Introduction
Breast cancer is the most common malignant disease
that mainly affects females. Recurrence and metastasis result in
unfavorable prognoses for breast cancer patients; up to 30% of
patients succumb to this disease due to relapse and metastasis
after having standard-of-care therapy (1). Therefore, novel therapies are urgently
required. Cancer metastases can be attributed to multiple factors
such as cancer cell biological processes that underlie the
dissemination and metastatic outgrowth of cancer cells, cancer stem
cells (CSCs) (2,3). MicroRNAs (miRs) are a class of small
noncoding RNAs (19-22 nt) that are involved in biological processes
such as proliferation, differentiation, apoptosis, and development
(4,5). miR-based therapeutic strategies are
promising for cancer therapy. MiR-7 is an intronic microRNA that
resides in the first intron of the heterogeneous ribonuclear
protein K gene on chromosome 9 and is downregulated in different
cancer types (6). Our previous
study had revealed that miR-7, which was downregulated in breast
CSCs (BCSCs; EpCAM+CD44+CD24-/low)
isolated from the human MCF-7 and MDA-MB-231 cell lines, inhibited
cell invasion and metastasis, decreased the BCSC population, and
partially reversed EMT in MDA-MB-231 cells by directly targeting
the oncogene, SETDB1(7). However,
the molecular mechanism by which miR-7 plays a role in BCSC subset
downregulation is not clear. Due to the existence of a BCSC subset,
chemotherapy, radiotherapy sensitivity, and therapeutic effects
have been revealed to be decreased. It is known that the quantity
of the BCSC subset is closely related to the survival of breast
cancer patients (8). Therefore, it
is of great significance to elucidate the molecular mechanism of
miR-7 to reduce the amount of the BCSC subset and to use miR-7 to
target BCSC in the treatment of breast cancer.
Although our latest work investigated the
relationship between miR-7 and ALDH1A3, a few questions remain
unanswered (9). For example, it is
not known how miR-7 downregulates CD44 and what role ALDH1A3 plays
in impacting CD44 expression in MDA-MB-231 cells, which leads to a
decrease of the BCSC subset. In the present study, the mechanisms
involved in small interfering (si)ALDH1A3 downregulation of CD44
via the TGF-β1 pathway were explored.
Materials and methods
Cell culture and reagents
Human breast cancer cell lines MDA-MB-231, SK-BR-3,
and MCF-7 were obtained from the Shanghai Institute of Biochemistry
and Cell Biology, Chinese Academy of Sciences. The LD cell line was
established by our laboratory (Department of Pathogenic Biology and
Immunology, School of Medicine, Southeast University, Nanjing,
China) from a human breast cancer postsurgery sample (8). All cells were cultured in Dulbecco's
modified Eagle's medium containing 10% fetal bovine serum (GibcoTM;
Thermo Fisher Scientific, Inc.). Lipofectamine™ 2000 reagent was
obtained from Invitrogen; Thermo Fisher Scientific, Inc. TGF-β1
(Novoprotein Scientific, Inc.) and SB431542 (selective inhibitor of
TGF-βRI) (cat. no. A8249; APeXBIO Technology LLC) were used to
treat cells.
Human breast cancer samples
The data of 12 clinical human breast cancer
postsurgery samples, were recorded in our previous study (8), and were obtained from the Department
of General Surgery of Zhongda Hospital at Southeast University
(Nanjing, China). The investigation was approved by the Ethics
Committee at Southeast University School of Medicine, and informed
consent for the use of the postsurgery samples was obtained from
the donors who were breast cancer patients.
Magnetic cell sorting (MACS) for
BCSCs
CD44 (cat. no. 130-095-194)/CD24 (cat. no.
130-095-951)/CD326 (cat. no. 130-061-101) antibodies conjugated to
magnetic microbeads (Miltenyi Biotec GmbH) were used to obtain the
BCSCs from MDA-MB-231 cell lines, respectively. The isolation
process was according to the manufacturer's instructions.
siRNA design and plasmid
transfection
siALDH1A3 was designed based on the ALDH1A3 DNA
sequence (GenBank no. NM_001128128.2) using the siDESIGN design
software (http://www.dharmacon.com/). siALDH1A3
(5'-GUUCAAAAGUAUCGAAGAA-3') and siRNA-NC
(5'-GGCUCUAGAAAAGCCUAUGCdTdT-3') sequences were synthesized by
Guangzhou RiboBio Co., Ltd. MDA-MB-231 cells were transiently
transfected using Lipofectamine® 2000 transfection
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 1
µl Lipofectamine was added to 50 µl serum-free and antibiotic-free
DMEM medium, mixed with 50 nM siRNA at room temperature for 20 min.
The mixture was added to 1x105 cells in 24 well-plate,
which were cultured at 37˚C with 5% CO2 for 4-6 h for
transfection. The subsequent experiments were performed 48 h after
transfection.
Flow cytometry (FCM)
CD44-APC antibodies (1:100 dilution; cat. no.
17-0441-82; eBioscience; Thermo Fisher Scientific, Inc.) diluted in
PBS were used to label 1x106 cells at 4˚C for 30 min and
the stained cells were analyzed using BD FACSAria (BD Biosciences)
according to the manufacturer's instructions. FlowJo v10 (FlowJo
LLC) was used for analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
To evaluate the expression of miR-7, Smad2, Smad3,
Smad4, transforming growth factor-β receptor 2 (TGFBR2), and CD44
respectively, total cellular RNA was isolated from each sample
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.) and used for the reverse transcription reactions
(PrimeScript™ RT Master Mix; cat. no. RR036A; Takara Bio, Inc.),
followed by qPCR (One Step TB Green® PrimeScript™ RT-PCR
kit; cat. no. RR066A; Takara Bio, Inc.) was performed on a
StepOnePlus™ System (AB Applied Biosystems; Thermo Fisher
Scientific, Inc.). The 2-ΔΔCq method was used for
quantification (10). GAPDH was
used as an internal control. The following primer sequences were
used: miR-7 (forward, 5'-ACACTCCAGCTGGGTGGAAGACTA GTGATTT-3';
reverse, 5'-CTCAACTGGTGTCGTGGAG TCGGCAATTCAGTTGAGACAACAAA-3'),
Smad2 (forward, 5'-GTCGTCCATCTTGCCATTCAC-3'; reverse,
5'-TTCCTGCCCATTCTGCTCTC-3'), Smad3 (forward,
5'-GTCGTCCATCCTGCCTTTCA-3'; reverse, 5'-GTTTCT
TGACCAGGCTCTTGACC-3'), Smad4 (forward, 5'-GCT
GCTGGAATTGGTGTTGATG-3'; reverse, 5'-AGGTGT TTCTTTGATGCTCTGTCT-3'),
TGFBR2 (forward, 5'-GCA CGTTCAGAAGTCGGATG-3'; reverse, 5'-CTGCACCGT
TGTTGTCAGTG-3'), CD44 (forward, 5'-GCCCAATGCCTT TGATGGAC-3';
reverse, 5'-CCCATGTGAGTGTCTGGT AGC-3'); GAPDH (forward,
5'-AGGTCGGTGTGAACG GATTTG-3'; reverse, 5'-GGGGTCGTTGATGGCAACA-3');
and U6 (forward, 5'-GCTTCGGCAGCACATATACTAA AAT-3'; reverse,
5'-CGCTTCACGAATTTGCGTGTCAT-3'). U6 was used as an internal control
for miR-7 and GAPDH was used as an internal control for mRNA
(Smad2, Smad3, Smad4, TGFBR2 and CD44) quantification.
