Introduction
The vascular endothelium is considered to be crucial
for maintaining physiological balance in the vascular system and is
therefore regarded as the ‘guardian’ of vascular health (1). Endothelial dysfunction has been
implicated in the pathogenesis of cardiovascular diseases (2). Functional changes in the endothelial
cells and vascular system have been reported to serve an important
role in the pathology of a range of diseases, including peripheral
vascular disease, stroke, heart disease, diabetes mellitus, insulin
resistance, chronic renal failure, tumor growth and metastasis,
venous thrombosis and severe viral infections (3). Endothelial cells can synthesize and
subsequently release a number of factors that are involved in
regulating local permeability, vascular tension, smooth muscle cell
proliferation and migration, inflammatory response and platelet
function (4). Perturbation of the
tightly regulated balance in the vasculature can result in the
development of atherosclerotic lesions of varying severity
(5). Therefore, approaches aimed
at improving vascular endothelial function can reduce the risk of
or alleviate cardiovascular disease (6). Nitric oxide (NO) is a key signaling
molecule that is produced by vascular endothelial cells and serves
an important role in maintaining vascular tone and antioxidant
stress (7). In addition, other
factors can also activate endothelial cells, including
lipopolysaccharide (LPS), IL-1 and TNF-α, all of which are
dependent on the activation status of the NF-κB pathway (8). Endothelial cell activation can result
in the reduction in NO bioavailability (6), which in turn weakens the regulatory
functions of the endothelium over vascular tone, proliferation,
thrombosis, immunocyte reaction and barrier activity (7). In this regard, this reduction in NO
production or bioavailability can be regarded to be a predictor of
endothelial dysfunction (9).
Heparin is a high-concentration sulfated
glycosaminoglycan with strong acidity and a molecular weight of
1,200-40,000 kDa (10). It is a
natural anticoagulant in mammalian mast cells and neutrophils
(11) and promotes transcription
and release of placental growth factor from endothelial cells
(12). As an anticoagulant,
heparin has anti-inflammatory properties (13). However, to the best of our
knowledge, the effect of heparin in LPS-induced endothelial injury
remains unclear. Therefore, in the present study, experiments were
performed to investigate the possible effects and related mechanism
of heparin on vascular inflammation-induced endothelial injury.
Materials and methods
Materials Reagents
Heparin solution, with a molecular weight of 1,200
Da, was obtained from Changzhou Qianhong Biochemical Pharmaceutical
Co., Ltd. High-glucose DMEM, newborn calf serum (NBCS) and trypsin
was purchased from Thermo Fisher Scientific, Inc. LPS was obtained
from EMD Millipore. The ECL detection kit, PI and DAPI staining
solutions were acquired from Beyotime Institute of Biotechnology.
GAPDH (cat. no. ab8245), toll-like receptor 4 (TLR4; cat. no.
ab13556), myeloid differentiation primary response 88 (MyD88; cat.
no. ab107585), p-NF-κB (p65; cat. ab222494) and phosphorylated
(p)-NF-κB (p65; cat. no. ab183559) primary antibodies were
purchased from Abcam. Goat anti-rabbit IgG HRP-conjugated (cat. no.
70748; Cell Signaling Technology, Inc.) and FITC-labeled secondary
antibodies (cat. no. A10530; ThermoFisher Scientific, Inc.) were
obtained from Bioworld Technology, Inc. Small interfering RNA
(si)-TLR4 (sense, 5'-GGGCUUAGAACAACUAGAATT-3'; antisense,
5'-UUCUAGUUGUUCUAAGCCCTT-3') and si-negative control (si-NC; sense,
5'-UUCUCCGAACGUGUCACGUTT-3'; antisense,
5'-ACGUGACACGUUCGGAGAATT-3') construction was performed by Nanjing
KeyGen Biotech. Co. Ltd.
Equipment. The inverted fluorescence
microscope was obtained from Olympus Corporation and the
chemiluminescence imaging system was from Bio-Rad Laboratories,
Inc.
Cell lines. HUVECs were purchased from The
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences.
Methods
Cell culture. HUVECs were cultured in
high-glucose DMEM supplemented with 15% NBCS in a cell incubator at
37˚C with 5% CO2, for a passage cycle of 2-3 days.
Construction of an inflammatory injury model of
HUVECs. HUVECs were inoculated into a six-well plate at a
concentration of 2x105 cells/ml. After the cells reached
70-80% confluence, they were starved in DMEM for 12 h. A cell model
of endothelial cell inflammatory injury was established using LPS
(100 µg/ml) for 6 h (7).
Cell transfection. si-TLR4 (the negative
control used was si-NC) was constructed and transfected into HUVECs
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at a final concentration of 50 nmol/l of the
transfection with 10 nM si-TLR4 or si-NC. Following 6 h of
transfection at room temperature, the DMEM medium containing 10%
FBS (Sigma-Aldrich; Merck KGaA) was replaced, followed by
continuous culture for 48 h at room temperature. The transfected
cells were then collected before the transfection efficiency was
evaluated using reverse transcription-quantitative PCR
(RT-qPCR).
Cell grouping. HUVECs were divided into the
following groups: i) Negative control (NC; cultured with DMEM
medium); ii) LPS (intervention with 1,000 µg/l LPS); iii) LPS + Low
(induction with 1,000 µg/l LPS and intervention with 10 U/l
heparin); iv) LPS + Middle (induction with 1,000 µg/l LPS and
intervention with 20 U/l heparin); v) LPS + High (induction with
1,000 µg/l LPS and intervention with 100 U/l heparin); vi) si-TLR4
(transfection with si-TLR4 and induction with 1,000 µg/l LPS); vii)
heparin (induction with 1,000 µg/l LPS and intervention with 100
U/l heparin which was the most effective concentration of heparin;
heparin and LPS + High were similar in treatment); and viii)
heparin + si-TLR4 (transfection with si-TLR4, induction with 1,000
µg/l LPS and intervention with 100 U/l heparin). Following 48 h at
room temperature of the corresponding treatments (heparin and LPS
were delivered together at the same time), cells from each group
were used for subsequent experiments.
ELISA. TNF-α (cat. no. KGEHC103α-1), IL-1β
(cat. no. KGEHC002b-1), IL-6 (cat. no. KGEHC007-1) and IFN-γ (cat.
no. KGERC101g-1) detection kits were purchased from Nanjing KeyGen
Biotech, Co., Ltd. Following centrifugation of the cell culture
medium in each group at 3,000 x g for 5 min at 4˚C, the supernatant
was collected for subsequent measurements of the concentration of
the inflammatory factors, according to the manufacturer's protocols
in each kit.
5-Ethynyl-2'-deoxyuridine (EdU) staining.
HUVECs in the logarithmic growth phase were seeded into a 24-well
plate at a density of 5x104 cells/well. Cells were
incubated with DMEM medium and then treated for 48 h at room
temperature, according to the treatment protocol of each group.
Next, 10 µmol/l EdU reagent was added to the cells and incubated
for 2 h at room temperature, according to the protocol of the EdU
fluorescence staining cell proliferation kit (cat. no. KGA331-1000;
Nanjing KeyGen Biotech, Co., Ltd.). The EdU solution was removed by
washing with PBS, without DNA penetration and the cells were fixed
with 4% paraformaldehyde for 30 min at room temperature. After
washing the fixation solution away with PBS, Apollo staining
solution (part of Keygen EdU staining kit) was added and incubated
in the dark at room temperature for 30 min. After the staining
solution was washed off with PBS, 10 µmol/l DAPI (per well) was
used to stain the nucleus for 5 min at room temperature.
Fluorescence images of five random fields of view per well were
obtained using an IX73 fluorescence microscope (Olympus
Corporation; magnification, x200) and EdU-positive cells were
counted using ImageJ software v1.8.0 (National Institutes of
Health).
Cell apoptosis detection. After 48 h at room
temperature of corresponding treatments, HUVECs (1x105
cells/ml) in each group were digested and collected, followed by
incubation with 5 µl Annexin V-FITC for 10 min at room temperature
and 5 µl PI (cat. no. KGAV113; Nanjing KeyGen Biotech, Co., Ltd.)
for 10 min at room temperature in the dark. Apoptotic cells were
then analyzed using flow cytometry. The analysis was performed
using a BD FACSAria™ II flow cytometer (Becton-Dickinson and
Company), and the data were analyzed using CellQuest Pro software
(version 5.1; Becton-Dickinson and Company).
