Introduction
Endothelial cells are highly plastic cells that may
rapidly switch to an activated state to mediate angiogenesis during
wound healing (1,2). Formed blood vessels supply oxygen and
nutrients, remove wastes and contribute to the regeneration of
various tissues and organs (3).
Considering the importance of blood supply, vascularization is
regarded as an essential factor during the fabrication and
construction of tissue-engineered products (4-6).
Angiogenesis is also critical for the regeneration
of injured peripheral nerves. Following peripheral nerve injury,
polarized blood vessel formation within the bridge guides the
migration of Schwann cells towards and across the injured site and
encourages axon elongation and nerve repair (7). In tissue-engineered nerve grafts,
sufficient angiogenesis benefits cell survival, promotes cell
integration with biomaterials, provides a favorable nutritional
microenvironment and improves the regeneration and functional
recovery of injured nerves (8,9).
Therefore, investigating certain approaches to modulate the
phenotype of endothelial cells and to advance angiogenesis after
peripheral nerve injury is important in theoretical and practical
aspects.
MicroRNAs (miRNAs/miRs) are small single-strand
non-coding RNAs of ~22 nucleotides in length. miRNAs bind to their
target mRNAs, induce mRNA degradation and/or translational
repression and negatively regulate their target mRNAs (10,11). A
large number of miRNAs were identified to be differentially
expressed in the injured peripheral nerve stumps after nerve injury
(12). Differentially expressed
miRNAs may affect the proliferation, migration and/or remyelination
of Schwann cells and regulate the nerve regeneration process
(13). However, in spite of the
essential roles of endothelial cells during peripheral nerve
regeneration, the regulatory effects of miRNAs on endothelial
cells, as compared with Schwann cells, remain largely elusive.
miR-328 has been involved in numerous biological
activities, such as cellular apoptosis (14), brown adipose tissue differentiation
(15) and glucose uptake (16). Emerging studies have associated
miR-328 with numerous diseases, including leukemic blasts (17), atrial fibrillation (18,19),
intervertebral disc degeneration (20) and hepatocellular carcinoma (21). Dysregulated miR-328 was also
observed in prion-induced neurodegeneration (22), implying that miR-328 may have
important roles in nervous system diseases. Furthermore, miRNA
sequencing data of rat sciatic nerve stumps after nerve transection
indicated that miR-328 expression was altered after peripheral
nerve injury (12), suggesting that
miR-328 may be essential for peripheral nerve injury and
regeneration.
miR-328a-3p was previously named as miR-328 or
miR-328a according to miRbase (http://mirbase.org/). In the present study, the
effects of miR-328a-3p on the biological behavior of human
umbilical vein endothelial cells (HUVECs) were examined. Potential
upstream long non-coding RNAs (lncRNAs) and downstream target mRNAs
of miR-328a-3p were bioinformatically analyzed to reveal the
regulatory cascade of miR-328a-3p after peripheral nerve
injury.
Materials and methods
Cell culture
HUVECs, a gift from Dr Mi Shen's laboratory at the
Co-Innovation Center of Neuroregeneration, Nantong University
(Nantong, China), were seeded into cell culture flasks (Corning
Inc.) and cultured in endothelial cell medium (ECM; ScienCell
Research Laboratories, Inc.) containing 5% fetal bovine serum (FBS;
ScienCell Research Laboratories, Inc.), 1% endothelial cell growth
supplement (ECGS; ScienCell Research Laboratories, Inc.) and 1%
penicillin and streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.) in a humidified atmosphere with 5% CO2 at 37˚C.
The cell culture medium was refreshed every other day.
Cell transfection
Cultured HUVECs were transfected with chemically
synthesized miR-328a-3p mimics, mimics control, miR-328a-3p
inhibitor or inhibitor control sequences (Guangzhou RiboBio Co.,
Ltd.) using Lipofectamine® RNAiMAX transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h at 37˚C
according to the manufacturer's protocol. The sequences of
miR-328a-3p mimics were 5'-CUGGCCCUCUCUGCCCUUCCGU-3' and
3'-ACGGAAGGGCAGAGAGGGCCAG-5', the sequence of miR-328a-3p inhibitor
was 5'-ACGGAAGGGCAGAGAGGGCCAG-3', the sequences of mimics control
were 5'-UUUGUACUACACAAAAGUACUG-3' and 3'-AAACAUGAUGUGUUUUCAUGAC-5',
and the sequence of inhibitor control was
5'-CAGUACUUUUGUGUAGUACAAA-3'. HUVECs were transfected with 50 nM
miRNA mimics and 250 nM miRNA inhibitor prior to functional
investigations.
Animal surgery and tissue
preparation
A total of 20 6- to 8-week adult male Sprague-Dawley
rats, weighing 180-220 g, were obtained from the Experimental
Animal Center of Nantong University. The rats were maintained in
standard laboratory conditions (23±2˚C room temperature, 55±5%
relative humidity and a 12-h light/dark cycle) with free access to
food and water. All rats were anesthetized by intraperitoneal
injection of complex narcotics (85 mg/kg of trichloroacetaldehyde
monohydrate, 42 mg/kg of magnesium sulfate and 17 mg/kg of sodium
pentobarbital) (23). The rat
sciatic nerve was exposed through an incision on the lateral aspect
of the mid-thigh of the left hind limb. A 3 mm long segment of
sciatic nerve was crushed two times (15 sec each time, 3-sec
interval) using hemostatic forceps. To minimize the discomfort and
possible painful mechanical stimulation, the rats were housed in
large cages with sawdust bedding post-surgery. Under anesthesia
with compound anesthetic, the previously crushed sciatic nerve with
both nerve ends (1 mm long) was harvested at 0 h and on days 1, 4,
7 and 14 after nerve crush and animals were then immediately
euthanized by cervical dislocation. All experimental and animal
handling procedures were performed according to the Institutional
Animal Care Guidelines of Nantong University and all animal
experiments were ethically approved by the Administration Committee
of Experimental Animals, Nantong University (Nantong, China).
RNA isolation and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted using TRIzol®
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The contaminating DNA was removed using RNeasy spin
columns (Qiagen GmbH). The quality of the isolated RNA samples was
evaluated using an Agilent Bioanalyzer 2100 (Agilent Technologies,
Inc.) and the quantity of RNA samples was determined using a
NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific,
Inc.). To determine miR-328a-3p expression, a total amount of 20 ng
RNAs was reversely transcribed using a TaqMan® MicroRNA
Reverse Transcription kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and subjected to RNA amplification using the
QuantiNova SYBR Green PCR kit (Qiagen GmbH) on an Applied
Biosystems StepOne real-time PCR system. Bulge-loop miRNA qRT-PCR
Primer Sets (one RT primer and a pair of qPCR primers for each set)
specific for miR-328a-3p were designed by Guangzhou RiboBio Co.,
Ltd. RT-qPCR was performed using SYBR Green Premix Ex Taq (Takara
Bio, Inc.) with primers of lncRNAs (Table S1) on an Applied Biosystems StepOne
real-time PCR system to evaluate the expression abundances of the
lncRNAs. The thermocycling program was as follows: 10 min at 95˚C;
40 cycles of 2 sec at 95˚C; 20 sec at 60˚C and 10 sec at 70˚C
(24). The abundance of miR-328a-3p
in HUVECs was compared with the internal control U6 and
quantification of miR-328a-3p was performed using the
2-ΔΔCq method (23).