Dual-luciferase reporter assay
Targeted binding sites of miR-7 and TGFBR2 3'UTR
were predicted through TargetScan website (http://www.targetscan.org/vert_72/) (11). The full-length TGFBR2 3'UTR was
amplified via PCR using PrimeSTAR® (Takara Bio, Inc.)
from human genomic DNA, and the mutant TGFBR2 3'UTR was generated
by Mut Express II Fast Mutagenesis kit V2 (VazymeBiotech Co.,
Ltd.). These DNA fragments were cloned into a psiCHECK™-2 Vector
(Promega Coproration). Plasmids were cut and joined by endonuclease
QuickCut™ NotI, XhoI, and T4 DNA Ligase (Takara Bio,
Inc.). 1x105 MDA-MB-231 cells were seeded in a 24-well
plate and transfected, respectively, with the reporter constructs
and miR-7 mimic (forward, 5'-UGGAAGACU AGUGAUUUUGUUGUU-3'; reverse,
5'-AACAACAAAAUC ACUAGUCUUCCA-3'), miR-7 mimic control (forward,
5'-UU UGUACUACACAAAAGUACUG-3'; reverse, 5'-CAGUAC
UUUUGUGUAGUACAAA-3'), miR-7 inhibitor (5'-AAC
AACAAAAUCACUAGUCUUCCA-3') and miR-7 inhibitor control
(5'-CAGUACUUUUGUGUAGUACAAA-3'; all from Guangzhou RiboBio Co.,
Ltd.) for 48 h. Then, the luciferase reporter assay was performed
using a Dual-Luciferase Reporter System (Promega Corporation).
Western blotting
Approximately 1x106 cells were harvested
and lysed in RIPA lysis buffer (cat. no. P0013B; Beyotime Institute
of Biotechnology), and the lysates were run on a western blot as
previously described (12). The
antibodies used for western blotting included CD44 (1:2,000
dilution; cat. no. 60224-1-Ig), Smad3 (1:2,000 dilution; cat. no.
66516-1-Ig), GAPDH (1:10,000 dilution; cat no. 60004-1-Ig) and
TGFBR2 (1:2,000 dilution; cat. no. 66636-1-Ig) from ProteinTech
Group, Inc. IRDye® 680RD donkey anti-mouse IgG secondary
antibody (1:10,000 dilution; cat. no. P/N 926-68072) was from
LI-COR Biosciences.
ChIP-PCR assays
The JASPAR software for bioinformatics prediction of
transcription factors was used (http://jaspar.genereg.net/). The ChIP assay was
performed according to the Chromatin Immunoprecipitation Kit
Instruction Manual (EZ-ChIP™ cat. no. 17-371; Merck Millipore;
Merck KGaA). The anti-Smad2 + Smad3 antibody (product code
ab207447) was used to precipitate the protein-DNA complexes (Abcam)
(13), and the DNA isolated through
ChIP reactions was subjected to PCR using primers specific to the
promoter of CD44 (forward, 5'-CCCAGATGGAGAAAGCTCTG-3'; reverse,
5'-ACTTGGCTTTCTGTCCTCCA-3').
Expression of lentivirus-infected
cells and siRNA silencing gene
The three-plasmid system consisted of pSPAX2 (10
µg), pMD2G (5 µg), and pHBLV-U6-ZsGreen (15 µg). miR-7 fragment
(245 bp) was inserted into the lentivirus vector and cultured 48-72
h in 2nd generation 293T cells (Shanghai Institute of Biochemistry
and Cell Biology), and the virus solution was obtained
(plaque-forming units/ml=0.033; MOI=1). MDA-MB-231 cells
(1x104) were infected for 4 h with 10 µl lenti-miR-7
virus solution and miR-7 overexpression monoclonal cells were
selected after 5 days. siRNA and miR-7 mimic were synthesized by
the Guangzhou RiboBio Co., Ltd. and used as previously described
(8). A total of 50 nM siRNA/miR-7
mimic/inhibitor and negative controls were used to transfect
cells.
Statistical analysis
SPSS Statistics 21.0 (IBM Corp.) and GraphPad Prism
8.0 (GraphPad Software, Inc.) were used for data analysis and
imaging. Values of interest were presented as the mean ± standard
deviation. Statistical analyses were performed using Tukey's post
hoc test after one-way analysis of variance (ANOVA) for multiple
comparisons, and Spearman's correlation analysis. Values shown are
from one representative experiment. P<0.05 was considered to
indicate a statistically significant difference.
