Introduction
Inflammatory bowel diseases (IBD) are immune
disorders associated with chronic inflammation in the intestinal
tract, and include Crohn's diseases and ulcerative colitis
(1-3).
Previous studies revealed that the microbiota, genetics, immune
dysregulation and stress participate in the development of IBD
(3-7).
The microbiota is thought to play a central role in the
pathogenesis of IBD, and multiple strategies have been developed to
treat IBD via targeting the gut flora (8-11).
Furthermore, previous studies have shown that certain microbiota or
gut dysbiosis can influence the outcomes of IBD, autoimmune
disorders and diabetes (12-14).
Moreover, segmented filamentous bacteria alone can have an
important impact on the development of IBD and also regulate the
immune responses in gut (15,16).
Another previous study showed that the ratio of Firmicutes
and Bacteroidetes can influence the sensitivity of mice to
IBD induction, and a lower Firmicutes/Bacteroidetes ratio
appears to be related to higher susceptibility to IBD in humans and
mice (17).
The regulation of gut microbiota is very complex
(18). Intestinal epithelial cells
(IECs) include different cell types, such as Paneth cells, goblet
cells and enterocytes, which produce antimicrobial peptides and
mucus, including regenerating islet-derived protein 3 γ, Reg3α,
mucin-1 and mucin-2, to regulate the constitution and homeostasis
of gut microbiota (19,20). Abnormal inflammatory cytokine
production in the intestine can also regulate the gut microbiota
directly or indirectly (21).
Immune cells can secret inflammatory cytokines such as tumor
necrosis factor α (TNF-α), interleukin 17 (IL-17) and IL-22 to
regulate the function of IECs, thereby influencing the balance of
intestinal flora (22,23). Moreover, stress can also affect the
homeostasis of gut microbiota, causing mice to be more sensitive to
IBD (24).
MicroRNAs (miRNA/miRs) are small non-coding RNA
molecules that exist in animals and plants, and play critical roles
in the regulation of gene expression (25,26).
Previous findings revealed that miRNAs can modulate gene expression
during or after transcription by targeting specific nucleotide
sequences (25). Moreover, miRNAs
participate in the development of different immune disorders and
cancer types (26-30).
The expression of miR-602 has been found to inhibit hepatitis C
virus genotype 1b infection and regulate the expression levels of
the tumor suppressor genes p53, p73 and Ras association domain
family 1 isoform A in a hepatoma cell line (31). miR-602 is also an important
signaling regulator in chondrocytes (32). Tumor necrosis factor
receptor-associated factor 6 (TRAF6) signaling has be found to be
involved in the development of IBD (33,34),
and previous studies showed that miR-146a can regulate the TRAF6
signaling pathway (34,35). However, it is not fully understood
whether miRNA, in particular miR-602, plays a role in IBD
pathogenesis and the regulation of gut microbiota.
The present results suggested that miR-602 is
downregulated in intestinal tissues from IBD mice. Furthermore,
overexpression of miR-602 may prevent the development of IBD,
increase the production of proinflammatory cytokines and decrease
intestinal integrity in an IBD mouse model. In addition, the
inhibitory effect of miR-602 was lost when the microbiota was
depleted using an antibiotic mix. It was also found that miR-602
can target the 3'-untranslated region (3'UTR) of TRAF6 and inhibit
the related signaling pathways in IECs.
Materials and methods
Bioinformatics assay
The expression level of miR-602 in intestinal
tissues under normal or ulcerative colitis condition was based on
GSE43009 and GSE53867 as previously described (36). GSE43009 and GSE53867 datasets were
downloaded from NCBI's Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo/). The
read count for miR-602 was selected and used for further
analysis.
Cell culture
Non-transformed rat small intestinal IEC-6 cells
(Bena Culture Collection) were cultured in DMEM (cat. no. 8112040;
Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(cat. no. 10099-141; Gibco; Thermo Fisher Scientific, Inc.), 100
U/ml penicillin and 100 µg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.) at 37˚C with 5% CO2. Coca-2 (American
Type Culture Collection), a human intestinal epithelial cell line,
was cultured in DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.)
containing 10% FBS and 1% penicillin-streptomycin at 37˚C with 5%
CO2.
Mice
Experiments were performed according to the
Institutional Guidelines for the Care and Use of Laboratory Animals
in Research and were approved by the Committee of Affiliated
Hospital of Nanjing University of Chinese Medicine. Male C57BL/6
mice (age, 6-8 weeks; weight, 20 g) were purchased from the
Shanghai SLAC Laboratory Animal Co., Ltd. and raised in the
Affiliated Hospital of Nanjing University of Chinese Medicine. The
mice were housed at a constant room temperature (23±2˚C) and
relative humidity (50±10%) with free access to food and water in a
fixed 12-h light/dark cycle.
Humane endpoint criteria were used in all
experiments involving mice, which were monitored daily: Euthanasia
was performed when mice exhibited signs of distress (weight loss
≥25%; hunched posture; diarrhea; loss of active movements), while
animals with diarrhea but with weight loss <25% received daily
intraperitoneal (i.p.) injections of 1 ml saline (x3, at 8 h
intervals) to avoid dehydration (37). At the end of the experiment or for
euthanasia, animals were euthanized with an overdose of
pentobarbital (i.p, 500 mg/kg supplemented as needed), deep
anesthetization was confirmed by loss of the pedal withdrawal
reflex. In total, 50 mice were used, and no mice were euthanized or
found dead over the course of the experiments.
Induction and evaluation of IBD
To induce IBD, mice were treated with drinking water
containing 3% dextran sulfate sodium (DSS; MP Biomedicals LLC). DSS
water was replaced every other day. There was no restriction
regarding the dose of DSS solution that was provided ad
libitum for 7 consecutive days, and control mice received
standard drinking water throughout the 7-day experiment. The
disease index of IBD consisted of the following factors: Body
weight loss, stool consistency and rectal bleeding (38). Body weight loss score was assessed
as followed: i) No weight loss =0; ii) weight loss from 1-5%
compared to baseline =1; iii) weight loss from 6-10% compared to
baseline =2; iv) weight loss from 11-20% compared to baseline =3;
and v) weight loss >20% compared to baseline =4. Stool
consistency was determined as follows: i) Well-formed pellets =0;
ii) pasty and semi-formed stools =2; and iii) liquid stools =4. For
bleeding, the following criteria were used: i) No blood =0; ii)
positive bleeding =2; and iii) gross bleeding =4.
Adenovirus (Ad) vector treatment
Adenovirus vectors (pAdEasy/Track-CMV) used in the
present study were purchased from Agilent Technologies Inc. Blank
vectors without miR-602 were used as negative controls. To
overexpress miR-602 (5'-GACACGGGCGACAGCUGCGGCCC-3'), 100 µl of
adenovirus suspension was injected intravenously into 8-week-old
mice (2x109 particles/g body weight) retro-orbitally
using a 0.5 insulin syringe (Becton, Dickinson and Company) on day
1 and 3. The treatment lasted for 7 days, during which the mice
were checked daily. After adenovirus treatment, mice were treated
with 3% DSS (w/v) in drinking water for 7 days to establish an IBD
mouse model.
Fecal microbiota depletion and fecal
microbiota transplantation (FMT)
For fecal microbiota depletion, antibiotics were
administered in autoclaved drinking water for 4 weeks in the
following concentrations: Vancomycin (1 g/l), ampicilin (1.5 g/l),
kanamycin (1 g/l) and metronidazole (1.5 g/l). This time course is
consistent with standard antibiotic administration used in multiple
studies for altering microbiota (39). After microbiota depletion, mice were
treated with 3% DSS (w/v) in drinking water for 7 days to establish
the IBD mouse model.
For mice with microbiota depletion, FMT was
performed as described previously (40). In the morning, feces were collected
and immediately diluted (1:20 w/vol) in sterile, anoxic Ringer
buffer (Beijing Solarbio Science & Technology, Co., Ltd.)
containing 1 g/l L-cysteine (Sigma-Aldrich; Merck KGaA) as a
reducing agent. This was used as inoculum. Colonization was
achieved by intragastric gavage with 200 µl inoculum once per day
for 5 consecutive days. A total of 10 days after FMT, mice were
treated with DSS for 7 days to establish the IBD mice model.
The mice were housed at a constant room temperature
(23±2˚C) and relative humidity (50±10%) under a fixed 12-h
light/dark cycle. They were given free access to food and water.