5-Ethynyl-2'-deoxyuridine (EdU)
proliferation assay
A total of 4x103 HUVECs were resuspended
in 100 µl culture medium and seeded onto poly-L-lysine-coated
96-well plates at a density of 4x104 cells/ml. HUVECs
were exposed to 100 µM EdU for 2 h and then fixed with 4%
paraformaldehyde at 25˚C for 15 min. Cell proliferation was
determined by using a Cell-Light EdU DNA Cell Proliferation Kit
(Guangzhou RiboBio Co., Ltd.) following the manufacturer's
protocol. Images were acquired using a DMR fluorescence microscope
(Leica Microsystems). The numbers of EdU-positive cells and total
cells were determined by Apollo 567 fluorescent dyes and Hoechst
33342 staining. The proliferation rates of HUVECs were calculated
by dividing the numbers of EdU-positive cells by the numbers of
total cells.
Wound-healing assay
HUVECs were seeded onto a mold chamber with a 1 mm
wide insert placed on the bottom of a 6-well plate and grown in ECM
medium containing 2.5% FBS, 1% ECGS and 1% penicillin and
streptomycin. After the cells had grown confluent, the insert was
removed, leaving a blank space. Images were acquired using a DMR
inverted microscope (Leica Microsystems) at 0 and 12 h after
removal of the placed insert. The relative gap closure was measured
at these time-points using Image-Pro Plus 6.0 (Media Cybernetics,
Inc.) from randomly selected image fields.
Transwell migration assay
A total of 1x104 HUVECs were resuspended
in 100 µl ECM medium and seeded onto the upper compartment of a 6.5
mm Transwell chamber with 8 µm pores (Costar; Corning Inc.) at a
density of 1x105 cells/ml. A total of 500 µl cell
culture medium (ECM medium containing 2.5% FBS) was added to the
lower compartment of the Transwell chamber and the HUVECs were
cultured for 24 h. The upper surface of the upper chamber was wiped
with a cotton swab and the bottom surface of the upper chamber was
stained with 0.1% crystal violet at 25˚C for 15 min. Images were
acquired using a DMR inverted microscope (Leica Microsystems). The
crystal violet-stained cells were dissolved in 33% acetic acid and
the optical densities of the solutions were measured at 570 nm
(Agilent Technologies, Inc.).
Matrigel tubulogenesis assay
HUVECs were seeded onto cell culture plates
pre-coated with Matrigel® (BD Biosciences) and incubated
for 6 h to allow the formation of tubules. Images were acquired
using a DMR inverted microscope (Leica Microsystems). The numbers
of nodes, meshes and branches in the formed tubules were
quantitated using the Angiogensis analyzer in ImageJ software
v1.8.0 (National Institutes of Health) (25).
Bioinformatics analysis
Upstream regulatory lncRNAs of miR-328a-3p were
predicted using TargetScan (http://www.targetscan.org/vert_71/). Downstream target
mRNAs of miR-328a-3p were predicted using miRWalk 3.0 (http://mirwalk.umm.uni-heidelberg.de/),
miRanda (http://www.microrna.org/microrna/home.do) and the
miRNA Target Prediction Database (miRdb; http://mirdb.org/). Upstream lncRNAs and downstream
mRNAs were linked to construct the miR-328a-3p-centered competing
endogenous RNA (ceRNA). Heatmaps of lncRNAs and mRNAs were
generated using meV software 4.9.0 (http://www.tm4.org/) according to previously obtained
sequencing data of sciatic nerve stumps at 0, 1, 4, 7 and 14 days
after nerve crush injury was described in the aforementioned
protocol (26). Sequencing data
were deposited in the National Center for Biotechnology Information
database with the accession number PRJNA394957 (SRP113121)
(27).
Statistical analysis
An unpaired Student's t-test was applied for
comparisons of data between two groups. One-way ANOVA was applied
for comparisons of data among multiple groups. The calculation of
P-values and the generation of graphs were performed with SigmaPlot
v 14.0 (Systat Software, Inc.).
Results
miR-328a-3p inhibits the proliferation
of HUVECs
HUVECs were cultured and transfected with the mimics
or inhibitor of miR-328a-3p to examine the effects of miR-328a-3p
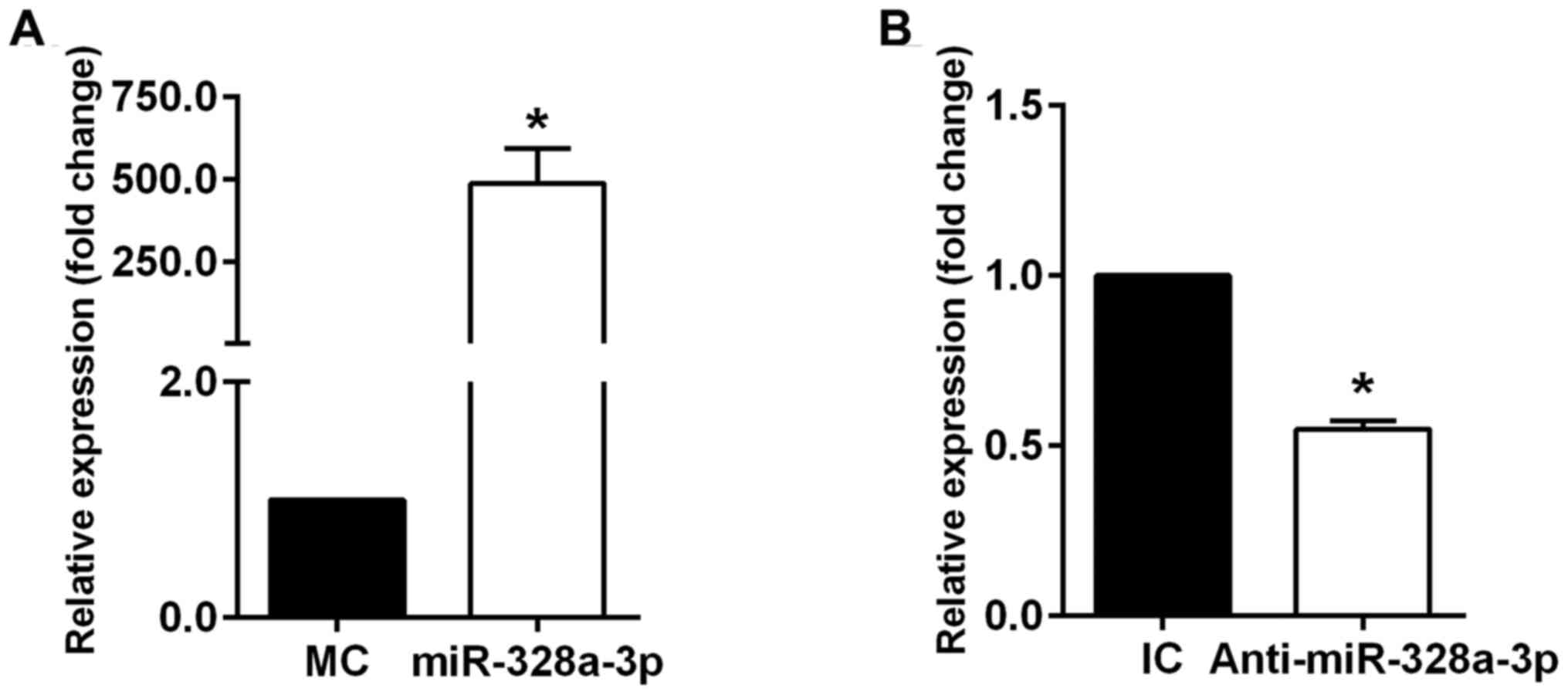
on endothelial cells. Efficient up- or downregulation of
miR-328a-3p in cultured HUVECs with miR-328a-3p mimics or inhibitor
was confirmed by RT-qPCR analysis. Transfection of HUVECs with
miR-328a-3p mimics significantly increased miR-328a-3p expression
(Fig. 1A), while transfection of
HUVECs with miR-328a-3p inhibitor significantly decreased
miR-328a-3p expression (Fig. 1B) as
compared with that in their respective negative controls.
