Introduction
Nasopharyngeal carcinoma (NPC) is a head and neck
malignant tumour derived from nasopharyngeal epithelium and has a
high incidence rate, with >20 cases per 100,000 individuals in
southern China (1). Several factors
contribute to NPC occurrence and development, including
Epstein-Barr virus (EBV) infection, chemical carcinogens, genetic
variation and spontaneous mutation (2). NPC is characterized by metastases via
both lymph and blood vessels at the early stage of the disease
(3), and subsequently induces the
development of distant metastases to the bone, lung and liver,
which are major causes of treatment failure after different
treatments (4). Although much
effort has been devoted, the mechanisms that regulate NPC malignant
behaviours, including proliferation, migration, invasion and colony
formation, remain poorly understood.
Small non-coding microRNAs (miRNAs/miRs) are a group
of small, non-coding RNA molecules that play critical roles in
regulating tumorigenesis processes. Moreover, accumulating evidence
has demonstrated that abnormalities in the expression profiles of
miRNAs are a hallmark of tumorigenesis and development (5). By acting as tumour promotors or
inhibitors, miRNAs tightly regulate the occurrence, development and
progression of cancer (6). miR-144
is an miRNA identified as a tumour regulator in several types of
cancer, including lung cancer, nasopharyngeal carcinoma,
hepatocellular carcinoma and clear cell renal cell carcinoma
(7-10).
However, miR-144 has been reported to act as either an oncogene or
tumour suppressor in different types of cells, which has raised an
interesting study topic on its regulatory role in NPCs.
Insulin receptor substrate (IRS) proteins are a
group of signalling intermediates downstream of cytoplasmic
molecules that play critical roles in several diseases, including
cancer (11). One member of IRS,
termed insulin receptor substrate-1 (IRS-1), was reported to be
associated with tumour initiation and progression. IRS-1, activated
by insulin/insulin-like growth factor I receptors, induces the
transduction of intracellular cascades, including activating
PI3K-AKT and Ras-MAPK pathways (12). As IRS-1 regulates several basal
processes, including inhibiting autophagy and decreasing oxidative
stress-induced apoptosis, the overexpression of IRS-1 promotes
malignant behaviours in several cancer cells (13,14). A
study reported that IRS-1 is downregulated by miR-145 and leads to
the inhibition of proliferation in colon cancer cells (15). Wu et al (16) reported that, in laryngeal squamous
cell carcinoma, miR-144 suppresses the growth and metastasis by
targeting IRS1, indicating that miRNAs, including miR-144, may
regulate malignant behaviours in cancer by regulating IRS-1.
Dishevelled (Dvl)2 is a cytoplasm-nucleus shuttling
protein and thus tightly regulates Wnt signaling (17), which is regulated by a variety of
factors, including ubiquitin-proteasome and autophagy-lysosomal
pathways (18,19). Zhang et al (18) reported that Dvl2 is bound by
GABARAPL1, a member of the ATG8 family, and results in degradation
through autophagy. Further research also revealed that IRS-1
interacts directly with Dvl2, and overexpression of IRS-1 increased
the protein level of Dvl2 by inhibiting ubiquitination and
subsequent degradation (20).
Subsequently, Dvl2 stabilized by IRS-1 promotes Wnt/β-catenin
signalling and thus promotes epithelial-mesenchymal transition
(EMT) and cell proliferation in response to Wnt stimulation
(20). These results indicate that
miRNAs may regulate Dvl2 signalling by post-transcriptional
regulation of IRS-1 and modifying physiological processes.
The present study focused on the biological
functions and underlying molecular mechanisms of miR-144 via its
regulation of IRS-1 and subsequent regulation of malignant
behaviors of NPCs.
Materials and methods
Cell culture
Nasopharyngeal carcinoma cell lines NPC-TW01 and
SUNE1 were bought from The Cell Bank of Type Culture Collection of
The Chinese Academy of Sciences. All cells were maintained in
RPMI-1640 medium (Thermo Fisher Scientific, Inc.) supplemented with
10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) in a
humidified atmosphere containing 5% CO2 at 37˚. Cells
were passaged every three days.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was isolated using TRIzol®
according to manufacturer's instruction and was reverse transcribed
into complementary DNA using Bio-Rad I Script cDNA Synthesis System
(Bio-Rad Laboratories, Inc.) at 42˚C for 60 min and denatured at
80˚C for 10 min. SYBR-Green PCR Master mix (Thermo Fisher
Scientific, Inc.) was then employed for qPCR according to
manufacturer's instruction. The primers used were as follows: IRS-1
forward, 5'-CAAGACCATCAGCTTCGTGA-3'; IRS-1 reverse, 5'-AGA
GTCATCCACCTGCATCC-3'; Dvl2 forward, 5'-GCCTATCCA GGTTCCTCCTC-3';
Dvl2 reverse, 5'-AGAGCCAGTCAACC ACATCC-3'; β-actin forward,
5'-CATGTACGTTGCTATCCA GGC-3'; and β-actin reverse,
5'-CTCCTTAATGTCACGCAC GAT-3'. For quantitative detection of miRNAs,
total RNA was revere transcribed using All-in-One™ miRNA qRT-PCR
Detection System (Guangzhou RiboBio Co., Ltd.) following the
manufacturer's instructions. Primer for miR-144 (cat. no.