TUNEL assay. Cells were treated according to
the protocols of the Fluorometric TUNEL System (cat. no. KGA7071;
Nanjing KeyGen Biotech, Co., Ltd.) after corresponding treatment of
HUVECs in each group for 48 h at room temperature. Cells were
seeded on coverslips, washed three times in PBS for 5 min each,
fixed in 4% formaldehyde for 20 min at room temperature and
incubated in 70% ethanol at -20˚C for 30 min. The coverslips were
washed a further three times and the cells were permeabilized. The
permeabilization was performed in 0.1% Triton X-100/0.1% sodium
citrate at room temperature for 10 min. After three 5-min washes in
PBS, the cells were incubated with 3% H2O2 at
room temperature for 10 min. After another three 5-min washes in
PBS, the cells were incubated with TdT enzyme at 37˚C for 90 min,
which was protected from light. After two 2-min washes in PBS, the
nuclei were stained with Hoechst 33258 at room temperature for 20
min in the dark. The cells were finally washed in the dark three
times in PBS containing 0.5% Tween-20 for 2 min each and mounted in
glycerol. Next, cells were observed under a fluorescence microscope
and images were captured (five fields; magnification, x200).
RT-qPCR. After 48 h of treatment at room
temperature, HUVECs in each group were collected and total RNA was
extracted using an RNAiso Plus kit (Takara Bio, Inc.). Next, cDNA
synthesis was performed with a PrimeScript™ RT kit (Takara Bio,
Inc.). The following thermocycling conditions were used: Initial
denaturation at 95˚C for 30 sec, then 55˚C for 30 sec and 72˚C 30
sec. The synthesized cDNA were collected for qPCR amplification in
a LightCycler 480 fluorescent PCR system (Roche Diagnostics),
according to the steps of SYBR Green RT-qPCR kit (cat. no. RR086B;
Takara Bio, Inc.). The reaction conditions were as follows:
Pre-denaturation at 95˚C for 15 min, followed by 40 cycles of
denaturation at 95˚C for 10 sec, annealing at 55˚C for 20 sec and
extension at 72˚C for 20 sec. The genes GAPDH was used for
normalization of mRNA expressions. Relative expression levels of
the respective target gene were calculated according to the
2-ΔΔCq method (14).
The primer sequences are shown in Table I.
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5'→3') |
|---|
| Toll-like | F:
TGGATACGTTTCCTTATAAG |
| receptor 4 | R:
GAAATGGAGGCACCCCTTC |
| Myeloid | F:
ACCTGGCTGGTTTACACGTC |
| differentiation
primary response 88 | R:
CTGCCAGAGACATTGCAGAA |
| NF-κB (p65) | F:
ATGCTTACTGGGTGCCAAAC |
| | R:
GGCAAGTCACTCAGCCTTTC |
| GAPDH | F:
AGGTCGGTGTGAACGGATTTG |
| | R:
TGTAGACCATGTAGTTGAGGTCA |
Western blot (WB) analysis. HUVECs were
collected following treatment in each group for 48 h at room
temperature. The collected cells were lysed on ice with RIPA lysis
buffer [10 mmol/l Tris (pH 8.0), 150 mmol/l NaCl, 1% Nonidet P-40,
0.1% SDS and 0.5% deoxycholate II] for 30 min. Cells were then
centrifuged at 14,000 x g for 30 min at 4˚C and the supernatant
containing the protein was obtained. Following protein
quantification using a BCA assay kit, an equal amount of protein
(30 µg/lane) was separated via 10% SDS-PAGE. Following
electrophoresis, the proteins were transferred onto a PVDF membrane
and blocked with a TBS-0.1% Tween-20 solution containing 5% skimmed
milk. Next, the membranes were incubated with anti-TLR4 (cat. no.
ab13556; 1:200), anti-MyD88 (cat. no. ab107585, 1:200),
anti-p-NF-κB (p65; cat. no. ab183559; 1:200), NF-κB (p65; cat. no.
ab32536; 1:200) and anti-GAPDH (cat. no. ab8245; 1:100) primary
antibodies at room temperature for 2 h. After the membranes were
washed, the HRP-conjugated secondary antibody was added for
subsequent incubation at a dilution of 1:4,000 at room temperature
for 1 h. ECL was used for development of the membrane to visualize
the bands. ImageJ software v1.8.0 (National Institutes of Health)
was used to analyze the gray values of the bands, where GAPDH was
used to normalize the results.
Immunofluorescence. After 48 h of treatment
in each group at room temperature, HUVECs were fixed with 3.5%
paraformaldehyde for 10 min at room temperature, permeabilized with
0.2% Triton X-100 on ice for 15 min and blocked with 3% BSA
(Sigma-Aldrich; Merck KGaA) for 30 min. Next, a p-NF-κB (p65; cat.
ab222494; 1:200) primary antibody was added to the cells and
incubated overnight at 4˚C. The next day, a FITC-labeled secondary
antibody was added according to the manufacturer's instructions and
incubated for 1 h at room temperature. Following 50 µl DAPI
staining for 5 min at room temperature, images of the stained cells
were captured using a laser confocal microscope (five files;
magnification, x200). This experiment was repeated three times.
Statistical analysis
SPSS 20.0 software (IBM Corp.) was used for
statistical analysis. To analyze data with a normal distribution
and homogeneity of variance, a one-way ANOVA was used followed by a
Tukey's post hoc test for pairwise comparisons. A two-tailed
hypothesis test was performed with α=0.05. P<0.05 was considered
to indicate a statistically significant difference. The experiments
were repeated three times.
Results
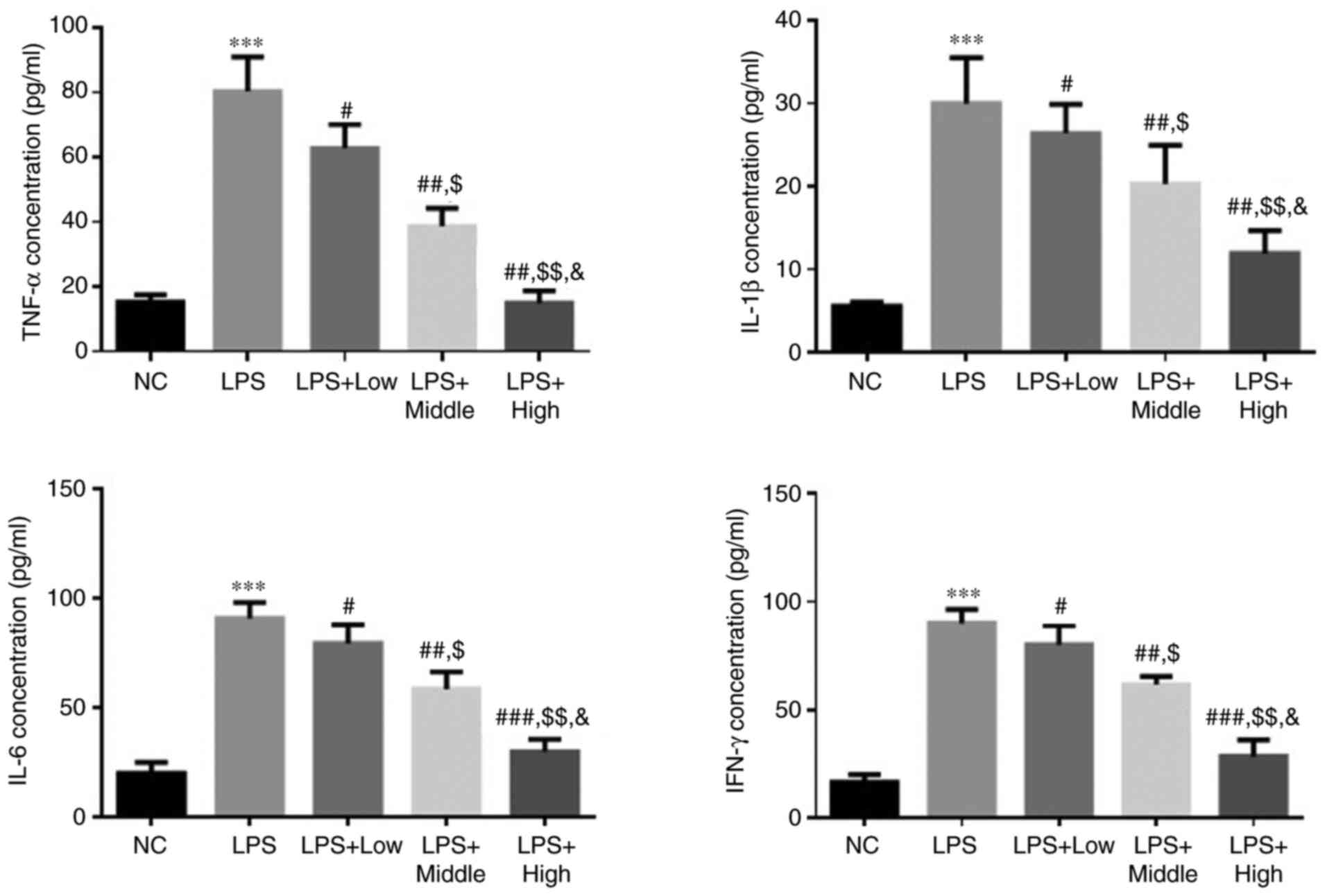
Effect of heparin on TNF-α, IL-1β,
IL-6 and IFN-γ levels in LPS-induced endothelial injury
Compared with those in the NC group, the levels of
TNF-α, IL-1β, IL-6 and IFN-γ in the LPS group were significantly
higher (all P<0.001; Fig. 1).
