Introduction
Ovarian cancer is the second most common
gynecological malignancy after uterine corpus cancer and it is the
eighth most common cause of cancer-associated death globally, with
a 5-year survival rate <45% (1,2).
However, mortality from ovarian cancer is the highest among all
gynecological malignancies (1). Due
to the lack of early-stage symptoms and reliable diagnostic
methods, it remains difficult to detect the occurrence of ovarian
cancer at the early stage (3).
Frequently, when the patient exhibits typical symptoms of ovarian
cancer, metastasis has already occurred, such that >70% patients
are diagnosed at the late stage (4). The main therapeutic method for ovarian
cancer includes surgery and adjuvant treatment for chemotherapy,
but these treatment strategies can also be combined with
radiotherapy and biological therapy (5). Although ovarian cancer can be
completely curable after initial surgery and chemotherapy, the
majority of patients with advanced disease tend to suffer tumor
recurrence, where the 10-year survival rate for patients with
ovarian cancer at stages III and IV is <30% (6). Therefore, identification of additional
early detection markers that are also sensitive is crucial for the
early diagnosis and treatment of ovarian cancer.
MicroRNAs (miRNAs or miRs) are non-coding RNA
molecules that are 17-27 nucleotides in length and serve a
regulatory role in various life processes (7). Numerous studies have shown that
miR-638 serves as a suppressive factor in various malignant tumors,
including gastric, breast and cervical cancer, where its expression
is reduced (8-10).
However, in other malignant tumors, such as esophageal squamous
cell carcinoma and melanoma, it is abundantly expressed and
functions as a promoter of malignant physiology (11,12).
The role of miR-638 in ovarian cancer cells remain poorly
understood. Bafilomycin A1 treatment was able to inhibit the
proliferation of the ovarian cancer cell line HO-8910, which was
accompanied by miR-638 upregulation (13), suggesting that increased miR-638
levels may inhibit the proliferation of ovarian cancer cells.
High mobility group A1 (HMGA1) is a chromosomal
binding protein that is involved in various cell processes by
regulating gene transcription (14). Recent evidence suggested that HMGA1
protein is expressed at high levels during embryonic development,
which is then reduced to markedly low levels or not detected after
aging (15). In 1983, the abnormal
expression of HMGA1 was first identified in aggressive cervical
cancer cells, following which the role of HMGA1 in malignant cancer
became gradually elucidated (16),
including in breast, gastric and thyroid cancer (17-20).
Furthermore, HMGA1 is also known to be highly expressed in ovarian
cancer tissues (21), where
downregulation of HMGA1 can inhibit the proliferation of ovarian
cancer cells in vitro and tumor formation in vivo
(22). Therefore, it was
hypothesized that miR-638 may be involved in the regulation of
malignancy in ovarian cancer cells by regulating HMGA1.
The present study aimed to explore the effects of
miR-638 on ovarian cancer cell proliferation, cell cycle and
apoptosis, in addition to investigating the possible underlying
mechanism. The results may facilitate the early detection and
treatment of ovarian cancer, thereby helping to reduce ovarian
cancer-related mortality.
Materials and methods
Clinical ovarian cancer sample
collection
A total of 30 paired ovarian cancer tissues and
adjacent normal tissues (2 cm from ovarian cancer tissues) were
collected from female patients with ovarian cancer (mean age,
41.5±4.9 years) following excision of the tumor. The cancer tissues
were collected in the Second Affiliated Hospital of Xi'an Jiaotong
University (Xi'an, China) between February 2018 and February 2019
with informed consent signed by the patients. All procedures were
performed in accordance with the principles outlined in the
Declaration of Helsinki. All the protocols were approved by the
Ethics Committee of the Second Affiliated Hospital of Xi'an
Jiaotong University.
Cell culture
The ovarian cancer cell lines A2780, ES-2, OVCAR-3
and Caov-3 were purchased from Procell Life Science &
Technology Co., Ltd. SKOV-3 and 293T cells were obtained from
Shanghai Zhongqiaoxinzhou Biotechnology Co., Ltd. A2780, Caov-3 and
293T cells were cultured in DMEM (HyClone; Cytiva) supplemented
with 10% FBS (Biological Industries). ES-2 cells were cultured in
McCoy's 5A medium (Procell Life Science & Technology Co., Ltd.)
supplemented with 10% FBS. OVCAR-3 cells were cultured in RPMI-1640
medium (HyClone; Cytiva) supplemented with 20% FBS and 0.01 mg/ml
bovine insulin (Beijing Solarbio Science & Technology Co.,
Ltd.). SKOV-3 cells were cultured in RPMI-1640 medium supplemented
with 15% FBS. All cells were incubated at 37˚C in an incubator with
5% CO2.
Cell transfection
A total of 100 pmol miR-638 mimics or 100 pmol
negative control (NC) mimics were transfected into OVCAR-3 cells,
whilst 100 pmol miR-638 inhibitor or 100 pmol NC inhibitor were
transfected into Caov-3 cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. miR-638 expression and cell viability were
detected 24 h after transfection. Apoptosis, cell cycle and protein
expression were measured 48 h after transfection.
To verify the association between miR-638 and HMGA1,
OVCAR-3 cells were co-transfected with 50 pmol miR-638 mimics and 1
µg HMGA1 overexpression plasmid (GenScript), and the detection was
performed 24 or 48 h after transfection. NC mimics and empty vector
served as the negative controls, respectively. The sequences used
were as follows: miR-638 mimics forward,
5'-AGGGAUCGCGGGCGGGUGGCGGCCU-3' and reverse,
5'-GCCGCCACCCGCCCGCGAUCCCUUU-3'; NC mimics forward,
5'-UUCUCCGAACGUGUCACGUTT-3' and reverse,
5'-ACGUGACACGUUCGGAGAATT-3'; miR-638 inhibitor,
5'-AGGCCGCCACCCGCCCGCGAUCCCU-3'; and NC inhibitor,
5'-UUGUACUACACAAAAGUACUG-3'.
Cell counting kit-8 (CCK-8) assay
OVCAR-3 and Caov-3 cells were transferred to 96-well
plates (3x103 cells/well), and cell viability was
detected using CCK-8 assay (Sigma-Aldrich; Merck KGaA) at 24, 48
and 72 h after transfection. Briefly, 10 µl CCK-8 solution was
added to each well containing 100 µl normal medium, followed by
incubation for 1 h at 37˚C with 5% CO2. The optical
density value, representing the viability of the cells was measured
using a microplate reader (ELX-800; BioTek Instruments, Inc.) at
450 nm.
Cell cycle assay
The aforementioned OVCAR-3 and Caov-3 cells were
first digested with 0.2% trypsin, harvested by centrifugation at
1,000 x g for 5 min at 4˚C and washed with PBS twice 48 h after
transfection. Subsequently, Cell Cycle Detection kit (Beyotime
Institute of Biotechnology) was used for cell cycle detection
according to the manufacturer's protocol. Briefly, the cells
(2x104) were fixed in pre-cooled 70% ethanol at 4˚C for
12 h, stained with 25 µl propidium iodide (PI) solution at 37˚C for
30 min in the dark, analyzed by flow cytometry (NovoCyte; ACEA
Bioscience, Inc.; Agilent Technologies, Inc.) and quantified using
NovoExpress v1.2.5 (Agilent Technologies, Inc.).
Cell apoptosis
Consistent with the procedure of cell cycle assay,
the OVCAR-3 and Caov-3 cells were harvested. Annexin-V/PI kit
(Beyotime Institute of Biotechnology) was used for apoptotic
detection according to the manufacturer's protocol. Briefly, 200 µl
Annexin V-FITC and 10 µl PI were added to resuspend the cells
(2x104). The cells were stained for 15 min in the dark
and then placed in an ice bath, followed by analysis using flow
cytometry (NovoCyte; ACEA Bioscience, Inc.; Agilent Technologies,
Inc.) and quantified using NovoExpress v1.2.5 (Agilent
Technologies, Inc.).