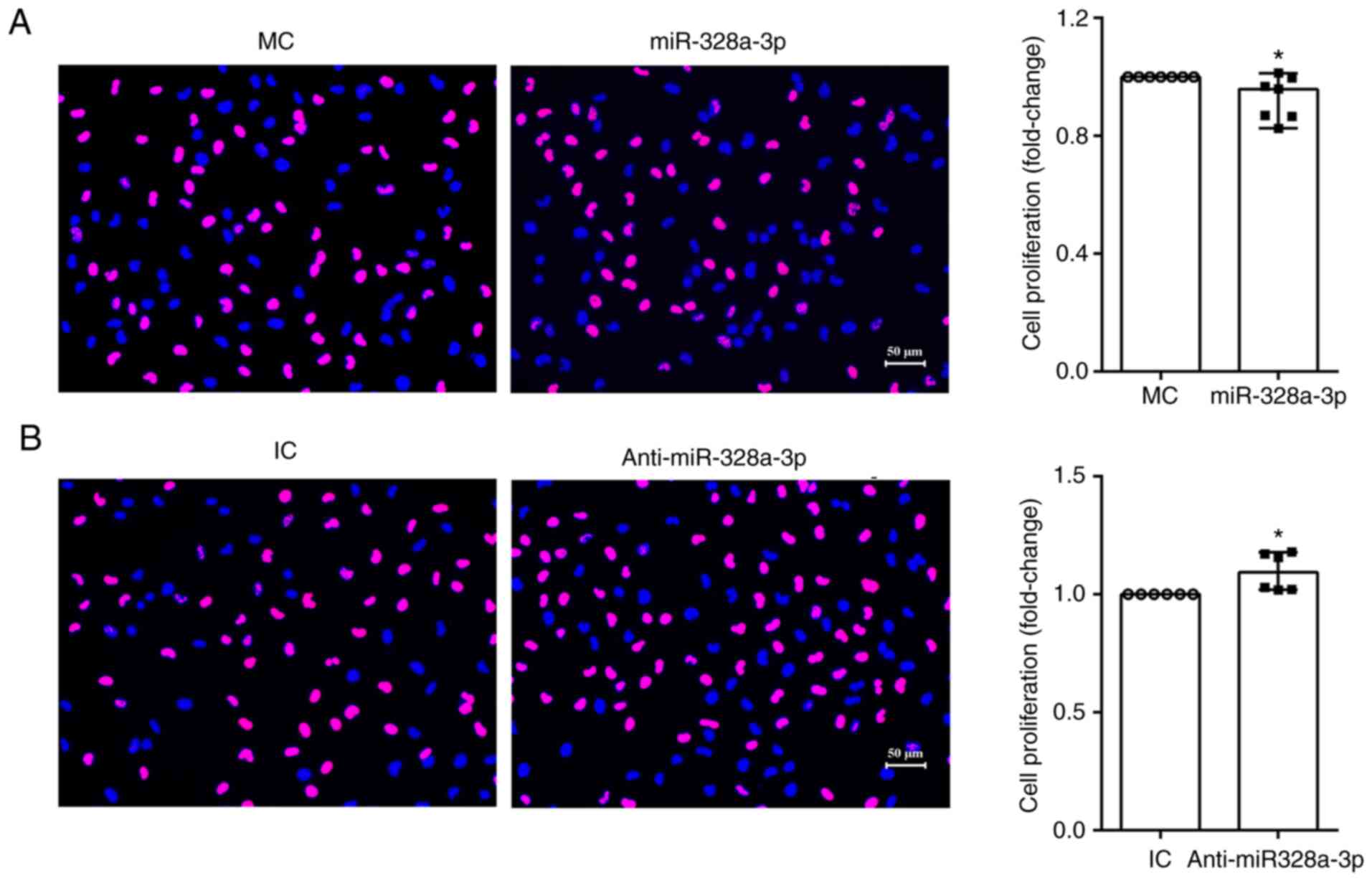
The EdU proliferation assay was performed to
determine the biological effects of miR-328a-3p on the
proliferation of HUVECs. The amount of proliferating cells in
HUVECs transfected with miR-328a-3p mimics, as compared with that
in HUVECs transfected with mimics control, was lower, while the
amount of total cells in both groups was similar. The ratio of
proliferating cells to total cells was lower after transfection
with miR-328a-3p mimics (Fig. 2A).
On the other hand, HUVECs transfected with miR-328a-3p inhibitor
had an increased amount of proliferating cells and a significantly
increased proliferation ratio (Fig.
2B). These outcomes indicated that miR-328a-3p slightly
impaired the proliferation of endothelial cells.
