Introduction
According to a recent study, bladder cancer (BCa) is
the most common type of malignancy of the urinary system and poses
a major threat to human health worldwide, 573,278 new cases of
bladder cancer and 212,536 mortalities from bladder cancer occurred
in 2021(1). Surgery and
chemotherapy are the predominant treatment strategies used for BCa;
however, the treatment efficacy is far from satisfactory (2). Thus, novel therapeutic strategies for
this disease are required.
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNA of >200 nucleotides in length (3,4). As
they are generated via RNA polymerase II-mediated transcription,
lncRNAs have been identified to play a crucial role in multiple
types of cancer, including colon and non-small cell lung cancer,
hepatocellular carcinoma and breast cancer (5-8).
According to recent studies, lncRNAs can inhibit or promote the
expression of certain genes at the transcriptional or
post-transcriptional level (9,10). For
instance, lncRNA small nucleolar RNA host gene 3 (SNHG3) is
significantly upregulated in BCa tissues and associated with
reduced overall survival rates of BCa patients. Silencing of SNHG3
expression suppresses BCa cell proliferation and metastasis by
increasing miR-515-5p expression (11). lncRNA KCNQ1 opposite
strand/antisense transcript 1 is notably upregulated in BCa tissues
and promote the progression of BCa through by modulating miR-218-5p
expression and the activity of Heparan Sulfate-Glucosamine
3-Sulfotransferase 3B1(12).
In the present study, abnormally expressed lncRNAs
in BCa tissues and adjacent normal tissues were examined using the
Cancer RNA-Seq Nexus (CRN) database, with a particular focus was on
the lncRNAs that were lowly expressed in BCa and had not yet been
reported, which may have more research value. To the best of our
knowledge, as the role of lncRNA LOC339524 in BCa has not been
studied before, it was selected for subsequent analysis. The aims
of the present study were to investigate the expression of lncRNA
LOC339524 in BCa, evaluate the effect of lncRNA LOC339524 on
regulating the malignant phenotypes of BCa cells in vitro
and explore the mechanism that underlies lncRNA LOC339524 in BCa
progression.
Materials and methods
Patient studies
A total of 52 clinical samples (26 BCa tissues and
26 para-carcinoma tissues) were collected from 26 patients (male,
n=17 and female, n=9; age range, 43-67 years) with BCa who
underwent surgery at The Third Affiliated Hospital, Army Medical
University (Chongqing, China) between September 2017 and February
2019. The tissues were immediately cut into small sections and
transferred to liquid nitrogen (-196˚C) for use in further
experiments. The tissues were all pathologically identified as BCa
or para-carcinoma tissues. Exclusion criteria included a prior
history of cancer, chemotherapy or radiotherapy prior to undergoing
surgery or a lack of written informed consent. The present study
was approved by the Ethics Committee of The Third Affiliated
Hospital, Army Medical University. All patients were informed of
the study protocol and signed informed consent forms prior to
participation.
Cell lines and culture
Human BCa cell lines (J82, T24, UM-UC-3 and 5637)
and the normal bladder epithelial cell line, SV-HUC-1, were
purchased from Shanghai Cell Bank of Chinese Academy of Sciences.
J82, T24 and 5637 cells were routinely cultured in RPMI-1640 medium
(HyClone; Cytiva) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.). The UM-UC-3 and SV-HUC-1 were routinely cultured
in DMEM (HyClone; Cytiva) supplemented with 10% FBS. All cells were
maintained in an incubator at 37˚C with 5% CO2. For 5 µl
5-Aza-2'-deoxycytidine (Sigma-Aldrich; Merck KGaA) treatment, the
5-Aza-2'-deoxycytidine compound was dissolved in DMSO and diluted
in cell culture medium to 2 µmol/l. Cells were cultured in the cell
culture supplemented with DMSO at 37˚C for three consecutive days
as the vehicle-treated cells.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the tissues and cells
using RNAiso Plus (Takara Bio, Inc.). Total RNA was reverse
transcribed into cDNA using a PrimeScript RT Master mix (Takara
Bio, Inc.) according to the manufacturer's protocol. The following
thermocycling conditions were used for reverse transcription: 16˚C
for 30 min, 42˚C for 30 min and 85˚C for 5 min. qPCR was
subsequently performed using SYBR Premix Ex Taq kit (Takara Bio,
Inc.). The thermocycling conditions for qPCR were: 95˚C for 2 min,
40 cycles of 95˚C for 15 sec and 60˚C for 30 sec. The primers used
for the qPCR are listed in Table
SI. GAPDH was selected as the internal reference gene for mRNA,
while U6 was selected as the internal reference gene for the
normalization of microRNA (miRNA/miR) expression. The
2-ΔΔCq method was used for analyzing the relative gene
expression (13).
Cell transfection
LOC339524 plasmid (pcDNA3.0), miR-875-5p mimic
(5'-UAUACCUCAGUUUUAUCAGGUG-3'), miR-875-5p inhibitor
(5'-CACCUGAUAAAACUGAGGUAUA-3'), mimic-NC
(5'-UUCUCCGAACGUGUCACGUTT-3'), inhibitor-NC
(5'-CAGUACUUUUGUGUAGUACAA-3') and NC empty plasmid (pcDNA3.0,
control for LOC339524) were obtained from Shanghai GeneChem Co.,
Ltd. The cells were seeded into 6-well plates at a density of
2x105 cells/well and prior to transfection, were washed
with PBS. Then, 40 nM plasmid, mimic, inhibitor, respective NC or 2
µl Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) were individually diluted in 50 µl Opti-MEM
(Invitrogen; Thermo Fisher Scientific, Inc.). Following 20 min of
incubation, plasmid/oligonucleotide Opti-MEM solution and
Lipofectamine 2000 Opti-MEM solution were mixed and incubated at
4˚C for a further 20 min. The complex was then added to each well.
The cells were collected for subsequent experimentation following
48 h of further culture.
Cell Counting Kit (CCK)-8 assay
Following transfection for 48 h, cells were seeded
into 96-well plates at a density of 1x103 cells/well
(six replicates/group). Following the incubation at 37˚C for 24 h,
10 µl CCK-8 reagent (Dojindo Molecular Laboratories, Inc.) was
added/well and incubated at 37˚C for 3 h. The absorbance of each
well was measured at a wavelength of 450 nm using a
spectrophotometer (BioTek Instruments, Inc.). The data were
collected for 5 days and the experiment was repeated four
times.
Colony formation assay
Following transfection for 48 h, cells were seeded
into 6-well plates at a density of 1x103 cells/well.
Following incubation at 37˚C for 12 days, the colonies in each
group were fixed with 4% paraformaldehyde at room temperature for
30 min and stained with 0.1% crystal violet at room temperature for
30 min. The colonies were visualized and counted. The experiment
was repeated four times.