HmiRQP0910) and RNU6-2 (cat. no. HmiRQP9001) were bought from
Guangzhou RiboBio Co., Ltd. Relative quantification of mRNA and
miRNA expression were calculated by using the 2-ΔΔCq
method (21). All RT-qPCR reactions
were performed in triplicates.
Western blotting
The cells were lysed using RIPPA buffer
(Sigma-Aldrich; Merck KGaA) and total protein concentration was
quantified using the BCA protein assay kit (Sigma-Aldrich; Merck
KGaA) following the manufacturer's instructions. Total protein (20
µg) was separated by 10% sodium dodecyl sulfate polyacrylamide gel
electrophoresis, transferred to PVDF membrane and blocked with
TBS-Tween (0.1%) containing 5% bovine serum albumin (Sigma-Aldrich;
Merck KGaA) at room temperature for 30 min, and probed with the
specific primary antibodies at room temperature for 60 min.
Anti-IRS-1 antibody (cat. no. ab52167), anti-Dvl2 antibody (cat.
no. ab22616) and anti-β-actin antibody (cat. no. ab8226) were all
bought from Abcam, and diluted in 1:2,000 before use. Then, the
blot was incubated with specific horseradish peroxidase
(HRP)-conjugated secondary antibodies (cat. nos. ab7090 or ab97040;
Abcam) at dilution of 1:1,000, and the immunoreactive bands were
imaged using enhanced chemiluminescence (Thermo Fisher Scientific,
Inc.). The blots were analyzed using ImageJ software (version-2.0;
National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
For cell proliferation assay, 5x103 cells
were seeded into 96-well plates and were analyzed at the indicated
time points (24, 48, 72, 96 and 120 h). Then, 10 µl CCK-8 reagent
(Sigma-Aldrich; Merck KGaA) was added into each well at 37˚C for
another 2-h incubation. The OD470 nm value was determined by a
microplate reader (Synergy 2 Multi-Mode Microplate Reader; BioTek
Instruments, Inc.).
Transfection
PcDNA3.1-Dvl2 plasmid was synthesized by Invitrogen
(Thermo Fisher Scientific, Inc.), and empty vector pcDNA3.1 was
employed as negative control. Small interfering (si)RNA target to
IRS-1 (5'-CAAUGAGUGUGCAUAAACUUC-3'), miR-144 mimic
(5'-UACAGUAUAGAUGAUGUACU-3'), miR-144 inhibitor
(5'-UACAGUAUAGAUGAUGUACU-3') and corresponding negative controls
were bought from Guangzhou RiboBio Co., Ltd.
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) was
employed for transfection. For plasmid transfection, 1.6 µg of
plasmid was used. For mimic, inhibitor and siRNA transfection, 50
µM of sample was employed following the manufacturer's instruction.
After transfection (4 h), the cells were employed for further
analyses.
Cell cycle analysis
The cells of interest were trypsinized and suspended
with PBS, and then pelleted. After discarding the supernatant,
cells were resuspended and fixed with ice-cold 70% ethanol. After 4
h, cells were pelleted and washed with ice-cold PBS, and suspended
with 5 µg/ml propidium iodide (PI; Sigma-Aldrich; Merck KGaA) for
10 min in darkness at room temperature. Then, stained cells were
analyzed using 3 laser Navios flow cytometers (Beckman Coulter,
Inc.) and results were analyzed using FlowJo software (FlowJo LLC;
version 9.7.4).
Scratch assay
When the cells reached 95% confluence, cells were
scratched with a sterile plastic micropipette tip. After wounding,
plates were washed three times with ice-cold PBS, and RIPM-1640
without FBS was added for 24-h incubation. After culturing for 24
h, images were captured by an inverted fluorescence microscope
(Nikon Corporation; magnification, x40) and the wound healing
ability of each cell line was analyzed. Distances between the edges
of the scratch were measured using ImageJ software (version-2.0;
National Institutes of Health) to quantitatively evaluate cell
migration. Cell migration was calculated according to the following
formula: Cell migration ratio = 100 x (0 h scratch width-24 h
scratch width)/0 h scratch width.
Transwell assay
In order to analyze invasion capacity,
1x104 cells were suspended and plated into the upper
chamber of Transwell inserts (8-µm pore size; Corning Inc.), coated
with 0.3% Matrigel (Sigma-Aldrich; Merck KGaA) in RPMI-1640 medium
without FBS addition, and maintained in an incubator at 37˚C. In
the lower chamber, 500 µl RPMI-1640 medium supplemented with 10%
FBS was added. After 24-h incubation, the invaded cells were fixed
with 4% paraformaldehyde at room temperature for 10 min and stained
with 0.25% crystal violet (Sigma-Aldrich; Merck KGaA) at room
temperature for 10 min. Invaded cells were then imaged under a X71
(U-RFL-T) fluorescence microscope (Olympus Corporation;
magnification, x40).
Colony formation in soft agar
Collected cells were counted and resuspended in 0.3%
low-melting soft agar in RPMI-1640 containing 10% FBS, then plated
onto 0.6% solidified agar in RPMI-1640 containing 10% FBS in 6-well
plates (5x103 cells/well). The samples were maintained
at 37˚C for 3 weeks. Colonies containing 50 cells or more were
counted under a X71 (U-RFL-T) fluorescence microscope (Olympus
Corporation; magnification, x40).