Results
Inhibition of ALDH1A3 decreases CD44
expression
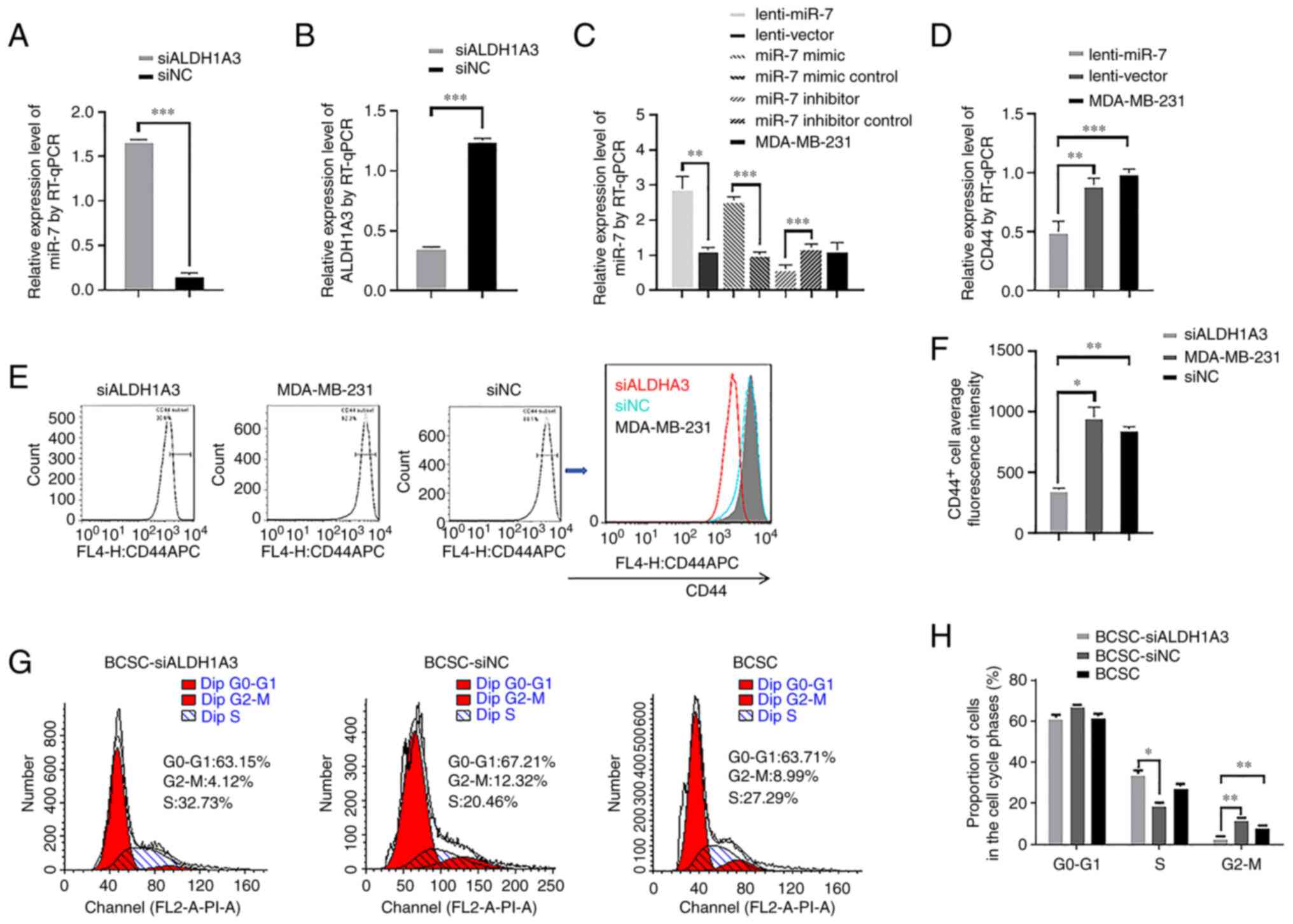
miR-7 and ALDH1A3 expression was detected by RT-qPCR
after knocking down ALDH1A3 with siRNA, and it was revealed that
miR-7 expression was significantly increased (Fig. 1A) and ALDH1A3 expression was
significantly decreased (Fig. 1B).
The results confirmed that not only could miR-7 regulate
ALDH1A3(9) but ALDH1A3 could also
reversely regulate miR-7 expression. To demonstrate transfection
efficiency, lenti-miR-7 and miR-7 mimic were transfected into
MDA-MB-231 cells, and RT-qPCR results revealed that they could
significantly increase the expression of miR-7, while the miR-7
inhibitor could inhibit the expression of miR-7 (Fig. 1C). Using a lentivirus to overexpress
miR-7 (lenti-miR-7) could significantly reduce the expression of
CD44 mRNA (Fig. 1D). Then, the
ratio of CD44+ cells in the MDA-MB-231 cells was
examined by FCM after knocking down ALDH1A3 with siRNA (Fig. 1E). As revealed in Fig. 1F, the ratio of CD44+
cells was significantly decreased compared to the control group. In
order to further evaluate the effect of siALDH1A3 on cell
proliferation, FCM was used to analyze the cell cycle of BCSCs
(Fig. 1G). Cell cycle analysis
revealed that BCSC-siALDH1A3 increased the S phase and reduced the
G2/M phase compared to the BCSC-siNC cell population (Fig. 1H). Collectively, the knockdown of
ALDH1A3 expression reduced the CD44 expression in MDA-MB-231 cells
and their in vitro proliferation in BCSCs.
miR-7 overexpression inhibits TGF-β1
signaling pathway and downregulates CD44 expression
To demonstrate whether miR-7 inhibits the TGF-β1
signaling pathway, lenti-miR-7 and lentivector cells were treated
with 10 ng/ml TGF-β1 and 100 ng/ml TGF-β1 type I receptor
antagonist SB431542. RT-qPCR results revealed that miR-7 inhibited
the effect of TGF-β1-upregulation of CD44. However, SB431542 could
enhance the effect of miR-7 concomitantly by inhibiting TGF-β1,
compared with the control group (Fig.
2A). Next, RT-qPCR was used to evaluate the effect of miR-7 on
the main signaling molecules of the TGF-β1 signaling pathway. As
revealed in Fig. 2B, miR-7
downregulated the expression levels of Smad2, Smad3, and Smad4. In
addition, as indicated in Fig. 2C,
miR-7 also downregulated the expression of TGFBR2. To demonstrate
the targeted regulatory relationship between miR-7 and TGFBR2, the
targeted binding sites of miR-7 and TGFBR2 3'UTR were predicted
through the bioinformatics website (Fig. 2D). For this reason, the MDA-MB-231
cells were transfected with psiCHECK-2-TGFBR2 and
psiCHECK-2-TGFBR2-Mut dual-luciferase recombinant plasmids using
Lipofectamine 2000, respectively. As revealed in Fig. 2E, the luciferase activity of the
dual-luciferase recombinant plasmid was not altered in the TGFBR2
3'UTR mutation group, while the luciferase activity of the
wild-type dual-luciferase recombinant plasmid exhibited a
significant decrease, indicating that miR-7 mimic could effectively
bind to TGFBR2 3'UTR and reduce the relative luciferase activity of
the wild-type vector. Western blot results further demonstrated
that miR-7 downregulated TGFBR2, Smad3, and CD44 (Fig. 2F). Furthermore, siALDH1A3 and
lenti-miR-7 were transfected with Lipofectamine 2000 separately to
verify the expression of TGFBR2 in the differently treated BCSCs.