Measurement of DNA content
Following transfection for 48 h, cells were
harvested (1,500 x g; 10 min; 4˚C) and suspended in propidium
iodide (RNaseA, 10 mg/l; Beyotime Institute of Biotechnology) at
4˚C for 40 min. Flow cytometric analysis (FACSCalibur flow
cytometer; BD Biosciences) was performed to assess the relative DNA
content in T24 and 5637 cells with a CellQuest Pro system software
package (version 5.1; BD Biosciences).
Bioinformatics analysis
Expression profiles for LOC339524 were obtained from
the Cancer RNA-seq Nexus (CRN) database (14) and the Gene Expression Profiling
Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/index.html) (15). The DIANA TOOLS database (http://diana.imis.athena-innovation.gr)
(16) was used to predict the
target miRNA of LOC339524. The MethHC database (https://awi.cuhk.edu.cn/~MethHC/methhc_2020/php/index.php)
(17) was used to determine the
degree of methylation of the LOC339524 promoter in BCa.
Dual luciferase reporter assay
The binding sites between LOC339524 and miR-875-5p
were predicted using the DIANA TOOLS database (http://diana.imis.athena-innovation.gr).
The wild-type (WT) LOC339524-WT luciferase plasmid containing the
binding sites for miR-875-5p and the LOC339524-mutant (MUT) plasmid
containing the mutated binding sites were constructed by Changzhou
Ruibo Bio-Technology Co., Ltd. The binding sites between miR-875-5p
and the COPS7A 3'-untranslated region (3'-UTR) were predicted using
miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2)
and TargetScan (http://www.targetscan.org) databases. The COPS7A
3'-UTR-WT luciferase plasmid containing the miR-875-5p binding
sites and the COPS7A 3'-UTR-MUT plasmid containing the MUT binding
sites were constructed. Cells (5,637) were seeded into 96-well
plates (1x104 cells/well). Luciferase plasmids were
co-transfected with miR-875-5p mimic or mimic-NC using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After incubated at 37˚C for 48 h, cells (5,637)
were harvested. The Firefly and Renilla luciferase
activities were analyzed using a Dual-Luciferase Reporter assay
system (Promega Corporation) according to the manufacturer's
instructions. The relative luciferase activity was normalized to
Renilla luciferase activity.
Western blotting
Cells were washed twice with PBS and total protein
was extracted using RIPA lysis buffer (Beyotime Institute of
Biotechnology) supplemented with PMSF protein inhibitor (Beyotime
Institute of Biotechnology). The protein concentration was
quantified using a standard BCA protein assay and 30 µg of
protein/lane was separated via 10% SDS-PAGE. The separated proteins
were subsequently transferred onto PVDF membranes (MilliporeSigma)
and blocked with 5% non-fat milk at 26˚C for 2 h. The membranes
were then incubated overnight at 4˚C with the following primary
antibodies: Rabbit anti-COPS7A (1:1,000; cat. no. ab124705; Abcam),
rabbit anti-CDK2 (1:1,000; cat. no. ab32147; Abcam), rabbit
anti-CDK4 (1:1,000; cat. no. ab108357; Abcam), rabbit anti-cyclin
D2 (1:1,000; cat. no. ab230883; Abcam) and rabbit anti-GAPDH
(1:2,500; cat. no. ab9485; Abcam). Following the primary antibody
incubation, the membranes were incubated with a goat anti-rabbit
IgG H&L secondary antibody (1:5,000; cat. no. ab6721; Abcam) at
room temperature for 2 h according to the protocol. The protein
bands were visualized using an ECL system (Thermo Fisher
Scientific, Inc.). GAPDH was used as the internal loading
control.
Biotin pull-down assay
The biotin-labeled LOC339524 and miR-875-5p probe
were synthesized by Changzhou Ruibo Bio-Technology Co., Ltd. The
LOC339524 or miR-875-5p probe was incubated with
streptavidin-coupled probe-bound DynaBeads (Invitrogen; Thermo
Fisher Scientific, Inc.) at 4˚C for 12 h. Following incubation, the
RNA complexes bound to the beads were eluted. RT-qPCR was used to
detect the purified RNAs.
Statistical analysis
Data are presented as the mean ± SD from at least
three independent experiments. SPSS software (version 20.0; IBM
Corp.) was used for statistical analysis. Comparisons among
multiple groups were analyzed using a one-way ANOVA followed by a
Tukey's post hoc test, while the statistical differences between
two unpaired groups were analyzed using an unpaired Student's
t-test. The statistical differences between tumor and adjacent
non-tumor samples of the same individuals were analyzed using a
paired Student's t-test. Correlations analysis were analyzed by the
Pearson's correlation test. P<0.05 was considered to indicate a
statistically significant difference.
Results
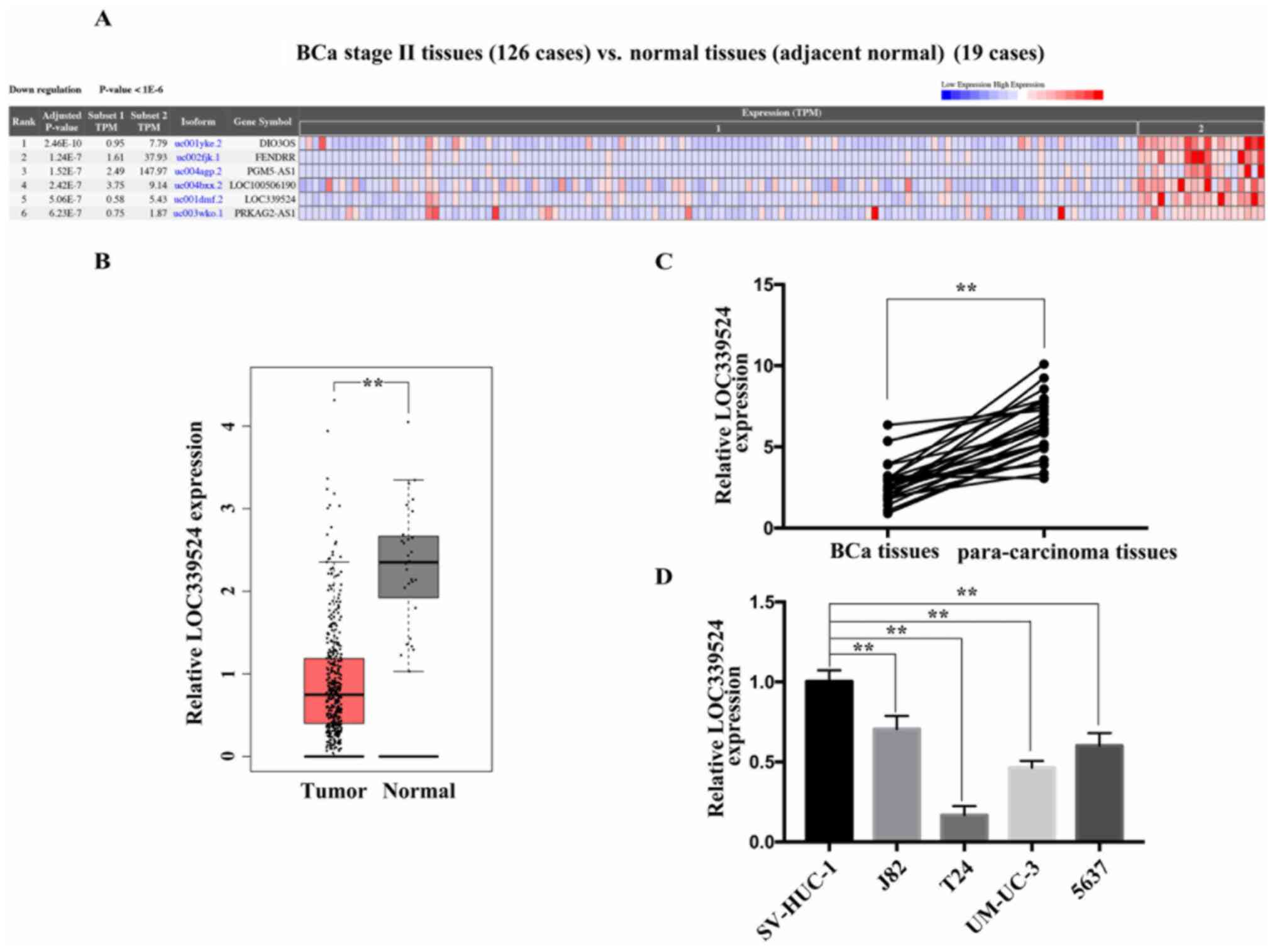
LOC339524 is expressed at low levels in BCa
tissues and cell lines. The expression profile of LOC339524 was
screened from the CRN database. As shown in Fig. 1A, the expression levels of LOC339524
were downregulated in BCa tissues compared with adjacent tissues.
