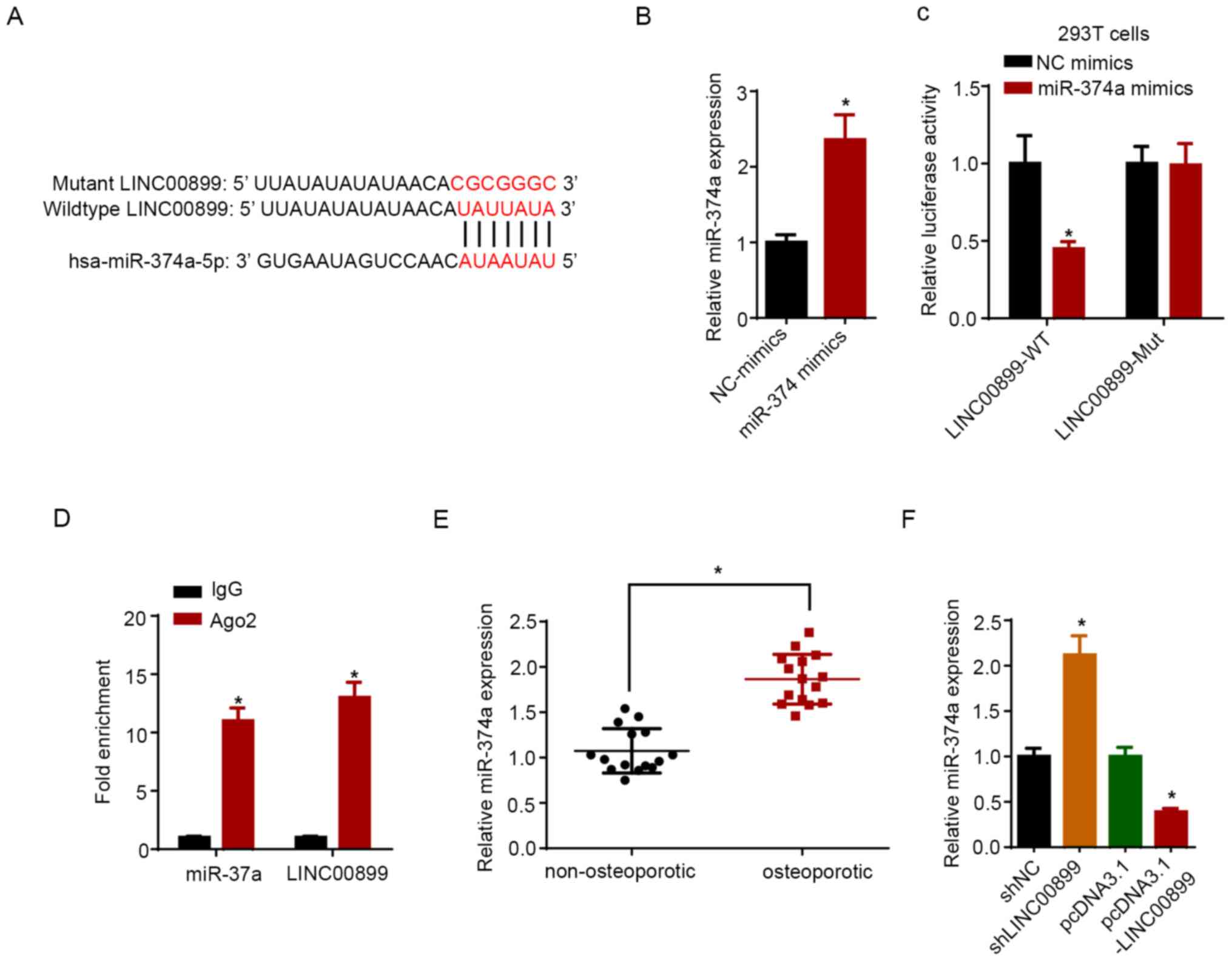
miR-374a is a target of LINC00899
A potential binding site for miR-374a on LINC00899
was predicted using the StarBase software (Fig. 3A). miR-374a expression was increased
in hBMSCs following transfection with miR-374a mimics compared with
that in cells transfected with NC mimics (Fig. 3B). Data from dual-luciferase
reporter assays revealed that miR-374a mimic transfection reduced
the luciferase activity of LINC00899-WT, but not on the
LINC00899-Mut plasmid (Fig. 3C).