Each cage housed a total of 5 mice. To investigate the potential
role of microbiota in miR-602 mediated therapy in IBD, mice
injected with adenovirus carrying negative control (Ad-NC) were
co-housed with mice injected with Ad-miR-602 for three weeks, after
which the IBD model was induced. A total of 5 mice were included in
each group.
Fecal microbiota was isolated from Ad-NC and
Ad-miR-602 mice, and these fecal microbiotas were separately
transferred into antibiotic-treated mice. After 10 days, IBD model
was induced in these microbiota-transferred mice and the
pathogenesis index was recorded.
Histological analysis
Hematoxylin and eosin (H&E) staining was used to
assess the induction of IBD. After euthanizing the mice, colon
tissues from control mice and IBD mice were collected and fixed at
room temperature in PBS containing 4% paraformaldehyde for 72 h.
The samples were sectioned to 6 µm thickness using a microtome
(cat. no. SM2500; Leica Microsystems GmbH). Then, the sections were
stained with hematoxylin and eosin (hematoxylin, 1.0 kg/l; eosin,
0.827 kg/l), and examined under light microscopy (Olympus
Corporation; magnification, x100).
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
To isolate RNA from colon tissues, a mortar and a
pestle was used to grind the frozen tissues which were pre-cooled
by liquid nitrogen. After euthanizing the mice, the colon tissue
samples were collected. After length measurement and weighing, the
tissue sample was put into liquid nitrogen and grinded. The treated
samples were lysed with TRIzol® (Invitrogen; Thermo
Fisher Scientific, Inc.) for extraction of mRNA. The generation of
cDNA from the extracted mRNA samples was performed using the
PrimeScript™ RT Reagent kit (cat. no. RR037B, Takara Bio, Inc.).
After mixing RNA with the kit components as described in the user
manual, the mixture was incubated at 37˚C for 15 min three times,
and then incubated at 85˚C for 5 sec to inactivate the reverse
transcriptase. RT-qPCR analysis was performed to assess gene
expression with Premix Ex Taq™ (Probe qPCR; cat. no. RR390W; Takara
Bio, Inc.). The thermocycling conditions were as follows: 15 min at
95˚C (initial denaturation), followed by 35 cycles of 15 sec at
94˚C (denaturation), 30 sec at 60˚C (annealing) and 30 sec at 72˚C
(extension). For gene expression in cells, IEC-6 and caco-2 cells
were collected, total RNA was isolated with TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.), and reverse
transcription and qPCR was performed as aforementioned. miRNA and
mRNA expression levels were quantified using the 2-ΔΔCq
method (41). The primers used for
PCR are listed in Table I.
 | Table IPrimer used for reverse
transcription-quantitative PCR. |
Table I
Primer used for reverse
transcription-quantitative PCR.
| Gene | Forward
(5'-3') | Reverse
(5'-3') |
|---|
| TNFα (m) |
ACTGAACTTCGGGGTGATCG |
GTTTGCTACGACGTGGGCTA |
| IL-1β (m) |
TGCCACCTTTTGACAGTGATG |
ATGTGCTGCTGCGAGATTTG |
| IL-6 (m) |
GTCCTTCCTACCCCAATTTCCA |
TAACGCACTAGGTTTGCCGA |
| TGF-β (m) |
GTGGACACTCGATCGCTACC |
GATGCGAGGGACTCAAGAGG |
| OCLN (m) |
TTTCAGGTGAATGGGTCACCG |
GCTCCCAAGATAAGCGAACCT |
| MUC-2 (m) |
TTGTCACCTTCGATGGGCTC |
TCTCGTGGCGCACAATAAGT |
| MUC-3 (m) |
CAAGAAGAGCGCAAAGCAGG |
CCTCCATCCCACACACTTCC |
| ZO-1 (m) |
AGAAAAAGAATGCACAGAGTTGT |
GAAATCGTGCTGATGTGCCA |
| TRAF6 (m) |
TCCAGAGTTTGCCGTCCAAG |
TGGGTCCCTTCAGAAGTTCA |
| GAPDH (m) |
TCTTTTGCGTCGCCAGCC |
CCATGGGTGGAATCATATTGGAAC |
| TRAF6 (h) |
GCGCACTAGAACGAGCAAG |
GGCAGTTCCACCCACACTAT |
| GAPDH (h) |
CTTTTGCGTCGCCAGGTGAAG |
ACCAAATCCGTTGACTCCGAC |
| miR-602 (h/m) |
GACACGGGCGACAGCUG |
CTCGCTTCGGCAGCACA |
| U6 (h/m) |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
Western blotting
Cells were transfected with Ad-NC and Ad-miR-602
using Lipofectamine® 2000 (Thermo Fisher Scientific,
Inc.). After 24 h incubation at 37˚C, cell samples were collected
and lysed with Cell lysis buffer (Beyotime Institute of
Biotechnology) containing the protease inhibitor cocktail (Beyotime
Institute of Biotechnology) for western blotting and IP. After the
protein concentration was determined using a bicinchoninic acid
protein assay kit (Beyotime Institute of Biotechnology), protein
extracts (10 µg) were analyzed via 12% SDS-PAGE. The proteins were
then transferred onto nitrocellulose membranes, which were
subsequently incubated with the following primary antibodies at 4˚C
overnight: TRAF6 (1:1,000; cat. no. ab227560; Abcam) and GAPDH
(1:2,000; cat. no. 60004; ProteinTech Group, Inc.). After washing
with PBST in triplicate, horseradish peroxidase-conjugated
anti-anti-rabbit (1:2,000; cat. no. SA00001-7L; ProteinTech Group,
Inc.) or anti-mouse (1:2,000; cat. no. SA00001-1; ProteinTech
Group, Inc.) secondary antibodies were added and incubated at room
temperature for 2 h. The membranes were then washed with
Tris-buffered saline and 0.1% Tween-20 in triplicate. Protein
levels were assessed using an enhanced chemiluminescence mix
(Thermo Fisher Scientific, Inc.).
Luciferase reporter assay
The luciferase reporter assay was performed as
described previously (42). MODE-K
and Caco-2 cells were co-infected with wild-type or mutant
TRAF6-3'UTR-Luc firefly luciferase constructs (100 ng) and 40
nmol/l miR-602 mimics using Lipofectamine 2000 reagent (Thermo
Fisher Scientific, Inc.). Then, 2 ng of pRL-SV40 plasmids (Addgene,
Inc.) were infected to assess the transfection efficiency. After
incubation for 24 h at 22˚C, luciferase activity was analyzed using
the Dual-Luciferase® Reporter (DLR™) Assay System
(Promega Corporation).