The data from the GEPIA database revealed the same trend (Fig. 1B). The expression of LOC339524 was
then examined in 26 pairs of BCa and para-carcinoma tissues using
RT-qPCR. The results revealed that LOC339524 expression levels were
significantly downregulated in BCa tissues compared with those in
para-carcinoma tissues (Fig. 1C).
In addition, the expression levels of LOC339524 in four BCa cell
lines (J82, T24, UM-UC-3 and 5637) were detected using RT-qPCR; the
SV-HUC-1 cell line served as a control. Compared with the SV-HUC-1
cells, the expression levels of LOC339524 in the BCa cell lines
were downregulated (Fig. 1D).
Notably, 5637 and T24 cells exhibited the lowest expression levels
of LOC339524 amongst the BCa cell lines.
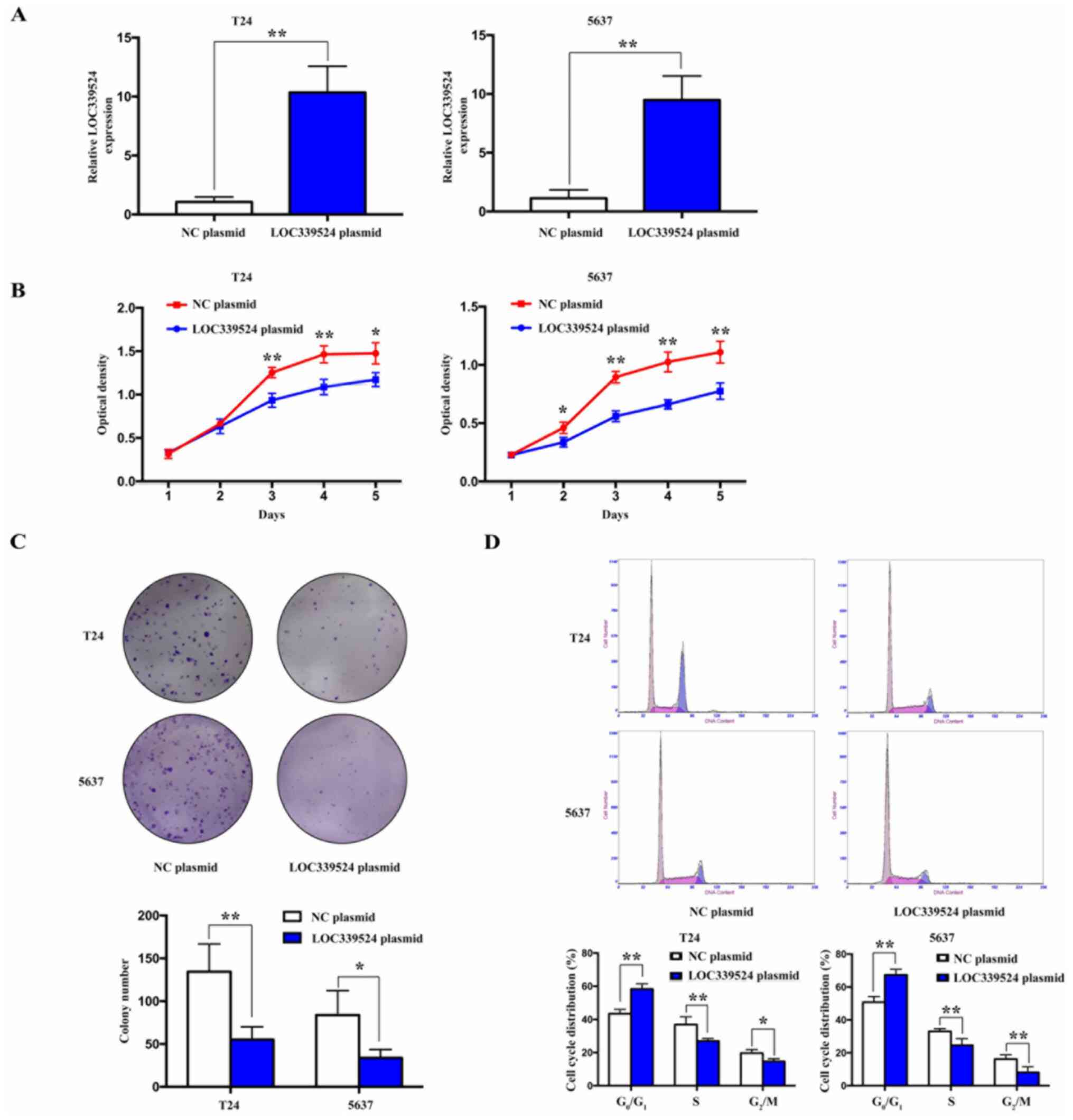
lncRNA LOC339524 is closely associated
with the proliferative ability of BCa cells
To investigate the role of LOC339524 in the
proliferation of BCa cells, LOC339524 was successfully
overexpressed in the 5637 and T24 cell lines (Fig. 2A). Cell proliferation was detected
using a CCK-8 assay and the results indicated that LOC339524
overexpression suppressed the proliferative ability of the BCa
cells (Fig. 2B). In addition, the
colony formation ability was significantly suppressed when
LOC339524 was overexpressed in both cell lines (Fig. 2C). Furthermore, cell cycle analysis
indicated that the number of cells in the
G0/G1 phase was increased following the
overexpression of LOC339524, while that in the S and
G2/M phases was decreased compared with the NC plasmid
group (Fig. 2D). Taken together,
these findings indicated that LOC339524 may significantly inhibit
the proliferation of BCa cells by blocking cell cycle
progression.