TUNEL assay
The transfected OVCAR-3 and Caov-3 cells were first
permeabilized by 0.1% Triton X-100. In Situ Cell Death
Detection kit (Roche Diagnostics) was used for labelling the
apoptotic cells according to the manufacturer's protocols. Briefly,
when the cell confluency reached 70%, the cells were permeabilized
with 200 µl 0.1% Triton X-100 for 15 min at room temperature, and
then incubated in TUNEL working solution (Enzyme solution:Label
Solution, 1:9) in the dark at 37˚C for 1 h. The cells were
counterstained by DAPI solution (Beyotime Institute of
Biotechnology) for 5 min at room temperature and then sealed with
anti-attenuation sealing reagent (Beijing Solarbio Science &
Technology Co., Ltd.). Finally, the apoptotic cells were observed
under a fluorescence microscope at x400 magnification (BX53; Olympus
Corporation). At least five fields of view were randomly observed
under the fluorescence microscope.
Dual-luciferase reporter assay
The putative binding site of miR-638 was predicted
by bioinformatics analysis (https://www.targetscan.org; release 7.2 March 2018).
According to the procedure of dual-luciferase reporter assay with
certain modifications (23), 293T
cells were plated in 12-well plates and incubated in DMEM
containing 10% FBS until 70% confluence, followed by incubation in
DMEM without FBS for 1 h at 37˚C. The wild-type or mutant 3'
untranslated region (UTR) sequences of HMGA1 with suspected miR-638
binding sites was then inserted into the luciferase reporter vector
pmiRGLO (Promega Corporation). 293T cells were co-transfected with
the constructed reporters (1.5 µg) and miR-638 mimics or NC mimics
(75 pmol) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). After 4 h of incubation at 37˚C, the
medium was discarded and replaced with normal DMEM for another 48
h. The luciferase activity was detected using the Dual-Luciferase
assay kit (Promega Corporation) according to the manufacturer's
protocol. The data were presented as Firefly/Renilla
luciferase activity.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from OVCAR-3 and Caov-3 cells was
extracted with Total RNA Isolating kit (BioTeke Corporation)
following the manufacturer's protocols. Complementary DNA was
obtained by RT using M-MLV reverse transcriptase kit (Takara
Biotechnology Co., Ltd.) and RT Primer (GenScript) according to the
manufacturer's protocol. The relative expression level of miR-638
was determined by qPCR amplification using TaKaRa Taq™ HS Perfect
Mix (Takara Biotechnology Co., Ltd.) with SYBR Green (BioTeke
Corporation). The thermocycling conditions consisted of:
Pre-denaturation at 94˚C for 30 sec, followed by 40 cycles at 94˚C
for 5 sec and 60˚C for 15 sec. miR-638 expression was normalized
against the expression of U6. The relative expression level of
miR-638 was converted to fold changes according to the
2-ΔΔCq method (24). The
primers used were as follows: miR-638 forward,
5'-AATAGGGATCGCGGGCGG-3' and reverse, 5'-GCAGGGTCCGAGGTATTC-3', and
U6 forward, 5'-GCTTCGGCAGCACATATACT-3' and reverse,
5'-GTGCAGGGTCCGAGGTATTC-3'.
Western blotting
Proteins were isolated from the aforementioned
OVCAR-3 and Caov-3 cells sing RIPA (Beyotime Institute of
Biotechnology) and PMSF solution (Beyotime Institute of
Biotechnology). The proteins were quantified using the BCA assay
(Beyotime Institute of Biotechnology) and then equal amounts of
protein (15-30 µg) were separated by 8-15% SDS-PAGE, transferred
onto PVDF membranes and blocked in 5% bovine serum albumin
(Biosharp Life Sciences) for 1 h at room temperature. After
blocking, the membranes were incubated with primary antibodies
against proliferating cell nuclear antigen (PCNA; dilution,
1:1,000; Affinity Biosciences; cat. no. AF0239), caspase-3
(dilution, 1:1,000; Cell Signaling Technology, Inc.; cat. no.
14220), poly(ADP-ribose) polymerase (PARP; dilution, 1:1,000; Cell
Signaling Technology, Inc.; cat. no. 9532), cyclin D1 (dilution,
1:500; ABclonal Biotech Co., Ltd.; cat. no. A0310), cyclin B1
(dilution, 1:2,000; ProteinTech Group, Inc.; cat. no. 55004-1-AP),
PTEN (dilution, 1:500; ProteinTech Group, Inc.; cat. no.
22034-1-AP), HMGA1 (dilution, 1:1,000; Affinity Biosciences; cat.
no. AF5218) or β-actin (dilution, 1:2,000; ProteinTech Group, Inc.;
cat. no. 60008-1-Ig) at 4˚C overnight, followed by incubation with
the corresponding anti-rabbit or anti-mouse IgG-horseradish
peroxidase secondary antibodies (dilution, 1:10,000; ProteinTech
Group, Inc.; cat. nos. SA00001-1 and SA00001-2) at 37˚C for 40 min.
The protein bands were visualized using enhanced chemiluminescence
(7 Sea Pharmatech Co., Ltd.) and quantitatively analyzed using
Gel-Pro Analyzer 4.0 (Media Cybernetics, Inc.).
Statistical analysis
Data are presented as the mean ± standard deviation.