Compared to the control (BCSC-lentivector), the expression of
TGFBR2 in BCSC-lenti-miR-7 group was decreased (Fig. 2G). Finally, the ChIP-PCR assay was
performed. Bioinformatics predicted that the transcription factor
Smad3 binds to the upstream region of the CD44 gene promoter
(14,15). Immunoprecipitation was performed
with the Smad3 antibody, and DNA fragments were eluted from the
immunoprecipitation complex. The identification of a positive
amplification product indicated that Smad3, a transcription factor,
could regulate gene expression by binding the upstream region of
the CD44 promoter (Fig. 2H).
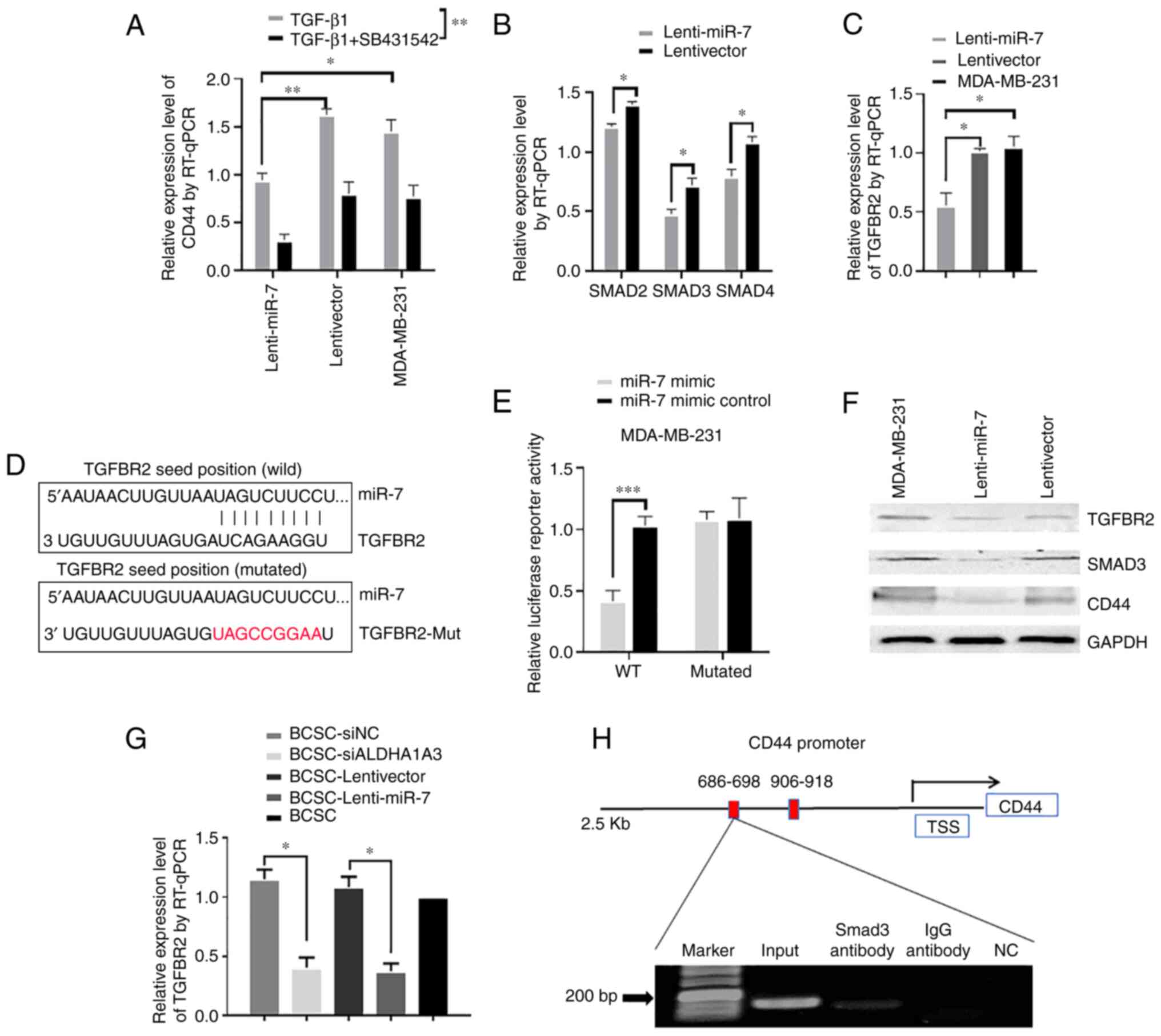 | Figure 2miR-7 directly targets TGFBR2 and
Smad3 binds to the CD44 promoter region. (A) Lenti-miR-7 and
lentivector MDA-MB-231 cells were respectively incubated with
TGF-β1 and TGF-β1 + SB431542, and then CD44 mRNA relative
expression levels were measured by RT-qPCR. (B) The relative
expression levels of Smad2, Smad3, and Smad4 were measured by
RT-qPCR in lenti-miR-7 and lentivector MDA-MB-231 cells. (C) TGFBR2
mRNA relative expression levels were measured by RT-qPCR. (D)
TGFBR2 3'UTR with the miR-7 binding site was predicted, and
complementary sequences of miR-7 to TGFBR2 3'UTR mutated are
presented in red. (E) Cells were harvested and luciferase
activities were measured after 48-h transfection. (F) The
expression levels of TGFBR2, Smad3, and CD44 were analyzed by
western blotting. (G) TGFBR2 mRNA relative expression levels were
measured by RT-qPCR in BCSC-siALDH1A3, BCSC-lenti-miR-7, and
control cells. (H) ChIP-PCR assay in MDA-MB-231 cells.
*P<0.05, **P<0.01 and
***P<0.001. miR-7, microRNA-7; TGFBR2, transforming
growth factor-β receptor 2; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; BCSC, breast
cancer stem cell; si, small interfering. |
Demonstration of
miR-7-TGFBR2-Smad3-CD44 axis
It was further evaluated whether there is a
miR-7-TGFBR2-Smad3-CD44 axis in both breast cancer cell lines and
breast cancer surgical specimens. First, surgical tissues were
obtained from 12 breast cancer patients and RT-qPCR was used to
detect the expression levels of miR-7, Smad3, CD44 and TGFBR2.