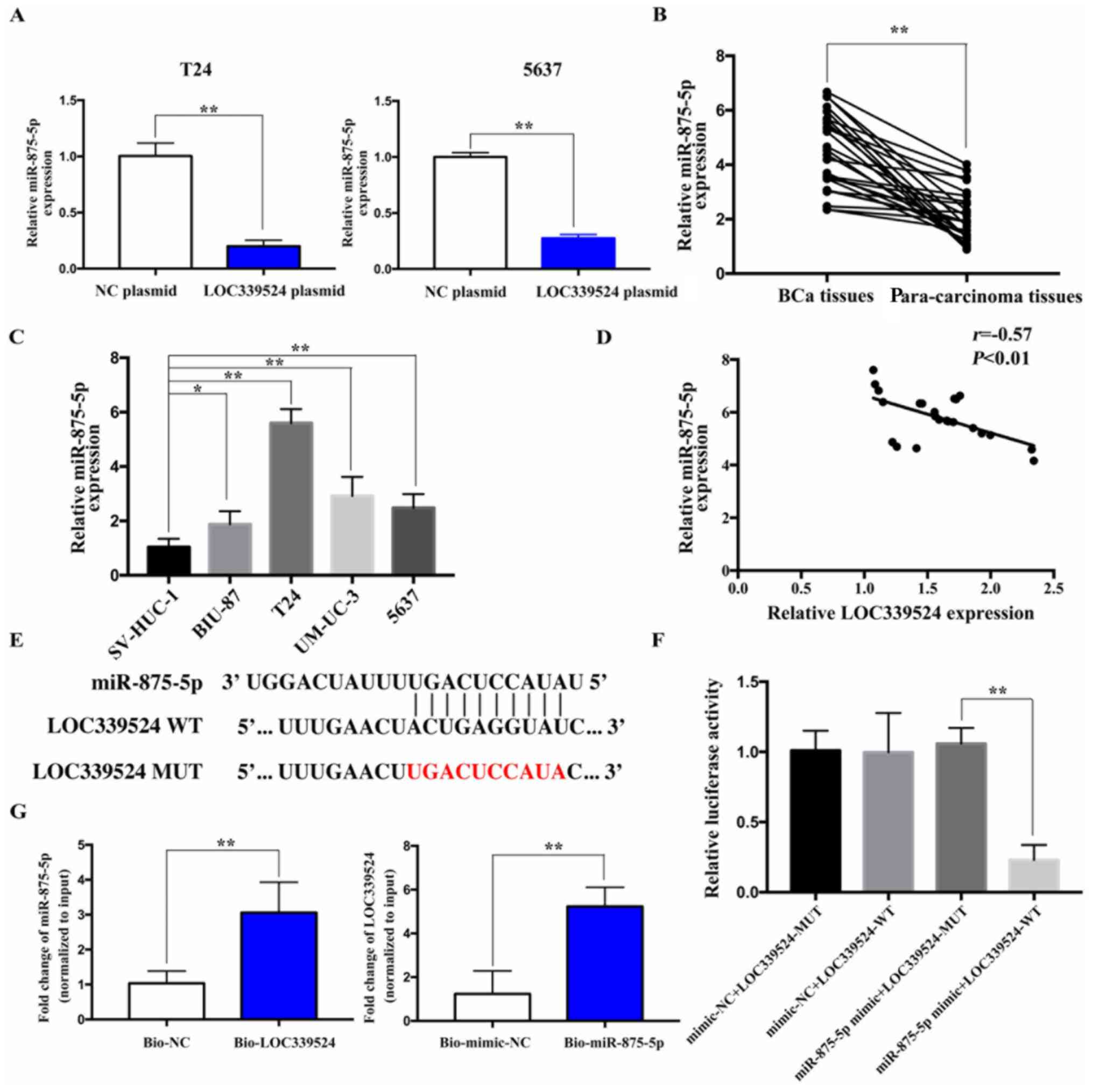
LOC339524 serves as a ceRNA by binding
to miR-875-5p
The DIANA TOOLS database revealed that LOC339524
could possibly bind to miR-875-5p via complementary base pairing.
The expression of miR-875-5p was detected by RT-qPCR after
LOC339524 was overexpressed. The overexpression of LOC339524
significantly downregulated the expression of miR-875-5p compared
with the NC plasmid group in both cell lines (Fig. 3A). Subsequently, the expression
levels of miR-875-5p were detected in 26 BCa and para-carcinoma
tissues using RT-qPCR. miR-875-5p expression was found to
significantly upregulated in BCa samples compared with
para-carcinoma samples (Fig. 3B).
In addition, the expression levels of miR-875-5p in BCa cell lines
were significantly upregulated compared with those in the SV-HUC-1
cells (Fig. 3C). Furthermore, the
expression levels of LOC339524 and miR-875-5p were negatively
correlated with each other in BCa tissues (Fig. 3D). Thus, LOC339524 was hypothesized
to function as a sponge for miR-875-5p.
The fluorescent reporter plasmids, LOC339524-WT and
LOC339524-MUT, containing the miR-875-5p binding site, were
constructed (Fig. 3E). miR-875-5p
overexpression significantly decreased the relative luciferase
activity following co-transfection with LOC339524-WT in 5637 cells,
whereas miR-875-5p overexpression exerted no significant effects on
the relative luciferase activity of the cells following
co-transfection with LOC339524-MUT (Fig. 3F). These findings indicated that
LOC339524 may directly bind to miR-875-5p. A biotin pull-down assay
was then conducted to investigate the endogenous interaction
between LOC339524 and miR-875-5p. The results indicated that
LOC339524 and miR-875-5p were significantly enriched in the
bio-miR-875-5p and bio-LOC339524 groups, respectively, compared
with the bio-NC/mimic-NC group (Fig.
3G).