RIP assay showed that LINC00899 and miR-374a were significantly
enriched in samples pulled down by Ago2-containing beads compared
with those pulled down by the IgG-containing beads (Fig. 3D). Furthermore, the expression
levels of miR-374a were measured in non-osteoporotic and
osteoporotic tissues, where the results showed that the miR-374a
expression level was significantly lower in non-osteoporotic
tissues compared with that in osteoporotic tissues (Fig. 3E). RT-qPCR confirmed that LINC00899
was upregulated in hBMSCs transfected with the LINC00899
overexpression plasmid compared with that in cells transfected with
the empty plasmid (Fig. 3F).
Subsequently, LINC00899 knockdown was found to significantly
increase the expression of miR-374a, whilst LINC00899
overexpression exerted the opposite effect (Fig. 3G). Overall, these results indicated
that LINC00899 could directly interact with and inhibit miR-374a
expression.
LINC00899 regulates osteogenic
differentiation by inhibiting miR-374a expression
Bioinformatics analysis using the Starbase software
predicted that RUNX2 could be a potential target of miR-374a
(Fig. 4A). Subsequently,
dual-luciferase reporter assay verified that transfection with the
miR-374a mimics significantly attenuated the luciferase activity of
RUNX2-WT (Fig. 4B). However,
miR-374a overexpression exerted no significant effect on the
luciferase activity of RUNX2-Mut (Fig.
4B). miR-374a expression was significantly downregulated
following the transfection of miR-374a inhibitor into hBMSCs
(Fig. 4C). In addition, silencing
of miR-374a significantly increased the expression of RUNX2
(Fig. 4D). To further investigate
whether LINC00899 could regulate the expression of RUNX2 via
miR-374a, hBMSCs were transfected with shNC, shLINC00899 or
shLINC00899 + miR-374a inhibitor. RT-qPCR and western blot analysis
showed that LINC00899 knockdown downregulated RUNX2 expression, but
the simultaneous inhibition of miR-374 expression significantly
reversed this effect (Fig. 4E and
F). Furthermore, co-transfection of
hBMSCs with the miR-374a inhibitor partially abolished the
suppressive effects of LINC00899 knockdown on the expression of OPN
and OCN (Fig. 4G-I). Results from
the ALP activity assay further verified that miR-374a silencing
could reverse the inhibitory effects of LINC00899 knockdown on ALP
activity (Fig. 4J). Additionally,
ARS staining showed that miR-374a inhibitor could markedly restore
the osteogenic capacity of hBMSCs transfected with shLINC00899
(Fig. 4K). These observations
suggest that LINC00899 can upregulate RUNX2 expression by sponging
miR-374a to alleviate osteoporosis.