RT-qPCR results revealed that the relative expression level of
miR-7 was expressed at a lower level in breast cancer tissues
compared with adjacent noncancerous tissues, while the relative
expression levels of TGFBR2, CD44, and Smad3 were significantly
higher in breast cancer tissues than in adjacent noncancerous
tissues, as revealed in Fig. 3A. It
was determined that miR-7 was negatively correlated with TGFBR2
(Fig. 3B), CD44 (Fig. 3E), and Smad3 (Fig. 3C). Smad3 was positively correlated
with CD44 (Fig. 3D). Second, the
relative expression levels of TGFBR2, Smad3, and CD44 were
concurrently analyzed in breast cancer cell lines SK-BR-3, MCF-7,
and LD using RT-qPCR after miR-7 mimic transfection. As revealed in
Fig. 4A-C, the relative expression
levels of TGFBR2, Smad3, and CD44 were all downregulated in the
miR-7-mimic-transfected cells. In contrast, the mRNA expression
levels of TGFBR2, Smad3, and CD44 were all upregulated after miR-7
inhibitor transfection (Fig. 4D-F).
These results suggested that the miR-7-TGFBR2-Smad3-CD44 axis
exists objectively in both breast cancer cell lines and breast
cancer surgical specimens.
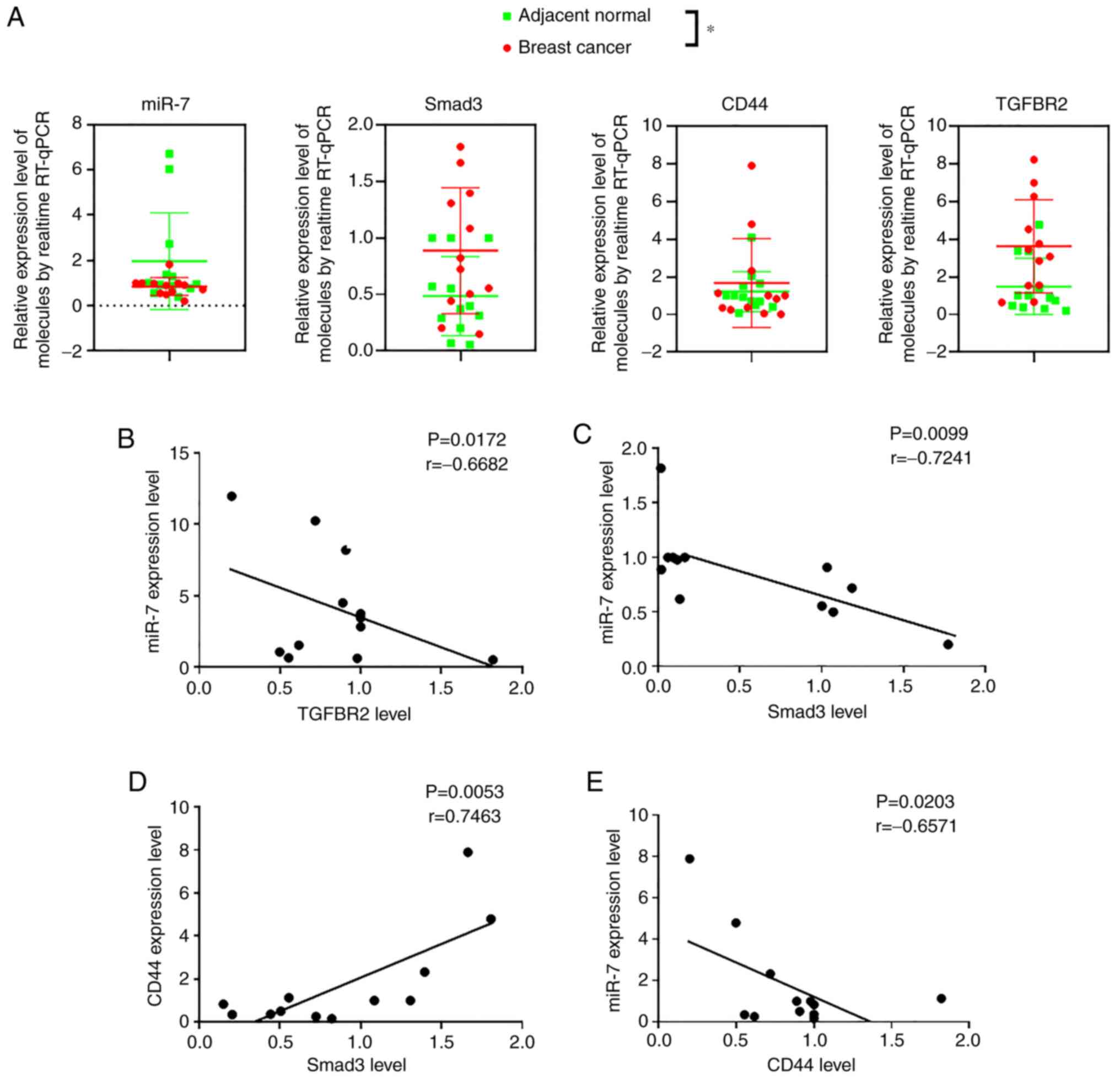 | Figure 3Detection of miR-7 and BCSC-related
molecular expression in breast cancer surgical specimens. (A)
Relative expression levels of miR-7, Smad3, CD44, and TGFBR2 in
breast cancer postsurgery samples analyzed by RT-qPCR. (B-E)
Relative expression levels of miR-7 and TGFBR2, miR-7 and Smad3,
Smad3 and CD44, and miR-7 and CD44, respectively. The green points
represent adjacent noncancerous tissues; the red points represent
tumor tissues (n=12). *P<0.05. miR-7, microRNA-7;
BCSC, breast cancer stem cell; TGFBR2, transforming growth factor-β
receptor 2; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction. |
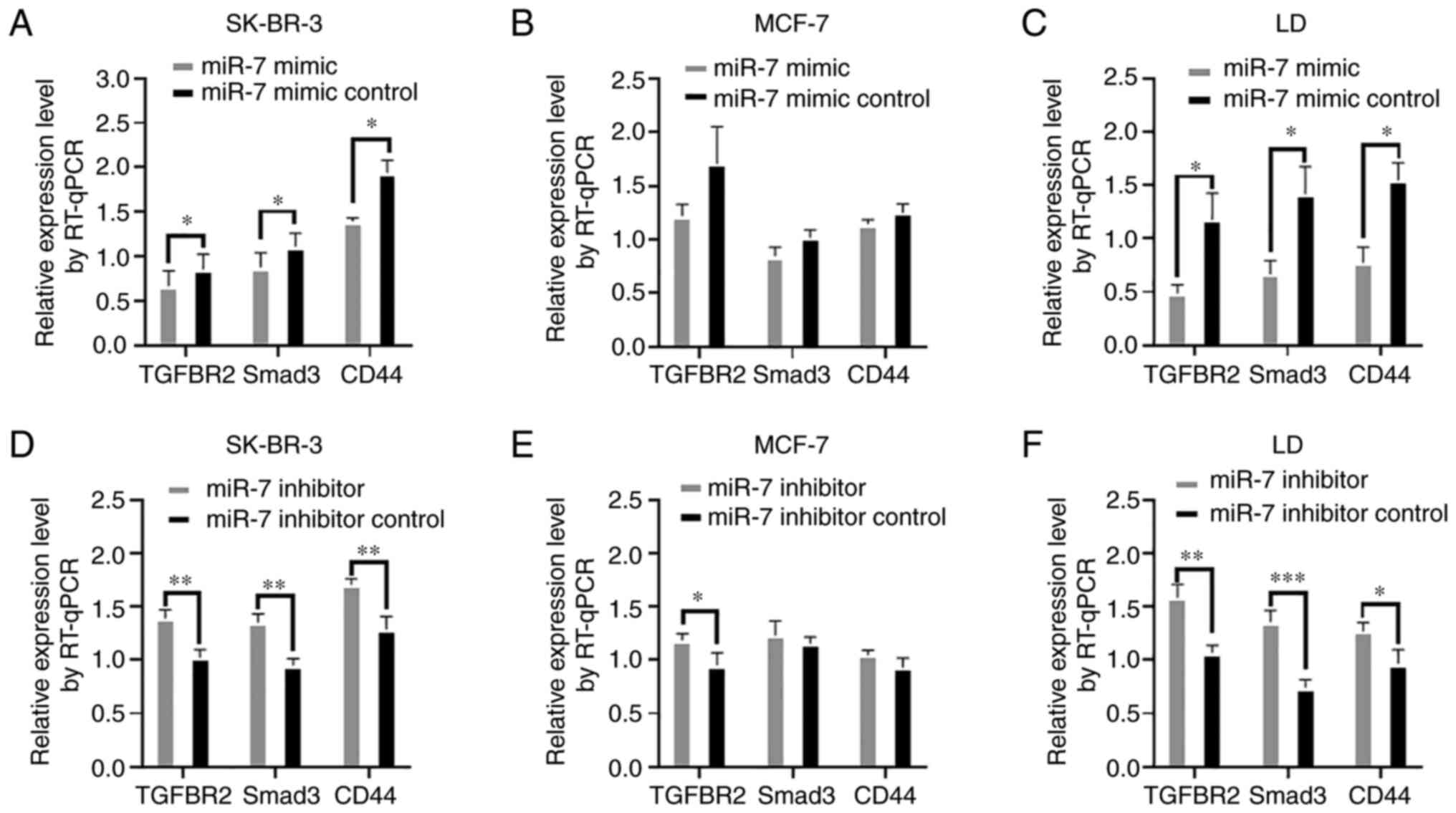 | Figure 4Detection of the molecular expression
in SK-BR-3, MCF-7, and LD breast cancer cell lines. (A-C) TGFBR2,
Smad3, and CD44 expression levels in SK-BR-3, MCF-7, and LD cells
were measured by RT-qPCR analysis after miR-7 mimic transfection.
(D-F) TGFBR2, Smad3, and CD44 relative expression levels in