All experiments were repeated at least three times. Comparisons
between ovarian cancer and adjacent normal tissues were performed
with paired Student's t-tests. Differences between two independent
groups were analyzed using unpaired Student's t-test. For
comparison of ≥3 groups, one-way analysis of variance followed by
Tukey's post hoc test was performed. Statistical analysis of data
was performed using GraphPad Prism 8.0 (GraphPad Software, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-638 is expressed in ovarian cancer
tissues and cell lines
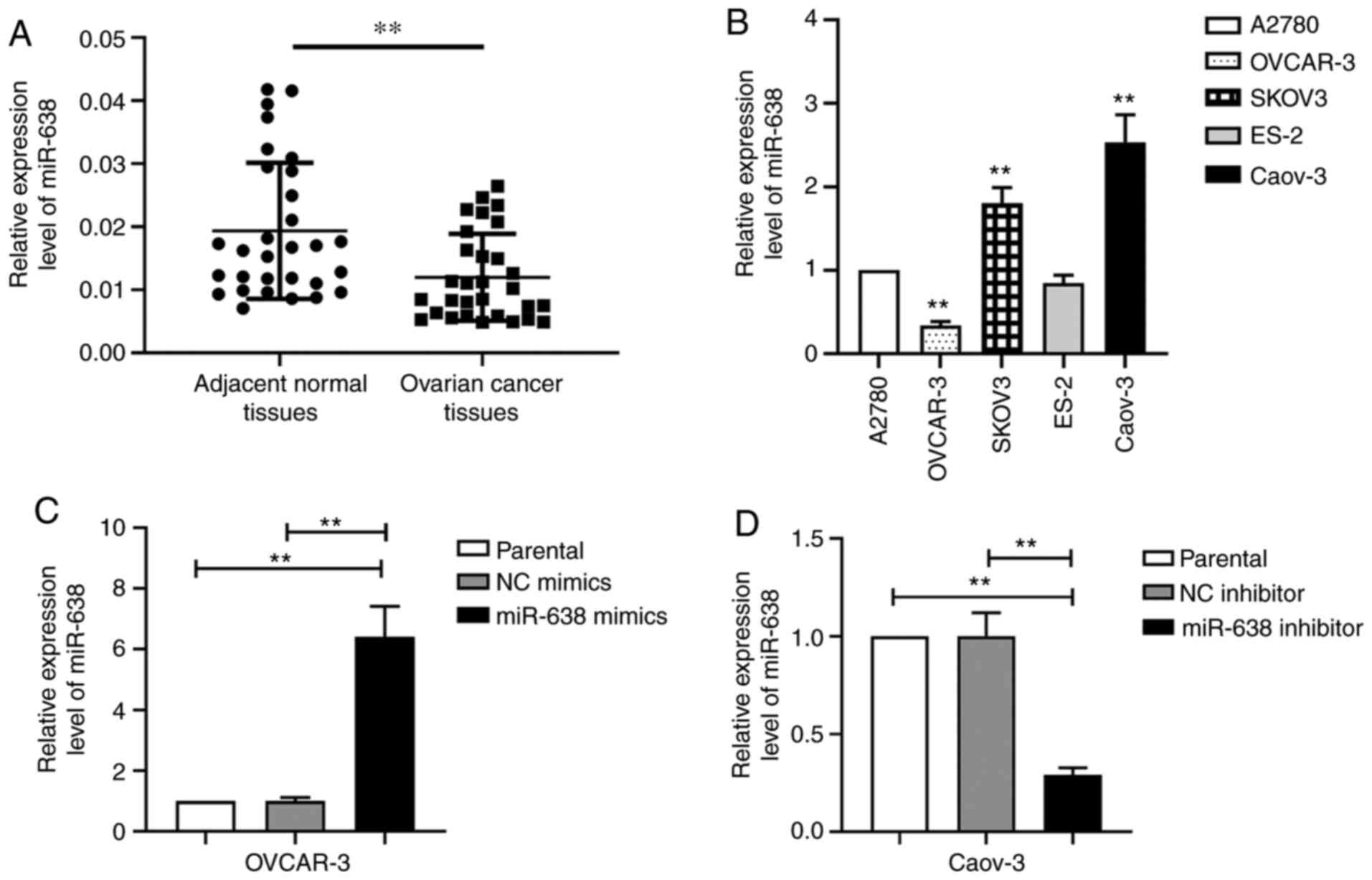
The expression levels of miR-638 in clinical ovarian
cancer samples were first measured. miR-638 levels were found to be
significantly lower in ovarian cancer samples compared with those
in adjacent normal tissues (Fig.
1A). The expression levels of miR-638 in different ovarian
cancer cell lines were subsequently measured by RT-qPCR. Caov-3
cells had the highest miR-638 expression level, followed by SKOV3,
A2780 and ES-2 cells (Fig. 1B).
OVCAR-3 cells exhibited the lowest miR-638 expression level
(Fig. 1B). Therefore, OVCAR-3 cells
were used for miR-638 overexpression and Caov-3 cells were used for
miR-638-knockdown in subsequent experimentation. After transfection
with miR-638 mimics, the miR-638 levels in OVCAR-3 cells were
significantly increased compared with those in parental or cells
transfected with NC mimics (Fig.
1C). After the Caov-3 cells were transfected with the miR-638
inhibitor, miR-638 expression was significantly decreased compared
with that in cells transfected with NC inhibitor (Fig. 1D). These results suggested that
miR-638 overexpression in OVCAR-3 cells and miR-638 knockdown in
Caov-3 cells were successful.
Effect of miR-638 on cell viability,
apoptosis and cell cycle in ovarian cancer cells
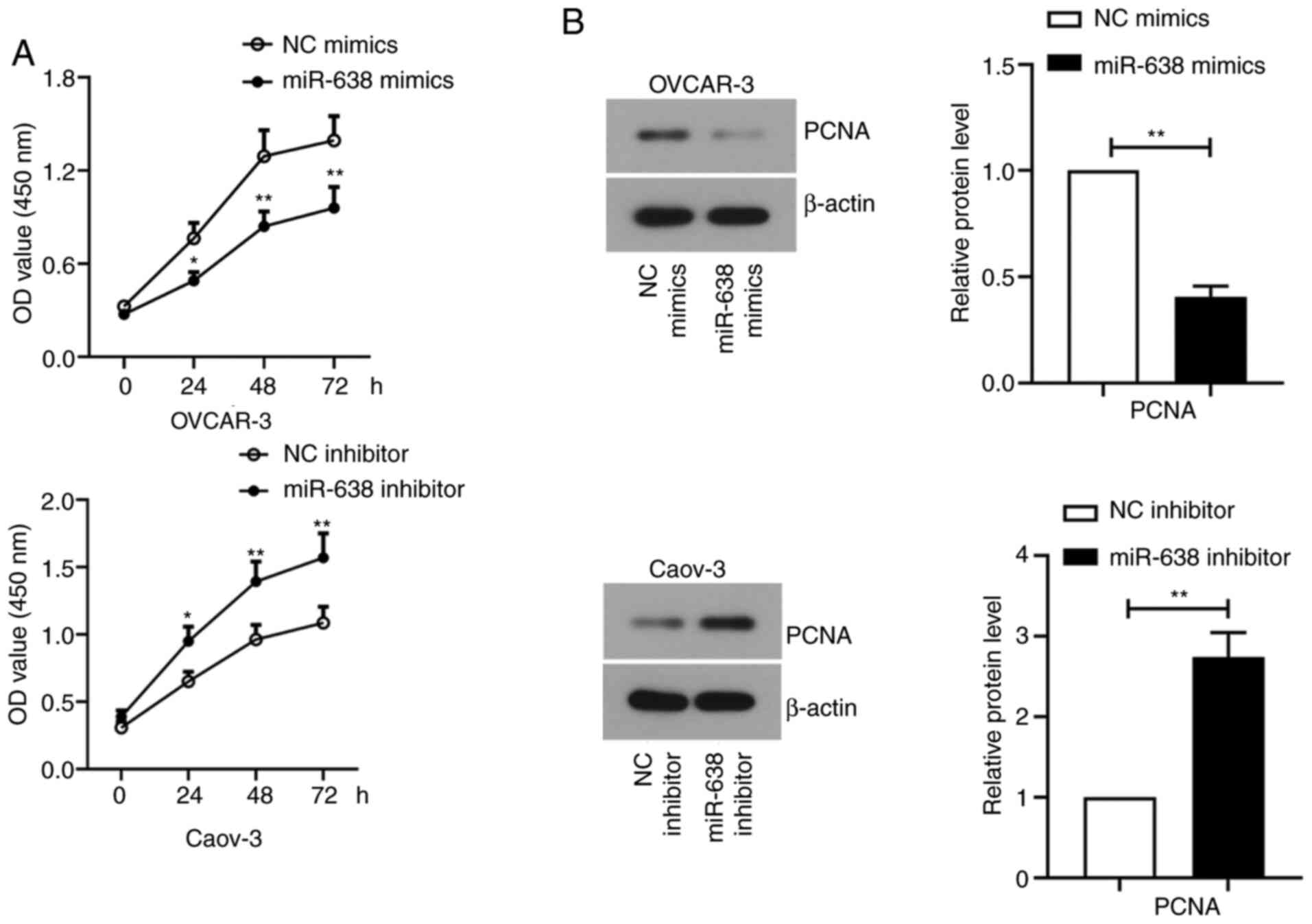