Statistical analysis
In the present study, all data are presented as the
mean ± SD. One-way analysis of variance (ANOVA) was performed to
compare multiple groups with one variable followed by Tukey's
post-hoc test. Significance between two groups was calculated by
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-144
post-transcriptionally regulates IRS-1 in NPC cells
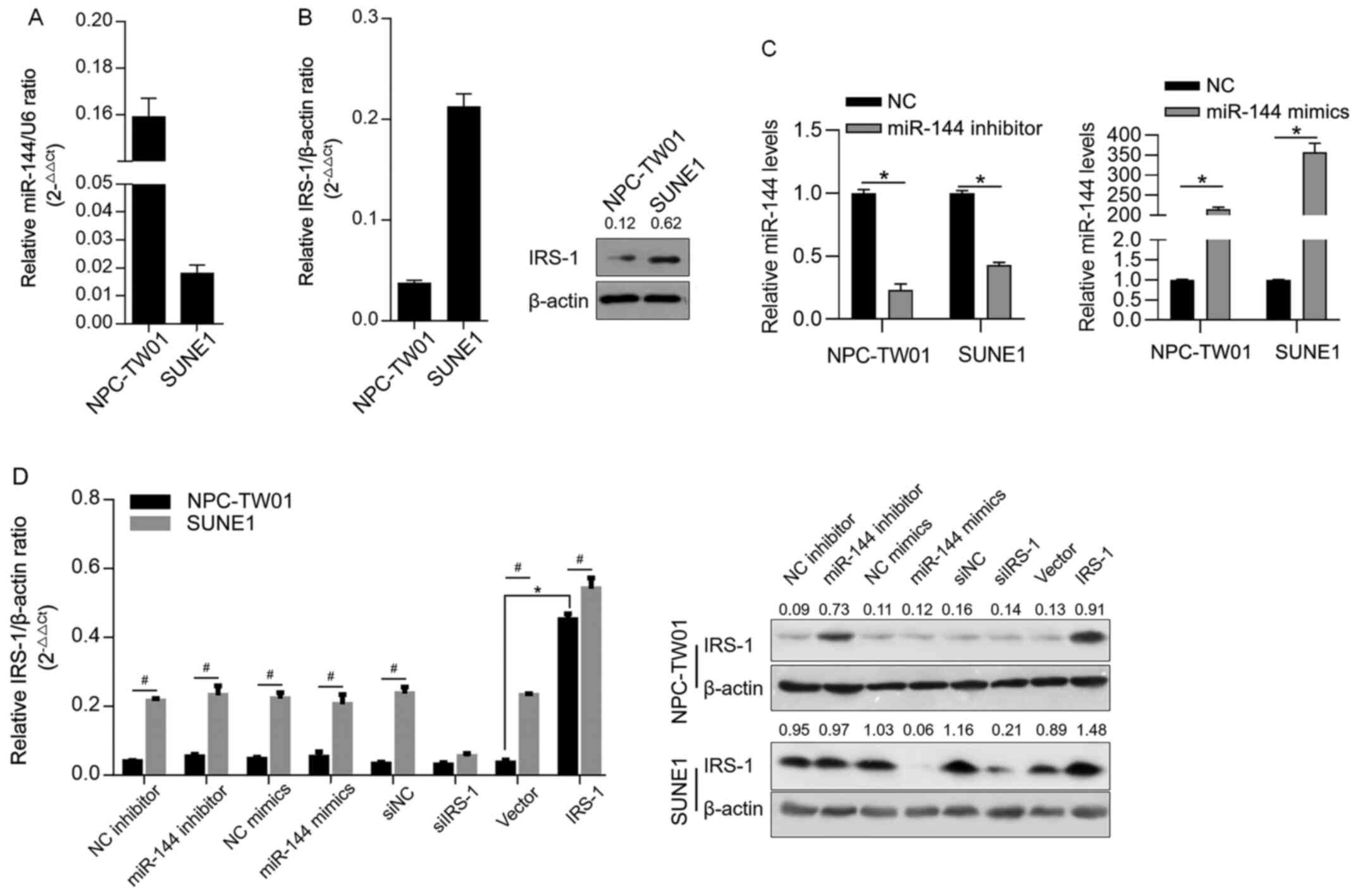
In order to determine the expression level of
miR-144 in NPC cell lines, RT-qPCR was performed to detect miR-144
level normalized to U6 RNA. It was observed that miR-144 was
detected in all tested NPC cell lines with different expression
profiles (Fig. 1A). The protein
levels of IRS-1 in the cell lines were then detected by
semi-quantitative western blotting. As shown in Fig. 1B, IRS-1 was detected in all these
cell lines. By considering that the expression levels of miR-144
and IRS-1 were contrasting in NPC-TW01 and SUNE1 cells, these two
cells were selected for further analysis. To confirm whether
miR-144 post-transcriptionally regulates IRS-1, miR-144 inhibitor,
miR-144 mimic, siIRS-1 or IRS-1 were efficiently transfected,
confirmed by RT-qPCR (Fig. 1C), and
IRS-1 protein was measured 48 h later. As shown in Fig. 1D, without affecting IRS-1 mRNA, the
introduction of miR-144 inhibitor increased IRS-1 protein, whereas
the miR-144 mimic downregulated the expression level of IRS-1
protein, which was similar to the observation following siIRS-1
transfection. Briefly, miR-144 post-transcriptionally regulates
IRS-1.