SK-BR-3, MCF-7, and LD cells were detected after the inhibitor
transfection by RT-qPCR. *P<0.05,
**P<0.01 and ***P<0.001. TGFBR2,
transforming growth factor-β receptor 2; RT-qPCR, reverse
transcription-quantitative polymerase chainreaction; miR-7,
microRNA-7. |
Discussion
The previous results of our research group revealed
that the overexpression of miR-7 caused the downregulation of
ALDH1A3 and CD44(9). However, how
miR-7 downregulates CD44 and what role ALDH1A3 plays in impacting
CD44 expression was not elucidated. Therefore, the molecular
mechanism that affects the expression of CD44 was explored by
regulating ALDH1A3 in the present study.
ALDH1A3 is the major isozyme that contributes to
ALDH enzyme activities in 58 human cancer cell lines. In breast
cancer, glioma, and melanoma, ALDH1A3 expression is regulated by
several mechanisms at epigenetic, transcriptional, and
posttranslational levels (16-18).
Attenuation of ALDH1A3 expression by RNA interference (RNAi)
significantly suppressed cell proliferation, reduced the number of
cancer cells that persisted after anticancer drug treatment, and
interfered with tumor growth in a mouse xenograft model (17). Our previous study (9) revealed that miR-7 expression could
downregulate ALDH1A3 expression. RT-qPCR experiments revealed that
miR-7 expression was upregulated by downregulating ALDH1A3
expression using siRNA. These experimental results suggested that
there is a mutual regulation between miR-7 and ALDH1A3. To further
understand and reveal the relationship between miR-7 and ALDH1A3,
further investigation was carried out.
The cell surface protein CD44 has been widely used
as a CSC marker in breast cancer and various other types of cancers
(19,20). CD44 is important for tumor
initiation in vivo and predominantly expressed in metastatic breast
cancer cells. Previous research results have also revealed that in
these metastatic breast cancer cell lines, knockdown of CD44
significantly inhibited breast cancer metastasis (21,22).
Therefore, it is of great significance to explore the mechanism of
miR-7 inhibition of CD44 and reduce the tumorigenicity of
BCSCs.