Statistical analysis
All experiments were performed ≥3 times. Data are
presented as the mean ± SEM. Data were analyzed using a one-way
ANOVA with Tukey's multiple comparison test or an unpaired
two-tailed Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
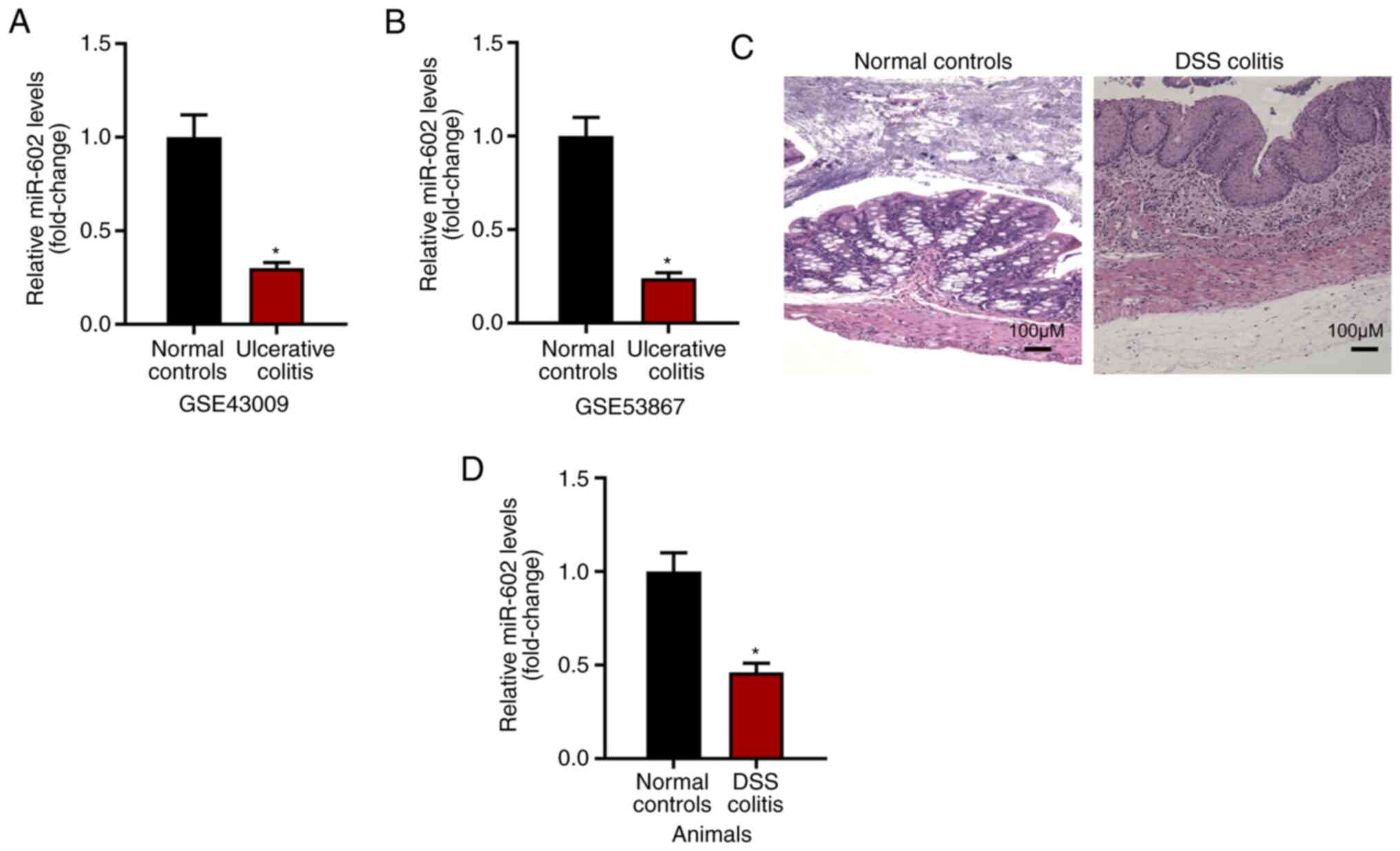
Expression level of miR-602 is
negatively related to IBD development
The present study investigated the expression level
of miR-602 in intestinal tissues under normal or ulcerative colitis
conditions based on GSE43009 and GSE53867. It was found that,
compared with normal intestinal tissues from healthy mice, tissues
with ulcerative colitis had a decreased expression level of miR-602
(Fig. 1A and B). To examine this result, IBD was induced
using DSS (Fig. 1C) and the
intestinal tissues were collected for RNA analysis. It was found
that intestinal tissues from IBD mice had decreased miR-602
expression compared with intestinal tissue from control mice
(Fig. 1D). Collectively, the
present results suggested that the expression level of miR-602 is
negatively related to IBD development.
Overexpression of miR-602 alleviates
IBD in mice
To investigate the role of miR-602 in IBD
development, miR-602 was overexpressed in mice by injecting
Ad-miR-602. After 5 days, it was found that the expression level of
miR-602 was increased in intestinal tissues in mice with Ad-miR-602
compared with those injected with Ad-NC (Fig. 2A). The present study established an
IBD mouse model using DSS, and then recorded body weight, occult
blood, and diarrhea score. The present results suggested that
overexpression of miR-602 increased body weight, but significantly
decreased occult blood, diarrhea score and disease activity index
(Fig. 2C-E). Compared with healthy
mice, Ad-NC treated mice showed a significantly higher colonic
weight/length ratio, while the colonic weight/length ratio of mice
overexpressing miR-602 was similar to healthy mice (Fig. 2F). Therefore, the present results
indicated that miR-602 may alleviate IBD.
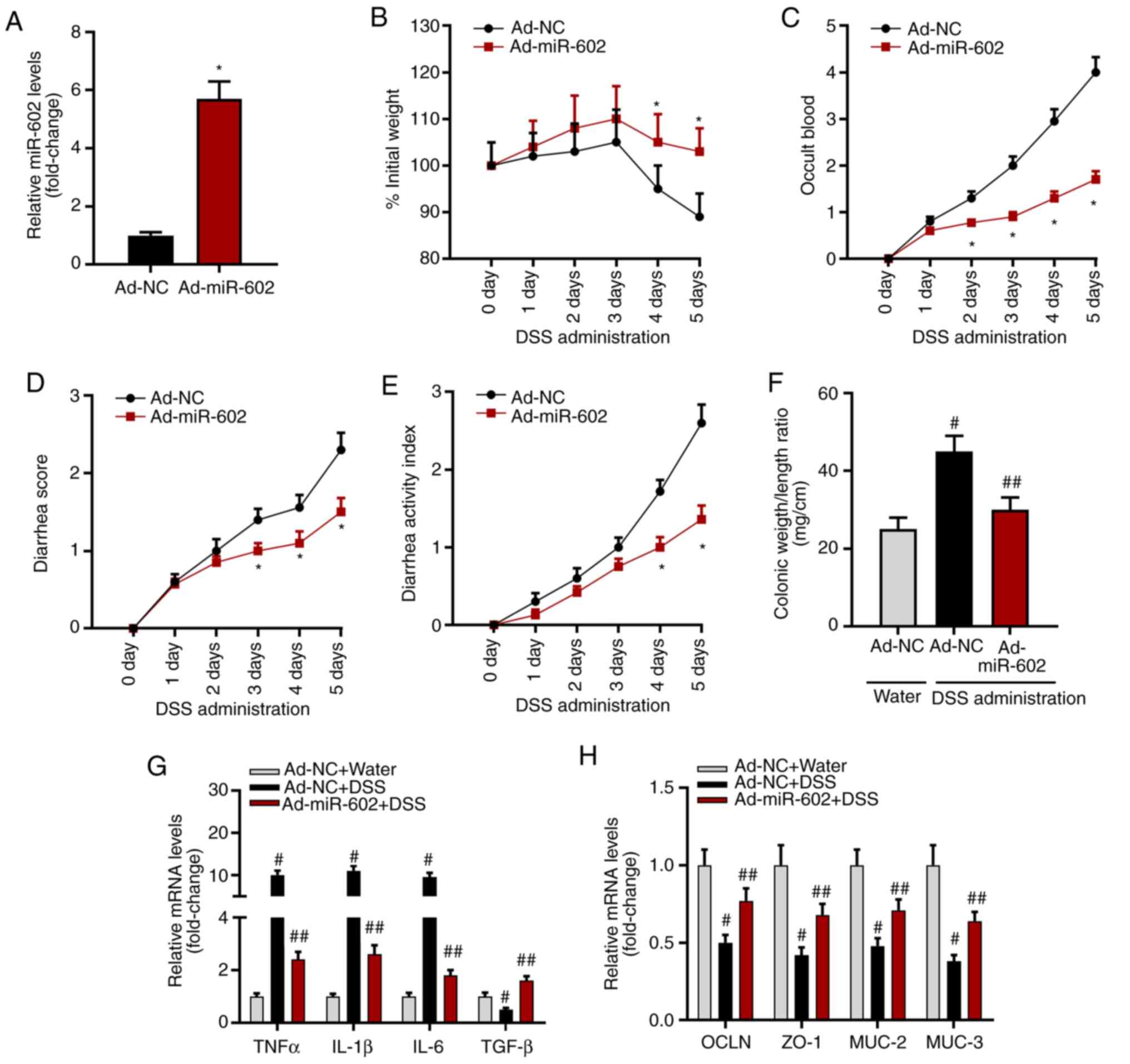 | Figure 2Overexpression of miR-602 alleviates
IBD. Mice were injected with Ad-NC or Ad-miR-602. (A) Expression
level of miR-602. Mice transfected with Ad-NC or Ad-miR-602 were
treated with DSS to induce IBD. *P<0.05 vs. the Ad-NC
group. (B) Body weight, (C) occult blood, (D) diarrhea score and
(E) diseases activity index were analyzed. All,
*P<0.05 vs. the Ad-NC group. (F) Colonic weight and
length were measured, and the ratio of weight to length was
calculated. #P<0.05 and ##P<0.01 vs.
the Ad-NC water group. mRNA expression levels of (G) TNF-α, IL-1β,
IL-6 and TGF-β. (H) mRNA expression levels of OCLN, ZO-1, MUC-2 and
MUC-3. n=5-6. #P<0.05 and ##P<0.01 vs.
the corresponding Ad-NC water group. TNFα, tumor necrosis factor α;
IL, interleukin; TGF-β, transforming growth factor β; OCLN,
occludin; MUC, Mucin; ZO-1, zonula occludens; TRAF6, Tumor necrosis
factor receptor-associated factor 6; IBD, inflammatory bowel
diseases; miR, microRNA; DSS, dextran sulfate sodium; Ad,
adenovirus; NC, negative control. |
Furthermore, the mRNA expression level of
inflammatory cytokines and the genes related to intestinal
integrity were investigated. It was found that miR-602
overexpression significantly inhibited mRNA expression levels of
the proinflammatory cytokines TNF-α, IL-1β and IL-6, but increased
the expression levels of the anti-inflammatory cytokine
transforming growth factor β (TGF-β; Fig. 2G). Moreover, miR-602 overexpression
promoted the expression of occludin, zonula occludens 1 (ZO-1),
Mucin 2 (MUC-2) and MUC-3 (Fig.