In the heparin groups, the levels of TNF-α, IL-1β, IL-6 and IFN-γ
were all significantly decreased compared with those in the LPS
group (all P<0.05; Fig. 1). In
addition, there was a significant dose-dependent effect among the
three heparin treatment groups (all P<0.05; Fig. 1).
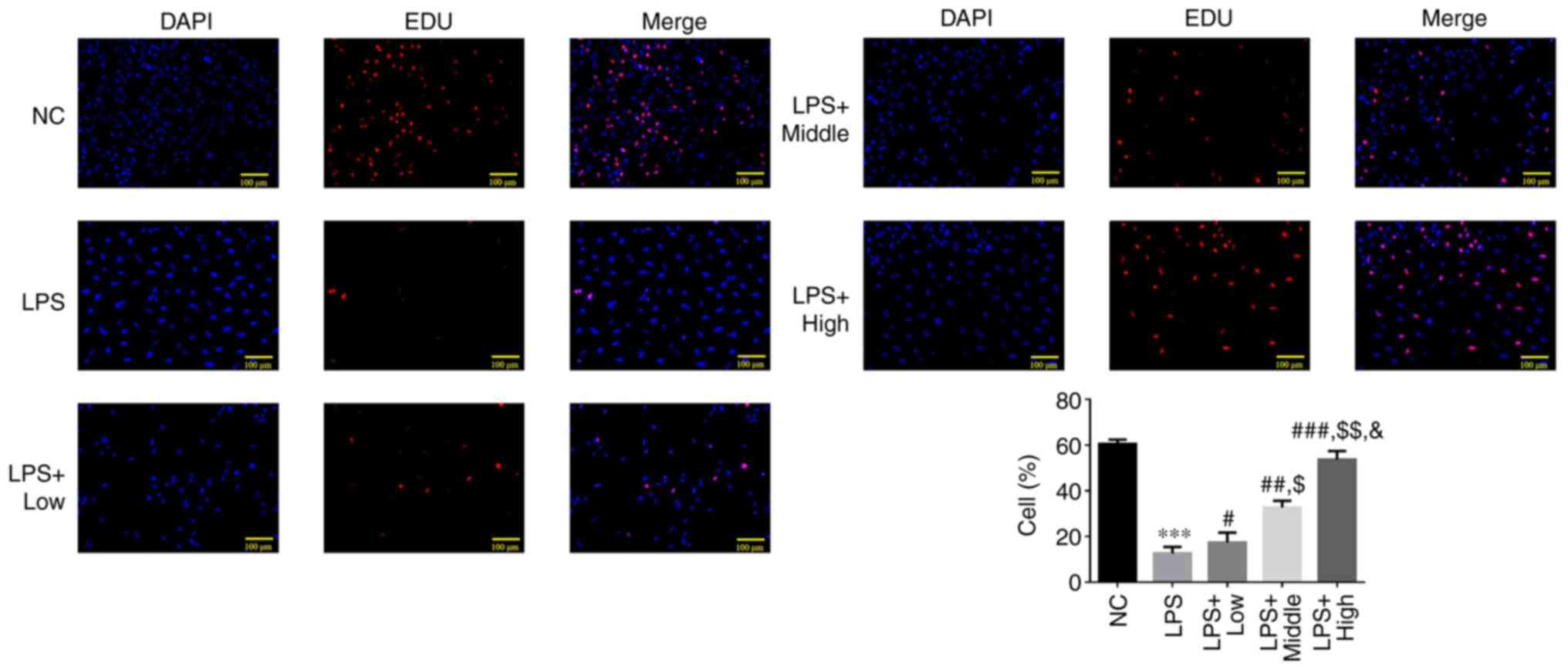
Effect of heparin on the proliferating
cell count after LPS-induced endothelial injury
A significant reduction in the EdU-positive cell
count was observed in the LPS group compared with that in the NC
group (P<0.001; Fig. 2). By
contrast, the EdU-positive cell count was significantly increased
in the three heparin groups compared with that in the LPS group
(P<0.05; Fig. 2), where a
significant dose-dependent effect was observed among the three
heparin treatment groups (all P<0.05; Fig. 2).
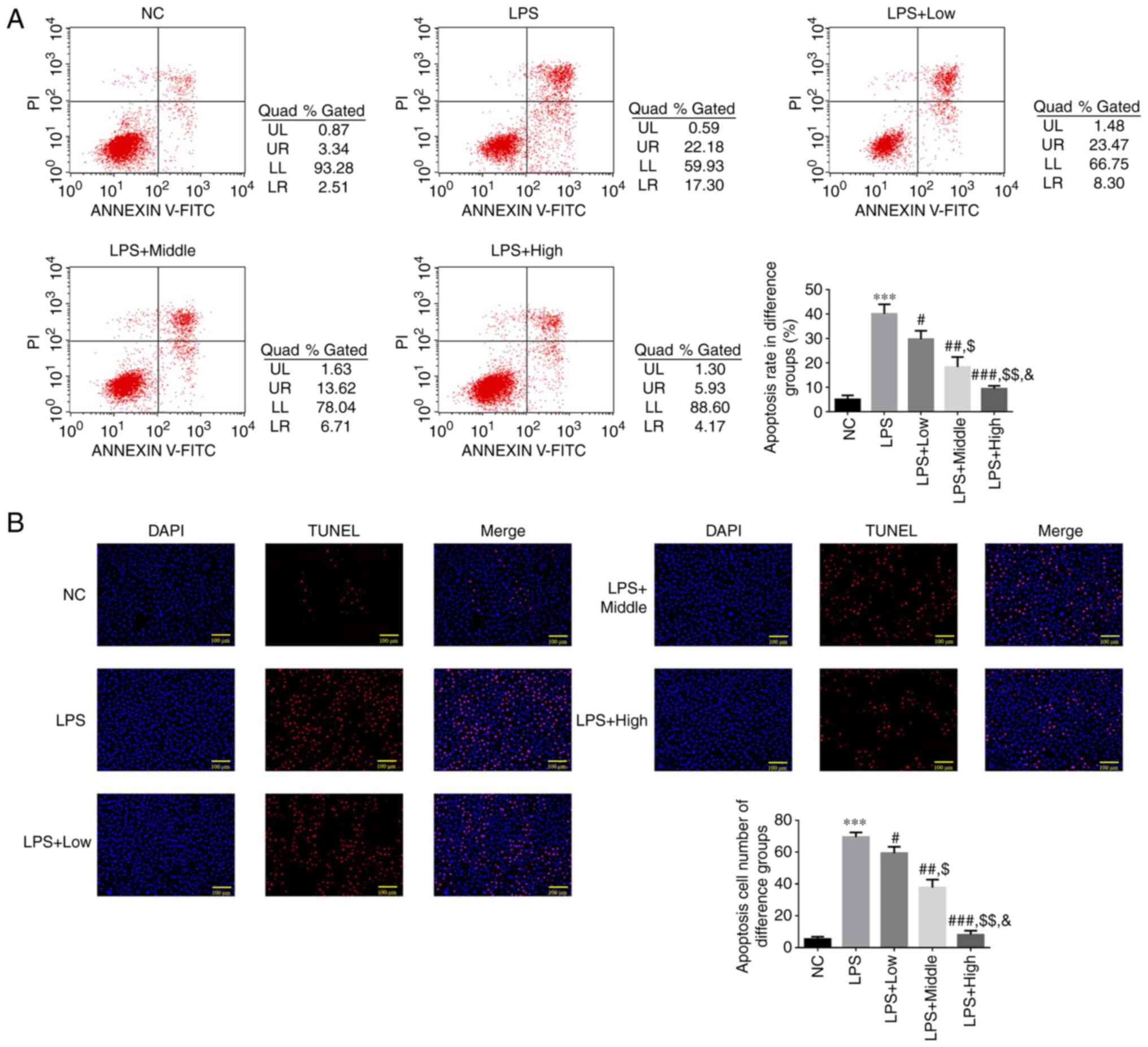
Flow cytometry analysis of
heparin-mediated regulation of apoptosis following LPS-induced
endothelial injury
According to the flow cytometry results, the
apoptotic rate in the LPS group was significantly higher compared
with that of the NC group (P<0.001; Fig. 3A). The apoptotic rate in all three
of the heparin groups was significantly lower compared with that in
the LPS group (P<0.05; Fig.
3A), with a significant dose-dependent effect observed among
the three heparin groups (all P<0.05; Fig. 3A).