Cell viability was measured at 24, 48 and 72 h after
transfection by CCK-8 assay. The viability of OVCAR-3 cells
transfected with miR-638 mimics was significantly decreased
compared with that in cells that were subjected to NC mimics
transfection. By contrast, miR-638 inhibitor transfection
significantly promoted Caov-3 cell viability compared with that in
cells transfected with the NC inhibitor (Fig. 2A). In addition, the expression of
PCNA, an indicator of cell proliferation (25), was found to be significantly
downregulated in OVCAR-3 cells transfected with miR-638 mimics
compared with that in cells transfected with NC mimics (Fig. 2B). By contrast, PCNA expression was
significantly upregulated after miR-638 inhibitor transfection in
Caov-3 cells compared with that in cells transfected with NC
inhibitor (Fig. 2B). To confirm
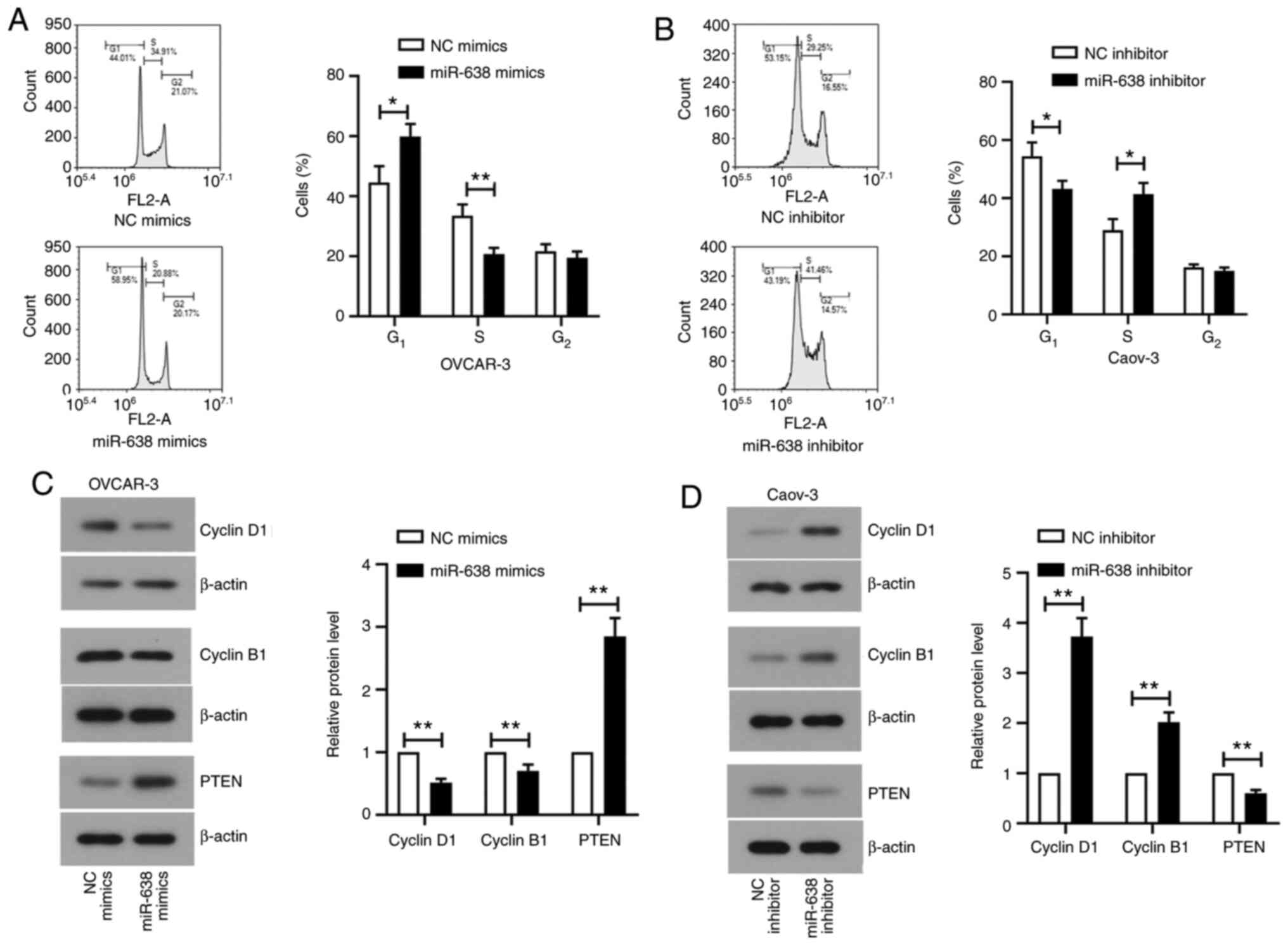
whether miR-638 was involved in cell cycle regulation, cell cycle
status was assessed after transfection by flow cytometry. There was
a significant increase in the number of cells at the G1
phase and a significant decrease in the number of cells at the S
phase in miR-638 mimics-transfected OVCAR-3 cells (Fig. 3A). Opposite trends were observed in
Caov-3 cells transfected with miR-638 inhibitor (Fig. 3B). miR-638 overexpression was found
to significantly suppress cyclins D1 and B1 expression whilst
significantly increasing PTEN expression in OVCAR-3 cells compared
with that in cells transfected with mimics NC (Fig. 3C). By contrast, opposite results
were observed in Caov-3 cells transfected with the miR-638
inhibitor (Fig. 3D).
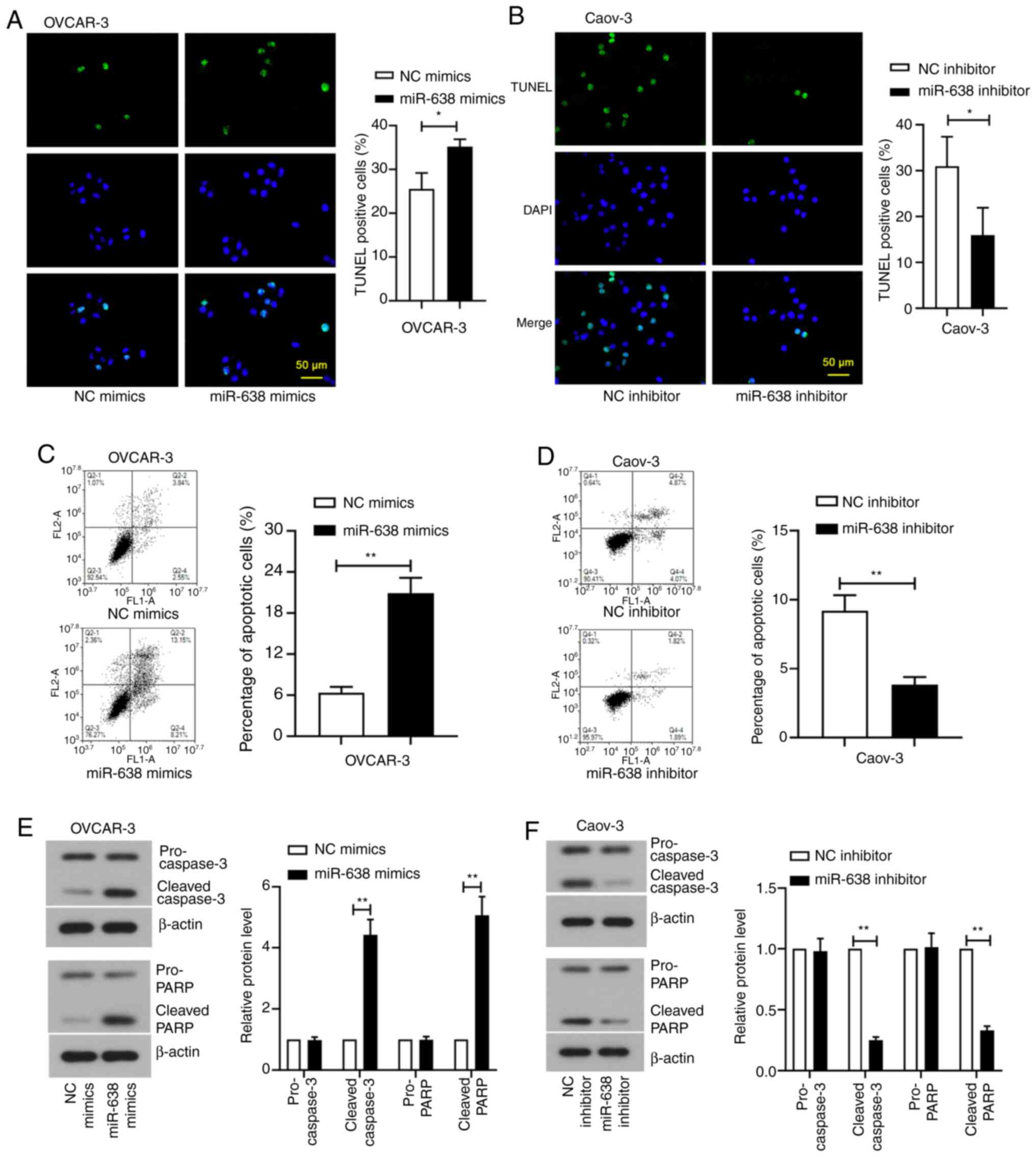
TUNEL staining and flow cytometry assay were next
used to measure apoptosis after transfection. The number of
TUNEL-positive OVCAR-3 cells was significantly increased after
transfection with miR-638 mimics compared with that in cells
transfected with mimics NC (Fig.