2H), indicating that miR-602 may prevent against the
destruction of intestinal tissue integrity. Collectively, the
present results suggested that miR-602 could have a protective role
in IBD.
Co-housing healthy,
miR-602-overexpressing mice with IBD mice showed beneficial effects
on IBD
Microbiota has been shown to play critical roles in
the development of IBD (43). To
investigate the potential role of microbiota in miR-602 mediated
therapy in IBD, mice injected with Ad-NC were co-housed with mice
injected with Ad-miR-602, after which IBD was induced 5 days later.
An IBD model was also induced in Ad-NC mice without co-housing,
which were used as control. It was found that those in the Ad-NC +
Ad-miR-602 group had little difference in body weight change
compared with the Ad-NC + Ad-NC group (Fig. 3A). Furthermore, the Ad-NC +
Ad-miR-602 group exhibited less severity in occult blood, diarrhea
and disease scores (Fig. 3B-D). It
was also demonstrated that, while co-housing with Ad-miR-602 mice
did not significantly influence the colonic weight/length ratio,
co-housing decreased the expression levels of TNF-α and increased
the expression of the cellular integrity-related gene ZO-1 in
intestinal tissues. Mice could transmit microbes to others upon
transient co-housing through coprophagy and grooming behaviors.
Therefore, the present results suggested that microbiota from
miR-602-overexpressing mice may be transferred to Ad-NC mice during
the co-housing process, and that the microbiota plays a protective
role in miR-602-mediated IBD.
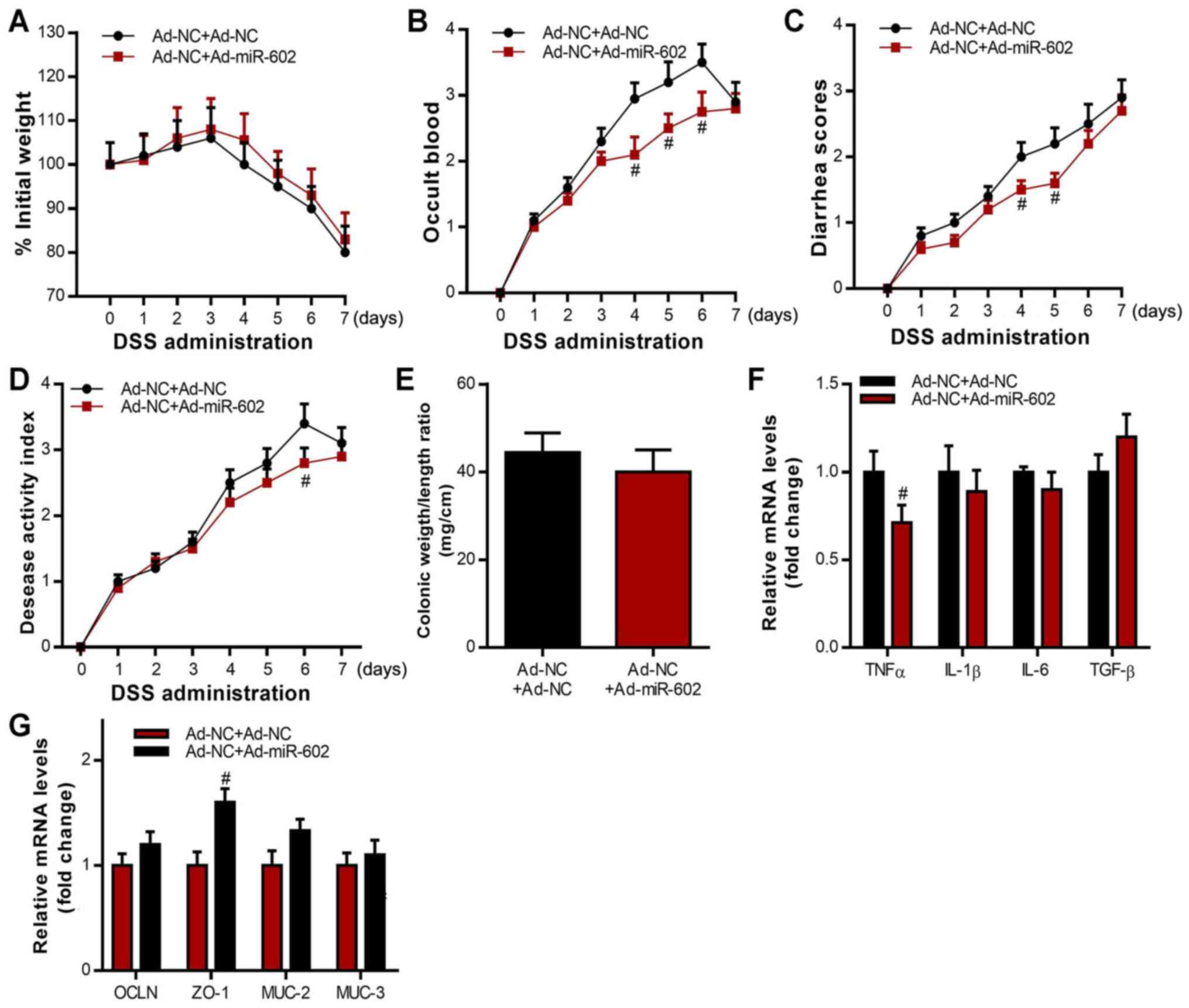 | Figure 3Co-housing Ad-miR-602-treated mice
attenuates IBD in Ad-NC-treated mice. Ad-NC-treated mice were
co-housed with or without Ad-miR-602-treated mice, then induced
with IBD. (A) Body weight, (B) occult blood, (C) diarrhea score and
(D) diseases activity index of mice were analyzed. (E) Colonic
weight and length were recorded, and the ratio of weight to length
was calculated. (F) mRNA expression levels of TNF-α, IL-1β, IL-6
and TGF-β. (G) mRNA expression levels of OCLN, ZO-1, MUC-2 and
MUC-3. TNFα, tumor necrosis factor α; IL, interleukin; TGF-β,
transforming growth factor β; OCLN, occludin; MUC, Mucin; ZO-1,
zonula occludens-1; TRAF6, Tumor necrosis factor
receptor-associated factor 6; IBD, inflammatory bowel diseases;
miR, microRNA; DSS, dextran sulfate sodium; Ad, adenovirus; NC,
negative control. n=5-6. #P<0.05 vs. the Ad-NC+Ad-NC
group in B-D, F, and G. |
Overexpression of miR-602 alleviates
IBD in a microbiota-dependent manner
To further investigate whether miR-602 alleviates
IBD via the microbiota, antibiotics were used to deplete the gut
microbiota in Ad-NC mice and Ad-miR-602 mice, and their responses
to DSS-induced IBD were assessed. The results revealed that the
inhibitory effect of miR-602 on body weight, occult blood, diarrhea
score, disease activity index and colonic weight/length ratio was
lost when gut microbiota were depleted (Fig. 4A-E). As presented in Fig. 4F, the inhibitory effect of miR-602
on the expression of various pro-inflammatory cytokines, including
TNF-α, IL-1β, IL-6 and TNF-β was also lost when gut microbiota were
depleted. In contrast, the stimulatory effect of miR-602 on the
expression of cellular integrity-related genes, including OCLN,
ZO-1, MUC-2 and MUC-3, was inhibited when gut microbiota were
depleted (Fig. 4G). These results
suggested that gut microbiota depletion abrogated the effects of
miR-602 overexpression by alleviating IBD and regulating gene
expression in the colon. Fecal microbiota was isolated from Ad-NC
and Ad-miR-602 mice, and these fecal microbiota were separately
transferred into antibiotic-treated mice. After 10 days, IBD was
induced in the microbiota-transferred mice and the pathogenesis was
recorded. The present results indicated that compared to Ad-NC-FMT
mice, those that received fecal microbiota from Ad-miR-602 mice had
a higher expression level of miR-602 in the colon (Fig. 5A), thus suggesting a bi-directional
interaction between miR-602 and gut microbiota. Furthermore, fecal
microbiota from Ad-miR-602 mice, as opposed to that from Ad-NC
mice, prevented weight loss, occult blood and diarrhea in IBD mice
(Fig. 5B-E). It was found that
Ad-miR-602 mouse-derived fecal microbiota also reduced the colonic
weight/length ratio (Fig. 5F). The
expression levels of inflammatory cytokines and genes related to
intestinal integrity were also examined, and it was demonstrated
that Ad-miR-602 mouse-derived fecal microbiota decreased the
expression levels of the proinflammatory genes TNF-α, IL-1β and
IL-6 (Fig. 5G). Moreover,
Ad-miR-602 mouse-derived fecal microbiota significantly increased
the expression level of TGF-β (Fig.