TUNEL detection analysis of
heparin-mediated regulation of cell apoptosis following LPS-induced
endothelial injury
The TUNEL assay results indicated that the LPS group
exhibited a significantly increased count of TUNEL-positive cells
compared with that in the NC group (P<0.001; Fig. 3B). However, the number of
TUNEL-positive cells in the three heparin groups was significantly
decreased compared with that in the LPS group (P<0.05; Fig. 3B), with a significant
dose-dependent effect observed among the three heparin groups (all
P<0.05; Fig. 3B).
Effect of heparin on TLR4, MyD88 and
NF-κB p65 expression
According to the RT-qPCR results, si-TLR4
significantly decreased TLR4 gene expression, as presented in
Fig. S1, the LPS group exhibited
significantly increased mRNA expression levels of TLR4, MyD88 and
NF-κB (p65) compared with those in the NC group (all P<0.001;
Fig. 4A). However, intervention
with all three doses of heparin significantly downregulated the
expression levels of TLR4, MyD88 and NF-κB (p65) compared with
those in the LPS group (all P<0.05; Fig. 4A), with a significant
dose-dependent effect observed among the three groups (all
P<0.05; Fig. 5). In addition,
WB results showed that compared with those in the NC group, the
protein expression levels of TLR4, MyD88 and p-NF-κB (p65) were all
significantly upregulated in the LPS group (all P<0.001;
Fig. 4B). A significant decrease
in the protein expression of TLR4, MyD88 and p-NF-κB (p65) was also
observed in the three heparin groups compared with that in the LPS
group (all P<0.05; Fig. 4B). In
addition, a significant dose-dependent effect was observed among
the three heparin groups (all P<0.05; Fig. 4B).
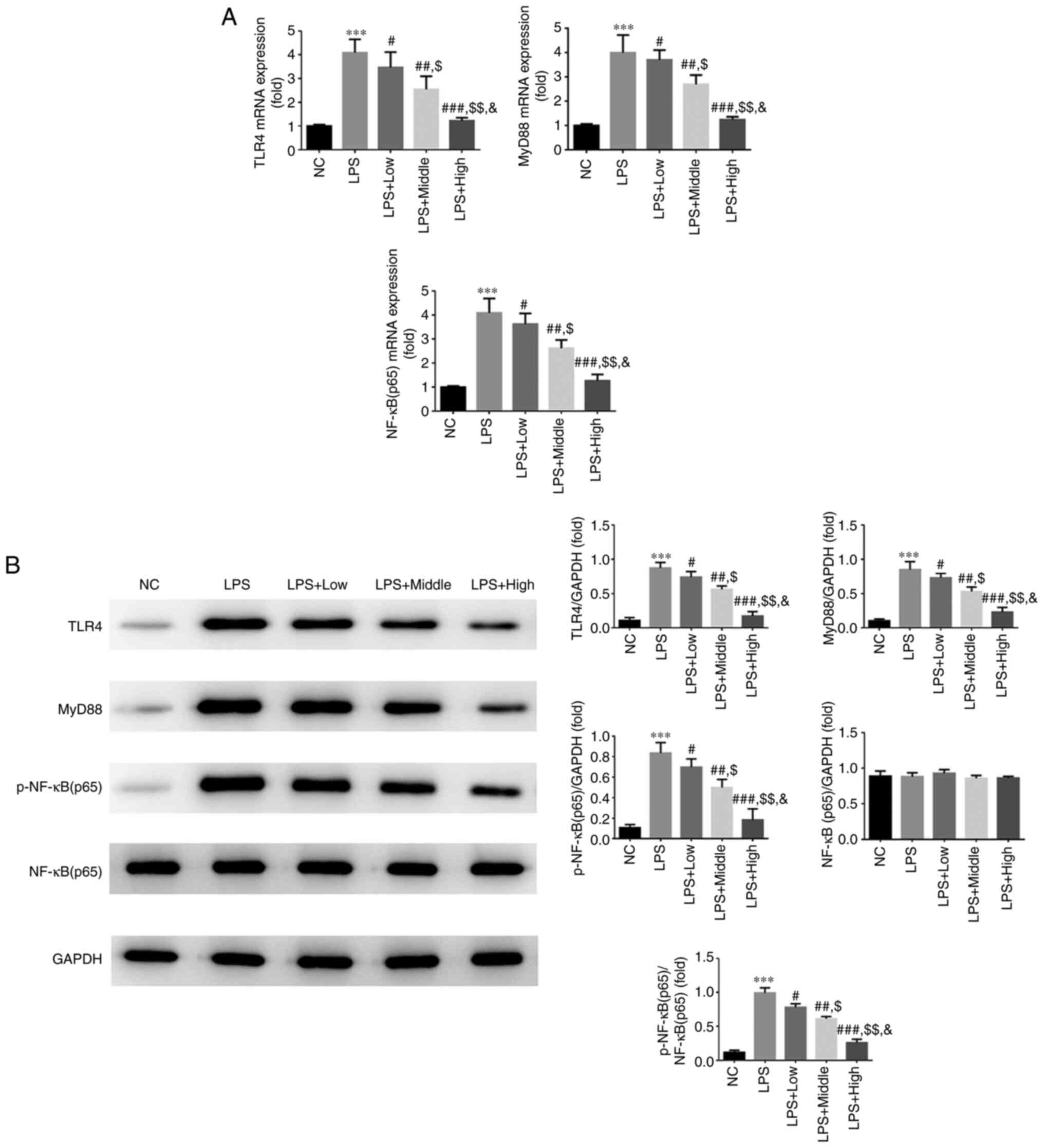 | Figure 4Effect of heparin on the expression
of TLR4, myD88 and NF-κB p65 in endothelial cells. (A) Relative
TLR4, myD88 and NF-κB p65 mRNA expression in the different
treatment groups. (B) Relative TLR4, myD88 and NF-κB p65 protein
expression in the different treatment groups.
***P<0.001 vs. NC; #P<0.05,
##P<0.01 and ###P<0.001 vs. LPS;
$P<0.05 and $$P<0.01 vs. LPS + Low
group; &P<0.05 vs. LPS + Middle. NC, normal
control group; LPS, lipopolysaccharide; LPS + Low, LPS-stimulated
cells were treated with low-dose heparin (10 U/l); LPS + Middle,
LPS-stimulated cells were treated with middle-dose heparin (20
U/l); LPS + High, LPS-stimulated cells were treated with high-dose
heparin (100 U/l); TLR4, toll-like receptor 4; myD88, myeloid
differentiation primary response 88. |
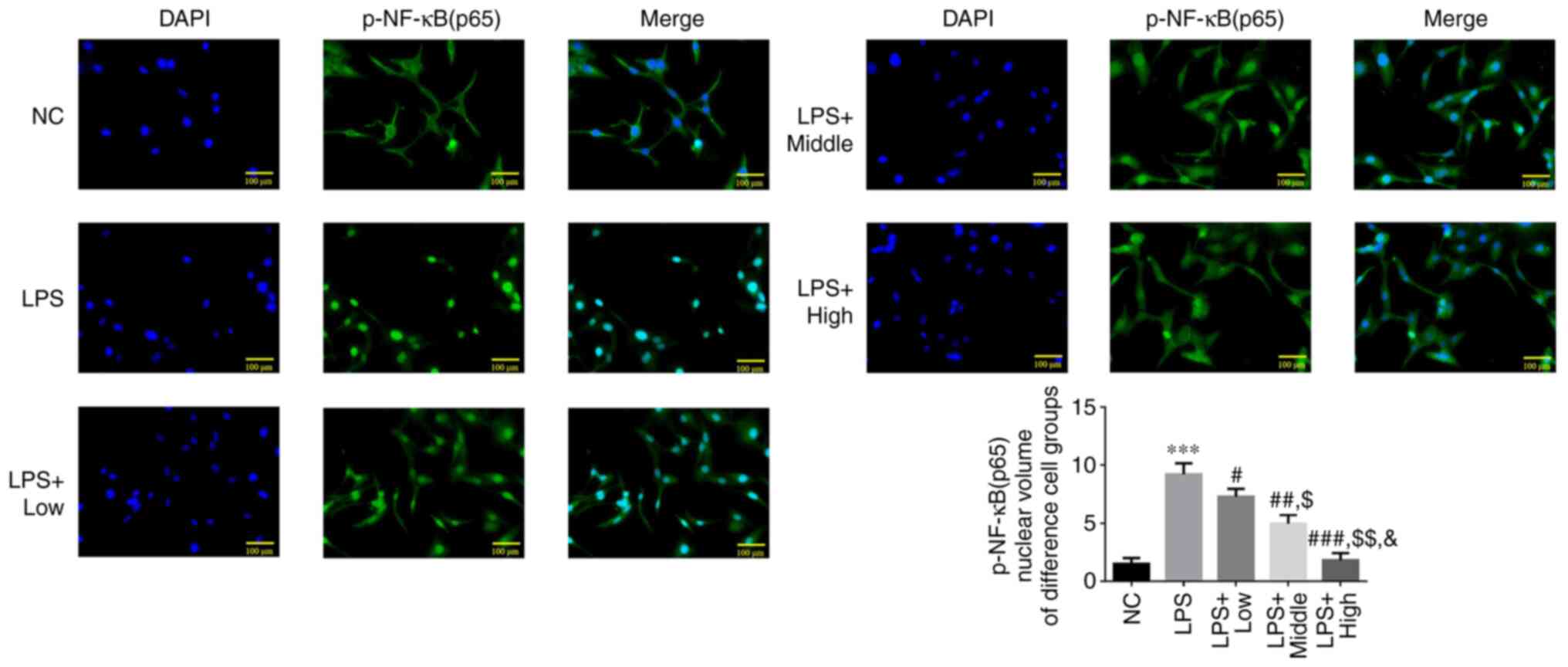
Effect of heparin on p-NF-κB (p65)
protein translocation into the nucleus
The results of the immunofluorescence assay showed
that the extent of p-NF-κB (p65) protein translocation into the
nucleus was significantly increased in the LPS group compared with
that in the NC group (P<0.001; Fig.