4A), whilst this number was significantly decreased after
transfection of Caov-3 cells with miR-638 inhibitor compared with
that in cells transfected with inhibitor NC (Fig. 4B). Flow cytometry assay also
subsequently showed that the percentage of apoptotic cells was
significantly increased in OVCAR-3 cells after miR-638 mimics
transfection but significantly decreased in Caov-3 cells after
miR-638 inhibitor transfection compared with those in their
respective NCs (Fig. 4C and
D). In addition, the expression
levels of apoptotic markers cleaved-caspase-3 and cleaved-PARP,
were found to be significantly increased with miR-638
overexpression in OVCAR-3 cells and significantly decreased with
miR-638 knockdown in Caov-3 cells compared with the levels
exhibited by their corresponding NCs (Fig. 4E and F). These data suggest that miR-638 is
involved in proliferation, cell cycle arrest and apoptosis in
ovarian cancer cells.
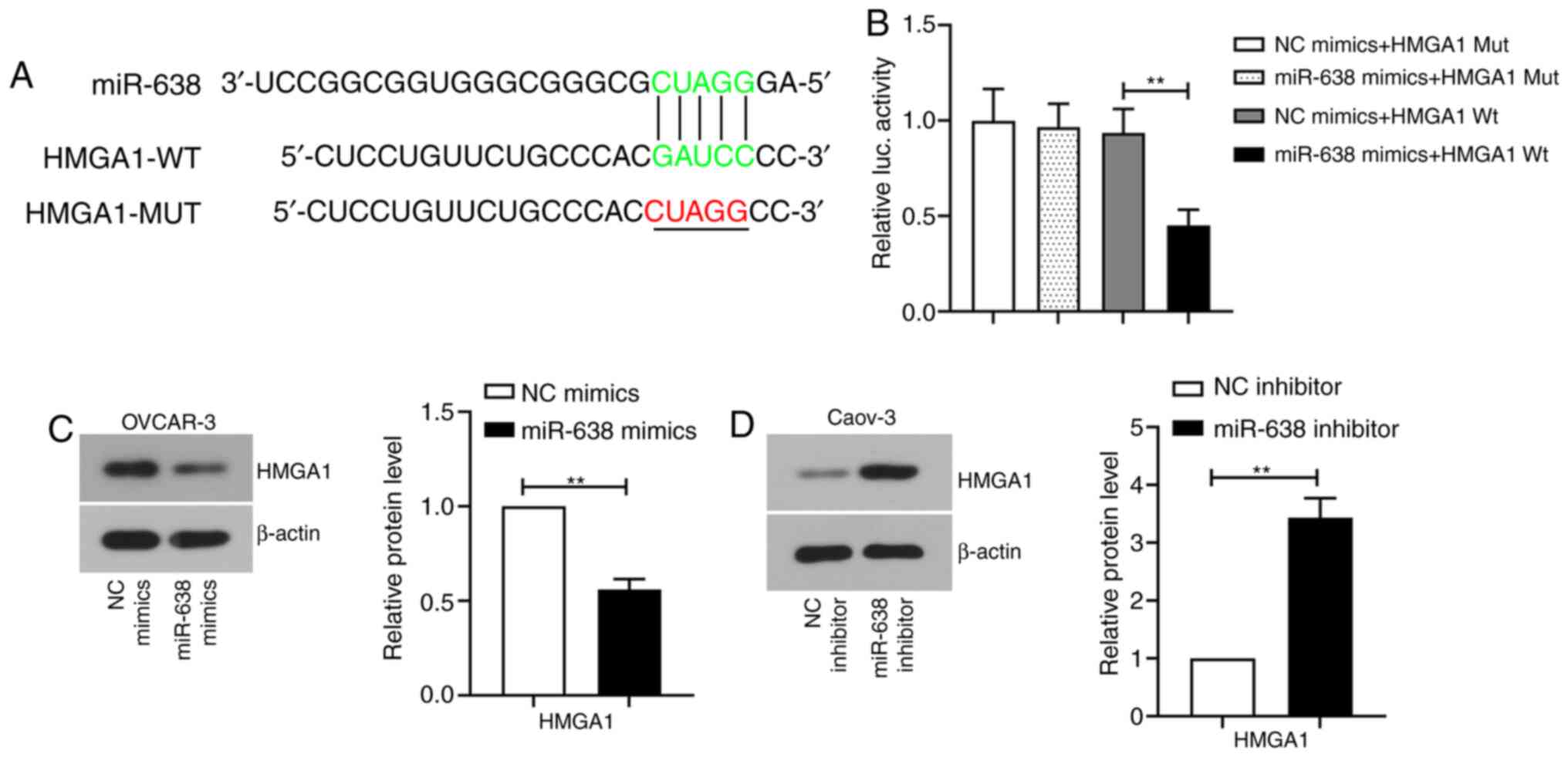
HMGA1 is the direct target of
miR-638
The putative binding site of miR-638 was predicted
by bioinformatics analysis. The results showed that there was a
potential association between miR-638 and HMGA1 (Fig. 5A). To verify this prediction,
luciferase reporter assay was performed in 293T cells. Transfection
with miR-638 mimics significantly inhibited the luciferase activity
of the wild-type reporter gene containing HMGA1-3'UTR compared with
that in cells co-transfected with the NC mimics (Fig. 5B), whilst miR-638 mimics did not
affect the luciferase activity of the mutated reporter gene
containing HMGA1-3'UTR (Fig. 5B).
These results suggested that miR-638 could specifically bind to
HMGA1 mRNA. To further verify the targeting association between
miR-638 and HMGA1, HMGA1 protein expression was detected by western
blotting. HMGA1 protein expression was significantly reduced in
OVCAR-3 cells transfected with miR-638 mimics and notably increased
in Caov-3 cells transfected with miR-638 inhibitor compared with
those in their corresponding NC (Fig.
5C and D). These results
suggest that HMGA1 is the direct target of miR-638 in ovarian
cancer cells.