To better understand the relationship between miR-7
and CD44, a bioinformatics approach was used to predict whether
miR-7 regulates CD44 cell surface expression via the TGF-β1
signaling pathway. Therefore, confirmation that the TGF-β1
signaling pathway is regulated by miR-7 and affects CD44 gene
expression was first required. TGF-β1 (10 ng/ml) and TGF-β1 type I
receptor antagonist SB431542 (100 ng/ml) were used to treat
lenti-miR-7 and lentivector cells by Lipofectamine 2000,
respectively (23). After 72 h, it
was revealed that TGF-β1 could significantly increase the
intracellular CD44 mRNA expression. On this basis, the inhibition
of TGF-β1 by SB431542 could significantly reduce CD44 mRNA
expression in MDA-MB-231 cells. Compared with lentivector cells,
the expression of CD44 mRNA was lower in the case of miR-7
overexpression in lenti-miR-7 cells. The aforementioned results
revealed that the TGF-β1 signaling pathway was involved in the
regulation of CD44. Next, the expression levels of Smad2, Smad3,
and Smad4 in lenti-miR-7 cells were examined. It was revealed that,
in addition to the inhibitory factor, Smad2, Smad3, and Smad4 all
exhibited decreased mRNA expression. This further suggested that
TGF-β1 signaling was regulated by miR-7 overexpression.
To explain how miR-7 inhibits the TGF-β1 signaling
pathway, bioinformatics were used to predict whether miR-7 has
binding targets for TGFBR2 3'UTR. It is well known that TGF-β
ligands assemble their corresponding receptors that contain two
type 1 components and two type 2 components. Type 2 receptors serve
as activators to phosphorylate type I receptors, whereas type 1
receptors function as propagators to transduce the downstream
signal to cytoplasmic proteins (24). The components of both receptors are
serine/threonine kinases. TGF-β type I receptors and activin type 1
receptors phosphorylate SMAD2/3(25). The expression of TGFBR2 by RT-qPCR
was first detected and it was revealed that, in the case of miR-7
overexpression, TGFBR2 mRNA expression was downregulated. The
results of RT-qPCR were further confirmed by western blotting,
suggesting miR-7 affects TGFBR2 expression.
The psiCHECK-2-TGFBR2 and psiCHECK-2-TGFBR2-Mut
dual-luciferase reporters were constructed, respectively. The
psiCHECK™-2 vector was designed by Promega Coproration to provide a
quantitative and rapid approach for the optimization of RNAi. The
vectors enable the monitoring of changes in the expression of a
target gene fused to the reporter gene, containing as the primary
reporter gene the synthetic version of Renilla luciferase,
hRluc (26). This synthetic gene is
engineered for more efficient expression in mammalian cells and for
reduced anomalous transcription (27). After transfecting miR-7 mimic and
dual-luciferase reporters into MDA-MB-231 cells, the luciferase
activity of wild-type cells significantly decreased, while the
luciferase activity of mutant cells was not significantly altered,
indicating that miR-7 can bind to TGFBR2 3'UTR. Concurrently, the
mRNA expression of TGFBR2 in BCSCs that were isolated from
MDA-MB-231 cells according to the phenotypes of
CD44+CD24-ESA+ BCSCs was also
observed (26). The cDNA products
of these BCSCs had also been used in a previously published study
(9). The results in the present
study revealed that the mRNA expression of TGFBR2 in BCSC in the
siALDH1A3 and lenti-miR-7 groups was visibly lower than that in the
control group.
Genome-wide identification of transcription factor
binding sites (TFBSs) is key to understanding transcriptional
regulation. The genomic locations where transcription factors bind
to DNA are typically short (6-20 bp) and exhibit sequence
variability (28). The DNA sequence
of the 2,500-bp region upstream of the CD44 promoter was obtained,
Smad3 was used as a transcription factor for motif binding
prediction, and the TFBSs for research based on the motif
conservation score were selected (14). The ChIP-PCR analysis strongly
revealed that the Smad3 protein binds to the 686-698 position
upstream of the CD44 promoter. The results demonstrated that miR-7
affects the TGF-β1 signaling pathway molecule Smad3 by
downregulating TGFBR2 and then inhibits CD44 gene transcription.
Collectively, the present study identified miR-7-TGFBR2-Smad3-CD44
as a regulatory axis of BCSC marker CD44 expression.
RT-qPCR experiments were performed on cancer tissues
and adjacent tissues from 12 clinical breast cancer surgical
samples. It was revealed that, compared with adjacent tissues,
miR-7 in cancer tissues exhibited low expression, and TGFBR2,
Smad3, and CD44 exhibited high expression. These molecular
relationships were analyzed, and it was determined that miR-7 was
negatively correlated with TGFBR2, Smad3 and CD44 respectively, and
that CD44 and Smad3 were positively correlated. Then, SK-BR-3,
MCF-7, and LD breast cancer cell lines were used to analyze the
effects of miR-7 on the miR-7-TGFBR2-Smad3-CD44 axis. In the case
of miR-7 overexpression, TGFBR2, Smad3, and CD44 were all
downregulated; while by inhibiting miR-7 expression, all three
molecules, TGFBR2, Smad3, and CD44 were upregulated in the breast
cancer cell lines.
Collectively, the present findings revealed that the
downregulation of ALDH1A3 can upregulate miR-7 and reduce the ratio
of CD44+ cells in breast cancer cells via the
miR-7-TGFBR2-Smad3-CD44 axis. Notably, these findings have
potential clinical importance for understanding the multiple
regulatory roles of miR-7. Inhibiting ALDH1A3 and/or miR-7
overexpression may be an important method for treating breast
cancer.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81572887), and partly
supported by the National Key Research and Development Program of
China (grant no. 2017YFA0205502), the Scientific Research
Foundation of Graduate School of Southeast University (grant no.
YBJJ1849), and the Postgraduate Research & Practice Innovation
Program of Jiangsu Province (grant no. KYCX18_0165).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JD designed the experiments and MP, ML, MG, HZ
carried out the experiments, data analysis and manuscript writing.
HX, FZ, FM and RX performed reverse transcription PCR, western
blotting and flow cytometry assays. MP and ML performed ChIP-PCR
and dual-luciferase assays. JD interpreted the data, edited and
corrected the manuscript and approved the final version to be
published. JD and MP confirmed the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The investigation was approved by the Ethics
Committee at Southeast University School of Medicine (Nanjing,
China), and informed consent for the use of the postsurgery samples
was obtained from the donors who were breast cancer patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gonzalez-Angulo AM, Morales-Vasquez F and
Hortobagyi GN: Overview of resistance to systemic therapy in
patients with breast cancer. Adv Exp Med Biol. 608:1–22.