5G) and intestinal integrity-related genes (Fig. 5H). Collectively, the present results
suggested that miR-602 may help to prevent the development of IBD
in a microbiota-dependent manner.
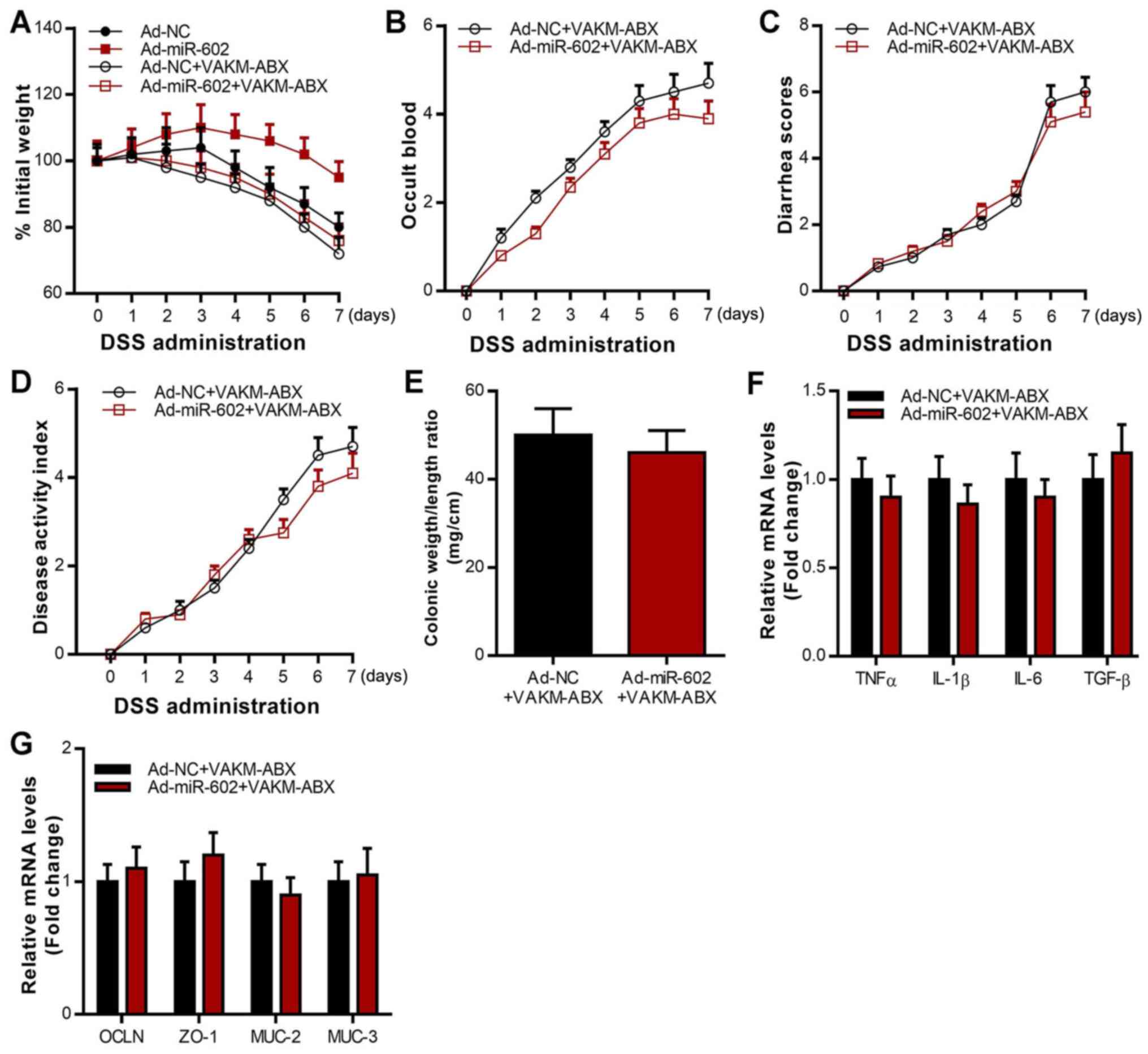 | Figure 4Antibiotics treatment abrogates the
beneficial effects of miR-602 overexpression in IBD mice. Mice were
treated with Ad-NC or Ad-miR-602, and then the gut microbiota was
depleted using an antibiotics mix. Then, DSS was used to induce
IBD. (A) Body weight, (B) occult blood, (C) diarrhea score and (D)
disease activity index. (E) Colonic weight and length were
measured, and the ratio of weight to length was calculated. (F)
mRNA expression of TNF-α, IL-1β, IL-6, TGF-β. (G) mRNA expression
level of OCLN, ZO-1, MUC-2 and MUC-3. n=5-6. TNFα, tumor necrosis
factor α; IL, interleukin; TGF-β, transforming growth factor β;
OCLN, occludin; MUC, Mucin; ZO-1, zonula occludens-1; TRAF6, Tumor
necrosis factor receptor-associated factor 6; IBD, inflammatory
bowel diseases; miR, microRNA; DSS, dextran sulfate sodium; Ad,
adenovirus; NC, negative control; VAKM, the combination of
vancomycin (1 g/l), ampicilin (1.5 g/l), kanamycin (1 g/l), and
metronidazole (1.5 g/l); ABX, antibiotics. n=5-6. |
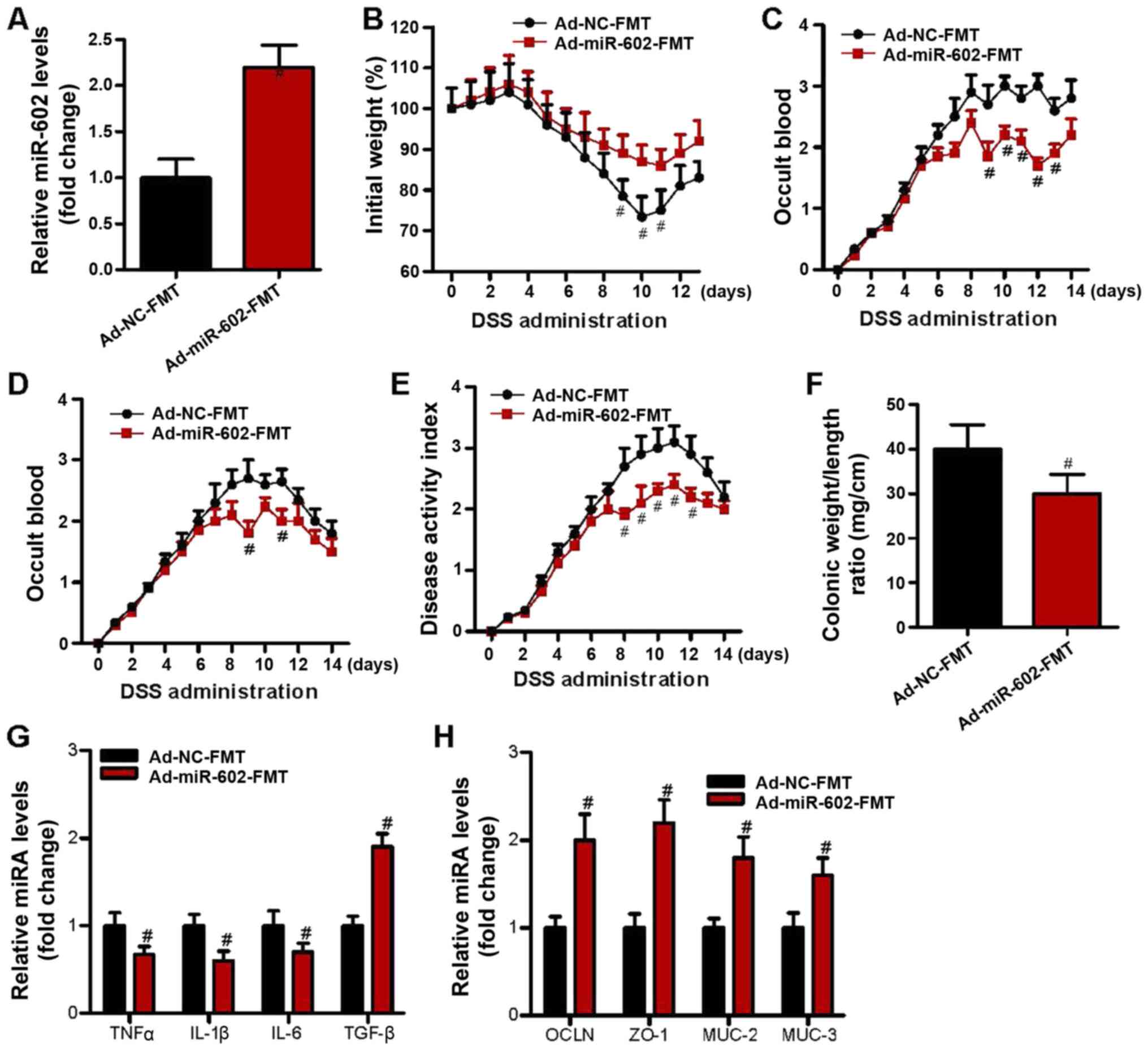 | Figure 5Fecal microbiota from
Ad-miR-602-treated mice attenuated IBD in mice. Fecal microbiota
was isolated from the Ad-NC or Ad-miR-602-treated mice derived
feces, and were transferred into antibiotic-treated mice. DSS
containing drinking water was used to induce IBD. (A) Expression
level of miR-602 in Ad-NC-FMT and Ad-miR-602-FMT groups. (B) Body
weight, (C) occult blood, (D) diarrhea score and (E) disease
activity index of the mice. (F) Colonic weight and length were used
to calculate the ratio of weight to length. (G) mRNA expression
levels of TNF-α, IL-1β, IL-6 and TGF-β. (H) mRNA expression levels
of OCLN, ZO-1, MUC-2 and MUC-3. FMT, fecal microbiota
transplantation; TNFα, tumor necrosis factor α; IL, interleukin;
TGF-β, transforming growth factor β; OCLN, occludin; MUC, Mucin;
ZO-1, zonula occludens-1; TRAF6, Tumor necrosis factor