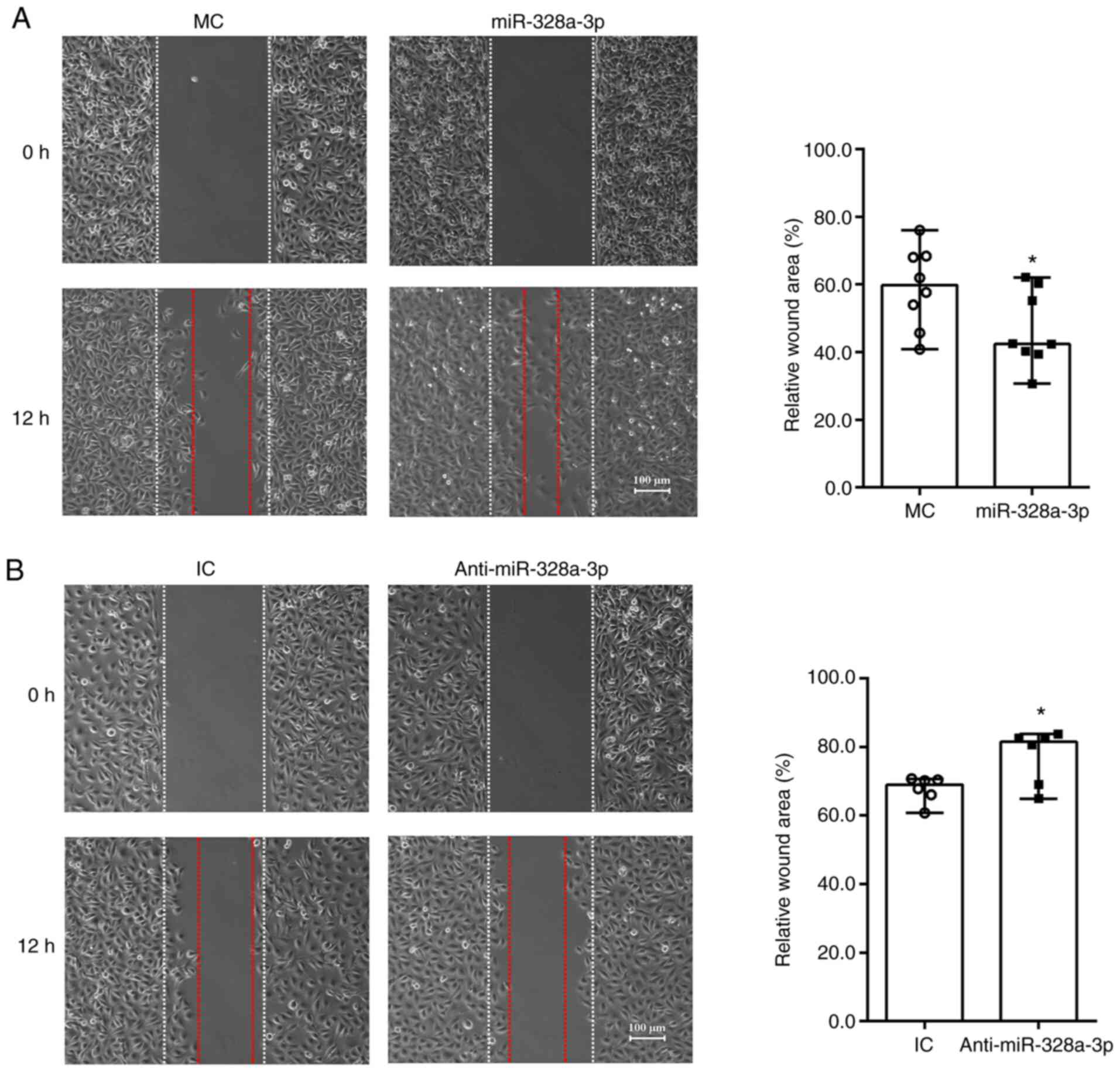
miR-328a-3p promotes the migration of
HUVECs
HUVECs transfected with miR-328a-3p mimics or
inhibitor were seeded onto mold chambers with inserts placed to
examine cell migration abilities. Equally wide blank spaces and
same sizes of cleared area were presented after insert removal (0
h) for both HUVECs transfected with miR-328a-3p mimics and mimics
control. Obvious cell migration was detected at 12 h after insert
removal. In HUVECs transfected with mimics control, numerous cells
migrated towards the wound area and filled the blank space.
Approximately 60% of cleared area remained after 12 h of insert
removal and cell culture, while ~40% of cleared area was covered by
migrated HUVECs (Fig. 3A). HUVECs
transfected with miR-328a-3p mimic exhibited a relatively greater
migration ability and occupied a larger space. In HUVECs
transfected with miR-328a-3p mimics, the remaining cleared area was
significantly smaller and only ~50% of cleared area was left after
12 h of cell culture (Fig. 3A). In
contrast to the decrease of relative cleared areas observed by
miR-328a-3p mimics transfection, miR-328a-3p inhibitor transfection
increased relative cleaned areas as compared with the control group
(Fig. 3B).
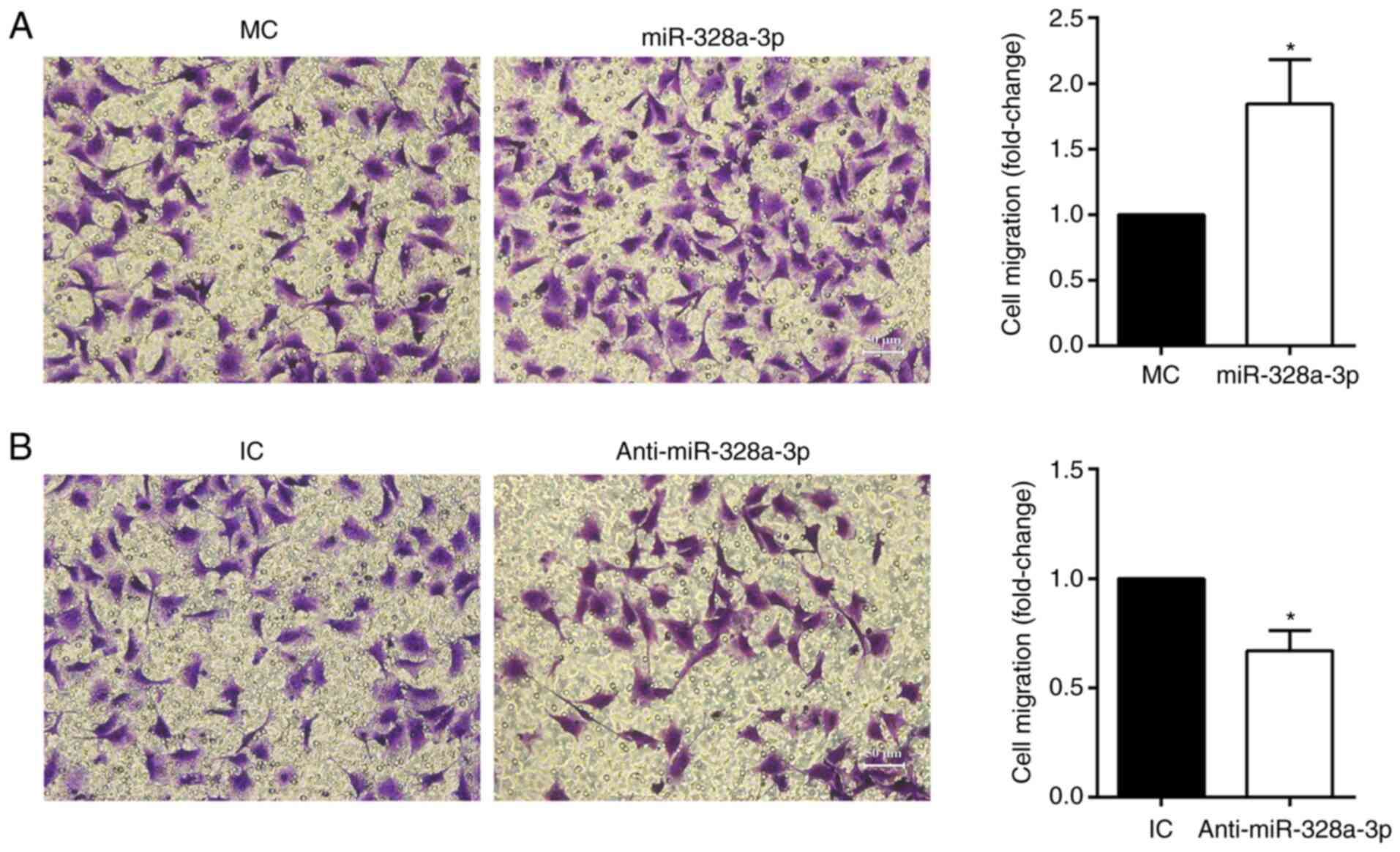
In addition to the wound-healing assay, a Transwell
assay was also applied to determine the effect of miR-328a-3p on
HUVEC migration. Transfection with miR-328a-3p mimics increased the
amount of migrated HUVECs (stained with crystal violet) to nearly
2-folds of that in the mimics control group (Fig. 4A), while transfection with
miR-328a-3p inhibitor reduced HUVEC migration, with a 31.07%
decrease as compared with the inhibitor control group (Fig. 4B).
miR-328a-3p promotes tubulogenesis of
HUVECs
HUVECs were seeded on Matrigel-coated culture plates
to examine their abilities to form capillary-like tubes. HUVECs
transfected with miR-328a-3p mimics, miR-328a-3p inhibitor and
corresponding non-targeting negative controls were all able to form
interconnecting tube networks (Fig.