miR-144 inhibits cell proliferation by
targeting IRS-1 in NPC cells
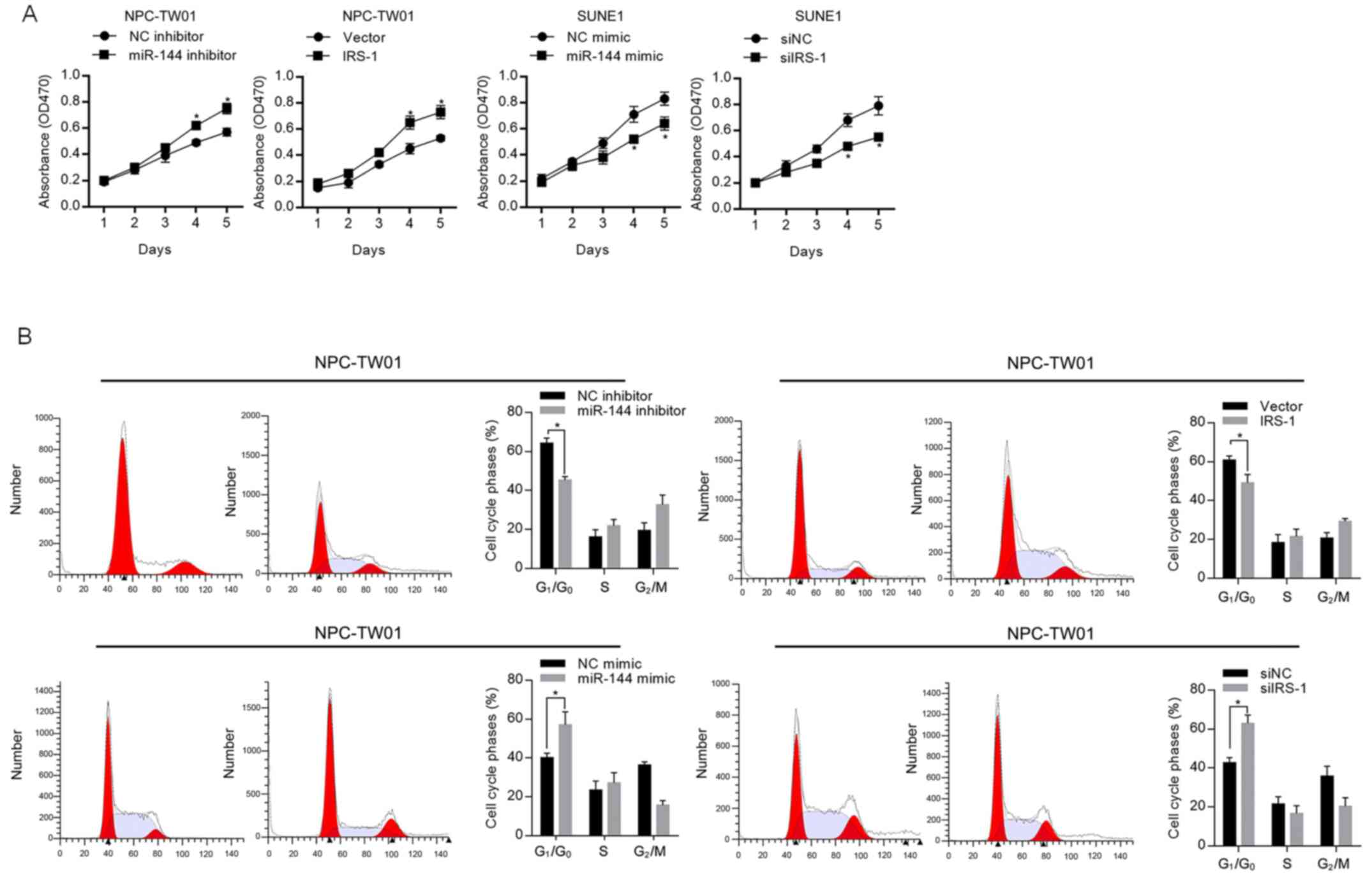
In order to evaluate the effect of miR-144 on
proliferation, the expression level of miR-144 in NPC-TW01 or SUNE1
was modified by introducing miR-144 inhibitor or miR-144 mimic.