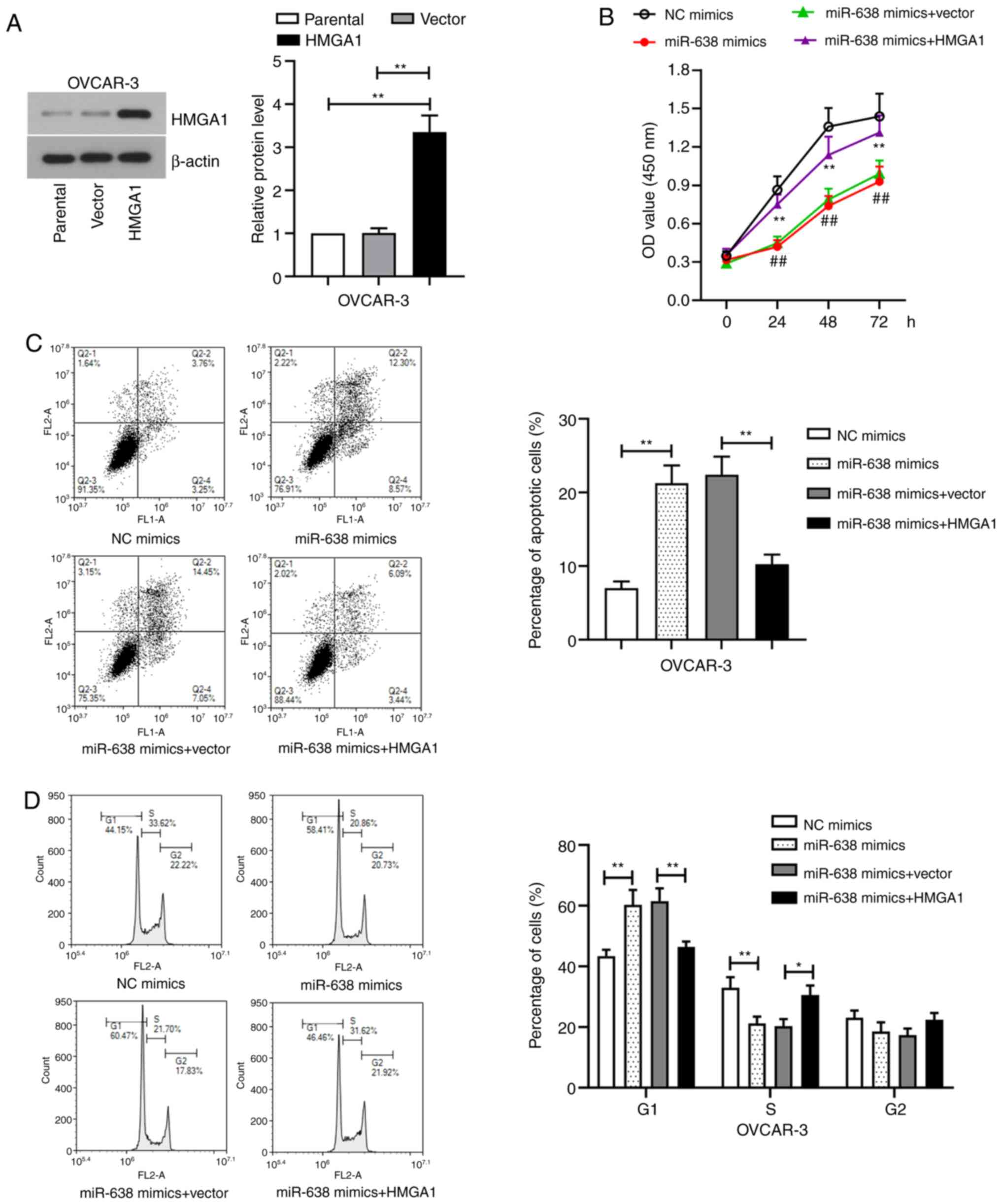
HMGA1 overexpression reverses miR-638
mimics-induced inhibition of proliferation and cell cycle
arrest
To verify whether miR-638 functions as a suppressor
gene by regulating HMGA1, OVCAR-3 cells were co-transfected with
miR-638 mimics and HMGA1 overexpression plasmid. HMGA1 expression
in OVCAR-3 cells was significantly enhanced after transfection with
the HMGA1 overexpression plasmid compared with that in cells
transfected with the empty vector (Fig.
6A), indicating that HMGA1 was successively transfected into
the cells. CCK-8 assay showed that the anti-proliferative effect of
miR-638 mimics was significantly reversed by HMGA1 overexpression
(Fig. 6B). Flow cytometry analysis
of apoptosis and cell cycle distribution showed that miR-638 mimics
and HMGA1 co-transfection significantly reduced apoptosis and cell
cycle arrest compared with those in cells transfected with miR-638
mimics alone in OVCAR-3 cells (Fig.
6C and D). These results
suggested that miR-638 served as a suppressor by targeting HMGA1 in
ovarian cancer cells.
Discussion
The present study primarily investigated the effect
and potential mechanism of the miR-638/HMGA1 axis on proliferation,
cell cycle and apoptosis in ovarian cancer cell lines, including
OVCAR-3 and Caov-3 cells. It was found that the expression of
miR-638 was relatively low in OVCAR-3 cells and relatively high in
Caov-3 cells. In addition, upregulating miR-638 expression
significantly inhibited the viability of OVCAR-3 cells, induced
cell cycle arrest at the G1 phase and promoted cell
apoptosis, all of which were prevented by miR-638 inhibition in
Caov-3 cells. Furthermore, results from the present study showed
that miR-638 could downregulate HMGA1 expression by binding to its
3'UTR, where HMGA1 overexpression could reverse the inhibitory
effects of miR-638 on ovarian cancer cell physiology. Therefore,
these data suggested that miR-638 may serve as a tumor suppressor
by targeting HMGA1 expression in ovarian cancer.
Since miRNAs are key factors in modulating tumor
occurrence and progression, identifying sensitive miRNAs and
understanding their functions may provide new strategies for
patients with cancer (26).
Previous reports have shown that the levels of multiple miRNAs are
altered in human ovarian cancer compared with those of normal
tissue (27). Previous studies have
reported that the expression levels of miR-603 and miR-31 were
decreased (28,29), whilst those of miR-552 and miR-182
were increased in ovarian cancer (30,31).
Furthermore, the miR-638 expression was found to be enhanced in
ovarian cancer cells after treatment with the antibiotic
bafilomycin A1(13). However, the
role of miR-638 in ovarian cancer remains poorly understood, which
provided a basis for the present study.
Unchecked cell proliferation is one of the hallmarks
of cancer, which may lead to high mortality among patients with
ovarian cancer, such that reversing aberrant miRNA expression can
effectively inhibit tumor cell proliferation, promote apoptosis and
arrest cell cycle progression (32). Zhao et al (33) found that miR-638 overexpression
could markedly suppress gastric cancer cell proliferation and
induce cell cycle arrest at the G1 phase in
vitro. By contrast, Cheng et al (34) observed that downregulating miR-638
could promote hepatocellular carcinoma cell growth in vitro
and tumor angiogenesis processes in vivo. Consistent with
these studies, the present study also found that upregulating
miR-638 levels could significantly inhibit ovarian cancer cell
viability, induce cell cycle arrest at the G1 phase and
promote cell apoptosis, suggesting that miR-638 is a tumor
suppressor in the ovarian cancer pathogenesis.