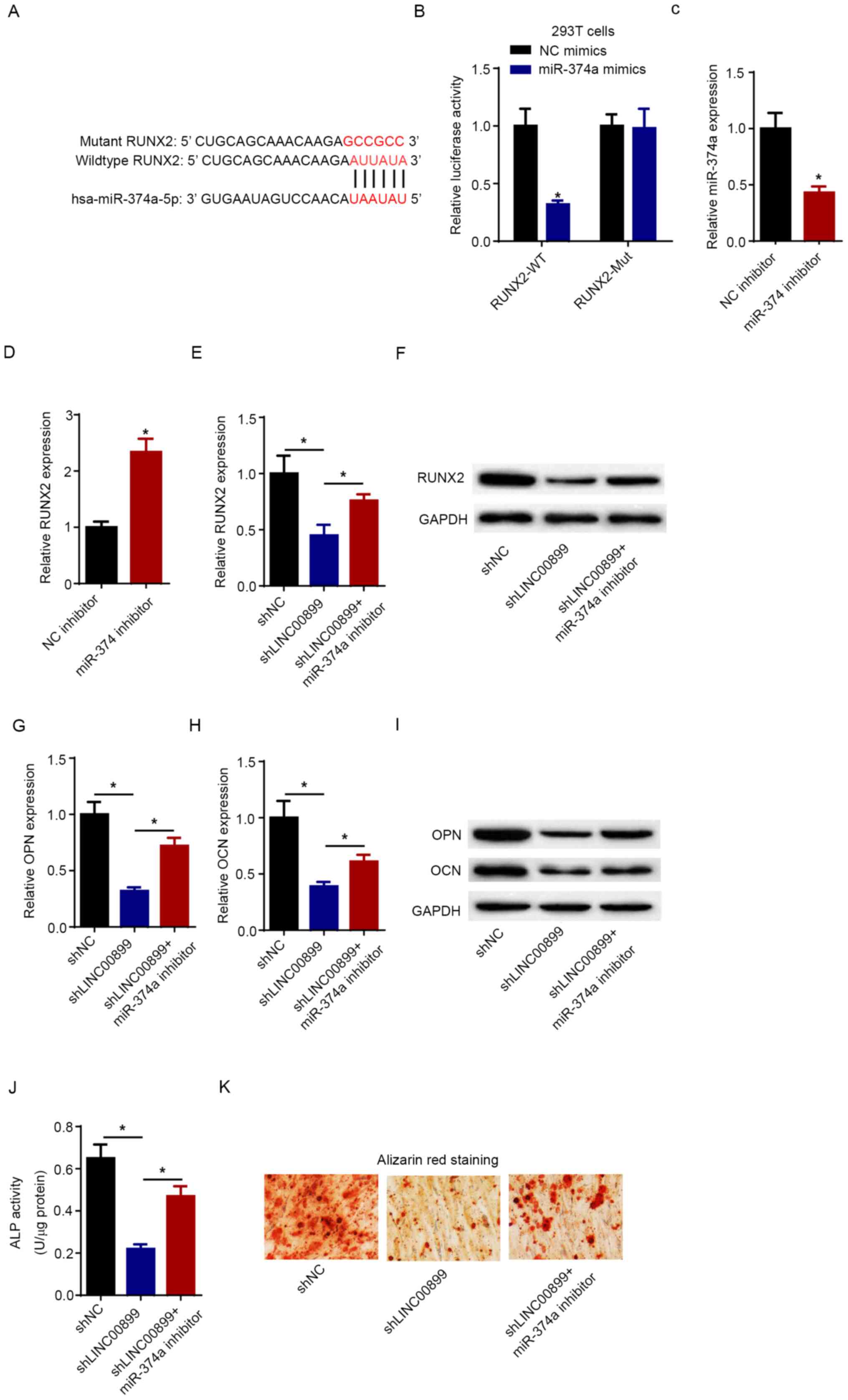 | Figure 4LINC00899 regulates osteogenic
differentiation by suppressing miR-374a expression. (A) The
predicted binding sites of miR-374a on the RUNX2 3'-untranslated
region. (B) Dual-luciferase reporter assay was used to determine
the luciferase activity of RUNX2-WT or RUNX2-Mut in 293T cells.
*P<0.05 vs. NC mimics. (C) Expression of miR-374a was
measured by RT-qPCR in hBMSCs transfected with NC inhibitor or
miR-374a inhibitor. *P<0.05 vs. NC inhibitor. (D) The
expression of RUNX2 in hBMSCs transfected with NC inhibitor and
miR-374a inhibitor was detected by RT-qPCR. *P<0.05
vs. NC inhibitor. (E) mRNA and (F) protein levels of RUNX2, mRNA
levels of (G) OPN and (H) OCN, (I) protein levels of OPN and OCN
were determined in hBMSCs transfected with shNC, shLINC00899,
shLINC00899 + miR-374a inhibitor by RT-qPCR and western blotting.