5A and B). Summarized data from
skeletonized images (Fig. 5C)
indicated that transfection with miR-328a-3p mimics largely
increased the number of formed tubes, with a 55.37% increase in the
number of nodes (Fig. 5E), a 34.97%
increase in the number of meshes (Fig.
5F); the increase number of branches was not significant
(Fig. 5G) as compared with the
mimics control. Curtailed formation of capillary-like tubes was
observed in HUVECs transfected with miR-328a-3p inhibitor (Fig. 5D). Transfection with miR-328a-3p
inhibitor, as compared with the inhibitor control, led to a 40.63%
decrease in the number of nodes (Fig.
5H), a 53.10% decrease in the number of meshes (Fig. 5I) and a slight decrease in the
number of branches (Fig. 5J).
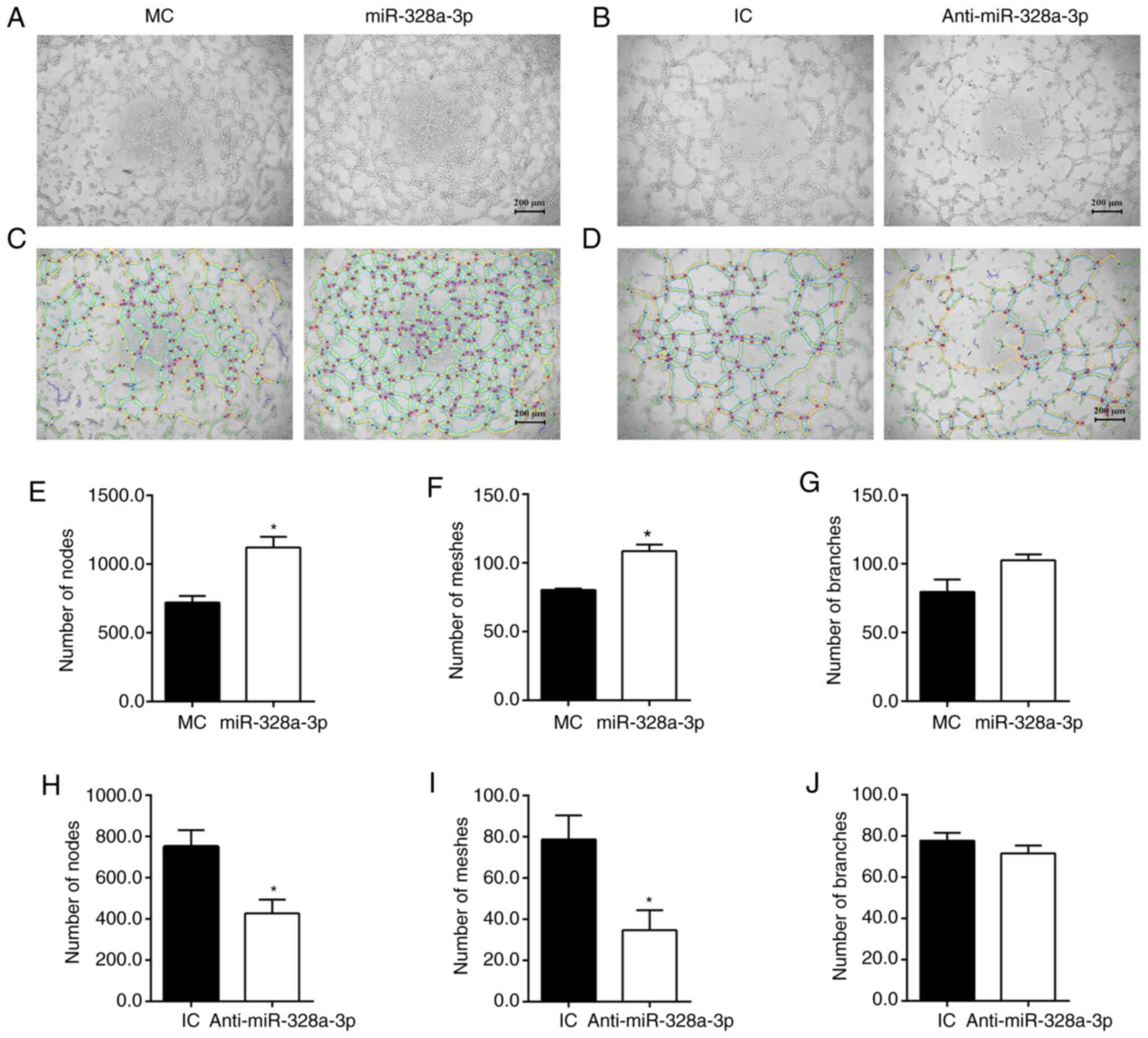 | Figure 5Effect of miR-328a-3p on
tubulogenesis of HUVECs. (A) Transfection with miR-328a-3p mimics
(miR-328a-3p), as compared with transfection with MC, increased
cell tubulogenesis. (B) Transfection with miR-328a-3p inhibitor
(Anti-miR-328a-3p), as compared with transfection with IC,
decreased cell tubulogenesis (scale bar, 200 µm). (C and D)
Skeletonized images of capillary-like tubes formed by HUVECs
transfected with (C) miR-328a-3p mimics or mimics control and (D)
miR-328a-3p inhibitor or IC. (E) The number of nodes in HUVECs
transfected with miR-328a-3p mimics or mimics control. (F) The
number of meshes in HUVECs transfected with miR-328a-3p mimics or
mimics control. (G) The number of branches in HUVECs transfected
with miR-328a-3p mimics or mimics control. (H) The number of nodes
in HUVECs transfected with miR-328a-3p inhibitor or IC. (I) The
number of meshes in HUVECs transfected with miR-328a-3p inhibitor
or IC. (J) The number of branches in HUVECs transfected with
miR-328a-3p inhibitor or inhibitor control. Values are expressed as
the mean with standard error of the mean from 3 or 4 experiments.
*P<0.05 vs. control. HUVEC, human umbilical vein
endothelial cell; miR, microRNA; MC, mimics control;
Anti-miR-328a-3p, miR-328a-3p inhibitor; IC, inhibitor control. |
Construction of miR-328a-3p-centered
ceRNA network
Upstream regulators and downstream effectors of
miR-328a-3p were analyzed using bioinformatics tools. Considering
that rat sciatic nerve crush injury has been commonly applied as an
animal model of peripheral nerve injury and gene expression
profiles of rat sciatic nerve stumps were previously determined,
upstream lncRNAs and downstream mRNAs of Rattus
Norvegicus-miR-328a-3p were investigated. TargetScan predicted
a total of 5 lncRNAs, XLOC_097278, XLOC_083369, XLOC_138151,
XLOC_174539 and XLOC_053870 that are able to bind to miR-328a-3p.
miRWalk 3.0, miRanda and miRdb prediction revealed various
potential target genes of miR-328a-3p. From these data, an
miR-328a-3p-centered lncRNA-miRNA-mRNA network was thus constructed
(Fig. 6A).