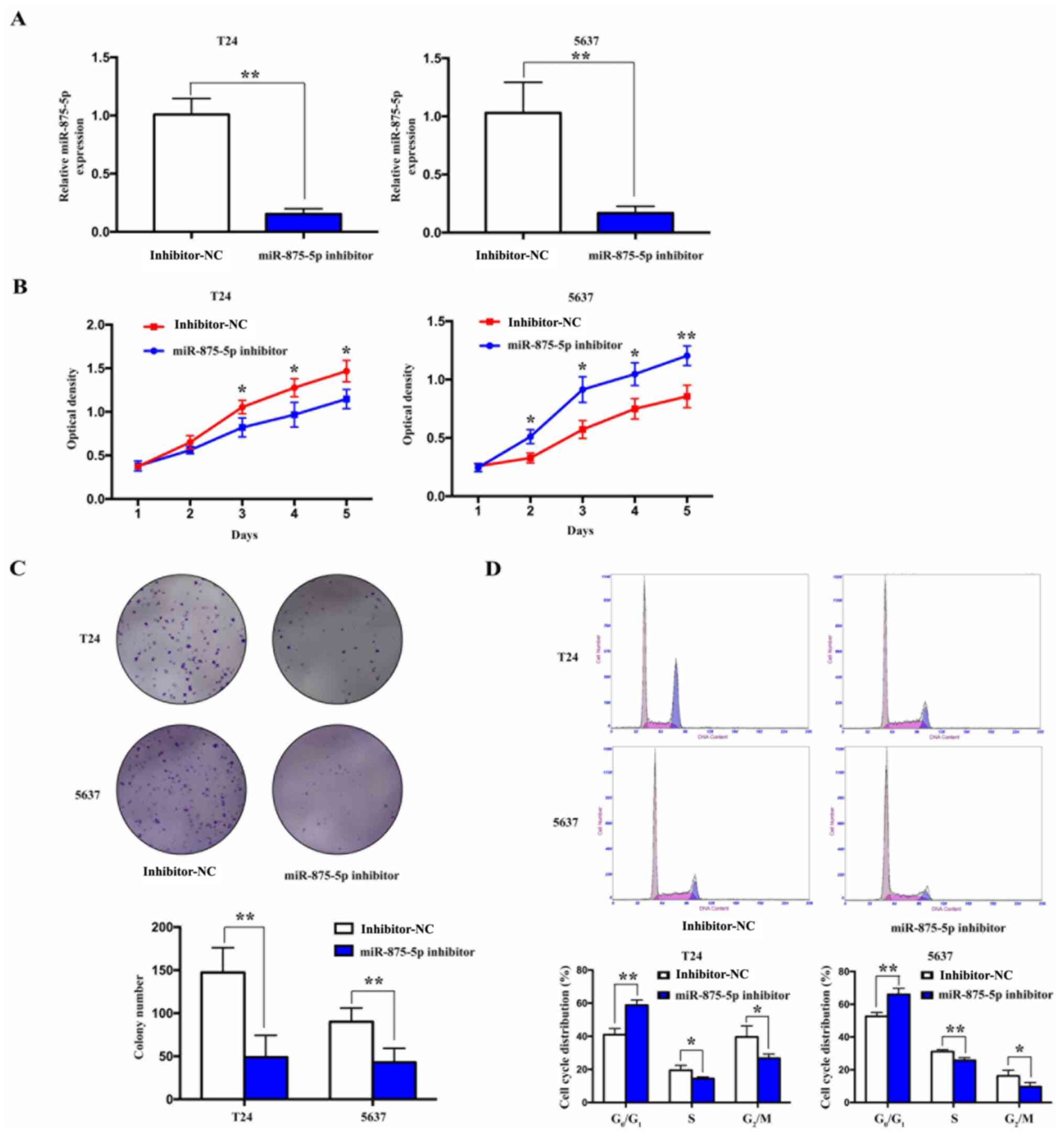
Knockdown of miR-875-5p suppresses the
proliferation of BCa cells
To examine the effects of miR-875-5p expression on
the proliferative ability of BCa cells, miR-875-5p expression was
knocked down in BCa 5637 and T24 cells via transfection with a
miR-875-5p inhibitor. Transfection with miR-875-5p inhibitor
significantly downregulated the expression levels of miR-875-5p
compared with the inhibitor-NC-transfected cells (Fig. 4A). Cell proliferation was then
detected using CCK-8 and colony formation assays. The results
revealed that the proliferation and colony formation ability of the
cells were significantly inhibited following the knockdown of
miR-875-5p compared with the inhibitor-NC transfected cells
(Fig. 4B and C). In addition, cell cycle analysis
revealed that, following the knockdown of miR-875-5p, the number of
cells in the S and G2/M phases decreased, while that in
the G0/G1 phase increased (Fig. 4D). These results indicated that the
knockdown of miR-875-5p may inhibit the proliferation of BCa
cells.
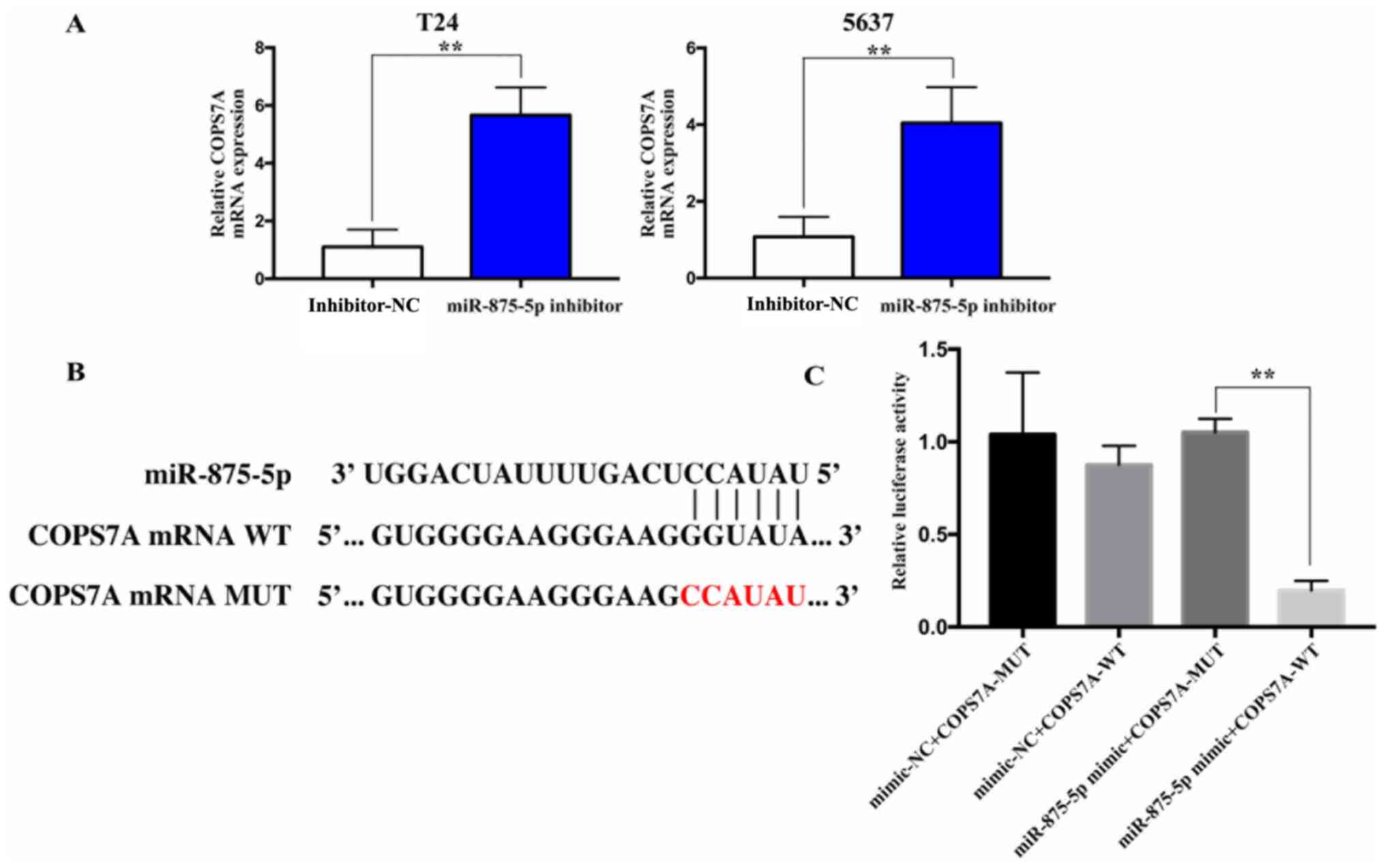
miRWalk and TargetScan databases were screened to
predict the target genes of miR-875-5p, and COPS7A was predicted as
the candidate target gene. The mRNA expression levels of COPS7A
were found to be upregulated following the inhibition of miR-875-5p
in 5637 and T24 cells compared with the inhibitor-NC group
(Fig. 5A). Subsequently, the
luciferase reporter plasmids, COPS7A 3'-UTR-WT and COPS7A
3'-UTR-MUT, containing the WT or MUT miR-875-5p binding site, were
respectively constructed (Fig. 5B).