PCNA is a standard marker of cell proliferation that
can be used to effectively evaluate the growth of malignant tumors
(35). PCNA dysfunction has been
widely used in the diagnosis of various cancer types, including
breast (36) and cervical cancer
(37). Consistent with a previous
study, results from the present study revealed that the cell
viability in OVCAR-3 cells was reduced with miR-638 overexpression,
which was accompanied with the decreased expression of PCNA. By
contrast, opposite results were observed following miR-638
knockdown in Caov-3 cells. Cyclins D1 and B1 are members of the
cyclin family that are associated with cell cycle progression and
can also be regulated by miRNA. It has been previously reported
that inhibiting cyclin D1 expression in epithelial ovarian cancer
cells by upregulating miR-211 can inhibit the progression of the
cell cycle through the G1 phase (38), whilst decreasing cyclin B1
expression in breast cancer cells by miR-379 transfection can
reduce proliferation (39). In the
present study, the S-phase cell fraction was decreased in OVCAR-3
cells overexpressing miR-638, but was increased in Caov-3 cells
following miR-638 knockdown. In line with the aforementioned data,
reduced cyclin D1 and cyclin B1 expression was observed in OVCAR-3
cells transfected with miR-638 mimics, whilst miR-638 inhibitor
transfection could reverse this phenomenon in Caov-3 cells. In
addition, cyclin D is one of the targets in the PTEN signaling
pathway, where PTEN overexpression can prevent the increase in
cyclin D1 during cell cycle progression from G1 to S
phase in U-87 and U-251 glioblastoma cells (40). Dysfunction of the suppressor PTEN is
a frequent phenomenon observed in numerous types of cancer, such as
ovarian cancer (31,41). Cleaved caspase-3 and cleaved PARP
are characteristic markers of cell apoptosis (42,43),
whereby elevating their expression levels may promote tumor
apoptosis. Zhang et al (44)
reported that miR-148a promoted apoptosis, which was characterized
by increased caspase-3 and PARP activation. In another study, Liu
et al (45) showed that
apoptosis, along with cleaved caspase-3 expression were decreased
in ovarian granulosa cells with miR-26b knockdown. Findings from
the present study suggest that miR-638 overexpression accelerated
apoptosis in OVCAR-3 cells by increasing cleaved caspase-3 and
cleaved PARP levels. These results indicate that miR-638
upregulation suppressed proliferation, induced cell cycle arrest
and promoted apoptosis in ovarian cancer cells.
Bioinformatics prediction indicated that miR-638 may
have a potential regulatory effect on HMGA1 by targeting its 3'UTR.
HMGA1 is a chromatin factor that is expressed at low levels in
normal human tissues but is overexpressed in certain malignant
tumors, including cervical, prostate and pancreatic cancer
(15). In addition, a previous
study has shown that HMGA1 overexpression is also a common feature
in ovarian cancer (46). However,
the mechanism involved in the regulation of HMGA1 expression has
yet to be fully elucidated. Although bioinformatics analysis showed
that miR-638 can directly target HMGA1, there is no previous report
on the association between miR-638 and HMGA1. Therefore, a
dual-luciferase assay was performed to verify the direct binding of
miR-638 to HMGA1 3'UTR, and the results confirmed this hypothesis.
Wei et al (47) found that
miR-296 diminished prostate cancer growth and invasion by directly
targeting HMGA1. In another study, Chen et al (48) found that HMGA1 was a target of the
miRNA let-7d-5p, where upregulating let-7d-5p expression could
suppress proliferation and facilitate apoptosis in ovarian cancer
via the p53 signaling pathway, which was mediated by HMGA1
expression. Based on these aforementioned studies, the present
study aimed to clarify whether miR-638/HMGA1 signaling was involved
in proliferation, apoptosis and cell cycle progression in ovarian
cancer cells. The inhibitory effects of miR-638 overexpression on
cell proliferation was reversed by HMGA1 overexpression, suggesting
that the growth-suppressive effect of miR-638 in ovarian cancer
cells was mediated by HMGA1 repression. However, this conclusion
was initially obtained through in vitro experiments.
Verifying the potentially anti-tumor effects of miR-638 in a
xenograft animal model would render results from the present the
study more convincing.
In conclusion, the findings of the present study
suggest that miR-638 expression is closely associated with ovarian
cancer. Upregulating miR-638 can inhibit proliferation, induce cell
cycle arrest and promote apoptosis in ovarian cancer cells.
Furthermore, the antitumor effects of miR-638 were mediated at
least partially by negatively regulating HMGA1 expression in
ovarian cancer cells. This finding may help to develop a novel
strategy for the prevention and treatment of ovarian cancer.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by a grant from the
Social Development Foundation of Department of Science and
Technology Shaanxi Province (grant no. 2017SF-202).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LM designed the study and drafted the manuscript. YJ
contributed to the experiments. WZ and XB analyzed the data. QY was
involved in conception and design, and drafting and revision of the
manuscript. LM and WZ confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All the protocols were approved by the Ethics
Committee of the Second Affiliated Hospital of Xi'an Jiaotong
University (Xi'an, China). Informed consent was signed by the
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Webb PM and Jordan SJ: Epidemiology of
epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol.
41:3–14. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bonifácio VDB: Ovarian cancer biomarkers:
Moving forward in early detection. Adv Exp Med Biol. 1219:355–363.
2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hennessy BT, Coleman RL and Markman M:
Ovarian cancer. Lancet. 374:1371–1382. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Doubeni CA, Doubeni AR and Myers AE:
Diagnosis and management of ovarian cancer. Am Fam Physician.
93:937–944. 2016.PubMed/NCBI
|
|
6
|
Vergote I, Tropé CG, Amant F, Kristensen
GB, Ehlen T, Johnson N, Verheijen RH, van der Burg ME, Lacave AJ,
Panici PB, et al: Neoadjuvant chemotherapy or primary surgery in
stage IIIC or IV ovarian cancer. N Engl J Med. 363:943–953.
2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Visone R and Croce CM: miRNAs and cancer.
Am J Pathol. 174:1131–1138. 2009.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Li M, Wang J and Liu H: Downregulation of
miR-638 promotes progression of breast cancer and is associated
with prognosis of breast cancer patients. Onco Targets Ther.
11:6871–6877. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Shen Y, Chen H, Gao L, Zhang W, He J, Yang
X, Qin L, Xue X and Guo Z: miR-638 acts as a tumor suppressor gene
in gastric cancer. Oncotarget. 8:108170–108180. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wei H, Zhang JJ and Tang QL: miR-638
inhibits cervical cancer metastasis through Wnt/β-catenin signaling
pathway and correlates with prognosis of cervical cancer patients.
Eur Rev Med Pharmacol Sci. 21:5587–5593. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bhattacharya A, Schmitz U, Raatz Y,
Schönherr M, Kottek T, Schauer M, Franz S, Saalbach A, Anderegg U,
Wolkenhauer O, et al: miR-638 promotes melanoma metastasis and
protects melanoma cells from apoptosis and autophagy. Oncotarget.
6:2966–2980. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ren Y, Chen Y, Liang X, Lu Y, Pan W and
Yang M: miRNA-638 promotes autophagy and malignant phenotypes of
cancer cells via directly suppressing DACT3. Cancer Lett.
390:126–136. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lu X, Chen L, Chen Y, Shao Q and Qin W:
Bafilomycin A1 inhibits the growth and metastatic potential of the
BEL-7402 liver cancer and HO-8910 ovarian cancer cell lines and
induces alterations in their microRNA expression. Exp Ther Med.
10:1829–1834. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Penzo C, Arnoldo L, Pegoraro S, Petrosino
S, Ros G, Zanin R, Wiśniewski JR, Manfioletti G and Sgarra R: HMGA1
modulates gene transcription sustaining a tumor signalling pathway
acting on the epigenetic status of triple-negative breast cancer
cells. Cancers (Basel). 11(1105)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang Y, Hu L, Zheng Y and Guo L: HMGA1 in
cancer: Cancer classification by location. J Cell Mol Med.