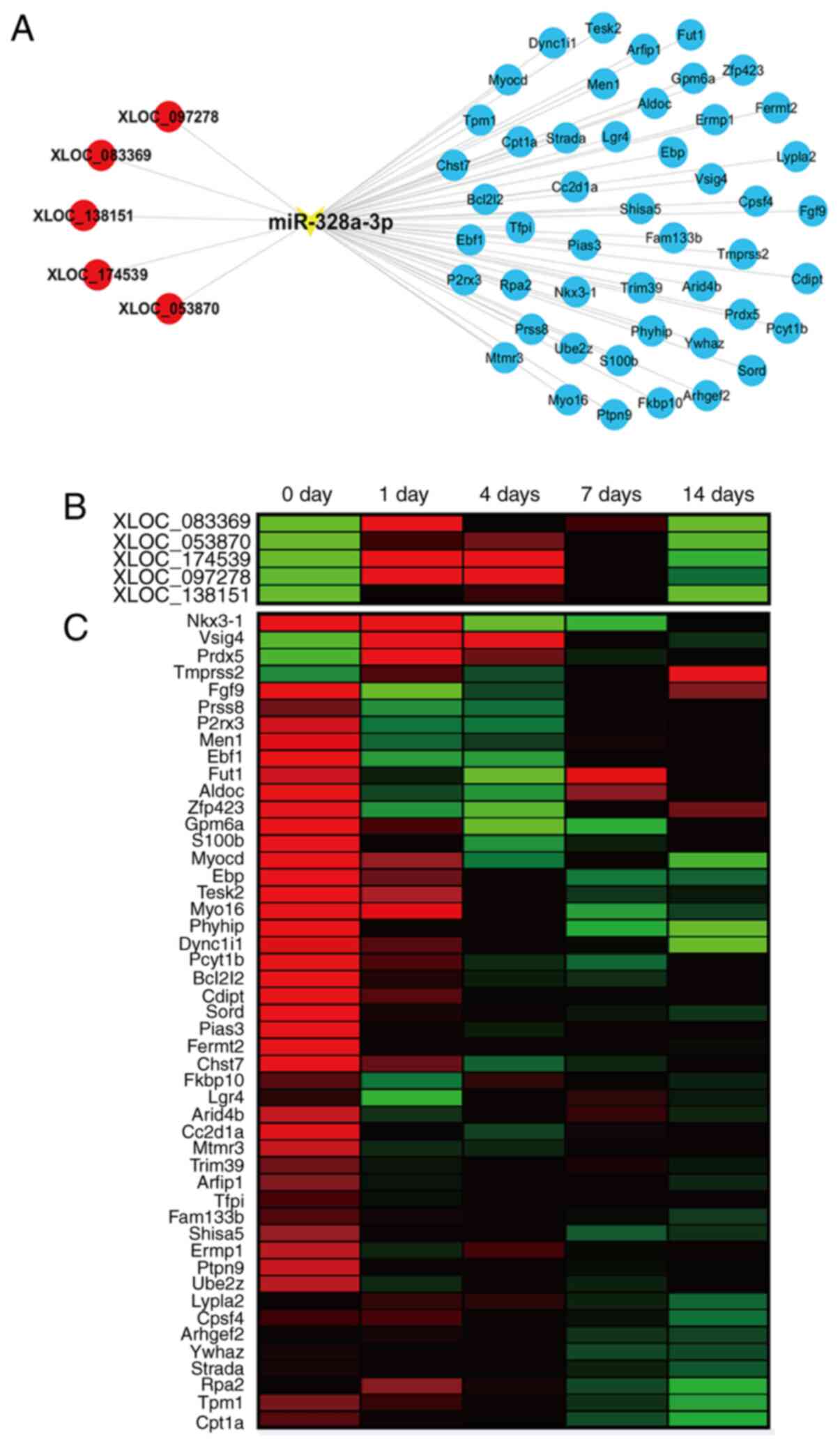 | Figure 6The ceRNA network of miR-328a-3p. (A)
Interactions of lncRNAs, miR-328a-3p and target mRNAs in the
miR-328a-3p-centered ceRNA network. lncRNAs were labeled in red
color, miRNA was labeled in yellow color and mRNAs were labeled in
blue color. (B) Heatmap of the temporal expression patterns of
lncRNAs in the miR-328a-3p-centered ceRNA network in the sciatic
nerve stumps at 0, 1, 4, 7 and 14 days after nerve crush injury.
Red color indicates upregulation and green color indicates
downregulation. (C) Heatmap of the temporal expression patterns of
mRNAs in the miR-328a-3p-centered ceRNA network in the sciatic
nerve stumps at 0, 1, 4, 7 and 14 days after nerve crush injury.
ceRNA, competing endogenous RNA; lncRNA, long non-coding RNA;
miRNA, microRNA. |
Previously generated sequencing data indicated that
all identified upstream lncRNAs were upregulated in sciatic nerve
stumps after nerve crush injury (Fig.
6B). The results of the RT-qPCR analysis demonstrated that the
expression levels of XLOC_097278, XLOC_083369, XLOC_138151,
XLOC_174539 and XLOC_053870 were elevated after sciatic nerve
injury (Fig. S1). However, the
majority of predicted potential target mRNAs were downregulated
after nerve injury, while certain mRNAs, such as Vsig4 and Prdx5,
had increased expression levels in the injured nerve stumps
(Fig. 6C).
Discussion
Peripheral nerve injury and repair is a complex
biological process that involves the participation of various
different cell types, such as Schwann cells, fibroblasts,
macrophages and endothelial cells. Following peripheral nerve
injury, miRNA-mediated phenotype modulation of Schwann cells has
been widely explored. However, the effects of miRNAs on endothelial
cells, which are cells that have important roles in the guidance of
Schwann cell migration after peripheral nerve injury, have not been
well demonstrated.
An earlier study by our group examined the
functional effects of let-7, a well-known miRNA that belongs to the
most abundant miRNA family in the genome (28), on endothelial cells (29). In the present study, the regulatory
roles of miR-328a-3p, another miRNA that was demonstrated to be
dysregulated in injured peripheral nerve stumps (12), were determined. The results of the
wound-healing assay, Transwell migration assay and Matrigel
tubulogenesis assay revealed that miR-328a-3p was able to
significantly promote the migration of endothelial cells and
stimulate the acquirement of functional capabilities. Considering
that proliferating cells are variables of these assays, an EdU
proliferation assay was also performed. In contrast to the effects
of miR-328a-3p on endothelial cell migration and tubulogenesis,
elevated miR-328a-3p slightly decreased EdU incorporation. These
results suggested that miR-328a-3p slightly inhibits the
proliferation but significantly promotes the migration of
endothelial cells. Numerous regulatory factors exert consistent
effects on cell proliferation and migration. However, numerous
factors may have distinct roles in cell proliferation and
migration. A previous study by our group indicated that MAPT, a
molecule whose expression was first downregulated after nerve
injury and then upregulated at later time-points, inhibits Schwann
cell proliferation but encourages Schwann cell migration (30). Therefore, it is likely that
miR-328a-3p has different roles to affect endothelial proliferation
and migration. These results excluded the possibility that enhanced
endothelial cell migration and tubulogenesis were caused by
increased numbers of cells and emphasized the promoting effects of
miR-328a-3p on endothelial cell migration and angiogenesis.
