Introduction
Alzheimer's disease (AD), the most prevalent form of
dementia in the elderly, is characterized by the progressive loss
of cognitive function. It is pathologically characterized by the
extracellular accumulation of amyloid β proteins and intracellular
formation of neurofibrillary tangles (1). Accumulating evidence has suggested an
important role of the genetic component, which is estimated to be
60-80%, in the pathogenesis of AD (2). Numerous susceptible loci containing
common variants [minor allele frequency (MAF) >5%] have been
identified in recent decades (3).
However, only the Ɛ4 allele of apolipoprotein E (APOE) is confirmed
to be the most important variant conferring a 3- to 4-fold risk to
AD (4). Furthermore, since most of
the loci reside in the intronic or intergenic regions, the
biological mechanisms remain difficult to interpret. With the
advent of next-generation sequencing, more and more rare coding
variants (MAF<1%) with a moderate effect on the risk of AD,
including those in phospholipase Cγ2 (PLCG2), ABI family member 3
(ABI3) and triggering receptor expressed on myeloid cells 2
(TREM2), have been identified (5).
TREM2 is located on chromosome 6p21.1 and encodes a
transmembrane protein on microglial cells. The protein is involved
in the innate immunity within the central nervous system (CNS) by
stimulating phagocytosis and inhibiting cytokine production
(6). The association between TREM2
and AD was first suggested by findings indicating that homozygous
loss-of-function variants of TREM2 caused autosomal recessive
Nasu-Hakola disease, which is characterized by polycystic
lipomembranous osteodysplasia with sclerosing leukoencephalopathy
and early-onset dementia (7).
Subsequent genome-wide and exome-wide sequencing revealed that rare
coding variants of TREM2 may confer a risk of AD (4,8). A
rare missense variant, p.Arg47His (R47H, rs75932628), has been
identified as a risk factor of AD in Caucasians (4,8), but
not in African-American or Asian populations (9,10).
Additional rare variants, including p.Arg62His (R62H, rs143332484),
p.Asp87Asn (D87N, rs142232675) and p.His157Tyr (H157Y, rs2234255),
have also been detected in AD subjects and indicated to increase
the susceptibility to AD (11,12).
Furthermore, certain TREM2 mutations, such as p.Gln33stop,
p.Tyr38Cys and Thr66Met, have been identified only in patients with
AD but not in normal controls, and appear to be causative (13), However, due to the low frequency of
these variants, studies with a small sample size may not have
adequate power to identify the genetic associations (14-16).
The results of association studies from different regions and
ethnicities are not always consistent. Therefore, a meta-analysis
for the genetic association between TREM2 rare coding variants and
AD risk involving 73,128 individuals was performed.
Materials and methods
Search strategy
The PubMed, Embase and Web of Science databases were
searched for articles investigating the association between TREM2
variants and the risk of AD using the following key words: (‘TREM2’
OR ‘triggering receptor expressed on myeloid cells 2’) and (‘AD’ OR
‘Alzheimer's disease’ OR ‘Alzheimer disease’) from inception to
December 31, 2019. Additional articles were obtained by manually
searching the reference lists of review and research articles.
Inclusion and exclusion criteria
For inclusion in the present meta-analysis, studies
were required to meet the following criteria: i) Investigation of
the association of TREM2 variants with the risk of AD in human
subjects; ii) reporting on genotype data of at least one of the
following four variants: R47H, R62H, D87N and H157Y; iii) studies
with a case-control design with unrelated samples; and iv)
providing sufficient data to calculate an odds ratio (OR) and
corresponding 95% CI. If studies had overlapping samples, only the
study with a larger sample size was included. Studies were excluded
if they were based on family samples, used imputed data or provided
insufficient genotype data.
Data extraction
A total of two independent researchers (RL and XW)
extracted the following information: First author, publication
year, ethnicity, sample size, genotype distribution in the case and
control group and the genotyping method. Discrepancies between
researchers were resolved by discussion.
Quality assessment
The present study used the Newcastle-Ottawa Scale
(NOS) (17), which includes 8 items
in 3 categories (selection, comparability and exposure), to assess
the quality of all included studies. There are 9 stars in total,
and studies having 7 or more stars were considered of high
quality.