CCK-8 cell viability assay results presented that, in NPC-TW01
cells, both the inhibition of miR-144 and overexpression of IRS-1
significantly promoted cell proliferation, and in SUNE1 cells, both
the introduction of miR-144 mimic and knockdown of IRS-1
significantly inhibited cell proliferation (Fig. 2A). The opposite effect of miR-144
and IRS-1 on cell proliferation indicated that miR-144 potentially
regulates cell proliferation via regulating IRS-1. To further
confirm the regulation of proliferation by miR-144, cell cycle was
then assessed by flow cytometry. As shown in Fig. 2B, miR-144 expression significantly
blocked the cell cycle at the G1/G0 phase,
which is in contrast to the observation in the presence of
IRS-1.
miR-144 decreases malignant behaviors
in NPC cells
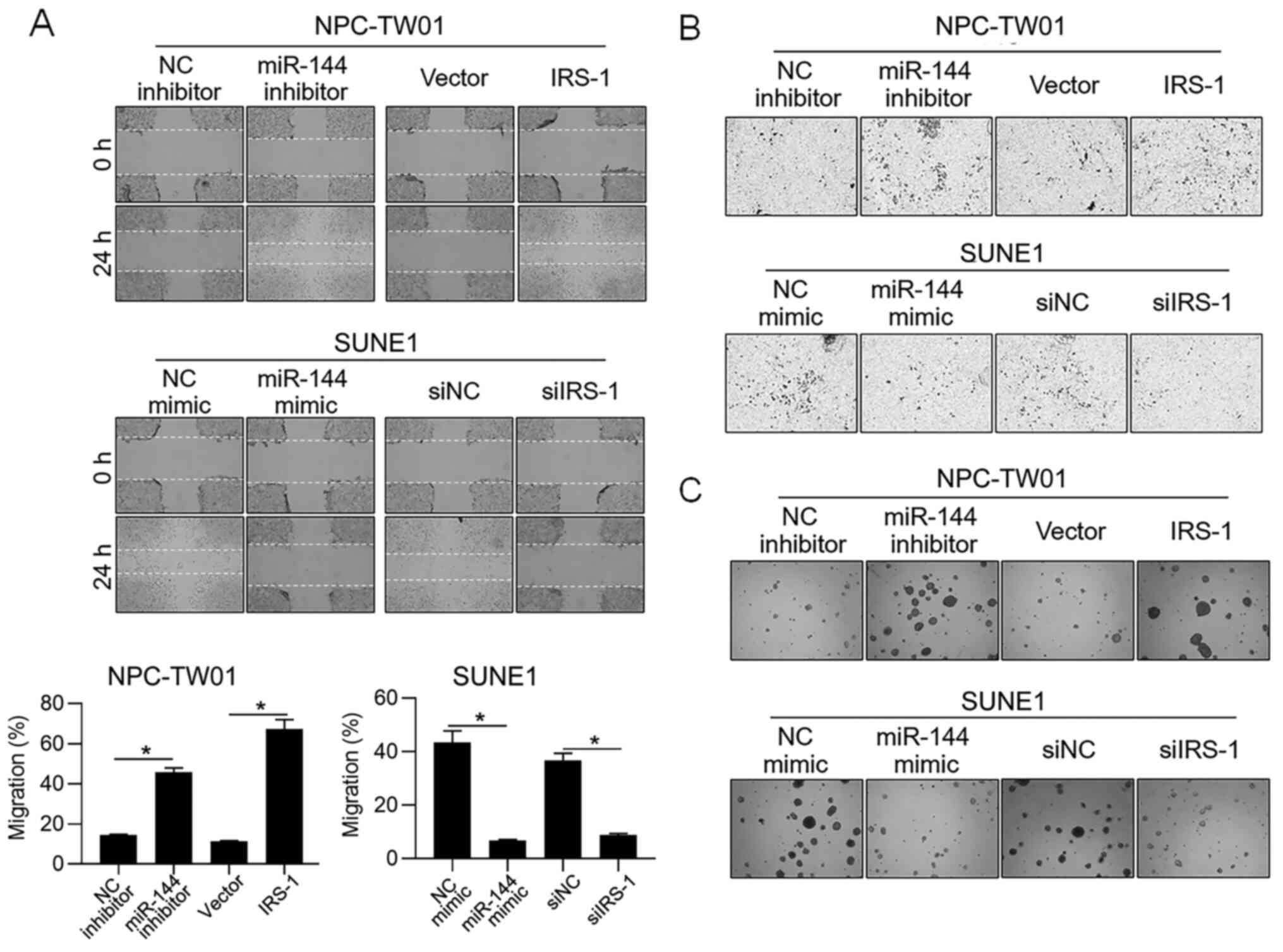
In order to further access whether miR-144 is
relevant in other malignant behaviors of NPC cells, cell migration,
invasion and tumor formation in soft agar were examined after
modification of miR-144 or IRS-1. As shown in Fig. 3A-C, in NPC-TW01 cells, inhibition of
miR-144 and overexpression of IRS-1 promoted migration, invasion
and colony formation in soft agar, and in SUNE1, the introduction
of miR-144 mimic and knockdown of IRS-1 obviously inhibited
migration, invasion and colony formation in soft agar. All these
observations are consistent with the effect of miR-144 on
proliferation, indicating that miR-144 exerted inhibitory effects
not only on proliferation.