receptor-associated factor 6; IBD, inflammatory bowel diseases;
miR, microRNA; DSS, dextran sulfate sodium; Ad, adenovirus; NC,
negative control. n=6. #P<0.05 vs. the Ad-NC-FMT
group. |
miR-602 inhibits TRAF6 signaling in
intestinal tissues
The present study investigated whether miR-602
regulates IBD via the TRAF6 signaling pathway in intestinal
tissues. It was demonstrated that TRAF6 expression level is
significantly increased in IBD mice compared with healthy control
mice (Fig. 6A). Moreover,
overexpression of miR-602 significantly inhibited TRAF6 mRNA and
protein expression levels in IEC-6 and Caco-2 cells (Fig. 6B-D). Furthermore, using a luciferase
reporter assay it was found that the mutation in 3'UTR miR-602
binding sites of TRAF6 (Fig. 6E)
abrogated the effects of wild-type miR-602 on TRAF6 expression
level (Fig. 6F and G).
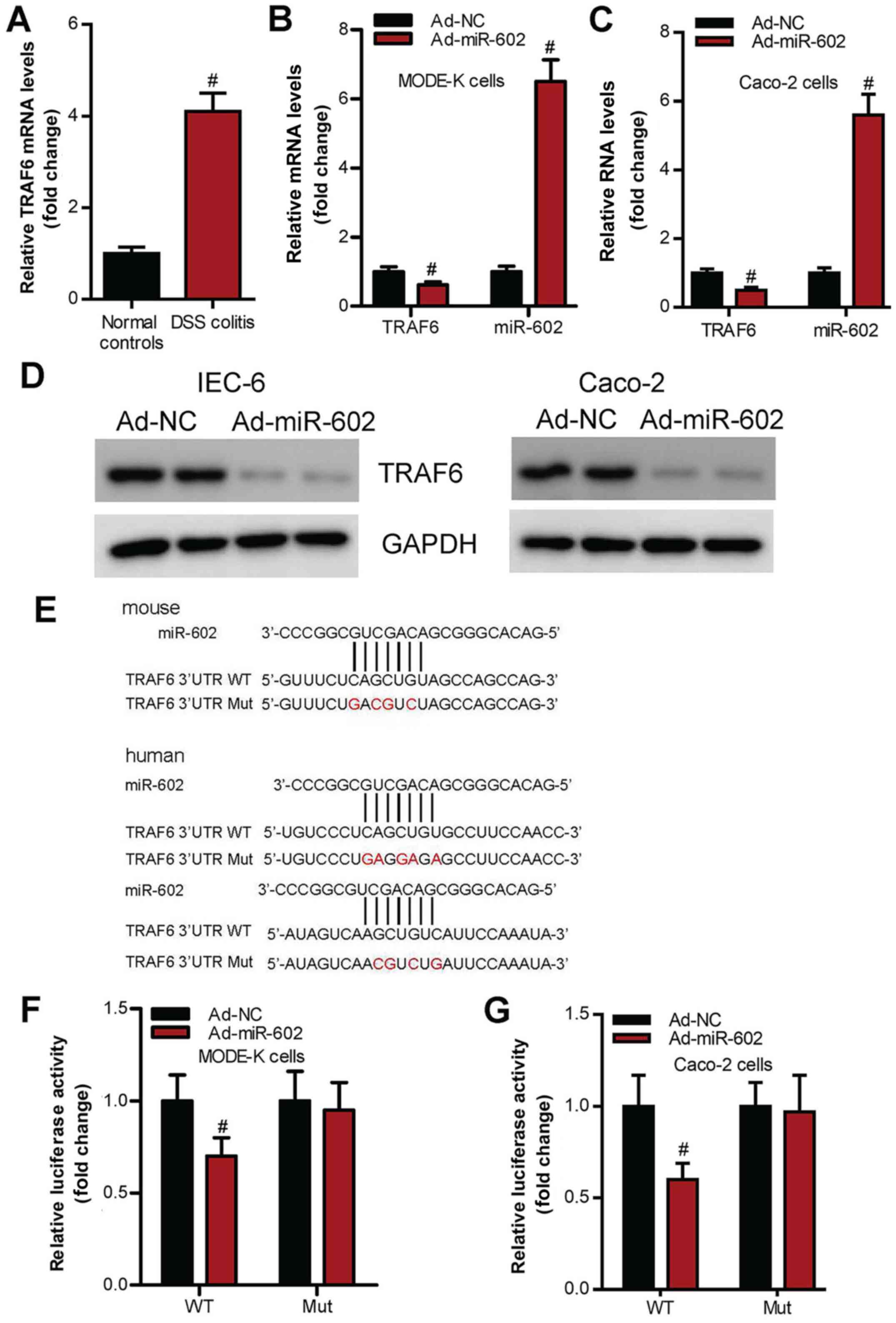 | Figure 6miR-602 targets TRAF6 signaling in
IEC. (A) Expression level of TRAF6 in intestinal tissues from
healthy mice and IBD mice. mRNA expression levels of TRAF6 and
miR-602 in (B) IEC-6 and (C) Caco-2 cells treated with Ad-NC or
Ad-miR-602. (D) Protein expression levels of TRAF6 in IEC-6 and
Caco-2 cells treated with Ad-NC or Ad-miR-602. (E) TRAF6 3'UTR Mut
site of mouse and human. There are two binding sites in the TRAF6
3'UTR, both of the binding sites were mutated. WT and TRAF6 3'UTR
Mut in (F) Mode-K and (G) Caco-2 cell lines treated with Ad-NC or
Ad-miR-602. The expression of TRAF6 was calculated by testing the
luciferase activities. TRAF6, Tumor necrosis factor
receptor-associated factor 6; IBD, inflammatory bowel diseases;
miR, microRNA; DSS, dextran sulfate sodium; Ad, adenovirus; NC,
negative control; WT, wild-type; Mut, mutant; 3'UTR, 3'untranslated
regions. n=5-6. #P<0.05 vs. Ad-NC group. |
Discussion
Genetic mutations and microenvironment exposure are
the most important factors for the occurrence of IBD (4,7),
however, recent studies have demonstrated that the microbiota also
plays a critical role in the progression of autoimmune disorders,
such as colitis (10,11,44).