Statistical analysis
Since the four variants examined are rare in the
general population, only the AD risk of carriers (heterozygous +
homozygous variants) vs. non-carriers (homozygous wild-type) was
compared. The OR was calculated as follows: OR=(no. of case
carriers/no. of case non-carriers)/(no. of control carriers/no. of
control non-carriers) and the 95% CI was determined to estimate the
association strength of each variant with AD risk. Between-study
heterogeneity was assessed by I2 statistics, which
indicated low, intermediate and high heterogeneity if
I2<25, 25-50 and >50%, respectively. The
random-effects model was applied if there was high heterogeneity;
otherwise, the fixed-effects model was used. Sensitivity analysis
was performed to evaluate the impact of each study on the pooled
effect size by excluding one study at a time. Publication bias was
detected by Begg's funnel plots and Egger's test. All statistical
analyses were performed by using STATA 12.0 (StataCorp LP).
P<0.05 was considered to indicate statistical significance.
Results
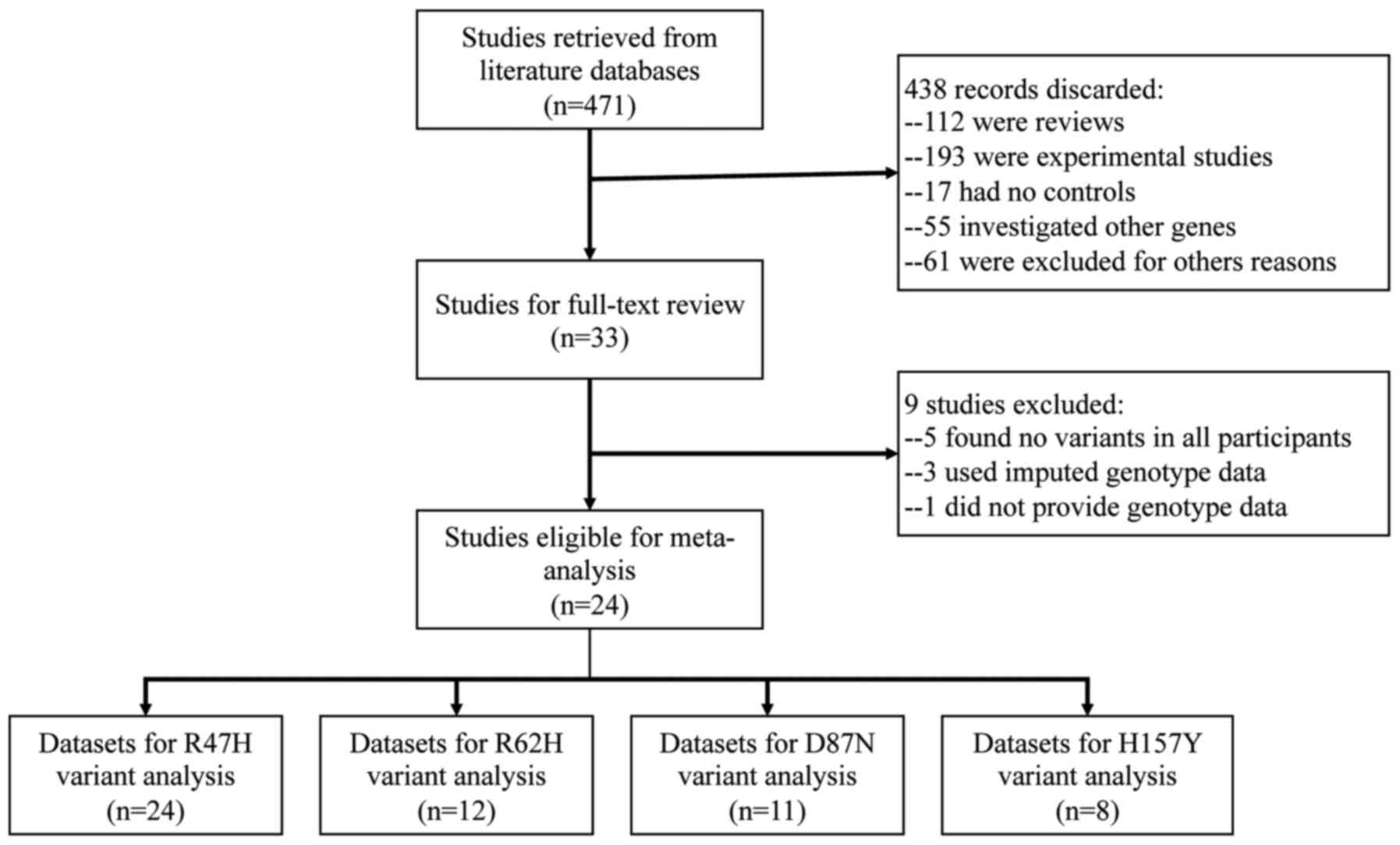
Study selection
A total of 33 studies investigating the genetic
association between rare TREM2 variants (R47H, R62H, D87N and
H157Y) and the risk of AD were identified. However, 9 studies were
excluded for the following reasons: A total of 5 did not provide