The functional roles of miR-328a-3p on cell
migration appeared to be contradictory in various different cell
types. It was reported that miR-328 functions as a tumor suppressor
gene to inhibit cell migration of gastric cancer cells (31) and osteosarcoma cells (32). On the contrary, in breast cancer
cells, elevated miR-328-3p was able to decrease the abundance of
glutamate metabotropic receptor 4 (GRM4) and counteract
GRM4-induced inhibition of breast cancer cell migration and
invasion (33). Of note, various
studies suggested that miR-328a-3p may even lead to different
cellular behaviors in endothelial cells exposed to diverse culture
conditions. Under high-glucose and low-serum conditions, the
migration and tube-like structure formation of HUVECs were
decreased, while the expression of miR-328 was increased.
Transfection of high-glucose and low-serum-cultured HUVECs with
miR-328 inhibitor stimulated cell migration and angiogenesis
(14). Another study suggested that
miR-328-3p promoted HUVEC migration and invasion and protected
HUVECs against oxidized low-density lipoprotein-induced injury
(34). These conflicting
observations implied that miR-328 may have diverse functions in
different microenvironments. In the present study, the biological
roles of miR-328a-3p in HUVECs cultured under standardized
conditions were examined. However, peripheral nerve injury
generally induced a hypoxia response at the injury site (7,35). In
future studies, it may be helpful to mimic the pathological
conditions of peripheral nerve injury, expose miR-328a-3p-treated
HUVECs to decreased oxygen levels and determine the modulating
effects of miR-328a-3p on the HUVEC phenotype under low oxygen
levels.
Besides the functional investigations of
miR-328a-3p, lncRNAs and mRNAs that may be associated with
miR-328a-3p were also analyzed. The sequences of miR-328a-3p were
highly conserved in different species, including human (H.
Sapiens) and rat (R. Norvegicus). Considering that the
transcriptome profiles were obtained and ceRNA networks of rats
after peripheral nerve injury were identified (36), the present study determined
lncRNA-miR-328a-3p-mRNA interactions, constructed a
miR-328a-3p-centered ceRNA network, and explored the temporal
expression patterns of miR-328a-3p-associated lncRNAs and mRNAs in
the injured sciatic nerves of rats. Following peripheral nerve
injury, the expression levels of miR-328a-3p first decreased,
reaching a valley level at 4 days after nerve injury, and then
gradually recovered to the uninjured level at 14 days (12). The heatmaps indicated inverse
correlations between the temporal expression profiles of
miR-328a-3p and all predicted upstream lncRNAs, as well as
miR-328a-3p and the potential target genes Vsig4 and Prdx5. Vsig4
encodes for a v-set and immunoglobulin domain-containing protein
and has been mainly considered as a negative regulator of T-cell
responses (37,38). Prdx5 encodes for peroxiredoxin 5, a
member of the peroxiredoxin family of antioxidant enzymes, and has
essential antioxidant roles (39).
The involvement of Vsig4 and Prdx5 in the peripheral nervous system
has remained to be fully elucidated, although Prdx5 was detected to
be expressed in peripheral nerves (40) and upregulated in dorsal root ganglia
neurons after peripheral nerve injury (41). In the present study, the sequencing
data suggested that the abundance of Vsig4 and Prdx5 was elevated
in sciatic nerve stumps after nerve injury and elevated Vsig4 and
Prdx5 may be induced by downregulation of miR-328a-3p. The binding
relationships of lncRNAs and mRNAs with miR-328a-3p may be further
validated by a luciferase assay and functional rescue assays.
Collectively, in the present study, the biological
effects of miR-328a-3p were investigated, revealing that
miR-328a-3p increased the migration and angiogenesis potential of
endothelial cells. These results provided a mechanism for the
regulation of endothelial cells during peripheral nerve
regeneration.
Supplementary Material
Analysis of the expression of
XLOC_083369, XLOC_053870, XLOC_174539, XLOC_097278 and XLOC_138151
in sciatic nerve segments at 0, 1, 4, 7 and 14 days after nerve
crush. Gene expression patterns of (A) XLOC_083369, (B)
XLOC_053870, (C) XLOC_174539, (D) XLOC_097278 and (E) XLOC_138151
were determined by reverse transcription-quantitative PCR with
normalization to GAPDH. Values are expressed as the mean with
standard error of the mean (n=3). *P<0.05 vs. day 0.
d, days.
Primer pairs for quantitative
real-time PCR of lncRNAs.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by the Natural Science
Foundation of Jiangsu Province, China (grant no. BK20200976), the
Priority Academic Program Development of Jiangsu Higher Education
Institutions (grant no. PAPD), and Postgraduate Research and
Practice Innovation Program of Jiangsu Province (grant no.
KYCX21_3076).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SY and XW conceived and designed the study. SC, YZ
and XC performed the experiments and acquired the experimental
data. SC and XW analyzed and interpreted the data. SY, XW and JZ
contributed reagents, materials and analysis tools. XW, SY and JZ
drafted the manuscript and revised it critically for important
intellectual content. SY and XW confirm the authenticity of all the
raw data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Experiments were ethically approved by the
Administration Committee of Experimental Animals, Nantong
University (Nantong, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Eelen G, de Zeeuw P, Treps L, Harjes U,
Wong BW and Carmeliet P: Endothelial cell metabolism. Physiol Rev.
98:3–58. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hassan M, Moghadamrad S, Sorribas M,
Muntet SG, Kellmann P, Trentesaux C, Fraudeau M, Nanni P, Wolski W,
Keller I, et al: Paneth cells promote angiogenesis and regulate
portal hypertension in response to microbial signals. J Hepatol.
73:628–639. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Velazquez OC: Angiogenesis and
vasculogenesis: Inducing the growth of new blood vessels and wound
healing by stimulation of bone marrow-derived progenitor cell
mobilization and homing. J Vasc Surg. 45 Suppl A:A39–A47.
2007.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Rossi L, Attanasio C, Vilardi E, De
Gregorio M and Netti PA: Vasculogenic potential evaluation of
bottom-up, PCL scaffolds guiding early angiogenesis in tissue
regeneration. J Mater Sci Mater Med. 27(107)2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wong HK, Ivan Lam CR, Wen F, Mark Chong
SK, Tan NS, Jerry C, Pal M and Tan LP: Novel method to improve
vascularization of tissue engineered constructs with biodegradable
fibers. Biofabrication. 8(015004)2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lloyd-Griffith C, McFadden TM, Duffy GP,
Unger RE, Kirkpatrick CJ and O'Brien FJ: The pre-vascularisation of
a collagen-chondroitin sulphate scaffold using human amniotic
fluid-derived stem cells to enhance and stabilise endothelial
cell-mediated vessel formation. Acta Biomater. 26:263–273.
2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Cattin AL, Burden JJ, Van Emmenis L,
Mackenzie FE, Hoving JJ, Garcia Calavia N, Guo Y, McLaughlin M,
Rosenberg LH, Quereda V, et al: Macrophage-induced blood vessels
guide schwann cell-mediated regeneration of peripheral nerves.
Cell. 162:1127–1139. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Muangsanit P, Shipley RJ and Phillips JB:
Vascularization strategies for peripheral nerve tissue engineering.