The results revealed that the 5637 cells co-transfected with
miR-875-5p mimic and the COPS7A 3'-UTR-WT plasmid exhibited a
significant decrease in the relative luciferase activity compared
with cells co-transfected with miR-875-5p mimic and the COPS7A
3'-UTR-MUT. However, the overexpression of miR-875-5p did not
affect the relative luciferase activity of 5637 cells
co-transfected with the COPS7A 3'-UTR-MUT plasmid. These findings
indicated that miR-875-5p may directly bind to COPS7A mRNA
(Fig. 5C).
COPS7A expression is downregulated in
BCa and the downregulation of LOC339524 occurs due to the
hypermethylation of its promoter
The mRNA expression levels of COPS7A were analyzed
in 26 BCa and para-carcinoma tissues using RT-qPCR. COPS7A mRNA
expression levels were discovered to be significantly downregulated
in BCa tissues compared with para-carcinoma tissues (Fig. 6A). Furthermore, a significant
negative correlation was identified between the expression of
COPS7A mRNA and miR-875-5p (Fig.
6B). Following LOC339524 overexpression in 5637 and T24 cells,
the protein expression levels of COPS7A were notably increased,
while the expression of CDK2, CDK4 and cyclin D1 proteins was
notably decreased (Fig. 6C). These
results indicated that LOC339524 may serve as a ceRNA in a network
involving miR-875-5p and COPS7A. The MethHC database revealed the
degree of methylation of the LOC339524 promoter in BCa (Fig. 6D). The results suggested that the
promoter was notably hypermethylated in BCa and that the low
expression of LOC339524 may be associated with the methylation
status. Following treatment with 5 µl 5-Aza-2'-deoxycytidine
(Sigma-Aldrich; Merck KGaA; 2 µmol/l), the expression levels of
LOC339524 were significantly upregulated in BCa cell lines compared
with the vehicle-treated cells (Fig.
6E).
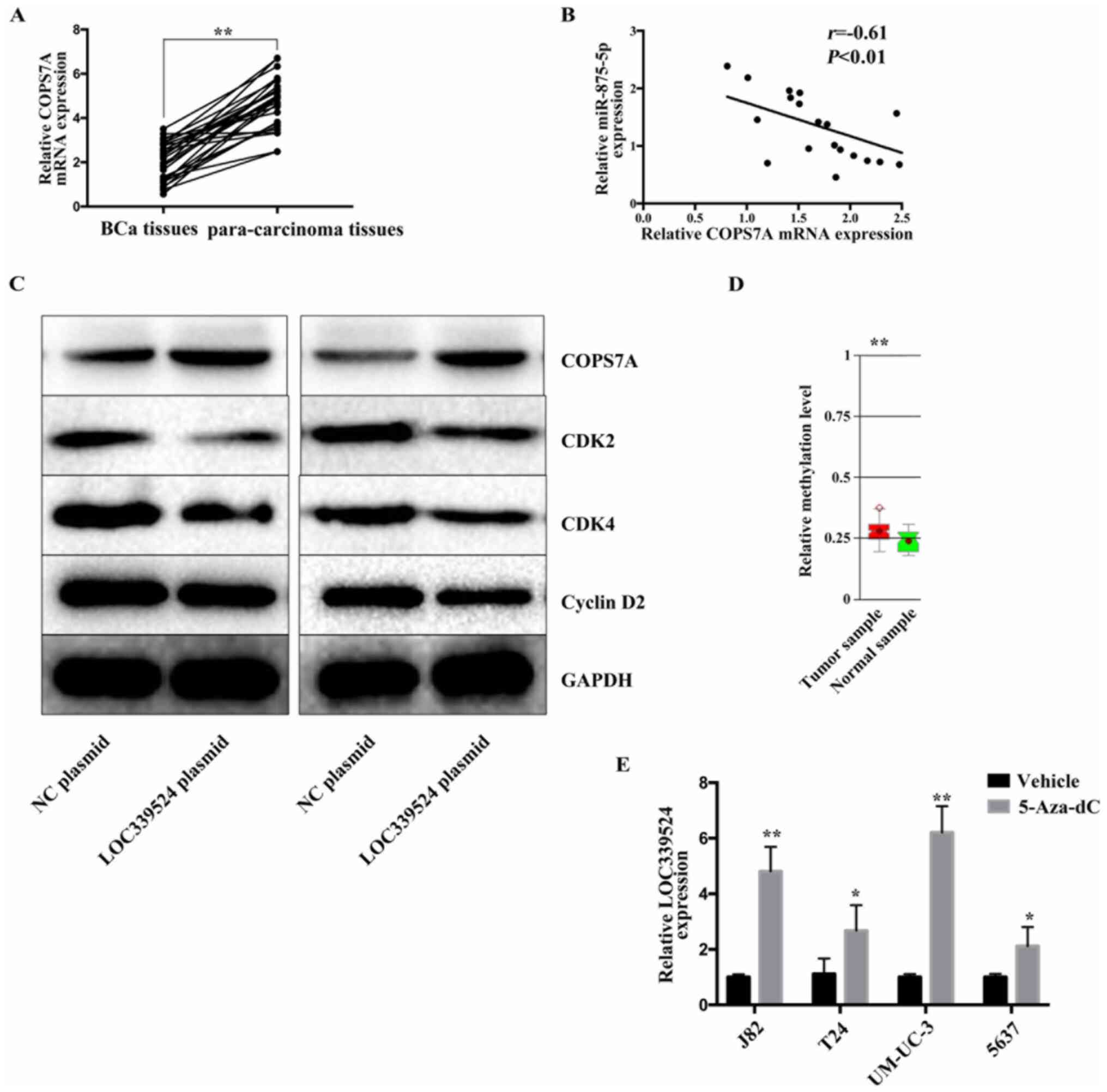 | Figure 6COPS7A is expressed at low levels in
BCa, and the downregulation of LOC339524 occurs due to the
increased methylation of its promoter. (A) Expression of COPS7A was
markedly decreased in BCa tissues compared with in para-carcinoma
tissues. (B) Expression of COPS7A mRNA and miR-875-5p were
negatively correlated in BCa tissues. (C) Following LOC339524
overexpression in 5637 and T24 cell lines, the expression of COPS7A
was notably increased at the protein level, the expression of CDK2,
CDK4 and Cyclin D2 proteins was significantly reduced. (D) MethHC
database was used to determine the DNA methylation status of
LOC339524 in BCa. (E) Expression of LOC339524 in BCa cell lines
following treatment with 5-Aza-CdR. All data are presented as the
mean ± SD. *P<0.05, **P<0.01. miR,
microRNA; BCa, bladder cancer; COPS7A, COP9 signalosome subunit 7A;
NC, negative control; 5-Aza-CdR, 5-Aza-2'-deoxycytidine. |
Discussion
BCa is the commonest malignant cancer of the urinary
system. Surgery and chemotherapy are the most common methods used
to treat BCa (18). Despite
advancements being made in the treatment of BCa, the mortality rate
of patients with advanced-stage BCa remains high, indicating
resistance to treatment (19). To
improve the treatment efficacy for BCa, novel therapeutic
strategies and targets are required. A multitude of lncRNAs have
been identified to serve as critical regulators of the development
of BCa (20). Dai et al
(21) reported that lncRNA integrin
subunit β1 (ITGB1) was upregulated in BCa tissues and affected the
malignant progression of BCa by regulating the expression of
miR-10a. The expression levels of ITGB1 were also found to be
associated with the pathological stage of BCa and were suggested to
be useful as a novel predictor of prognosis. Fang et al
(22) demonstrated that lncRNA
distal-less homeobox 6 antisense 1 (DXL6-AS1) was highly expressed
in BC tissues and acted as a sponge for miR-223. DXL6-AS1 has been
shown to reduce miR-223 expression, thereby enhancing BCa cell
proliferation and invasion. Yang et al (23) reported that lncRNA long intergenic
non-protein coding RNA 319 (LINC00319) was upregulated in BCa
tissues. LINC00319 enhanced cell proliferation and invasion by