Thus, targeting microbiota or developing microbiota-related
medicine may be an effective strategy to facilitate the treatment
of IBD. However, it is not fully understood how the constitution
and homeostasis of gut microbiota is regulated.
Previous studies have shown that miRNAs play an
important role in the initiation and development of IBD (28). It has been previously shown that
miR-148a is downregulated in human IBD and in tissues of patient
with colorectal cancer, and that restoration of miR-148a expression
inhibits colitis and colitis-associated tumorigenesis in mice
(45). By contrast, miR-155 is
increased in the colonic mucosa and peripheral blood in patients
with IBD, suggesting that miR-155 may act as a biomarker for the
diagnosis of IBD (46). Moreover, a
recent study showed that the microbiota decreases the expression
level of host miR-665, which then increases the expression level of
ATP Binding Cassette Subfamily C Member 3 in the host (46). In contrast to these previous
studies, the present results suggested that miR-602 regulated the
microbiota, which then alleviated IBD development. In addition, the
present study found that the regulation of gut flora by miR-602 may
be achieved via the targeting of TRAF6 in IECs, which play critical
roles in the regulation of gut microbiota (47). Therefore, further studies on the
exact mechanisms of miR-602 on IECs may facilitate the development
of a pharmacological target for the prevention and treatment of
IBD.
Abnormal production of inflammatory cytokines is
considered to be related to the development of colitis (48,49),
however, neutralization of inflammatory cytokines does not always
result in the alleviation of colitis (50), suggesting that the dysregulation of
inflammatory cytokines may be caused by other upstream events
(50). The present results
suggested that miR-602 downregulated the production of certain
proinflammatory cytokines, while upregulating the production of the
anti-inflammatory cytokine TGF-β. Furthermore, these effects were
dependent on the gut microbiota, as overexpression of miR-602 was
not seen to have inhibitory effects in microbiota-depleted mice.
Thus, dysbiosis in the gut may cause hyperactivation of the immune
system, and therapies targeting microbiota may be an effective
method to control immune responses during colitis.
Adoptive transfer of microbiota or the use of
microbial preparation are considered to be ideal strategies for the
treatment of autoimmune disorders and cancer types (51,52).
The present study found that miR-602 may shape the gut microbiota
balance, even under a co-housing situation. Furthermore, the
present results suggested that overexpression of miR-602 exerted
beneficial effects on IBD mice, indicating this miR-602 may
regulate the constitution and colonization of gut microbiota.
Moreover, microbiota has been found to influence the development of
different kinds of immune disorders, such as experimental
autoimmune encephalomyelitis and cancer types (53,54).
Thus, future studies could investigate the combination of
microbiota adoptive transfer and miR-602 overexpression, which may
be a potential therapeutic method for colitis.
Furthermore, the present study identified that
miR-602 exerted its inflammatory effect via the inhibition of TRAF6
expression. Previous studies have shown that TRAF6 plays a critical
role in IBD (33). It has been
shown that TRAF6 neddylation drives inflammatory arthritis by
increasing NF-κB activation, which then activate the expression of
various inflammatory cytokines including TNF-α, IL-6 and
IL-8(45).
Collectively, the present results suggested that
miR-602 may alleviate IBD in a microbiota-dependent manner and that
IECs play important roles in such processes. The present results
provide further evidence on the regulation of the gut microbiota
and facilitate the development of strategies for the treatment of
IBD.
Acknowledgements
Not applicable.
Funding
This project was sponsored by the Program of The State
Administration of Traditional Chinese Medicine of China (grant no.
JDZX2015083) and the National Natural Science Foundation of China
(grant no. 81774243).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HS contributed to the design and conception of the
study and revised it carefully for important intellectual content.
SZ, LeZ, WF, DDC and LuZ acquired the data by screening papers
identified on PubMed. DDC and YCH revised the study critically for
important intellectual content. SZ, LeZ, WF and YCH were involved
in drafting the manuscript. SZ and YCH analyzed and interpreted the
data. SZ also contributed to the conception and design of the study
and revised the manuscript. All authors agreed to be accountable
for all aspects of the work in ensuring that questions related to
the accuracy or integrity of any part of the work are appropriately
investigated and resolved. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Committee of
Affiliated Hospital of Nanjing University of Chinese Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hodson R: Inflammatory bowel disease.
Nature. 540(S97)2016.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Chu H, Khosravi A, Kusumawardhani IP, Kwon
AH, Vasconcelos AC, Cunha LD, Mayer AE, Shen Y, Wu WL, Kambal A, et
al: Gene-microbiota interactions contribute to the pathogenesis of
inflammatory bowel disease. Science. 352:1116–1120. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Neurath MF: Cytokines in inflammatory
bowel disease. Nat Rev Immunol. 14:329–342. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Kaser A, Zeissig S and Blumberg RS:
Inflammatory bowel disease. Annu Rev Immunol. 28:573–621.
2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Maloy KJ and Powrie F: Intestinal
homeostasis and its breakdown in inflammatory bowel disease.
Nature. 474:298–306. 2011.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Cho JH: The genetics and
immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol.
8:458–466. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Khor B, Gardet A and Xavier RJ: Genetics
and pathogenesis of inflammatory bowel disease. Nature.
474:307–317. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liang J, Sha SM and Wu KC: Role of the
intestinal microbiota and fecal transplantation in inflammatory
bowel diseases. J Dig Dis. 15:641–646. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lopetuso LR, Petito V, Graziani C,
Schiavoni E, Paroni Sterbini F, Poscia A, Gaetani E, Franceschi F,
Cammarota G, Sanguinetti M, et al: Gut microbiota in health,
diverticular disease, irritable bowel syndrome, and inflammatory
bowel diseases: Time for microbial marker of gastrointestinal
disorders. Dig Dis. 36:56–65. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wehkamp J and Frick JS: Microbiome and
chronic inflammatory bowel diseases. J Mol Med (Berl). 95:21–28.
2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Miyoshi J and Chang EB: The gut microbiota
and inflammatory bowel diseases. Transl Res. 179:38–48.
2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Baumgart DC and Carding SR: Inflammatory
bowel disease: Cause and immunobiology. Lancet. 369:1627–1640.
2007.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Campbell AW: Autoimmunity and the gut.
Autoimmune Dis. 2014(152428)2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Musso G, Gambino R and Cassader M:
Obesity, diabetes, and gut microbiota: The hygiene hypothesis
expanded? Diabetes Care. 33:2277–2284. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yi J, Jung J, Han D, Surh CD and Lee YJ:
Segmented filamentous bacteria induce divergent populations of
Antigen-Specific CD4 T cells in the small intestine. Mol Cells.
42:228–236. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ivanov II, Atarashi K, Manel N, Brodie EL,
Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al:
Induction of intestinal Th17 cells by segmented filamentous
bacteria. Cell. 139:485–498. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Molist F, Manzanilla EG, Perez JF and
Nyachoti CM: Coarse, but not finely ground, dietary fibre increases
intestinal Firmicutes:Bacteroidetes ratio and reduces
diarrhoea induced by experimental infection in piglets. Br J Nutr.
108:9–15. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Boulangé CL, Neves AL, Chilloux J,
Nicholson JK and Dumas ME: Impact of the gut microbiota on
inflammation, obesity, and metabolic disease. Genome Med.
8(42)2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu X, Lu J, Liu Z, Zhao J, Sun H, Wu N,
Liu H, Liu W, Hu Z, Meng G, et al: Intestinal Epithelial
Cell-Derived LKB1 suppresses colitogenic microbiota. J Immunol.