5). Following heparin treatment at all three doses, the amount
of p-NF-κB (p65) protein translocated into the nucleus was
significantly decreased compared with that in the LPS group (all
P<0.05; Fig. 5). In addition, a
significant dose-dependent effect was observed among the three
heparin groups (all P<0.05; Fig.
5).
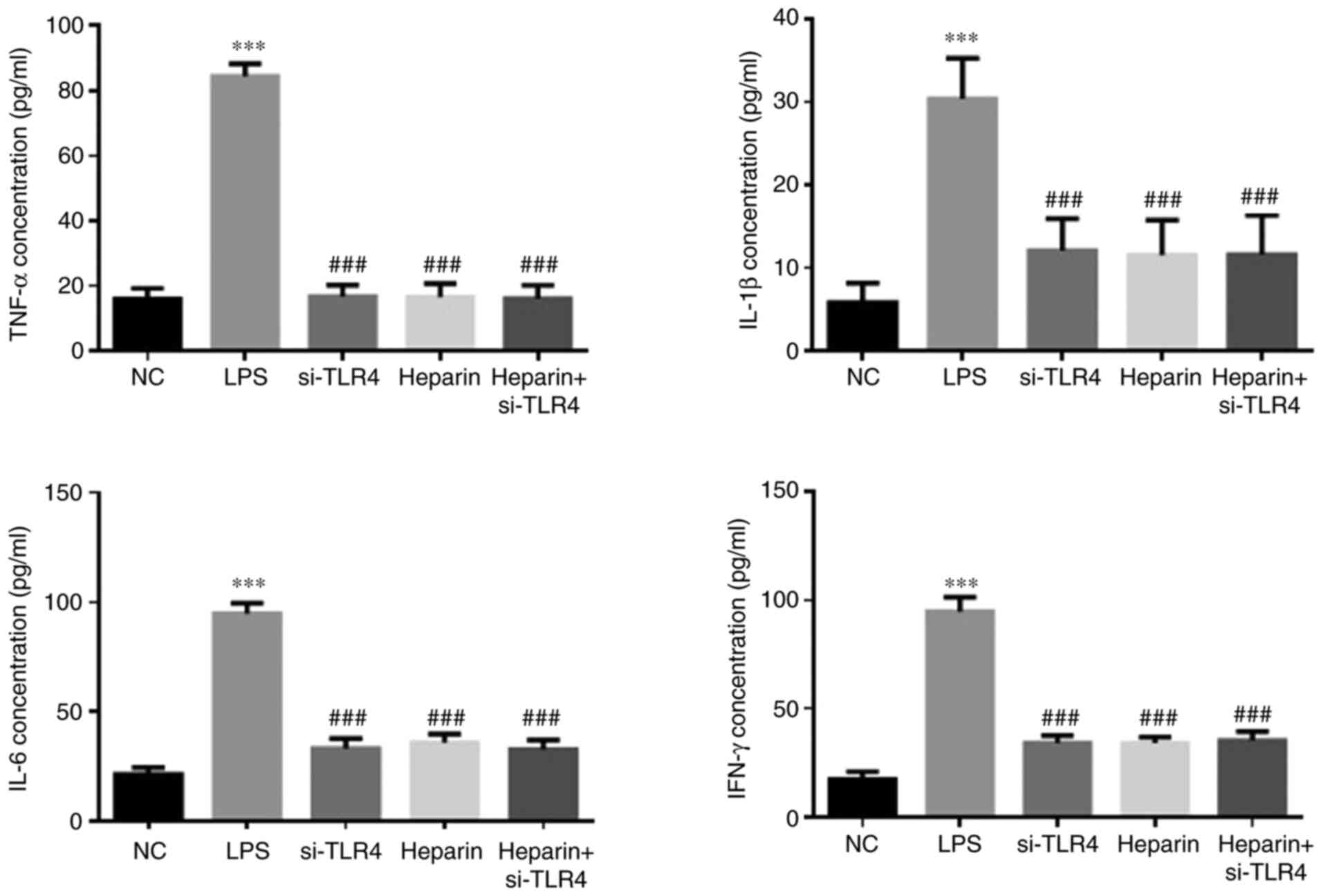
ELISA detection of TNF-α, IL-1β, IL-6
and IFN-γ levels in each group
The LPS group exhibited significantly higher levels
of TNF-α, IL-1β, IL-6 and IFN-γ (P<0.001; Fig. 6) compared with those in the NC
group. By contrast, the si-TLR4, heparin and heparin + si-TLR4
groups all exhibited significantly lower concentrations of TNF-α,
IL-1β, IL-6 and IFN-γ compared with those in the LPS group (all
P<0.001; Fig. 6). However,
there was no significant difference in the levels of these factors
among the si-TLR4, heparin and heparin + si-TLR4 groups (Fig. 6).
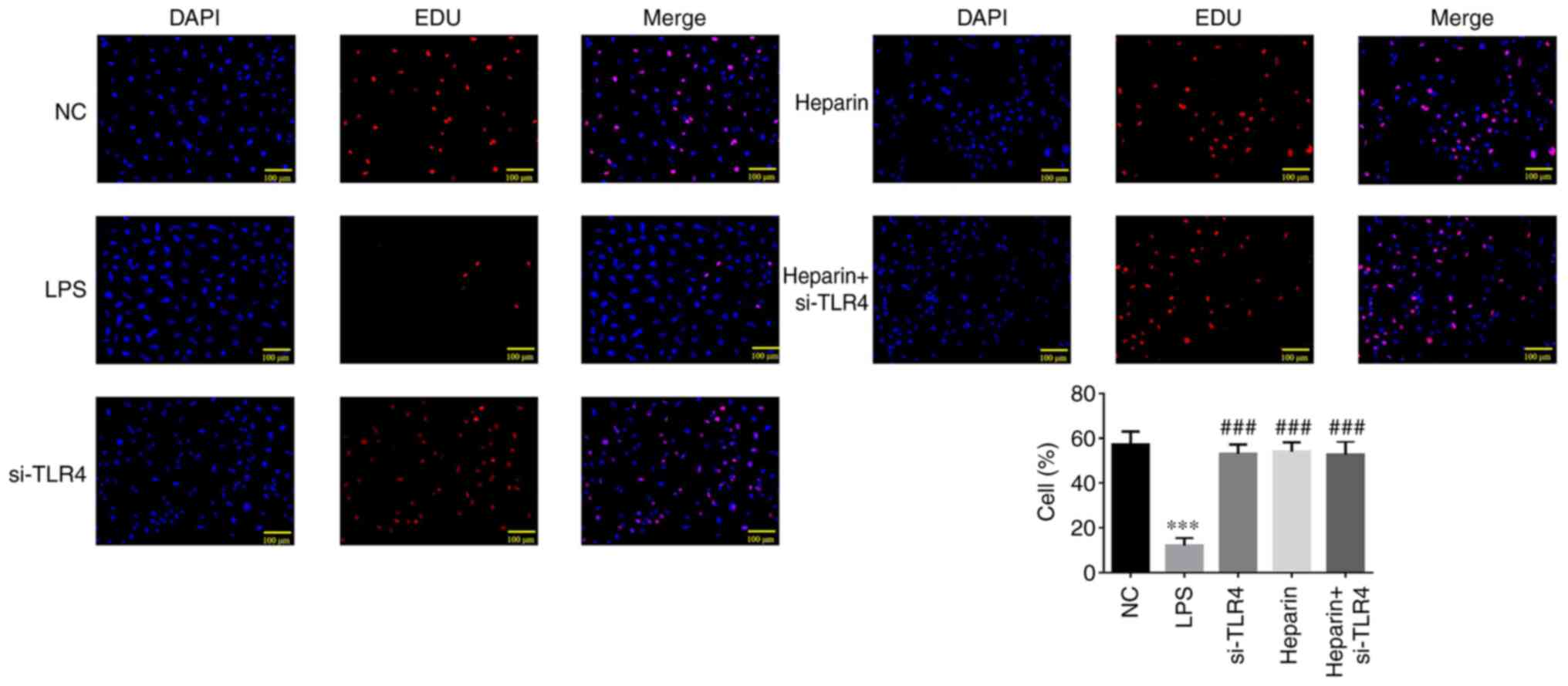
EdU detection of cell proliferation in
each treatment group
A significantly decreased number of EdU-positive
cells was observed in the LPS group compared with that in the NC
group (P<0.001; Fig. 7).