any variant of interest in neither the case nor control groups
(18-22),
3 used imputed data (5,23,24)
and 1 did not provide genotype data (25). A total of two studies used a
multi-stage strategy (8,26), and the datasets from discovery and
replication stages were independently included in the quantitative
synthesis. Finally, a total of 26 datasets from 24 studies
(4,8-12,14-16,26-40)
comprising 28,007 cases and 45,121 controls were eligible for the
meta-analysis. Among these studies, 24 were for R47H (4,8-11,14-16,26-37,39,40),
12 for R62H (8,9,11,14-16,27,28,31,32,38,39),
11 for D87N (8,9,11,15,26-28,31,32,36)
and eight for H157Y (8-12,26,31,36).
A flowchart of the literature search is provided in Fig. 1. Regarding ethnicity, the majority
of screened individuals were Caucasians (20 datasets) and the
others were from African (2 datasets), Asian (2 datasets) or mixed
populations (2 datasets). All studies were of high quality (≥7
stars) according to the NOS. The characteristics of all datasets
included in the quantitative synthesis are listed in Table I.
 | Table ICharacteristics of all datasets
included in the quantitative synthesis. |
Table I
Characteristics of all datasets
included in the quantitative synthesis.
| First author | Year | Ethnicity | Genotyping
method | Sample size
(cases/controls) | Variant | NOS score | Refs. |
|---|