200:1889–1900. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sekirov I, Russell SL, Antunes LC and
Finlay BB: Gut microbiota in health and disease. Physiol Rev.
90:859–904. 2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ding Y, Yanagi K, Cheng C, Alaniz RC, Lee
K and Jayaraman A: Interactions between gut microbiota and
non-alcoholic liver disease: The role of microbiota-derived
metabolites. Pharmacol Res. 141:521–529. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ramirez-Carrozzi V, Sambandam A, Luis E,
Lin Z, Jeet S, Lesch J, Hackney J, Kim J, Zhou M, Lai J, et al:
IL-17C regulates the innate immune function of epithelial cells in
an autocrine manner. Nat Immunol. 12:1159–1166. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Munoz M, Eidenschenk C, Ota N, Wong K,
Lohmann U, Kühl AA, Wang X, Manzanillo P, Li Y, Rutz S, et al:
Interleukin-22 induces interleukin-18 expression from epithelial
cells during intestinal infection. Immunity. 42:321–331.
2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gao X, Cao Q, Cheng Y, Zhao D, Wang Z,
Yang H, Wu Q, You L, Wang Y, Lin Y, et al: Chronic stress promotes
colitis by disturbing the gut microbiota and triggering immune
system response. Proc Natl Acad Sci USA. 115:E2960–E2969.
2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Mehta A and Baltimore D: MicroRNAs as
regulatory elements in immune system logic. Nat Rev Immunol.
16:279–294. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li Z and Rana TM: Therapeutic targeting of
microRNAs: Current status and future challenges. Nat Rev Drug
Discov. 13:622–638. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Whiteoak SR, Felwick R, Sanchez-Elsner T
and Fraser Cummings JR: MicroRNAs in inflammatory bowel diseases:
Paradoxes and possibilities. Inflamm Bowel Dis. 21:1160–1165.
2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kalla R, Ventham NT, Kennedy NA, Quintana
JF, Nimmo ER, Buck AH and Satsangi J: MicroRNAs: New players in
IBD. Gut. 64:504–517. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wu F, Zikusoka M, Trindade A, Dassopoulos
T, Harris ML, Bayless TM, Brant SR, Chakravarti S and Kwon JH:
MicroRNAs are differentially expressed in ulcerative colitis and
alter expression of macrophage inflammatory peptide-2 alpha.
Gastroenterology. 135:1624–1635.e24. 2008.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Neudecker V, Haneklaus M, Jensen O,
Khailova L, Masterson JC, Tye H, Biette K, Jedlicka P, Brodsky KS,
Gerich ME, et al: Myeloid-derived miR-223 regulates intestinal
inflammation via repression of the NLRP3 inflammasome. J Exp Med.
214:1737–1752. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yang L, Ma Z, Wang D, Zhao W, Chen L and
Wang G: MicroRNA-602 regulating tumor suppressive gene RASSF1A is
overexpressed in hepatitis B virus-infected liver and
hepatocellular carcinoma. Cancer Biol Ther. 9:803–808.
2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Akhtar N, Makki MS and Haqqi TM:
MicroRNA-602 and microRNA-608 regulate sonic hedgehog expression
via target sites in the coding region in human chondrocytes.
Arthritis Rheumatol. 67:423–434. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Shen J, Qiao Y, Ran Z and Wang T:
Different activation of TRAF4 and TRAF6 in inflammatory bowel
disease. Mediators Inflamm. 2013(647936)2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wu H, Fan H, Shou Z, Xu M, Chen Q, Ai C,
Dong Y, Liu Y, Nan Z, Wang Y, et al: Extracellular vesicles
containing miR-146a attenuate experimental colitis by targeting
TRAF6 and IRAK1. Int Immunopharmacol. 68:204–212. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Stickel N, Prinz G, Pfeifer D, Hasselblatt
P, Schmitt-Graeff A, Follo M, Thimme R, Finke J, Duyster J, Salzer
U and Zeiser R: miR-146a regulates the TRAF6/TNF-axis in donor T
cells during GVHD. Blood. 124:2586–2595. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Xia Z, Huang L, Yin P, Liu F, Liu Y, Zhang
Z, Lin J, Zou W and Li C: L-Arginine alleviates heat stress-induced
intestinal epithelial barrier damage by promoting expression of
tight junction proteins via the AMPK pathway. Mol Biol Rep.
46:6435–6451. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chen Z, Yu K, Zhu F and Gorczynski R:
Over-Expression of CD200 protects mice from dextran sodium sulfate
induced colitis. PLoS One. 11(e0146681)2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zaki MH, Boyd KL, Vogel P, Kastan MB,
Lamkanfi M and Kanneganti TD: The NLRP3 inflammasome protects
against loss of epithelial integrity and mortality during
experimental colitis. Immunity. 32:379–391. 2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Rodrigues RR, Greer RL, Dong X, Dsouza KN,
Gurung M, Wu JY, Morgun A and Shulzhenko N: Antibiotic-Induced
alterations in gut microbiota are associated with changes in
glucose metabolism in healthy mice. Front Microbiol.
8(2306)2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Le Roy T, Debédat J, Marquet F, Da-Cunha
C, Ichou F, Guerre-Millo M, Kapel N, Aron-Wisnewsky J and Clément
K: Comparative evaluation of microbiota engraftment following fecal
microbiota transfer in mice models: Age, kinetic and microbial
status matter. Front Microbiol. 9(3289)2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yu X, Wang D, Wang X, Sun S, Zhang Y, Wang
S, Miao R, Xu X and Qu X: CXCL12/CXCR4 promotes inflammation-driven
colorectal cancer progression through activation of RhoA signaling
by sponging miR-133a-3p. J Exp Clin Cancer Res.
38(32)2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Khan I, Ullah N, Zha L, Bai Y, Khan A,
Zhao T, Che T and Zhang C: Alteration of gut microbiota in
Inflammatory Bowel Disease (IBD): Cause or consequence? IBD
treatment targeting the gut microbiome. Pathogens.
8(126)2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Dalal SR and Chang EB: The microbial basis
of inflammatory bowel diseases. J Clin Invest. 124:4190–4196.
2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zhu Y, Gu L, Li Y, Lin X, Shen H, Cui K,
Chen L, Zhou F, Zhao Q, Zhang J, et al: miR-148a inhibits colitis
and colitis-associated tumorigenesis in mice. Cell Death Differ.
24:2199–2209. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Ye YL, Pang Z, Gu W and Zheng JJ:
Expression of microRNA-155 in inflammatory bowel disease and its
clinical significance. Zhonghua Yi Xue Za Zhi. 97:3716–3719.
2017.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
47
|
Han D, Walsh MC, Cejas PJ, Dang NN, Kim
YF, Kim J, Charrier-Hisamuddin L, Chau L, Zhang Q, Bittinger K, et
al: Dendritic cell expression of the signaling molecule TRAF6 is
critical for gut microbiota-dependent immune tolerance. Immunity.
38:1211–1222. 2013.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zundler S and Neurath MF: Pathogenic T
cell subsets in allergic and chronic inflammatory bowel disorders.
Immunol Rev. 278:263–276. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Strober W and Fuss IJ: Proinflammatory
cytokines in the pathogenesis of inflammatory bowel diseases.
Gastroenterology. 140:1756–1767. 2011.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Perrier C and Rutgeerts P: Cytokine
blockade in inflammatory bowel diseases. Immunotherapy.
3:1341–1352. 2011.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Roy S and Trinchieri G: Microbiota: A key
orchestrator of cancer therapy. Nat Rev Cancer. 17:271–285.
2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Zhu W, Winter MG, Byndloss MX, Spiga L,
Duerkop BA, Hughes ER, Büttner L, de Lima Romão E, Behrendt CL,
Lopez CA, et al: Precision editing of the gut microbiota
ameliorates colitis. Nature. 553:208–211. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Chu F, Shi M, Lang Y, Shen D, Jin T, Zhu J
and Cui L: Gut microbiota in multiple sclerosis and experimental
autoimmune encephalomyelitis: Current applications and future
perspectives. Mediators Inflamm. 2018(8168717)2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Vivarelli S, Salemi R, Candido S, Falzone
L, Santagati M, Stefani S, Torino F, Banna GL, Tonini G and Libra
M: Gut microbiota and cancer: From pathogenesis to therapy. Cancers
(Basel). 11(38)2019.PubMed/NCBI View Article : Google Scholar
|