Compared with that in the LPS group, si-TLR4, heparin and heparin +
si-TLR4 groups exhibited significantly increased EdU-positive cell
counts (all P<0.001; Fig. 7).
No differences could be observed in the number of proliferative
cells among the si-TLR4, heparin and heparin + si-TLR4 groups
(Fig. 7).
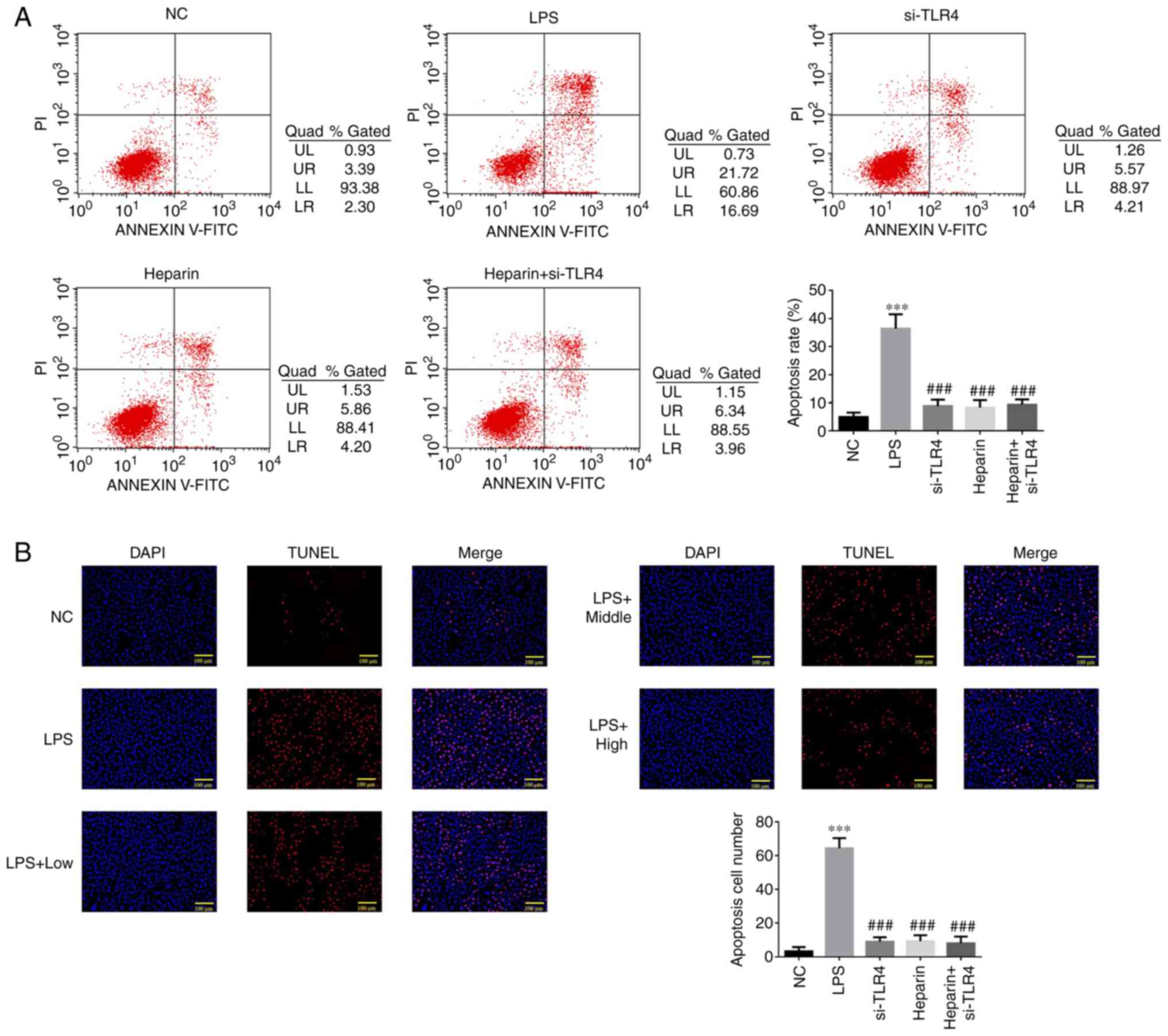
Flow cytometric detection of the cell
apoptotic rate in each treatment group
The LPS group exhibited a significantly increased
cell apoptotic rate compared with that in the NC group (P<0.001;
Fig. 8A). Compared with that in
the LPS group, the apoptotic rate in the si-TLR4, heparin and
heparin + si-TLR4 groups was significantly decreased (all
P<0.001; Fig. 8A). No
statistical differences were observed in the apoptotic rate among
the si-TLR4, heparin and heparin + si-TLR4 groups (Fig. 8A).
TUNEL detection of apoptotic cell
count in each group
A significantly increased TUNEL-positive cell count
was observed in the LPS group compared with that in the NC group
(P<0.001; Fig. 8B).
Furthermore, the TUNEL-positive cell count was significantly
decreased in the si-TLR4, heparin and heparin + si-TLR4 groups
compared with that in the LPS group (all P<0.001; Fig. 8B). No statistical differences were
observed in the TUNEL-positive cell count among the si-TLR4,
heparin and heparin + si-TLR4 groups (all P>0.05; Fig. 8B).
RT-qPCR and WB measurements of TLR4,
myD88 and NF-κΒ p65 expression
As shown in Fig.
9A, the LPS group exhibited significantly increased mRNA
expression levels of TLR4, MyD88 and NF-κB p65 (all P<0.001)
compared with those in the NC group. Furthermore, when compared
with those in the LPS group, significantly decreased mRNA
expression levels of TLR4, MyD88 and NF-κB p65 were observed in the
si-TLR4, heparin and heparin + si-TLR4 groups (all P<0.001). As
shown in Fig. 9B, the protein
expression levels of TLR4, MyD88 and NF-κB p65 were significantly
increased in the LPS group compared with those in the NC group (all
P<0.001). In addition, the si-TLR4, heparin and heparin +
si-TLR4 groups all exhibited significantly lower protein expression
levels of TLR4, MyD88 and NF-κB (p65) compared with those in the
LPS group (all P<0.001). No significant differences were
observed in the gene and protein expression levels of TLR4, MyD88
and NF-κB p65 among the si-TLR4, heparin and heparin + si-TLR4
groups (Fig. 9).