functioning as a ceRNA to sponge miR-4492, which attenuated its
downstream target, reactive oxygen species modulator 1. Zhuang
et al (24) found that
lncRNA gastric cancer-associated lncRNA1 (GClnc1) promoted the
proliferation, migration and invasiveness of BCa cells by
regulating the lin-28 homolog B/let-7a/Myc signaling pathway. These
findings indicated that lncRNAs may be used as potential biomarkers
and targets for BCa. According to current research (24), the majority of lncRNAs in BCa serve
as oncogenes. To the best of our knowledge, lncRNAs functioning as
tumor suppressor genes in BCa are rarely reported.
In the present study, abnormally expressed lncRNAs
were screened in BCa using the CRN database and lncRNA LOC339524
was selected for subsequent analysis. The roles of LOC339524 in
types of cancer have not been previously reported, to the best of
the authors' knowledge. The expression levels of LOC339524 were
found to be markedly downregulated in BCa tissues compared with
para-carcinoma tissues. Similarly, compared with SV-HUC-1 cells,
lncRNA LOC339524 expression was low in BCa cells, particularly in
5637 and T24 cells. Thus, 5637 and T24 cells were selected for use
in subsequent analyses. The in vitro experiments identified
that LOC339524 inhibited the proliferation of BCa cells. Previous
studies have demonstrated that lncRNAs can serve as ceRNAs that
sponge miRNAs, thereby affecting the expression of target genes
mediated by miRNAs and regulating the malignant activity of cancer
cells (25,26). Therefore, we hypothesized that
LOC339524 may affect the biological behavior of BCa cells through a
ceRNA mechanism. The present study confirmed that LOC339524 and
miR-875-5p could bind to each other, as demonstrated by
bioinformatics analysis and dual luciferase reporter assays. Wang
et al (27) found that
miR-875-5p was upregulated in non-small cell lung cancer tissues,
and attenuated SATB homeobox 2 expression to the promote
proliferation and invasion of non-small cell lung cancer cells.
Notably, miR-875-5p has been reported to exert tumor-promoting
effects in lung cancer (27).
However, to the best of our knowledge, the role of miR-875-5p in
BCa has not been studied to date. The results of the present study
also revealed that miR-875-5p expression was significantly
upregulated in BCa tissues and cells. The results of the current
study demonstrated that miR-875-5p served as an oncogene in BCa.
COPS7A belongs to the COP9 signalosome complex (CSN) complex
(28). CSN expression has been
shown to control cell cycle progression and to be associated with
carcinogenesis (29). COPS7A
expression was also found to be downregulated in gastric cancer and
suppressed gastric cancer cell proliferation by inactivating the
NF-κB signaling pathway (30). In
the present study, COPS7A expression was found to be downregulated
in BCa tissues. To the best of our knowledge, the present study was
the first to demonstrate that COPS7A is a direct target gene of
miR-875-5p via bioinformatics analysis and dual luciferase reporter
assays. Furthermore, it was identified that the promoter of
LOC339524 in BCa was markedly hypermethylated, and the decreased
expression of LOC339524 was associated with promoter
hypermethylation.
However, there are several limitations in this
study. The expression of LOC339524 were analyzed using limited
sample sizes. The functional experiments in the current study were
conducted at the cellular level and only proved the function of
LOC339524 in BCa cells instead of animals. Further studies are
needed to confirm the expression of LOC339524 in a large sample
size and observe the roles of LOC339524 in animal experiments.
In conclusion, the findings of the present study
demonstrated that the expression levels of LOC339524 were
downregulated in BCa tissues and cells. It was also found that
LOC339524 suppressed the proliferation of BCa cells in
vitro. Therefore, LOC339524 was proposed to function as a ceRNA
that promotes the expression of COPS7A by binding and sponging
miR-875-5p.