| Jonsson | 2013 | Caucasian | Assay, Taqman | 2037/9727 | R47H | 9 | (4) |
| Guerreiro
(discovery) | 2013 | Caucasian | NGS,
Sequencing | 1091/1105 | R47H, R62H, D87N,
H157Y | 9 | (8) |
| Guerreiro
(replication) | 2013 | Caucasian | Taqman | 1887/4061 | R47H | 9 | (8) |
| Pottier | 2013 | Caucasian | Sequencing | 726/783 | R47H, R62H,
D87N | 7 | (27) |
| Bertitez | 2013 | Caucasian | Sequencing | 504/550 | R47H, R62H,
D87N | 7 | (28) |
| Gonzalez | 2013 | Caucasian | Taqman | 427/2540 | R47H | 7 | (29) |
| Ruiz | 2014 | Caucasian | Taqman, HRM,
KASPar | 3172/2169 | R47H | 8 | (30) |
| Cuyvers | 2014 | Caucasian | Sequencing | 1216/1094 | R47H, R62H, D87N,
H157Y | 7 | (31) |
| Miyashita | 2014 | Asian | Taqman | 2190/2498 | R47H, H157Y | 8 | (10) |
| Jin | 2014 | Caucasian | Taqman | 2082/1648 | R47H, R62H, D87N,
H157Y | 7 | (11) |
| Slattery | 2014 | Caucasian | Sequencing | 971/534 | R47H, R62H,
D87N | 8 | (32) |
| Finelli | 2015 | Caucasian | Sequencing | 474/608 | R47H | 7 | (33) |
| Roussos | 2015 | Caucasian | Sequencing | 265/225 | R47H | 7 | (34) |
| Jin | 2015 | African | Sequencing | 906/2487 | R47H, R62H, D87N,
H157Y | 8 | (9) |
| Mehrjoo | 2015 | Caucasian | Sequencing | 131/157 | R47H, R62H | 7 | (14) |
| Rosenthal | 2015 | Caucasian | Taqman | 1613/2927 | R47H | 8 | (35) |
| Jiang | 2016 | Asian | NGS | 988/1354 | H157Y | 8 | (12) |
| Ghani | 2016 | Caucasian | NGS | 210/233 | R47H, D87N,