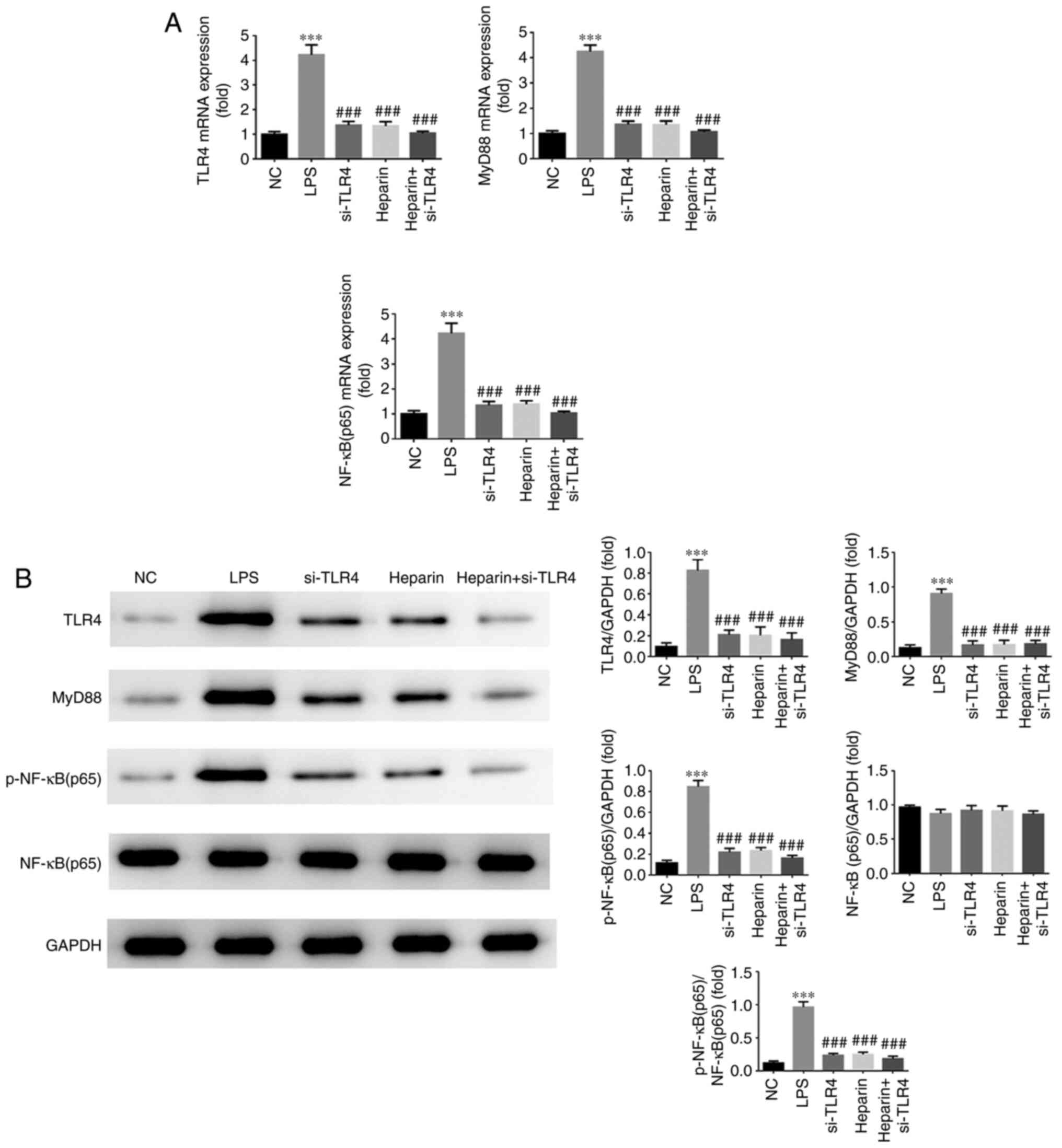 | Figure 9Reverse transcription-quantitative
PCR and western blot detection of TLR4, myD88 and NF-κB p65 gene
and protein expression. (A) Relative TLR4, myD88 and NF-κB p65 mRNA
expression in the different treatment groups. (B) Relative TLR4,
myD88 and NF-κB p65 protein expression in the different treatment
groups. ***P<0.001 vs. NC; ###P<0.001
vs. LPS. NC, normal control group; TLR4, toll-like receptor 4; LPS,
lipopolysaccharide; si, small interfering RNA; myD88, myeloid
differentiation primary response 88; p-, phosphorylated; si-TLR4,
LPS-stimulated cells were transfected with si-TLR4; heparin,
LPS-stimulated cells were treated with 100 U/l heparin; heparin +
si-TLR4, LPS-stimulated cells were transfected with si-TLR4 and
treated with 100 U/l heparin. |
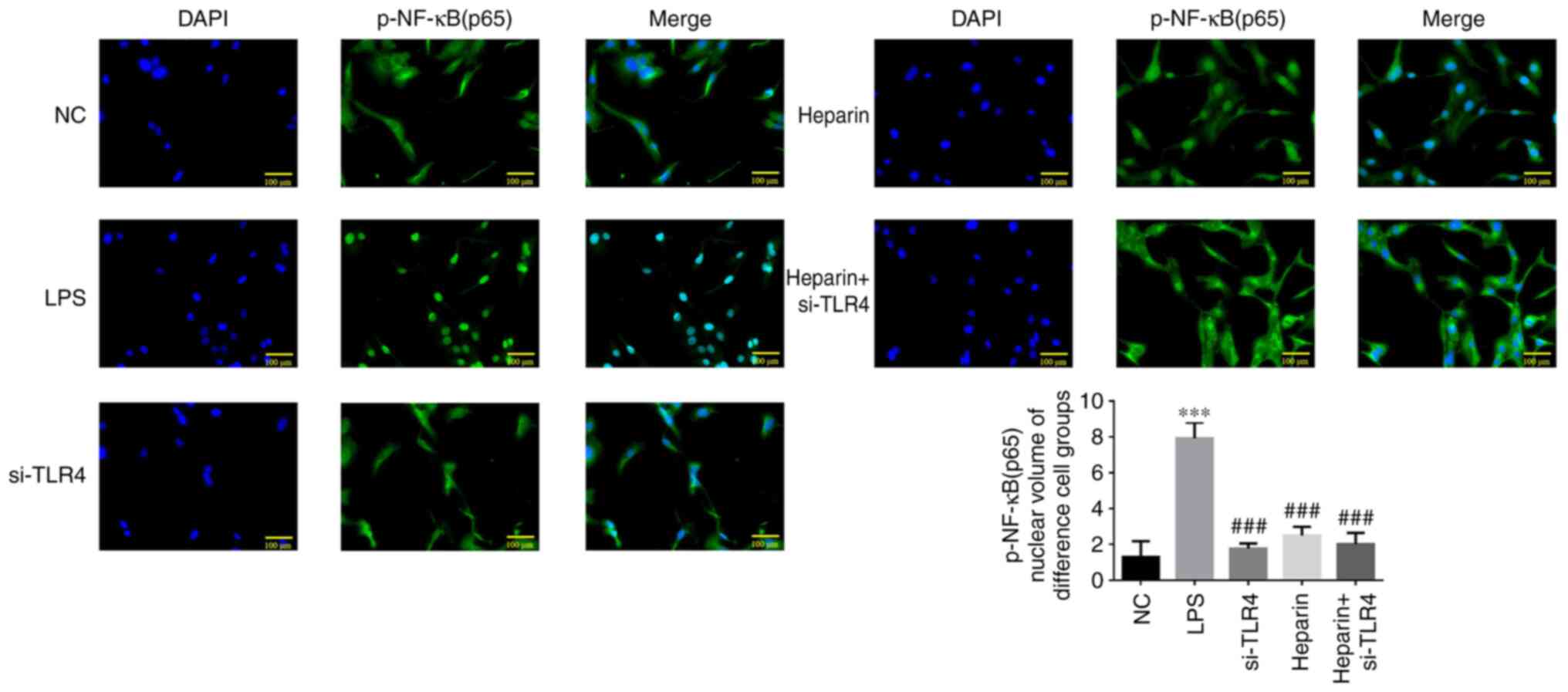
Immunofluorescence analysis of p-NF-κB
p65 protein translocation into the nucleus
Compared with that in the NC group, the degree of
p-NF-κB (p65) protein translocation into the nucleus was increased
in the LPS group (all P<0.001; Fig. 10). However, compared with that in
the LPS group, p-NF-κB p65 protein translocation into the nucleus
was decreased in the si-TLR4, heparin and heparin + si-TLR4 groups
(all P<0.001). No statistical differences were observed in
p-NF-κB p65 protein translocation into the nucleus among the
si-TLR4, heparin and heparin + si-TLR4 groups (Fig. 10).
Discussion
LPS has been identified to be the main component of
the cell wall of gram-negative bacteria (15). After being transported by LPS
binding protein (LBP), LPS binds to CD14 expressed on various
cytoplasmic membranes (16). After
binding with the LPS-LBP complex, CD14 activates the NF-κB
signaling pathway through TLR4(17). The resulting signaling cascade
activated can then promote the release of inflammatory cytokines,
including IL-6 and TNF-α (13,18,19).
TLRs are key components of the innate immune system
(20). Following activation, TLRs
relay the inflammatory signaling information through a
MyD88-dependent pathway to activate the expression and secretion of
inflammatory factors, resulting in inflammatory lesions (21-23).
Downstream, NF-κB (p65) is an important inflammatory regulator
(24). As a transcription factor,
it can activate the expression of a number of inflammatory
cytokines, including TNF-α, IL-1β, IFN-γ and IL-6 (25-27).
The expression of inflammatory factors induced by NF-κB p65 can
lead to potentiation of NF-κB activation by positive feedback,
which is mediated by the continuous translocation of p-NF-κB p65
into the nucleus, aggravating inflammatory injury (28). Consequently, the TLR4/MyD88/NF-κB
p65 signaling pathway serves a key role in the inflammatory
response.
Vascular endothelial cells at the inflammatory site
can serve a dual role, either as a participant or a regulator in
the inflammatory process (29).
Incalza et al (30) found
that long-term or repeated exposure to risk factors of
cardiovascular diseases can damage the endogenous anti-inflammatory
system within endothelial cells. Consequently, the endothelium can
lose not only its function, but endothelial cells can also detach
from the endothelium and enter the circulatory system, which can
induce an inflammatory reaction (31). Therefore, repairing endothelial
cell injury can serve an important role in preserving vascular
function (32).
A previous study (32) reported that heparin had
anti-inflammatory effects. In the present study, heparin exerted an
inhibitory effect on LPS-induced HUVEC apoptosis, secretion of the
inflammatory cytokines TNF-α, IL-1β, IL-6 and IFN-γ, in addition to
reducing the protein levels of TLR4, MyD88 and p-NF-κB p65.