23:2293–2302. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lund T, Holtlund J, Fredriksen M and
Laland SG: On the presence of two new high mobility group-like
proteins in HeLa S3 cells. FEBS Lett. 152:163–167. 1983.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Méndez O, Pérez J, Soberino J, Racca F,
Cortés J and Villanueva J: Clinical implications of extracellular
HMGA1 in breast cancer. Int J Mol Sci. 20(5950)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zanin R, Pegoraro S, Ros G, Ciani Y,
Piazza S, Bossi F, Bulla R, Zennaro C, Tonon F, Lazarevic D, et al:
HMGA1 promotes breast cancer angiogenesis supporting the stability,
nuclear localization and transcriptional activity of FOXM1. J Exp
Clin Cancer Res. 38(313)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Cao XP, Cao Y, Zhao H, Yin J and Hou P:
HMGA1 promoting gastric cancer oncogenic and glycolytic phenotypes
by regulating c-myc expression. Biochem Biophys Res Commun.
516:457–465. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Jing Z, Liu C, Zhang QH, Chen L, Shen YY,
Chen YJ, Zeng X, Zu XY and Cao RX: TGF-β1 induces HMGA1 expression:
The role of HMGA1 in thyroid cancer proliferation and invasion. Int
J Oncol. 50:1567–1578. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Peters DG, Kudla DM, DeLoia JA, Chu TJ,
Fairfull L, Edwards RP and Ferrell RE: Comparative gene expression
analysis of ovarian carcinoma and normal ovarian epithelium by
serial analysis of gene expression. Cancer Epidemiol Biomarkers
Prev. 14:1717–1723. 2005.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liu Y, Wang Y, Zhang Y, Fu J and Zhang G:
Knockdown of HMGA1 expression by short/small hairpin RNA inhibits
growth of ovarian carcinoma cells. Biotechnol Appl Biochem. 59:1–5.
2012.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Ma T, Zhao Z, Wang Z, Wang C and Zhang L:
miR-940 inhibits migration and invasion of tongue squamous cell
carcinoma via regulatingCXCR2/NF-κB system-mediated
epithelial-mesenchymal transition. Naunyn Schmiedebergs Arch
Pharmacol. 392:1359–1369. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Schmittgen TD and Livak KJ: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
25
|
González-Magaña A and Blanco FJ: Human
PCNA structure, function and interactions. Biomolecules.
10(570)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Heneghan HM, Miller N and Kerin MJ: miRNAs
as biomarkers and therapeutic targets in cancer. Curr Opin
Pharmacol. 10:543–550. 2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lu J, Wang L, Chen W, Wang Y, Zhen S, Chen
H, Cheng J, Zhou Y, Li X and Zhao L: miR-603 targeted hexokinase-2
to inhibit the malignancy of ovarian cancer cells. Arch Biochem
Biophys. 661:1–9. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Creighton CJ, Fountain MD, Yu Z, Nagaraja
AK, Zhu H, Khan M, Olokpa E, Zariff A, Gunaratne PH, Matzuk MM and
Anderson ML: Molecular profiling uncovers a p53-associated role for
microRNA-31 in inhibiting the proliferation of serous ovarian
carcinomas and other cancers. Cancer Res. 70:1906–1915.
2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chao A, Lin CY, Lee YS, Tsai CL, Wei PC,
Hsueh S, Wu TI, Tsai CN, Wang CJ, Chao AS, et al: Regulation of
ovarian cancer progression by microRNA-187 through targeting
Disabled homolog-2. Oncogene. 31:764–775. 2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhao W, Han T, Li B, Ma Q, Yang P and Li
H: miR-552 promotes ovarian cancer progression by regulating PTEN
pathway. J Ovarian Res. 12(121)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev. 9:287–314. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhao LY, Yao Y, Han J, Yang J, Wang XF,
Tong DD, Song TS, Huang C and Shao Y: miR-638 suppresses cell
proliferation in gastric cancer by targeting Sp2. Dig Dis Sci.
59:1743–1753. 2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Cheng J, Chen Y, Zhao P, Liu X, Dong J, Li
J, Huang C, Wu R and Lv Y: Downregulation of miRNA-638 promotes
angiogenesis and growth of hepatocellular carcinoma by targeting
VEGF. Oncotarget. 7:30702–30711. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Juríková M, Danihel L', Polák Š and Varga
I: Ki67, PCNA, and MCM proteins: Markers of proliferation in the
diagnosis of breast cancer. Acta Histochem. 118:544–552.
2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhao H, Ho PC, Lo YH, Espejo A, Bedford
MT, Hung MC and Wang SC: Interaction of proliferation cell nuclear
antigen (PCNA) with c-Abl in cell proliferation and response to DNA
damages in breast cancer. PLoS One. 7(e29416)2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Madhumati G, Kavita S, Anju M, Uma S and
Raj M: Immunohistochemical expression of cell proliferating nuclear
antigen (PCNA) and p53 protein in cervical cancer. J Obstet Gynecol
India. 62:557–561. 2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Xia B, Yang S, Liu T and Lou G: miR-211
suppresses epithelial ovarian cancer proliferation and cell-cycle
progression by targeting Cyclin D1 and CDK6. Mol Cancer.
14(57)2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Khan S, Brougham CL, Ryan J, Sahrudin A,
O'Neill G, Wall D, Curran C, Newell J, Kerin MJ and Dwyer RM:
miR-379 regulates Cyclin B1 expression and is decreased in breast
cancer. PLoS One. 8(e68753)2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Radu A, Neubauer V, Akagi T, Hanafusa H
and Georgescu MM: PTEN induces cell cycle arrest by decreasing the
level and nuclear localization of cyclin D1. Mol Cell Biol.
23:6139–6149. 2003.PubMed/NCBI View Article : Google Scholar
|
|
41
|
He L, Zhu W, Chen Q, Yuan Y, Wang Y, Wang
J and Wu X: Ovarian cancer cell-secreted exosomal miR-205 promotes
metastasis by inducing angiogenesis. Theranostics. 9:8206–8220.
2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hong SJ, Dawson TM and Dawson VL: Nuclear
and mitochondrial conversations in cell death: PARP-1 and AIF
signaling. Trends Pharmacol Sci. 25:259–264. 2004.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Porter AG and Janicke RU: Emerging roles
of Caspase-3 in. Cell Death Differ. 6:99–104. 2015.
|
|
44
|
Zhang H, Li Y, Huang Q, Ren X, Hu H, Sheng
H and Lai M: miR-148a promotes apoptosis by targeting Bcl-2 in
colorectal cancer. Cell Death Differ. 18:1702–1710. 2011.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Liu J, Tu F, Yao W, Li X, Xie Z, Liu H, Li
Q and Pan Z: Conserved miR-26b enhances ovarian granulosa cell
apoptosis through HAS2-HA-CD44-Caspase-3 pathway by targeting HAS2.
Sci Rep. 6(21197)2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Masciullo V, Baldassarre G, Pentimalli F,
Berlingieri MT, Boccia A, Chiappetta G, Palazzo J, Manfioletti G,
Giancotti V, Viglietto G, et al: HMGA1 protein over-expression is a
frequent feature of epithelial ovarian carcinomas. Carcinogenesis.
24:1191–1198. 2003.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wei JJ, Wu X, Peng Y, Shi G, Basturk O,
Yang X, Daniels G, Osman I, Ouyang J, Hernando E, et al: Regulation
of HMGA1 expression by MicroRNA-296 affects prostate cancer growth
and invasion. Clin Cancer Res. 17:1297–1305. 2011.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Chen YN, Ren CC, Yang L, Nai MM, Xu YM,
Zhang F and Liu Y: MicroRNA let-7d-5p rescues ovarian cancer cell
apoptosis and restores chemosensitivity by regulating the p53
signaling pathway via HMGA1. Int J Oncol. 54:1771–1784.
2019.PubMed/NCBI View Article : Google Scholar
|