miR-144 acts as a tumor suppressor
potentially via downregulating Dvl2 indirectly
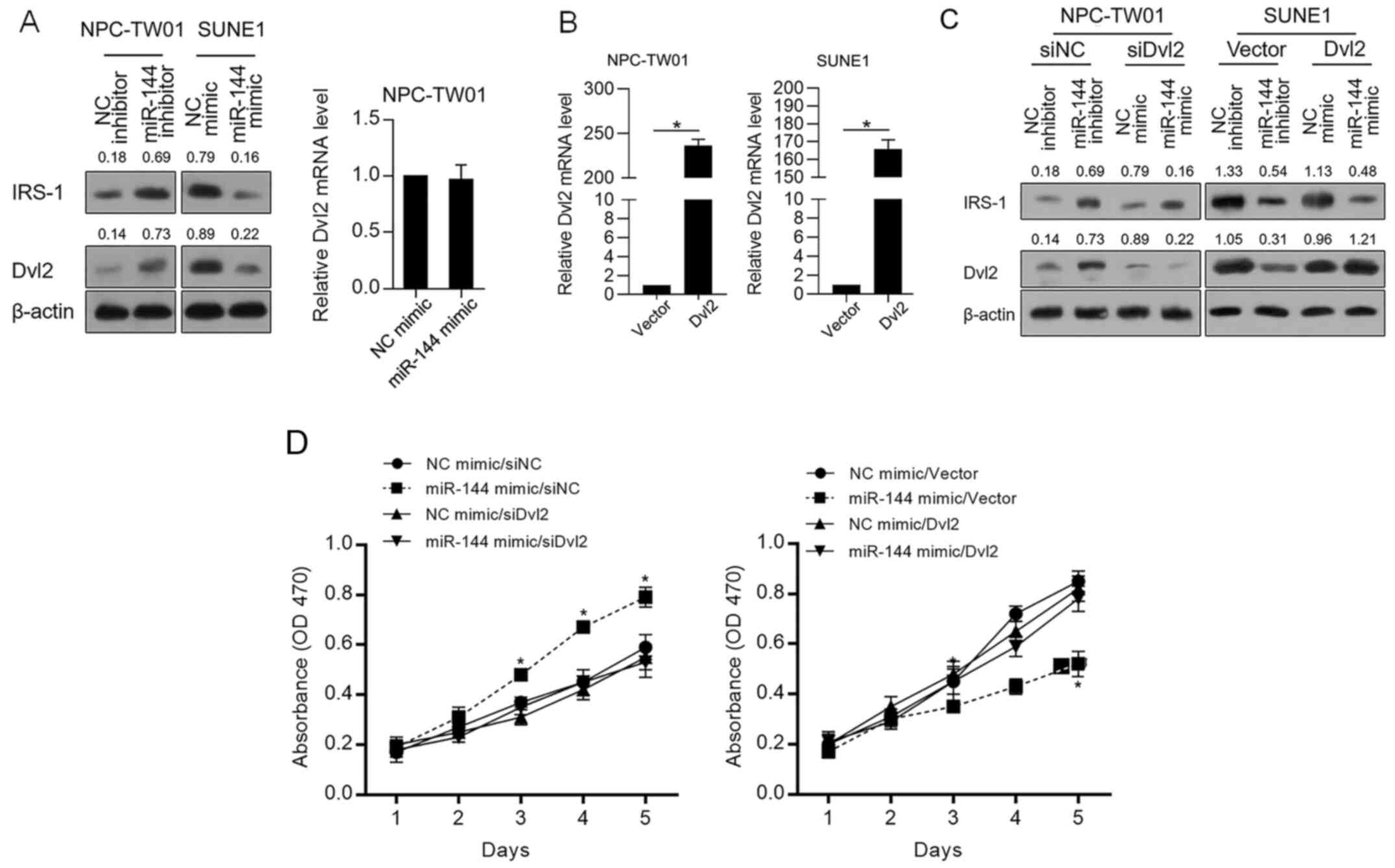
Accordingly, IRS-1 stabilizes Dvl2 and thus promotes
malignant behaviors in cancer cells (20). Thus, whether miR-144 regulates Dvl2
protein level was further confirmed. As shown in Fig. 4A, miR-144 negatively regulated IRS-1
and Dvl2 without disturbing Dvl2 mRNA, indicating that miR-144 may
regulate the proliferation of NPC cells via indirectly regulating
Dvl2. Thus, Dvl2 was modified in NPC-TW01 and SUNE1 cells by
efficiently introducing a Dvl2-coding vector (Fig. 4B). As shown in Fig. 4C, Dvl2 protein expression was
successfully modified despite the level of IRS-1. Then, cell
proliferation was analyzed. In Fig.
4D, it was observed that the inhibitory effect of miR-144 on
proliferation was significantly reversed by overexpression of Dvl2,
demonstrating that the inhibitory effect of miR-144 on
proliferation depends on the decrease in Dvl2.
Discussion
Accumulating evidence indicates that miRNAs exert
significant roles in the pathogenesis of NPC by regulating tumour
cell proliferation, metastasis, invasion and chemosensitivity
(22). For example, Qiu et
al (23) reported that in NPC
cells, miR-29a/b targeted the COL3A1 and SPARC genes and thus
enhanced cell migration, invasion and proliferation. Verhoeven
et al (24) reported that
miRNA and long non-coding RNA are regulated by high levels of EBV,
and overexpressed miRNAs then promote malignant behaviors in NPC
cells. It has also been reported that miR-24 promotes DNA
methylation and then acts as a radiosensitizer in NPC cells.