However, no significant enhancements were observed when heparin and
TLR4 knockdown were combined. Therefore, it was concluded that
heparin may serve an anti-inflammatory and protective role in
vascular endothelial injury by downregulating the TLR4/MyD88/NF-κB
(p65) signaling pathway.
In the present study, in vitro experiments
were conducted, where the results showed that heparin may exert a
protective effect on LPS-induced acute vascular endothelial injury.
The specific mechanism can be explained by its role in reducing the
inflammatory reaction and inhibiting the TLR4/MyD88/NF-κB (p65)
signaling pathway. The findings of the present study may provide a
foundation for further investigations into the protective effect of
heparin on the cardiovascular system.
Supplementary Material
TLR4 mRNA expressionafter si-TLR4
transfection. ***P<0.001 vs. si-NC. TLR4, toll-like
receptor 4; si-, small interfering RNA; NC, negative control;
si-NC, the cells were transfected with the non-targeting
si-negative control sequence; si-TLR4, the cells were transfected
with si-TLR4.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science
Foundation of China (grant no. 81760339), Ningxia Natural Science
Foundation of China (grant no. 2020AAC03331) and the Fourth Batch
of Ningxia Youth Talents Supporting Program (grant no.
TJGC2019087).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WL and QY designed the study. WL, YW and YL
performed the ELISA, flow cytometry and TUNEL experiments, ZW, XZ
and KH performed the rest of the experiments. WL, YW and XZ wrote
the manuscript. All authors read and approved the final version of
the manuscript. WL and QY confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Stanhewicz AE, Wenner MM and Stachenfeld
NS: Sex differences in endothelial function important to vascular
health and overall cardiovascular disease risk across the lifespan.
Am J Physiol Heart Circ Physiol. 315:H1569–H1588. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Widlansky ME, Gokce N, Keaney JF Jr and
Vita JA: The clinical implication of endothelial dysfunction. J Am
Coll Cardiol. 42:1149–1160. 2003.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rajendran P, Rengarajan T, Thangavel J,
Nishigaki Y, Sakthisekaran D, Sethi G and Nishigaki I: The vascular
endothelium and human diseases. Int J Biol Sci. 9:1057–1069.
2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sturtzel C: Endothelial cells. Adv Exp Med
Biol. 1003:71–91. 2007.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hergenreider E, Heydt S, Tréguer K,
Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS,
Yin X, Mayr M, et al: Atheroprotective communication between
endothelial cells and smooth muscle cells through miRNAs. Nat Cell
Biol Feb. 14:249–256. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Huang AL and Vita JA: Effects of systemic
inflammation on endothelium dependent vasodilation. Trends
Cardiovasc Med. 16:15–20. 2006.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kolluru GK, Bir SC and Kevil CG:
Endothelial dysfunction and diabetes: Effects on angiogenesis,
vascular remodeling, and wound healing. Int J Vasc Med.
2012(918267)2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Park KH and Park WJ: Endothelial
dysfunction: Clinical implication in cardiovascular disease and
therapeutic approaches. J Korean Med Sci. 30:1213–1225.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhong L, Simard MJ and Huot J: Endothelial
microRNAs regulating the NF-κB pathway and cell adhesion molecules
during inflammation. FASEB J. 32:4070–4084. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Onishi A, St Ange K, Dordick JS and
Linhardt RJ: Heparin and anticoagulation. Front Biosci (Landmark
Ed). 21:1372–1392. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Ong CS, Marcum JA, Zehr KJ and Cameron DE:
A century of heparin. Ann Thorac Surg. 108:955–958. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
McLaughlin K, Nadeem L, Wat J, Baczyk D,
Lye SJ and Kingdom JC: Low molecular weight heparin promotes
transcription and release of placental growth factor from
endothelial cells. Am J Physiol Heart Circ Physiol.
318:H1008–H1017. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen J, Wang H, Gao C, Liu D, Fan Y, Li W,
Chen Y and Pan S: Tetramethylpyrazine alleviates LPS-induced
inflammatory injury in HUVECs by inhibiting Rho/ROCK pathway.
Biochem Biophys Res Commun. 514:329–335. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Haarmann R, Ibrahim M, Stevanovic M,
Bredemeier R and Schleiff E: The properties of the outer membrane
localized Lipid A transporter LptD. J Phys Condens Matter.
22(454124)2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wan X, Wang PX, Zhou L and Xiang Q: Gene
expression of toll-like receptors in the liver, lungs and spleen in
mice after endotoxin challenge. Zhongguo Wei Zhong Bing Ji Jiu Yi
Xue. 16:73–76. 2004.PubMed/NCBI(In Chinese).
|
|
17
|
Zhang J, Zheng Y, Luo Y, Du Y, Zhang X and
Fu J: Curcumin inhibits LPS-induced neuroinflammation by promoting
microglial M2 polarization via TREM2/ TLR4/ NF-κB pathways in BV2
cells. Mol Immunol. 116:29–37. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xiao Q, Zhu X, Yang S, Wang J, Yin R, Song
J, Ma A and Pan X: LPS induces CXCL16 expression in HUVECs through
the miR-146a-mediated TLR4 pathway. Int Immunopharmacol.
69:143–149. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li Y, Zhu H, Wei X, Li H, Yu Z, Zhang H
and Liu W: LPS induces HUVEC angiogenesis in vitro through
miR-146a-mediated TGF-β1 inhibition. Am J Transl Res. 15:591–600.
2017.PubMed/NCBI
|
|
20
|
Cox D, Kerrigan SW and Watson SP:
Platelets and the innate immune system: Mechanisms of
bacterial-induced platelet activation. J Thromb Haemost.
9:1097–1107. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Cheng X, Yang YL, Yang H, Wang YH and Du
GH: Kaempferol alleviates LPS-induced neuroinflammation and BBB
dysfunction in mice via inhibiting HMGB1 release and
down-regulating TLR4/MyD88 pathway. Int Immunopharmacol. 6:29–35.
2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Gu J, Su S, Guo J, Zhu Y, Zhao M and Duan
JA: Anti-inflammatory and anti-apoptotic effects of the combination
of Ligusticum chuanxiong and Radix Paeoniae against focal cerebral
ischaemia via TLR4/MyD88/MAPK/NF-κB signalling pathway in MCAO
rats. J Pharm Pharmacol. 70:268–277. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li C, Ai G, Wang Y, Lu Q, Luo C, Tan L,
Lin G, Liu Y, Li Y, Zeng H, et al: Oxyberberine, a novel gut
microbiota-mediated metabolite of berberine, possesses superior
anti-colitis effect: Impact on intestinal epithelial barrier, gut
microbiota profile and TLR4-MyD88-NF-κB pathway. Pharmacol Res.
152(104603)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li H, Zhong X, Li W and Wang Q: Effects of
1,25-dihydroxyvitamin D3 on experimental periodontitis and
AhR/NF-κB/NLRP3 inflammasome pathway in a mouse model. J Appl Oral
Sci. 27(e20180713)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kim SY, Jin CY, Kim CH, Yoo YH, Choi SH,
Kim GY, Yoon HM, Park HT and Choi YH: Isorhamnetin alleviates
lipopolysaccharide-induced inflammatory responses in BV2 microglia
by inactivating NF-κB, blocking the TLR4 pathway and reducing ROS
generation. Int J Mol Med. 43:682–692. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kim DC, Quang TH, Oh H and Kim YC:
Steppogenin isolated from Cudrania tricuspidata shows
Antineuroinflammatory effects via NF-κB and MAPK pathways in
LPS-Stimulated BV2 and primary rat microglial cells. Molecules.
22(2130)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yu X, Lan P, Hou X, Han Q, Lu N, Li T,
Jiao C, Zhang J, Zhang C and Tian Z: HBV inhibits LPS-induced NLRP3
inflammasome activation and IL-1β production via suppressing the
NF-κB pathway and ROS production. J Hepatol. 66:693–702.
2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kauppinen A, Suuronen T, Ojala J,
Kaarniranta K and Salminen A: Antagonistic crosstalk between NF-κB
and SIRT1 in the regulation of inflammation and metabolic
disorders. Cell Signal. 25:1939–1948. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Pober JS and Sessa WC: Evolving functions
of endothelial cells in inflammation. Nature Rev Immunol.
7:803–815. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Incalza MA, D'Oria R, Natalicchio A,
Perrini S, Laviola L and Giorgino F: Oxidative stress and reactive
oxygen species in endothelial dysfunction associated with
cardiovascular and metabolic diseases. Vascul Pharmacol. 100:1–19.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Bischoff J: Endothelial-to-mesenchymal
transition. Circ Res. 124:1163–1165. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Deanfield JE, Halcox JP and Rabelink TJ:
Endothelial function and dysfunction: Testing and clinical
relevance. Circulation. 115:1285–1295. 2007.PubMed/NCBI View Article : Google Scholar
|