*P<0.05. (J) ALP activity was detected in hBMSCs
transfected with shNC, shLINC00899 and shLINC00899 + miR-374a
inhibitor. *P<0.05. (K) Alizarin red S staining was
detected in hBMSCs transfected with shNC, shLINC00899 and
shLINC00899 + miR-374a inhibitor (magnification, x200). The data
were presented as mean ± SD. miR, microRNA; LINC00899, long
intergenic non-protein coding RNA 899; RT-qPCR, reverse
transcription-quantitative PCR; hBMSCs, human bone mesenchymal stem
cells; sh, short hairpin; NC, negative control; WT, wild-type; Mut,
mutant; ALP, alkaline phosphatase; RUNX2, runt-related
transcription factor 2; OPN, osteopontin; OCN, osteocalcin. |
Discussion
The present study demonstrated that LINC00899 could
facilitate osteogenic differentiation and prevent osteoporosis by
regulating the miR-374a/RUNX2 axis. Specifically, LINC00899
downregulated miR-374a expression, which could in turn directly
target the 3'-UTR of RUNX2, thus downregulating its expression.
Emerging evidence has suggested that lncRNAs can
serve an important role in bone diseases, such as osteoporosis,
osteoarthritis, and osteosarcoma (28,29).
Han et al (30) revealed
that downregulation of lncRNA taurine upregulated 1 could
effectively inhibit osteoclast proliferation and serve as a
potential target for the treatment of osteoporosis. Shen et
al (31) demonstrated that
lncRNA hox transcript antisense RNA (HOTAIR) was highly expressed
in patients with osteoporosis, which prevents osteogenic
differentiation by regulating the Wnt/β-catenin signaling pathway.
In terms of LINC00899, Zhou et al (32) previously showed that the
upregulation of LINC00899 suppressed breast cancer progression by
regulating the expression of miR-425. Additionally, Dong et
al (33) revealed that the
upregulation of LINC00899 can promote the progression of acute
myeloid leukemia. However, the biological effects of LINC00899 in
osteogenic differentiation and osteoporosis remain poorly
understood. To the best of our knowledge, the present study was the
first to report that LINC00899 expression is downregulated in the
bone tissues of patients with osteoporosis. Furthermore, knocking
down LINC00899 expression was found to decrease the expression of
osteogenesis-related genes, inhibit ALP activity whilst reducing
ARS accumulation, eventually leading to the inhibition of
osteogenic differentiation and progression of osteoporosis.
Accumulating evidence has suggested that lncRNAs can
serve as miRNA sponges to regulate osteogenic differentiation and
osteoporosis progression. For example, Wang et al (13) demonstrated that the upregulation of
lncRNA KCNQ1OT1 promoted osteogenic differentiation by sponging
miR-214 to upregulate BMP2. Zhang et al (34) reported that the upregulation of
nuclear paraspeckle assembly transcript 1 promoted BMP1 expression
to regulate osteogenic differentiation in hBMSCs by sponging
miR-29b-3p. In addition, it has been suggested that miRNAs are
involved in the occurrence of bone diseases, such as osteoporosis
(35), osteoarthritis (36), and osteosarcoma (37). For example, Fan et al
(38) demonstrated that miR-532-3p
attenuated osteogenic differentiation by downregulating ETS-1.
Additionally, Xiaoling et al (39) previously reported that the
upregulation of miR-19b-3p could promote osteogenic differentiation
in BMSCs. In the present study, miR-374a was predicted to be a
potential target of LINC00899, which was verified by
dual-luciferase reporter and RIP assays. Furthermore, the dynamic
expression profile of LINC00899 and miR-374a during osteogenic
differentiation was also determined. The results revealed that the
expression of LINC00899 was gradually increased, whilst that of
miR-374a was decreased, during osteogenic differentiation. These
results suggest that miR-374a could be associated with osteoporosis
progression by regulating osteogenic differentiation.
RUNX2 is widely recognized as an important
transcription factor during osteogenic differentiation (40). Therefore, the regulatory mechanism
of RUNX2 during osteogenic differentiation has attracted the
attention of numerous research groups. Fu et al (41) showed that the upregulation of
HOTAIRM1 promoted osteogenesis by modulating the activity of JNK
and c-Jun to promote RUNX2 gene transcription. Another study by
Chen et al (42)
demonstrated that the upregulation of lncRNA AWPPH contributed to
non-traumatic osteonecrosis by upregulating RUNX2. However, the
mechanism associated with RUNX2 in osteoporosis has not been fully
elucidated. Therefore, the present study aimed to actively explore
the novel mechanisms by which RUNX2 may regulate osteogenic
differentiation. RUNX2 was predicted as the direct target of
miR-374a. RT-qPCR showed that RUNX2 was significantly upregulated
during osteogenic differentiation and in hBMSCs transfected with
the miR-374a inhibitor. In addition, depletion of LINC00899
inhibited the expression of osteogenesis-related markers RUNX2, OPN
and OCN. This effect was counteracted following the co-transfection
of hBMSCs with the miR-374a inhibitor. Furthermore, co-transfection
with the miR-374a inhibitor neutralized the decrease in ALP
activity and ARS accumulation induced by LINC00899 knockdown.