Further analysis revealed that miR-24 is downregulated in NPC
tissues compared with adjacent tissues (25). The present study found that miR-144
expression is differentially expressed in different NPC cell lines,
and its expression is negatively correlated with the IRS-1 level,
which was reported to be a target that is downregulated
post-transcriptionally. Overexpression of the miR-144 mimic
remarkably decreased the malignant behaviors of NPCs, including
proliferation, migration, invasion and tumour formation in soft
agar. Conversely, the downregulation of miR-144, by introducing a
specific inhibitor, promoted the aforementioned malignant
behaviors. Zhang et al (8)
reported that miR-144 promoted NPC malignancy by inhibiting PTEN
expression, which is in contradiction with the present study
results. The difference may be due to the different expression
patterns of PTEN in different NPC cell lines, and it is worth
investigating the roles of PTEN in additional NPC cell lines.
Dvl2 is mainly involved in the regulation of
Wnt/β-catenin signaling (17). In
recent research, Dvl2 was found to directly bind to the IRS-1
protein (20). IRS-1 binds to Dvl2
with or without insulin stimulation, and its overexpression
increases the protein level of Dvl2, thus promoting canonical Wnt
signalling and increasing the expression of CYCLIN D1 and C-MYC,
which are recognized as oncogenes (20). Further research showed that the
binding of IRS-1 to Dvl2 decreased ubiquitination and induced the
stabilization of Dvl2. According to the present study results and
consistent with previous findings, downregulation of IRS-1 by
miR-144 introduction decreased the protein level of Dvl2. Although
the binding of IRS-1 and Dvl2 was not investigated further, the
presented results also demonstrated that the regulation of IRS-1 by
miR-144 negatively regulated Dvl2. Moreover, the inhibitory effect
of the miR-144 mimic on proliferation was significantly abolished
by the exogenous expression of Dvl2, indicating that the regulatory
effects of miR-144 are potentially, at least in part, dependent on
the regulation of Dvl2.
IRS has been identified as a key player in the
regulation of cell proliferation and epithelial mesenchymal
transition (EMT), which is characterized by the expression levels
of E-cadherin and N-cadherin. It has been reported that the
overexpression of IRS-1 regulates the expression of E-cadherin
during EMT (26,27) and cyclinD1 during cell proliferation
(28). Although the hallmarks of
EMT or the effects of miR-144 on EMT were not detected, by
performing scratch and transwell assays, it was confirmed that
miR-144 inhibited the migration and invasion of NPC cells, which
was similar to the effect of IRS-1 knockdown being reversed by the
introduction of a miR-144 inhibitor, indicating that miR-144
potentially regulates EMT. By considering that miR-144
post-transcriptionally regulates IRS-1 by direct binding (29), dual-luciferase reporter gene assay
was not performed to prove that miR-144 post-transcriptionally
regulates IRS-1, which is a limitation of the present study.
In conclusion, the present study findings show that
miR-144 regulates IRS-1 and Dvl2. miR-144 inhibited the malignant
behavior of NPC cells, including proliferation, migration, invasion
and tumour formation, mainly via indirect regulation of Dvl2. The
findings improve the understanding of the molecular mechanisms of
miR-144 in the physiological processes of NPC cells. In future
research, it is worth confirming whether miR-144 regulates EMT and
the Wnt/β-catenin signaling pathway.
Acknowledgements
The authors would like to thank Professor Tao Hong
(Sichuan University, Chengdu, China) for language editing.
Funding
Funding: This work was supported by Scientific Fund of the First
People's Hospital of Neijiang (grant no. 18ZF0399).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
XA and JC designed the experiments, assessed the raw
data and are responsible for confirming the authenticity of the
data. XA, YJ and DC performed cell culture-associated experiments.
DC and JC are responsible for data collection and performed
statistical analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cho WC: Nasopharyngeal carcinoma:
Molecular biomarker discovery and progress. Mol Cancer.
(1)2007.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chou J, Lin YC, Kim J, You L, Xu Z, He B
and Jablons DM: Nasopharyngeal carcinoma - review of the molecular
mechanisms of tumorigenesis. Head Neck. 30:946–963. 2008.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Blanchard P, Lee A, Marguet S, Leclercq J,
Ng WT, Ma J, Chan AT, Huang PY, Benhamou E, Zhu G, et al: MAC-NPC
Collaborative Group. Chemotherapy and radiotherapy in
nasopharyngeal carcinoma: An update of the MAC-NPC meta-analysis.
Lancet Oncol. 16:645–655. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lai SZ, Li WF, Chen L, Luo W, Chen YY, Liu
LZ, Sun Y, Lin AH, Liu MZ and Ma J: How does intensity-modulated
radiotherapy versus conventional two-dimensional radiotherapy
influence the treatment results in nasopharyngeal carcinoma
patients? Int J Radiat Oncol Biol Phys. 80:661–668. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
6
|
Iorio MV and Croce CM: Causes and
consequences of microRNA dysregulation. Cancer J. 18:215–222.
2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chen S, Li P, Li J, Wang Y, Du Y, Chen X,
Zang W, Wang H, Chu H, Zhao G, et al: MiR-144 inhibits
proliferation and induces apoptosis and autophagy in lung cancer
cells by targeting TIGAR. Cell Physiol Biochem. 35:997–1007.
2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang LY, Ho-Fun Lee V, Wong AM, Kwong DL,
Zhu YH, Dong SS, Kong KL, Chen J, Tsao SW, Guan XY, et al:
MicroRNA-144 promotes cell proliferation, migration and invasion in
nasopharyngeal carcinoma through repression of PTEN.