H157Y | 7 | (36) |
| Sirkis
(discovery) | 2016 | Caucasian | NGS | 31/245 | R47H, D87N | 7 | (26) |
| Sirkis
(replication) | 2016 | Caucasian | NGS | 2927/2633 | R47H, D87N,
H157Y | 8 | (26) |
| Bellenguez | 2017 | Caucasian | NGS | 1779/1273 | R47H | 8 | (37) |
| Peplonska | 2018 | Caucasian | Sequencing | 274/208 | R47H, R62H,
D87N | 7 | (15) |
| Landoulsi | 2018 | African | Sequencing | 172/158 | R62H | 7 | (38) |
|
Arboleda-Bustos | 2018 | Caucasian | KASPar | 358/329 | R47H, R62H | 7 | (16) |
| Dalmasso | 2019 | Mixed | Taqman | 419/486 | R47H, R62H | 7 | (39) |
| Ayer | 2019 | Mixed | Taqman | 1157/5089 | R47H | 8 | (40) |
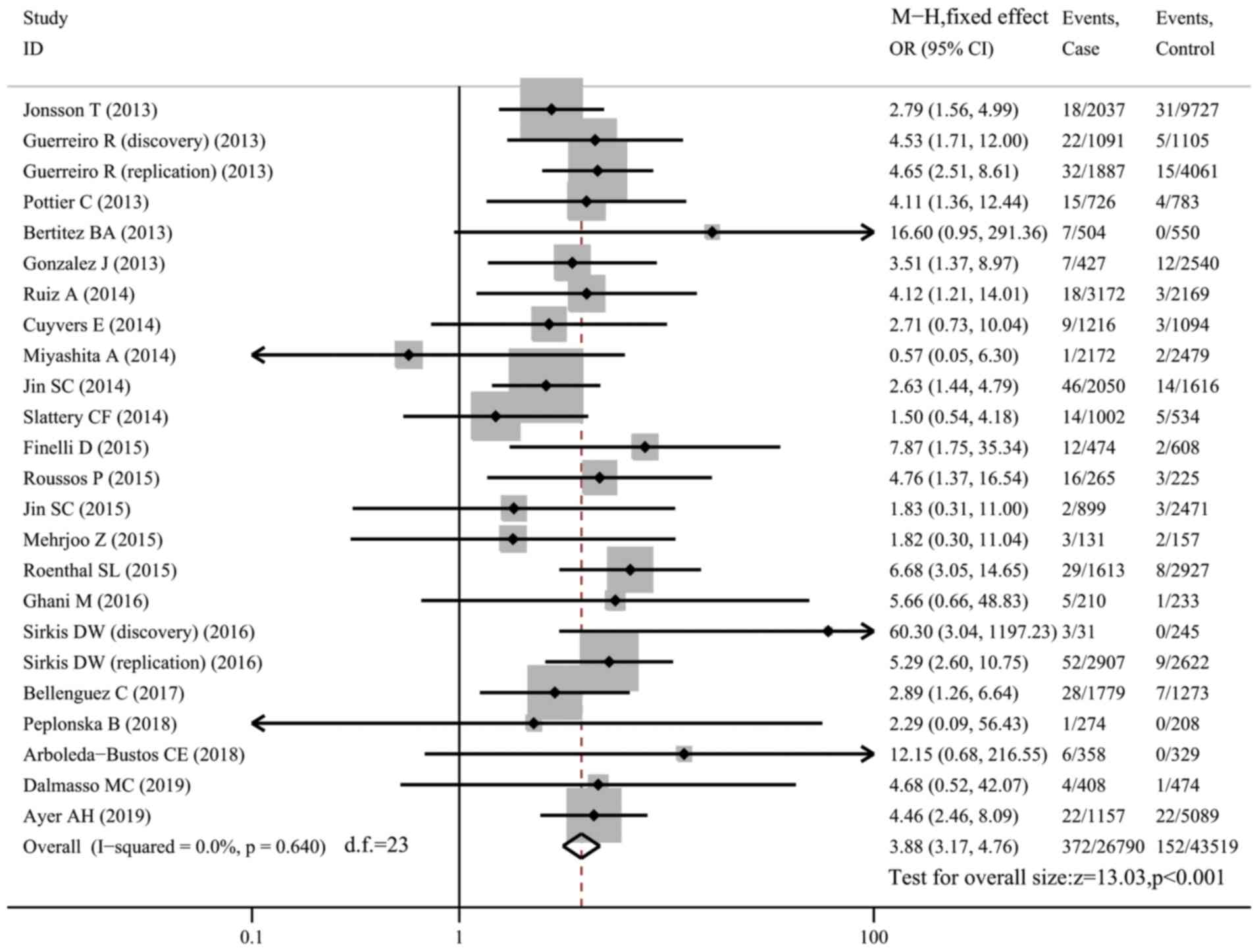
R47H and risk of AD
A total of 24 datasets (26,847 cases and 43,609
controls) investigated the association between the R47H variant and
risk of AD. Carriers of R47H accounted for 1.39% of AD patients and
0.35% of controls. There was no between-study heterogeneity
(I2=0) and the fixed-effects model was used. Compared
with non-carriers, R47H carriers were more susceptible to AD
(OR=3.88, 95% CI: 3.17-4.76, P<0.001; Fig. 2). Subgroup analysis of the datasets
of Caucasian populations (20 datasets with 22,175 cases and 33,049
controls) revealed an increased risk of AD in R47H carriers
(OR=3.93, 95% CI: 3.15-4.90, P<0.001, I2=0; Fig. S1).
There were three studies reporting on the