Anat Rec (Hoboken). 301:1657–1667. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Saffari TM, Bedar M, Hundepool CA, Bishop
AT and Shin AY: The role of vascularization in nerve regeneration
of nerve graft. Neural Regen Res. 15:1573–1579. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yu B, Zhou S, Wang Y, Ding G, Ding F and
Gu X: Profile of microRNAs following rat sciatic nerve injury by
deep sequencing: Implication for mechanisms of nerve regeneration.
PLoS One. 6(e24612)2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yu B, Zhou S, Yi S and Gu X: The
regulatory roles of non-coding RNAs in nerve injury and
regeneration. Prog Neurobiol. 134:122–139. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zou Y, Wu F, Liu Q, Deng X, Hai R, He X
and Zhou X: Downregulation of miRNA328 promotes the angiogenesis of
HUVECs by regulating the PIM1 and AKT/mTOR signaling pathway under
high glucose and low serum condition. Mol Med Rep. 22:895–905.
2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Oliverio M, Schmidt E, Mauer J, Baitzel C,
Hansmeier N, Khani S, Konieczka S, Pradas-Juni M, Brodesser S, Van
TM, et al: Dicer1-miR-328-Bace1 signalling controls brown adipose
tissue differentiation and function. Nat Cell Biol. 18:328–336.
2016.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Yi W, Tu MJ, Liu Z, Zhang C, Batra N, Yu
AX and Yu AM: Bioengineered miR-328-3p modulates GLUT1-mediated
glucose uptake and metabolism to exert synergistic
antiproliferative effects with chemotherapeutics. Acta Pharm Sin B.
10:159–170. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Eiring AM, Harb JG, Neviani P, Garton C,
Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, et al:
MiR-328 functions as an RNA decoy to modulate hnRNP E2 regulation
of mRNA translation in leukemic blasts. Cell. 140:652–665.
2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang
F, Zhang Y, Shan H, Luo X, Bai Y, et al: MicroRNA-328 contributes
to adverse electrical remodeling in atrial fibrillation.
Circulation. 122:2378–2387. 2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ruan ZB, Wang F, Bao TT, Yu QP, Chen GC
and Zhu L: Genome-wide analysis of circular RNA expression profiles
in patients with atrial fibrillation. Int J Clin Exp Pathol.
13:1933–1950. 2020.PubMed/NCBI
|
|
20
|
Guo W, Mu K, Zhang B, Sun C, Zhao L, Li
HR, Dong ZY and Cui Q: The circular RNA circ-GRB10 participates in
the molecular circuitry inhibiting human intervertebral disc
degeneration. Cell Death Dis. 11(612)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye
X, Zhang H, Yang P, Cui X, Ren Y, et al: A Noncoding regulatory
RNAs network driven by Circ-CDYL acts specifically in the early
stages hepatocellular carcinoma. Hepatology. 71:130–147.
2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Saba R, Goodman CD, Huzarewich RL,
Robertson C and Booth SA: A miRNA signature of prion induced
neurodegeneration. PLoS One. 3(e3652)2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang X, Chen Q, Yi S, Liu Q, Zhang R, Wang
P, Qian T and Li S: Correction: The microRNAs let-7 and miR-9
down-regulate the axon-guidance genes Ntn1 and Dcc during
peripheral nerve regeneration. J Biol Chem.
294(6695)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li S, Wang X, Gu Y, Chen C, Wang Y, Liu J,
Hu W, Yu B, Wang Y, Ding F, et al: Let-7 microRNAs regenerate
peripheral nerve regeneration by targeting nerve growth factor. Mol
Ther. 23:423–433. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zudaire E, Gambardella L, Kurcz C and
Vermeren S: A computational tool for quantitative analysis of
vascular networks. PLoS One. 6(e27385)2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yi S, Zhang H, Gong L, Wu J, Zha G, Zhou
S, Gu X and Yu B: Deep sequencing and bioinformatic analysis of
lesioned sciatic nerves after crush injury. PLoS One.
10(e0143491)2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhao L and Yi S: Transcriptional landscape
of alternative splicing during peripheral nerve injury. J Cell
Physiol. 234:6876–6885. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Roush S and Slack FJ: The let-7 family of
microRNAs. Trends Cell Biol. 18:505–516. 2008.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ji X, Hua H, Shen Y, Bu S and Yi S: Let-7d
modulates the proliferation, migration, tubulogenesis of
endothelial cells. Mol Cell Biochem. 462:75–83. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Yi S, Liu Q, Wang X, Qian T, Wang H, Zha
G, Yu J, Wang P, Gu X, Chu D and Li S: Tau modulates Schwann cell
proliferation, migration and differentiation following peripheral
nerve injury. J Cell Sci. 132(jcs222059)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yan BL, Li XL and An JY: MicroRNA-328 acts
as an anti-oncogene by targeting ABCG2 in gastric carcinoma. Eur
Rev Med Pharmacol Sci. 23:6148–6159. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Shi J, An G, Guan Y, Wei T, Peng Z, Liang
M and Wang Y: MiR-328-3p mediates the anti-tumor effect in
osteosarcoma via directly targeting MMP-16. Cancer Cell Int.
19(104)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Xiao B, Chen D, Zhou Q, Hang J, Zhang W,
Kuang Z, Sun Z and Li L: Glutamate metabotropic receptor 4 (GRM4)
inhibits cell proliferation, migration and invasion in breast
cancer and is regulated by miR-328-3p and miR-370-3p. BMC Cancer.
19(891)2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Qin X and Guo J: MicroRNA-328-3p protects
vascular endothelial cells against oxidized low-density lipoprotein
induced injury via targeting forkhead box protein O4 (FOXO4) in
atherosclerosis. Med Sci Monit. 26(e921877)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Smaila BD, Holland SD, Babaeijandaghi F,
Henderson HG, Rossi FMV and Ramer MS: Systemic hypoxia mimicry
enhances axonal regeneration and functional recovery following
peripheral nerve injury. Exp Neurol. 334(113436)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Qian T, Fan C, Liu Q and Yi S: Systemic
functional enrichment and ceRNA network identification following
peripheral nerve injury. Mol Brain. 11(73)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Jung K, Seo SK and Choi I: Endogenous
VSIG4 negatively regulates the helper T cell-mediated antibody
response. Immunol Lett. 165:78–83. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Vogt L, Schmitz N, Kurrer MO, Bauer M,
Hinton HI, Behnke S, Gatto D, Sebbel P, Beerli RR, Sonderegger I,
et al: VSIG4, a B7 family-related protein, is a negative regulator
of T cell activation. J Clin Invest. 116:2817–2826. 2006.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Hanschmann EM, Godoy JR, Berndt C,
Hudemann C and Lillig CH: Thioredoxins, glutaredoxins, and
peroxiredoxins-molecular mechanisms and health significance: From
cofactors to antioxidants to redox signaling. Antioxid Redox
Signal. 19:1539–1605. 2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lu JL, Vallat JM, Pollard JD, Knoops B and
Ouvrier R: Expression of the antioxidant enzyme peroxiredoxin 5 in
the human peripheral nervous system. J Peripher Nerv Syst.
11:318–324. 2006.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Valek L, Kanngießer M, Haussler A, Agarwal
N, Lillig CH and Tegeder I: Redoxins in peripheral neurons after
sciatic nerve injury. Free Radic Biol Med. 89:581–592.
2015.PubMed/NCBI View Article : Google Scholar
|