Carcinogenesis. 34:454–463. 2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bao H, Li X, Li H, Xing H, Xu B, Zhang X
and Liu Z: MicroRNA-144 inhibits hepatocellular carcinoma cell
proliferation, invasion and migration by targeting ZFX. J Biosci.
42:103–111. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Liu F, Chen N, Xiao R, Wang W and Pan Z:
miR-144-3p serves as a tumor suppressor for renal cell carcinoma
and inhibits its invasion and metastasis by targeting MAP3K8.
Biochem Biophys Res Commun. 480:87–93. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mardilovich K, Pankratz SL and Shaw LM:
Expression and function of the insulin receptor substrate proteins
in cancer. Cell Commun Signal. 7(14)2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Taniguchi CM, Emanuelli B and Kahn CR:
Critical nodes in signalling pathways: Insights into insulin
action. Nat Rev Mol Cell Biol. 7:85–96. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Chan SH, Kikkawa U, Matsuzaki H, Chen JH
and Chang WC: Insulin receptor substrate-1 prevents
autophagy-dependent cell death caused by oxidative stress in mouse
NIH/3T3 cells. J Biomed Sci. 19(64)2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bergmann U, Funatomi H, Kornmann M, Beger
HG and Korc M: Increased expression of insulin receptor substrate-1
in human pancreatic cancer. Biochem Biophys Res Commun.
220:886–890. 1996.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Shi B, Sepp-Lorenzino L, Prisco M, Linsley
P, deAngelis T and Baserga R: Micro RNA 145 targets the insulin
receptor substrate-1 and inhibits the growth of colon cancer cells.
J Biol Chem. 282:32582–32590. 2007.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wu X, Cui CL, Chen WL, Fu ZY, Cui XY and
Gong X: miR-144 suppresses the growth and metastasis of laryngeal
squamous cell carcinoma by targeting IRS1. Am J Transl Res. 8:1–11.
2016.PubMed/NCBI
|
|
17
|
Gao C and Chen YG: Dishevelled: The hub of
Wnt signaling. Cell Signal. 22:717–727. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang Y, Wang F, Han L, Wu Y, Li S, Yang
X, Wang Y, Ren F, Zhai Y, Wang D, et al: GABARAPL1 negatively
regulates Wnt/β-catenin signaling by mediating Dvl2 degradation
through the autophagy pathway. Cell Physiol Biochem. 27:503–512.
2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T,
Fu W, Zhang J, Wu W, Zhang X, et al: Autophagy negatively regulates
Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol.
12:781–790. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Geng Y, Ju Y, Ren F, Qiu Y, Tomita Y,
Tomoeda M, Kishida M, Wang Y, Jin L, Su F, et al: Insulin receptor
substrate 1/2 (IRS1/2) regulates Wnt/β-catenin signaling through
blocking autophagic degradation of dishevelled2. J Biol Chem.
289:11230–11241. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tan G, Tang X and Tang F: The role of
microRNAs in nasopharyngeal carcinoma. Tumour Biol. 36:69–79.
2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Qiu F, Sun R, Deng N, Guo T, Cao Y, Yu Y,
Wang X, Zou B, Zhang S, Jing T, et al: miR-29a/b enhances cell
migration and invasion in nasopharyngeal carcinoma progression by
regulating SPARC and COL3A1 gene expression. PLoS One.
10(e0120969)2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Verhoeven RJ, Tong S, Zhang G, Zong J,
Chen Y, Jin DY, Chen MR, Pan J and Chen H: NF-κB signaling
regulates expression of Epstein-Barr virus BART MicroRNAs and long
noncoding RNAs in nasopharyngeal carcinoma. J Virol. 90:6475–6488.
2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wang S, Zhang R, Claret FX and Yang H:
Involvement of microRNA-24 and DNA methylation in resistance of
nasopharyngeal carcinoma to ionizing radiation. Mol Cancer Ther.
13:3163–3174. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Carew RM, Browne MB, Hickey FB and Brazil
DP: Insulin receptor substrate 2 and FoxO3a signalling are involved
in E-cadherin expression and transforming growth factor-β1-induced
repression in kidney epithelial cells. FEBS J. 278:3370–3380.
2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sorokin AV and Chen J: MEMO1, a new
IRS1-interacting protein, induces epithelial-mesenchymal transition
in mammary epithelial cells. Oncogene. 32:3130–3138.
2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Dearth RK, Cui X, Kim HJ, Kuiatse I,
Lawrence NA, Zhang X, Divisova J, Britton OL, Mohsin S, Allred DC,
et al: Mammary tumorigenesis and metastasis caused by
overexpression of insulin receptor substrate 1 (IRS-1) or IRS-2.
Mol Cell Biol. 26:9302–9314. 2006.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Karolina DS, Armugam A, Tavintharan S,
Wong MT, Lim SC, Sum CF and Jeyaseelan K: MicroRNA 144 impairs
insulin signaling by inhibiting the expression of insulin receptor
substrate 1 in type 2 diabetes mellitus. PLoS One.
6(e22839)2011.PubMed/NCBI View Article : Google Scholar
|