association between the R47H variant and the susceptibility to
early-onset AD (EOAD), which onsets in patients below the age of 65
years. These 3 studies comprised a total of 1,758 cases of EOAD and
2,606 controls (26,27,36).
The variant occurred in 2.22% of EOAD cases but only in 0.42% of
controls, thus conferring a 4.86-fold increased risk (95% CI:
2.52-9.36, P<0.001, I2=0; Fig. S2) to EOAD.
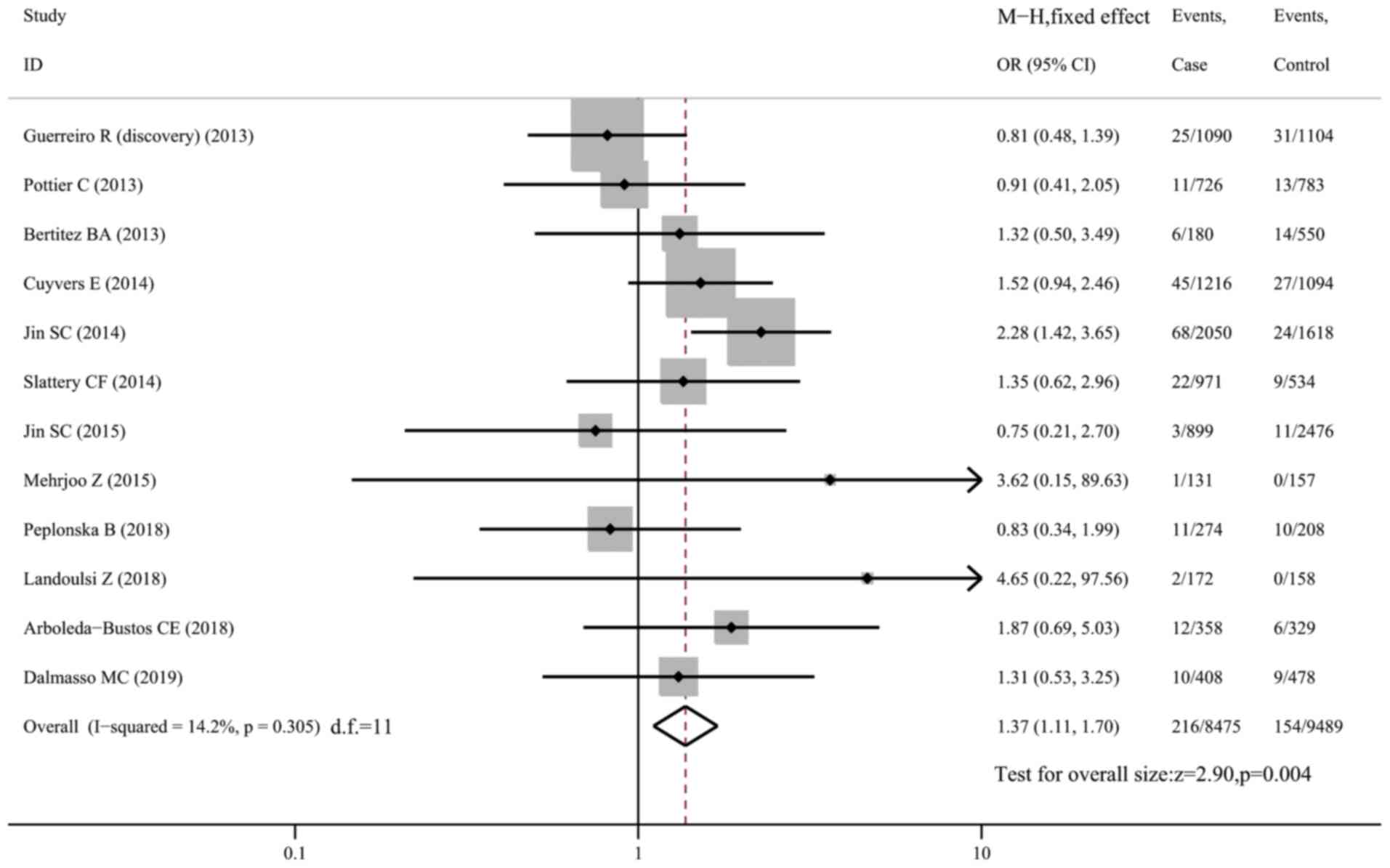
R62H and risk of AD
The R62H variant was genotyped in 12 datasets
comprising 8,525 AD cases and 9,539 controls. Pooled analysis using
the fixed-effects model demonstrated a higher risk of AD in
carriers than in non-carriers of R62H in the whole sample (OR=1.37,
95% CI: 1.11-1.70, P=0.004, I2=14.2%; Fig. 3) and in Caucasians only (OR=1.39,
95% CI: 1.11-1.74, P=0.004; Fig.
S3). However, sensitivity analysis revealed that the
association became insignificant (OR=1.17, 95% CI: 0.92-1.50,
P=0.192, I2=0; Fig. S4)
when the study by Jin et al (11) was excluded, indicating that the
study had a significant impact on the pooled effect size and was
the main source of heterogeneity. In a subgroup analysis of
Caucasian subjects after excluding the study by Jin et al
(eight datasets with 13,436 subjects), no association was
identified between the R62H variant and AD susceptibility (OR=1.17,
95% CI: 0.90-1.52, P=0.231, I2=0; Fig. S5).