Supplementary Material
Primers used for reverse
transcription-quantitative PCR.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BW and XH designed the experiments; HX, RY and HC
performed the experiments; BW and XH analyzed and interpreted the
data and drafted the manuscript. All authors read and approved the
final manuscript. BW and XH confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Third Affiliated Hospital, Army Medical University
(Chongqing, China). All patients were informed of the study
protocol and signed written informed consent forms.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN Estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Abdolmaleki F, Ghafouri-Fard S, Taheri M
and Omrani DM: P21-Associated ncRNA DNA damage-activated expression
in bladder cancer. Klin Onkol. 32:277–280. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Jiang D, Zhang Y, Yang L, Lu W, Mai L, Guo
H and Liu X: Long noncoding RNA HCG22 suppresses proliferation and
metastasis of bladder cancer cells by regulation of PTBP1. J Cell
Physiol. 2:1711–1722. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hu J, Shan Y, Ma J, Pan Y, Zhou H, Jiang L
and Jia L: lncRNA ST3Gal6-AS1/ST3Gal6 axis mediates colorectal
cancer progression by regulating alpha-2,3 sialylation via PI3K/Akt
signaling. Int J Cancer. 145:450–460. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bai J, Xu J, Zhao J and Zhang R:
Downregulation of lncRNA AWPPH inhibits colon cancer cell
proliferation by downregulating GLUT-1. Oncol Lett. 18:2007–2012.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li Y, Zhao L, Zhao P and Liu Z: Long
non-coding RNA LINC00641 suppresses non-small-cell lung cancer by
sponging miR-424-5p to upregulate PLSCR4. Cancer Biomark. 26:79–91.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lee YR, Kim G, Tak WY, Jang SY, Kweon YO,
Park JG, Lee HW, Han YS, Chun JM, Park SY and Hur K: Circulating
exosomal noncoding RNAs as prognostic biomarkers in human
hepatocellular carcinoma. Int J Cancer. 144:1444–1452.
2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Li J, Wang W, Xia P, Wan L, Zhang L, Yu L,
Wang L, Chen X, Xiao Y and Xu C: Identification of a five-lncRNA
signature for predicting the risk of tumor recurrence in patients
with breast cancer. Int J Cancer. 143:2150–2160. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yao F, Wang Q and Wu Q: The prognostic
value and mechanisms of lncRNA UCA1 in human cancer. Cancer Manag
Res. 11:7685–7696. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang Y, Liao G, Bai J, Zhang X, Xu L,
Deng C, Yan M, Xie A, Luo T, Long Z, et al: Identifying cancer
driver lncRNAs bridged by functional effectors through integrating
multi-omics data in human cancers. Mol Ther Nucleic Acids.
17:362–373. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Dai G, Huang C, Yang J, Jin L, Fu K, Yuan
F, Zhu J and Xue B: lncRNA SNHG3 promotes bladder cancer
proliferation and metastasis through miR-515-5p/GINS2 axis. J Cell
Mol Med. 24:9231–9243. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li Y, Shi B, Dong F, Zhu X, Liu B and Liu
Y: lncRNA KCNQ1OT1 facilitates the progression of bladder cancer by
targeting miR-218-5p/HS3ST3B1. Cancer Gene Ther. 28:212–220.
2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li JR, Sun CH, Li W, Chao RF, Huang CC,
Zhou XJ and Liu CC: Cancer RNA-Seq Nexus: A database of
phenotype-specific transcriptome profiling in cancer cells. Nucleic
Acids Res. 44:D944–D951. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM and Hatzigeorgiou
AG: DIANA-lncBase: Experimentally verified and computationally
predicted microRNA targets on long non-coding RNAs. Nucleic Acids
Res. 41:D239–D245. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Huang HY, Li J, Tang Y, Huang YX, Chen YG,
Xie YY, Zhou ZY, Chen XY, Ding SY, Luo MF, et al: MethHC 2.0:
Information repository of DNA methylation and gene expression in
human cancer. Nucleic Acids Res. 49:D1268–D1275. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yuan S, Luan X, Han G, Guo K, Wang S and
Zhang X: LINC01638 lncRNA mediates the postoperative distant
recurrence of bladder cancer by upregulating ROCK2. Oncol Lett.
18:5392–5398. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Hu X, Feng H, Huang H, Gu W, Fang Q, Xie
Y, Qin C and Hu X: Downregulated long noncoding RNA PART1 inhibits
proliferation and promotes apoptosis in bladder cancer. Technol
Cancer Res Treat. 18(1533033819846638)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Lyu L, Xiang W, Zhu JY, Huang T, Yuan JD
and Zhang CH: Integrative analysis of the lncRNA-associated ceRNA
network reveals lncRNAs as potential prognostic biomarkers in human
muscle-invasive bladder cancer. Cancer Manag Res. 11:6061–6077.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Dai L, Chai CM, Shen TY, Tian Y, Shang ZQ
and Niu YJ: lncRNA ITGB1 promotes the development of bladder cancer
through regulating microRNA-10a expression. Eur Rev Med Pharmacol
Sci. 23:6858–6867. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Fang C, Xu L, He W, Dai J and Sun F: Long
noncoding RNA DLX6-AS1 promotes cell growth and invasiveness in
bladder cancer via modulating the miR-223-HSP90B1 axis. Cell Cycle.
18:3288–3299. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yang Y, Zhang F, Huang H, Xie Z, Huang W,
Xie H and Wang F: Long noncoding RNA LINC00319 regulates ROMO1
expression and promotes bladder cancer progression via
miR-4492/ROMO1 axis. J Cell Physiol. 4:3768–3775. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhuang C, Ma Q, Zhuang C, Ye J, Zhang F
and Gui Y: lncRNA GClnc1 promotes proliferation and invasion of
bladder cancer through activation of MYC. FASEB J. 33:11045–11059.
2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Peng H and Li H: The encouraging role of
long noncoding RNA small nuclear RNA host gene 16 in
epithelial-mesenchymal transition of bladder cancer via directly
acting on miR-17-5p/metalloproteinases 3 axis. Mol Carcinog.
58:1465–1480. 2019.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Peng Y, Leng W, Duan S and Hong M: Long
noncoding RNA BLACAT1 is overexpressed in hepatocellular carcinoma
and its downregulation suppressed cancer cell development through
endogenously competing against hsa-miR-485-5p. Biomed Pharmacother.
116(109027)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang J, Lu Y, Ding H, Gu T, Gong C, Sun J,
Zhang Z, Zhao Y and Ma C: The miR-875-5p inhibits SATB2 to promote
the invasion of lung cancer cells. Gene. 644:13–19. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Davidsson J and Johansson B: Methylation
and expression analyses of Pallister-Killian syndrome reveal
partial dosage compensation of tetrasomy 12p and hypomethylation of
gene-poor regions on 12p. Epigenetics. 11:194–204. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yoshida A, Yoneda-Kato N, Panattoni M,
Pardi R and Kato JY: CSN5/Jab1 controls multiple events in the
mammalian cell cycle. FEBS Lett. 584:4545–4552. 2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zheng J, Zhang H, Ma R, Liu H and Gao P:
Long non-coding RNA KRT19P3 suppresses proliferation and metastasis
through COPS7A-mediated NF-κB pathway in gastric cancer. Oncogene.
45:7073–7088. 2019.PubMed/NCBI View Article : Google Scholar
|