LINC00899 also regulated RUNX2 expression by targeting miR-374a,
supporting the regulatory effects of the LINC00899/miR-374a/RUNX2
axis during osteogenic differentiation.
In summary, the present study uncovered a role of
LINC00899 in promoting osteogenic differentiation through the
miR-374a/RUNX2 axis. Therefore, these findings may provide a novel
theoretical basis for developing a treatment strategy for
osteoporosis.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
XG and KY designed the study. XG, YX and KY
performed the experiments, analyzed the data and prepared the
figures. XG and KY drafted the initial manuscript. XG and KY
authenticated the raw data in this study. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from the Ethics
Committee of the Affiliated Changzhou No. 2 People's Hospital of
Nanjing Medical University (Nanjing, China). All procedures
performed in studies involving human participants were in
accordance with the ethical standards of the institutional and
national research committee. Written informed consent was obtained
from all individual participants included in this study.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Montalcini T, Romeo S, Ferro Y, Migliaccio
V, Gazzaruso C and Pujia A: Osteoporosis in chronic inflammatory
disease: The role of malnutrition. Endocrine. 43:59–64.
2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kobayashi M, Sawada K, Yoshimura A,
Yamamoto M, Shimizu A, Shimura K, Komura N, Miyamoto M, Ishida K
and Kimura T: Clinical effects of switching from minodronate to
denosumab treatment in patients with postmenopausal osteoporosis: A
retrospective study. BMC Womens Health. 20(48)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bianco P, Sacchetti B and Riminucci M:
Stem cells in skeletal physiology and endocrine diseases of bone.
Endocr Dev. 21:91–101. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jing H, Liao L, An Y, Su X, Liu S, Shuai
Y, Zhang X and Jin Y: Suppression of EZH2 prevents the shift of
osteoporotic MSC fate to adipocyte and enhances bone formation
during osteoporosis. Mol Ther. 24:217–229. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Li CJ, Cheng P, Liang MK, Chen YS, Lu Q,
Wang JY, Xia ZY, Zhou HD, Cao X, Xie H, et al: MicroRNA-188
regulates age-related switch between osteoblast and adipocyte
differentiation. J Clin Invest. 125:1509–1522. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Li Y, Fan L, Hu J, Zhang L, Liao L, Liu S,
Wu D, Yang P, Shen L, Chen J and Jin Y: miR-26a rescues bone
regeneration deficiency of mesenchymal stem cells derived from
osteoporotic mice. Mol Ther. 23:1349–1357. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hu HL, Liu KY, Ram YI, Gao JL and Cao YM:
Long noncoding RNA MIRG induces osteoclastogenesis and bone
resorption in osteoporosis through negative regulation of miR-1897.
Eur Rev Med Pharmacol Sci. 23:10195–10203. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zeng X, Wang Y, Dong Q, Ma MX and Liu XD:
DLX2 activates Wnt1 transcription and mediates Wnt/β-catenin signal
to promote osteogenic differentiation of hBMSCs. Gene.