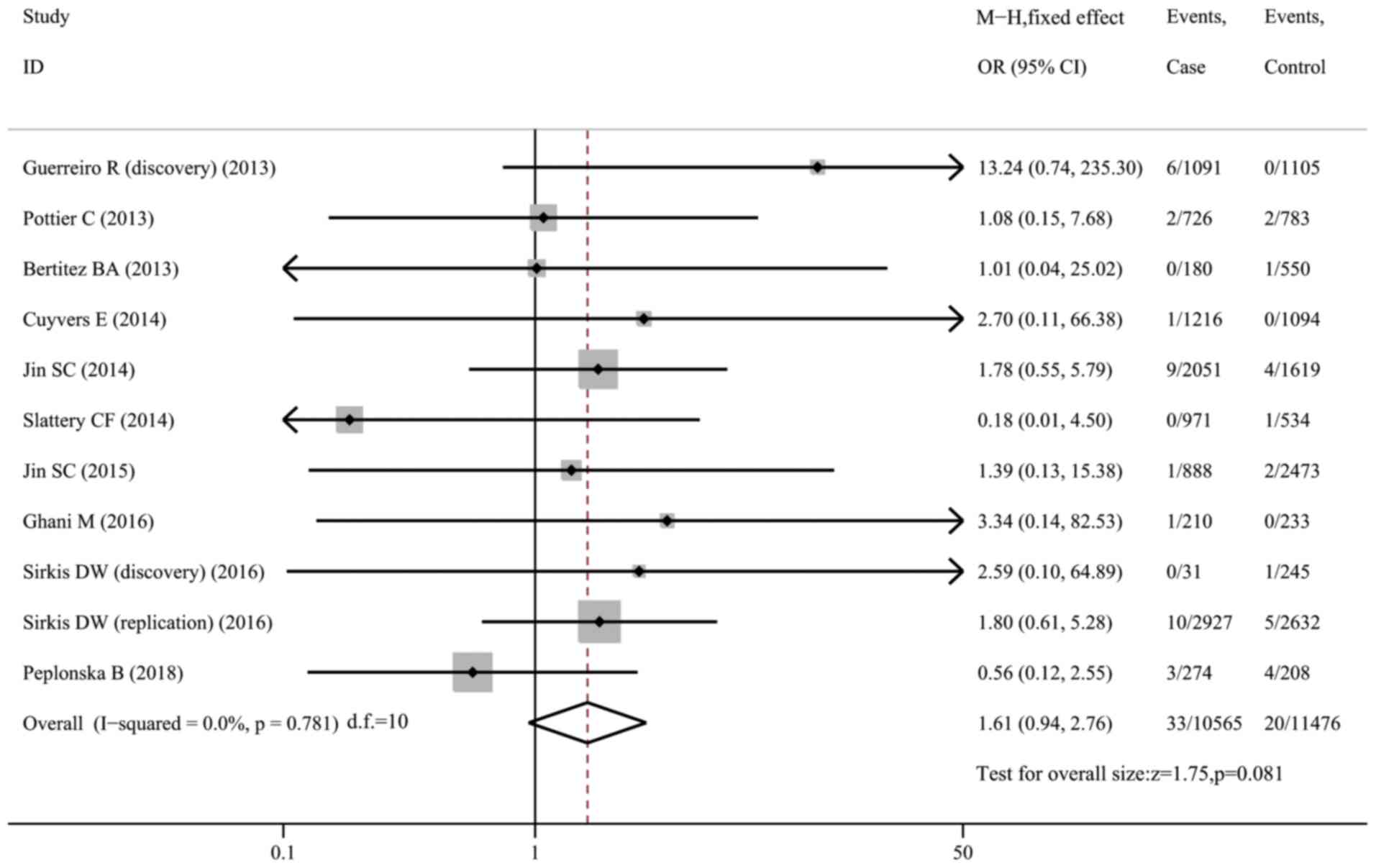
D87N and risk of AD
A total of 11 datasets (10,614 cases and 11,520
controls) were included in the meta-analysis on the association
between the D87N variant and AD susceptibility. However, no
significant associations were observed in the whole sample
(OR=1.61, 95% CI: 0.94-2.76, P=0.081, I2=0; Fig. 4) or in the Caucasian subgroup
(OR=1.62, 95% CI: 0.94-2.82, P=0.084, I2=0; Fig. S6).