2007.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Dou J, Li Y, Zhao F, Hu W, Wen P, Tang Q,
Chu L, Wang Y, Cao M, Jiang C and Gu N: Identification of tumor
stem-like cells in a mouse myeloma cell line. Cell Mol Biol
(Noisy-le-grand). 55 (Suppl):OL1151–OL1160. 2009.PubMed/NCBI
|
|
4
|
Bravo-Egana V, Rosero S, Molano RD,
Pileggi A, Ricordi C, Domínguez-Bendala J and Pastori RL:
Quantitative differential expression analysis reveals miR-7 as
major islet microRNA. Biochem Biophys Res Commun. 366:922–926.
2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Junn E, Lee KW, Jeong BS, Chan TW, Im JY
and Mouradian MM: Repression of alpha-synuclein expression and
toxicity by microRNA-7. Proc Natl Acad Sci USA. 106:13052–13057.
2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li X and Carthew RW: A microRNA mediates
EGF receptor signaling and promotes photoreceptor differentiation
in the Drosophila eye. Cell. 123:1267–1277. 2005.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhang H, Cai K, Wang J, Wang X, Cheng K,
Shi F, Jiang L, Zhang Y and Dou J: miR-7, inhibited indirectly by
lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of
breast cancer stem cells by downregulating the STAT3 pathway. Stem
Cells. 32:2858–2868. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Li M, Pan M, You C, Zhao F, Wu D, Guo M,
Xu H, Shi F, Zheng D and Dou J: miR-7 reduces the BCSC subset by
inhibiting XIST to modulate the miR-92b/Slug/ESA axis and inhibit
tumor growth. Breast Cancer Res. 22(26)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pan M, Li M, You C, Zhao F, Guo M, Xu H,
Li L, Wang L and Dou J: Inhibition of breast cancer growth via
miR-7 suppressing ALDH1A3 activity concomitant with decreasing
breast cancer stem cell subpopulation. J Cell Physiol.
235:1405–1416. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mon-López D and Tejero-González CM:
Validity and reliability of the TargetScan ISSF Pistol & Rifle
application for measuring shooting performance. Scand J Med Sci
Sports. 29:1707–1712. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Dou J, Wang Y, Yu F, Yang H, Wang J, He X,
Xu W, Chen J and Hu K: Protection against Mycobacterium
tuberculosis challenge in mice by DNA vaccine
Ag85A-ESAT-6-IL-21 priming and BCG boosting. Int J Immunogenet.
39:183–190. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cibi DM, Mia MM, Guna Shekeran S, Yun LS,
Sandireddy R, Gupta P, Hota M, Sun L, Ghosh S and Singh MK: Neural
crest-specific deletion of Rbfox2 in mice leads to craniofacial
abnormalities including cleft palate. Elife.
8(e45418)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fornes O, Castro-Mondragon JA, Khan A, van
der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M,
Baranašić D, et al: JASPAR 2020: Update of the open-access database
of transcription factor binding profiles. Nucleic Acids Res. 48
(D1):D87–D92. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Khan A, Fornes O, Stigliani A, Gheorghe M,
Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni
SR, Tan G, et al: JASPAR 2018: Update of the open-access database
of transcription factor binding profiles and its web framework.
Nucleic Acids Res. 46 (D1):D260–D266. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kawakami R, Mashima T, Kawata N, Kumagai
K, Migita T, Sano T, Mizunuma N, Yamaguchi K and Seimiya H:
ALDH1A3-mTOR axis as a therapeutic target for anticancer
drug-tolerant persister cells in gastric cancer. Cancer Sci.
111:962–973. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yamashita D, Minata M, Ibrahim AN,
Yamaguchi S, Coviello V, Bernstock JD, Harada S, Cerione RA,
Tannous BA, La Motta C, et al: Identification of ALDH1A3 as a
viable therapeutic target in breast cancer metastasis-initiating
cells. Mol Cancer Ther. 19:1134–1147. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang S, Zhou X, Liang C, Bao M, Tian Y,
Zhu J, Zhang T, Yang J and Wang Z: ALDH1A3 serves as a predictor
for castration resistance in prostate cancer patients. BMC Cancer.
20(387)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003.PubMed/NCBI View Article : Google Scholar : Erratum in: Proc
Natl Acad Sci USA 100: 6890, 2003.
|
|
20
|
Fillmore C and Kuperwasser C: Human breast
cancer stem cell markers CD44 and CD24: Enriching for cells with
functional properties in mice or in man? Breast Cancer Res.
9(303)2007.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Zhao P, Xu Y, Wei Y, Qiu Q, Chew TL, Kang
Y and Cheng C: The CD44s splice isoform is a central mediator for
invadopodia activity. J Cell Sci. 129:1355–1365. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang H, Brown RL, Wei Y, Zhao P, Liu S,
Liu X, Deng Y, Hu X, Zhang J, Gao XD, et al: CD44 splice isoform
switching determines breast cancer stem cell state. Genes Dev.
33:166–179. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sun C, Sun L, Jiang K, Gao DM, Kang XN,
Wang C, Zhang S, Huang S, Qin X, Li Y, et al: NANOG promotes liver
cancer cell invasion by inducing epithelial-mesenchymal transition
through NODAL/SMAD3 signaling pathway. Int J Biochem Cell Biol.
45:1099–1108. 2013.PubMed/NCBI View Article : Google Scholar : Erratum in: Int J
Biochem Cell Biol 105: 144, 2018.
|
|
24
|
Itatani Y, Kawada K and Sakai Y:
Transforming growth factor-β signaling pathway in colorectal cancer
and its tumor microenvironment. Int J Mol Sci.
20(E5822)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Vitiello GAF, Amarante MK, Banin-Hirata
BK, Campos CZ, de Oliveira KB, Losi-Guembarovski R and Watanabe
MAE: Transforming growth factor beta receptor II (TGFBR2) promoter
region polymorphism in Brazilian breast cancer patients:
Association with susceptibility, clinicopathological features, and
interaction with TGFB1 haplotypes. Breast Cancer Res Treat.
178:207–219. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kumar R, Conklin DS and Mittal V:
High-throughput selection of effective RNAi probes for gene
silencing. Genome Res. 13:2333–2340. 2003.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhao L and Zhang Y and Zhang Y: Long
noncoding RNA CASC2 regulates hepatocellular carcinoma cell
oncogenesis through miR-362-5p/Nf-κB axis. J Cell Physiol.
233:6661–6670. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha
V, Lindblad-Toh K, Lander ES and Kellis M: Systematic discovery of
regulatory motifs in human promoters and 3' UTRs by comparison of
several mammals. Nature. 434:338–345. 2005.PubMed/NCBI View Article : Google Scholar
|