744(144564)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gao GC, Yang DW and Liu W: LncRNA TERC
alleviates the progression of osteoporosis by absorbing miRNA-217
to upregulate RUNX2. Eur Rev Med Pharmacol Sci. 24:526–534.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang Y, Wu W, Jiao G, Chen Y and Liu H:
LncRNA SNHG1 modulates p38 MAPK pathway through Nedd4 and thus
inhibits osteogenic differentiation of bone marrow mesenchymal stem
cells. Life Sci. 228:208–214. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen RS, Zhang XB, Zhu XT and Wang CS:
LncRNA Bmncr alleviates the progression of osteoporosis by
inhibiting RANML-induced osteoclast differentiation. Eur Rev Med
Pharmacol Sci. 23:9199–9206. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang Y, Li Y, Song HQ and Sun GW: Long
non-coding RNA LINC00899 as a novel serum biomarker for diagnosis
and prognosis prediction of acute myeloid leukemia. Eur Rev Med
Pharmacol Sci. 22:7364–7370. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang CG, Liao Z, Xiao H, Liu H, Hu YH,
Liao QD and Zhong D: LncRNA KCNQ1OT1 promoted BMP2 expression to
regulate osteogenic differentiation by sponging miRNA-214. Exp Mol
Pathol. 107:77–84. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang N, Hu X, He S, Ding W, Wang F, Zhao
Y and Huang Z: LncRNA MSC-AS1 promotes osteogenic differentiation
and alleviates osteoporosis through sponging microRNA-140-5p to
upregulate BMP2. Biochem Biophys Res Commun. 519:790–796.
2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ding J, Sha L, Shen P, Huang M, Cai Q and
Li J: MicroRNA-18a inhibits cell growth and induces apoptosis in
osteosarcoma by targeting MED27. Int J Oncol. 53:329–338.
2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bellavia D, De Luca A, Carina V, Costa V,
Raimondi L, Salamanna F, Alessandro R, Fini M and Giavaresi G:
Deregulated miRNAs in bone health: Epigenetic roles in
osteoporosis. Bone. 122:52–75. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang T, Zhang C, Wu C, Liu J, Yu H, Zhou
X, Zhang J, Wang X, He S, Xu X, et al: miR-765 inhibits the
osteogenic differentiation of human bone marrow mesenchymal stem
cells by targeting BMP6 via regulating the BMP6/Smad1/5/9 signaling
pathway. Stem Cell Res Ther. 11(62)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang Y, Liu Y, Wu M, Wang H, Wu L, Xu B,
Zhou W, Fan X, Shao J and Yang T: MicroRNA-664a-5p promotes
osteogenic differentiation of human bone marrow-derived mesenchymal
stem cells by directly downregulating HMGA2. Biochem Biophys Res
Commun. 521:9–14. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li ZH, Hu H, Zhang XY, Liu GD, Ran B,
Zhang PG, Liao MM and Wu YC: miR-291a-3p regulates the BMSCs
differentiation via targeting DKK1 in dexamethasone-induced
osteoporosis. Kaohsiung J Med Sci. 36:35–42. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li W, Meng Z, Zou T, Wang G, Su Y, Yao S
and Sun X: miR-374a activates Wnt/β-catenin signaling to promote
osteosarcoma cell migration by targeting WIF-1. Pathol Oncol Res.
26:533–539. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Komori T: Requisite roles of Runx2 and
Cbfb in skeletal development. J Bone Miner Metab. 21:193–197.
2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhao Z, Zhao M, Xiao G and Franceschi RT:
Gene transfer of the Runx2 transcription factor enhances osteogenic
activity of bone marrow stromal cells in vitro and in vivo. Mol
Ther. 12:247–253. 2005.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li L and Jiang D: Hypoxia-responsive
miRNA-21-5p inhibits Runx2 suppression by targeting SMAD7 in
MC3T3-E1 cells. J Cell Biochem. 120:16867–16875. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Cheng F, Yang MM and Yang RH:
miRNA-365a-3p promotes the progression of osteoporosis by
inhibiting osteogenic differentiation via targeting RUNX2. Eur Rev
Med Pharmacol Sci. 23:7766–7774. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yang L, Zeng Z, Kang N, Yang JC, Wei X and
Hai Y: Circ-VANGL1 promotes the progression of osteoporosis by
absorbing miRNA-217 to regulate RUNX2 expression. Eur Rev Med
Pharmacol Sci. 23:949–957. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kosmacheva SM, Volk MV, Yeustratenka TA,
Severin IN and Potapnev MP: In vitro growth of human umbilical
blood mesenchymal stem cells and their differentiation into
chondrocytes and osteoblasts. Bull Exp Biol Med. 145:141–145.
2008.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tong X, Gu PC, Xu SZ and Lin XJ: Long
non-coding RNA-DANCR in human circulating monocytes: A potential
biomarker associated with postmenopausal osteoporosis. Biosci
Biotechnol Biochem. 79:732–737. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang Q, Li Y and Zhang Y, Ma L, Lin L,
Meng J, Jiang L, Wang L, Zhou P and Zhang Y: LncRNA MEG3 inhibited
osteogenic differentiation of bone marrow mesenchymal stem cells
from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed
Pharmacother. 89:1178–1186. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Han Y, Liu C, Lei M, Sun S, Zheng W, Niu Y
and Xia X: LncRNA TUG1 was upregulated in osteoporosis and
regulates the proliferation and apoptosis of osteoclasts. J Orthop
Surg Res. 14(416)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Shen JJ, Zhang CH, Chen ZW, Wang ZX, Yang
DC, Zhang FL and Feng KH: LncRNA HOTAIR inhibited osteogenic
differentiation of BMSCs by regulating Wnt/beta-catenin pathway.
Eur Rev Med Pharmacol Sci. 23:7232–7246. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhou W, Gong J, Chen Y, Chen J, Zhuang Q,
Cao J, Mei Z and Hu B: Long noncoding RNA LINC00899 suppresses
breast cancer progression by inhibiting miR-425. Aging (Albany NY).
11:10144–10153. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Dong X, Xu X and Guan Y: LncRNA LINC00899
promotes progression of acute myeloid leukaemia by modulating
miR-744-3p/YY1 signalling. Cell Biochem Funct. 38:955–964.
2020.PubMed/NCBI View
Article : Google Scholar
|
|
34
|
Zhang Y, Chen B, Li D, Zhou X and Chen Z:
LncRNA NEAT1/miR-29b-3p/BMP1 axis promotes osteogenic
differentiation in human bone marrow-derived mesenchymal stem
cells. Pathol Res Pract. 215:525–531. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kelch S, Balmayor ER, Seeliger C, Vester
H, Kirschke JS and van Griensven M: miRNAs in bone tissue correlate
to bone mineral density and circulating miRNAs are gender
independent in osteoporotic patients. Sci Rep.
7(15861)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Park S, Lee M, Chun CH and Jin EJ: The
lncRNA, nespas, is associated with osteoarthritis progression and
serves as a potential new prognostic biomarker. Cartilage.
10:148–156. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Delsin LEA, Roberto GM, Fedatto PF, Engel
EE, Scrideli CA, Tone LG and Brassesco MS: Downregulated
adhesion-associated microRNAs as prognostic predictors in childhood
osteosarcoma. Pathol Oncol Res. 25:11–20. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Fan Q, Li Y, Sun Q, Jia Y, He C and Sun T:
miR-532-3p inhibits osteogenic differentiation in MC3T3-E1 cells by
downregulating ETS1. Biochem Biophys Res Commun. 525:498–504.
2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Xiaoling G, Shuaibin L and Kailu L:
MicroRNA-19b-3p promotes cell proliferation and osteogenic
differentiation of BMSCs by interacting with lncRNA H19. BMC Med
Genet. 21(11)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yang GZ, Zhang WJ, Ding X, Zhang XK, Jiang
XQ and Zhang ZY: Effect of overexpression of transcription factor
Runx2 and Osterix on osteogenic differentiation of endothelial
cells. Shanghai Kou Qiang Yi Xue. 26:353–357. 2017.PubMed/NCBI(In Chinese).
|
|
41
|
Fu L, Peng S, Wu W, Ouyang Y, Tan D and Fu
X: LncRNA HOTAIRM1 promotes osteogenesis by controlling JNK/AP-1
signalling-mediated RUNX2 expression. J Cell Mol Med. 23:7517–7524.
2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen X, Li J, Liang D, Zhang L and Wang Q:
LncRNA AWPPH participates in the development of non-traumatic
osteonecrosis of femoral head by upregulating Runx2. Exp Ther Med.
19:153–159. 2020.PubMed/NCBI View Article : Google Scholar
|