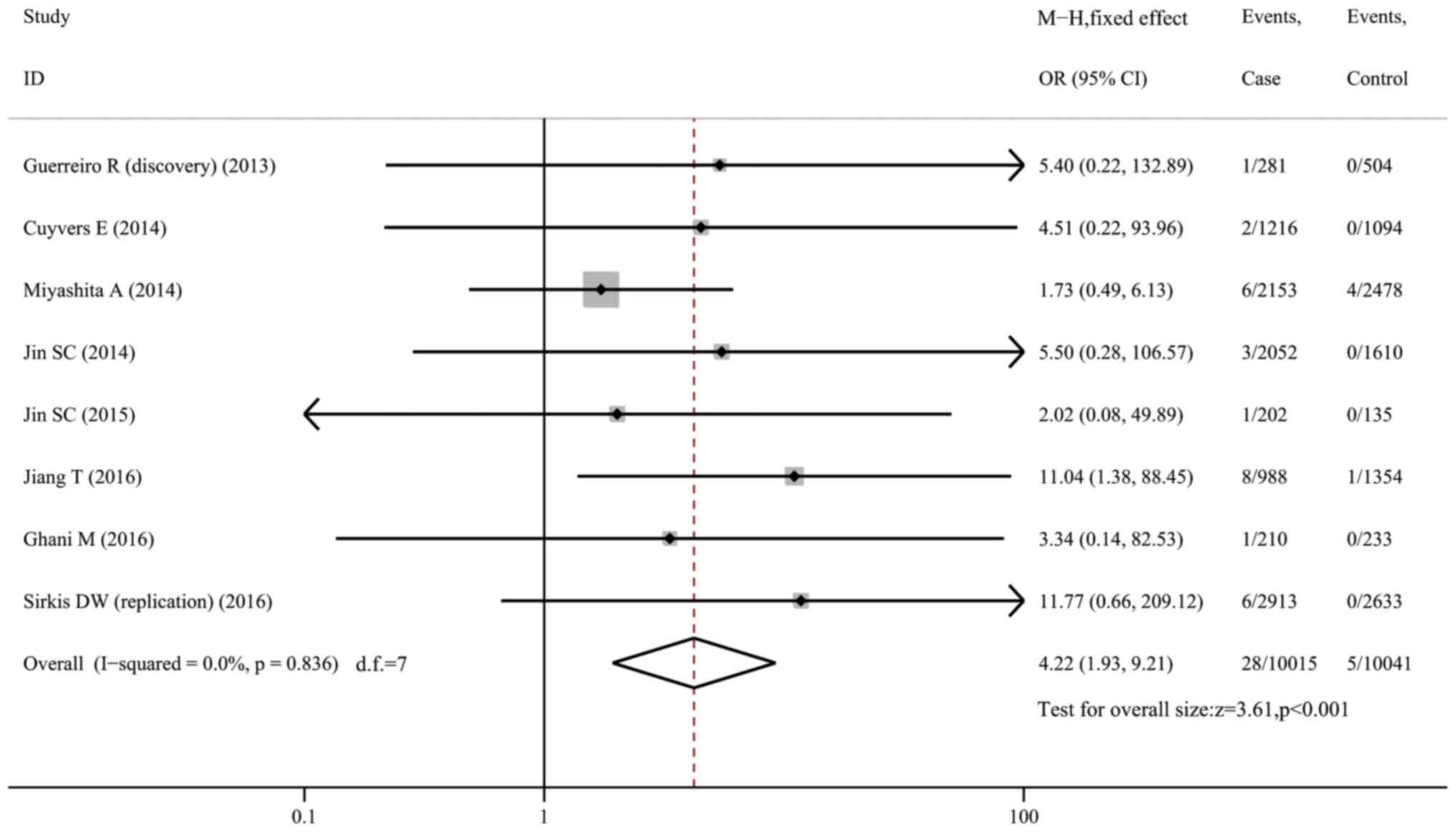
H157Y and risk of AD
The H157Y variant was rare in AD cases (0.28%) and
controls (0.05%). When pooling 8 datasets (10,096 cases and 10,099
controls) together, the analysis indicated that carriers of the
H157Y variant were more predisposed to AD (OR=4.22, 95% CI:
1.93-9.21, P<0.001, I2=0; Fig. 5). The variant conferred a 6.20-fold
risk to Caucasians (95% CI: 1.60-24.04, P=0.008, I2=0;
Fig. S7) from 5 datasets, but was
not associated with AD risk in Asians (OR=3.61, 95% CI: 0.61-21.37,
P=0.157, I2=55.1%; Fig.
S8) from two datasets. Of note, in Caucasians, the variant was
only found in AD patients (13/6,672) but was completely absent in
normal controls (0/6,074).
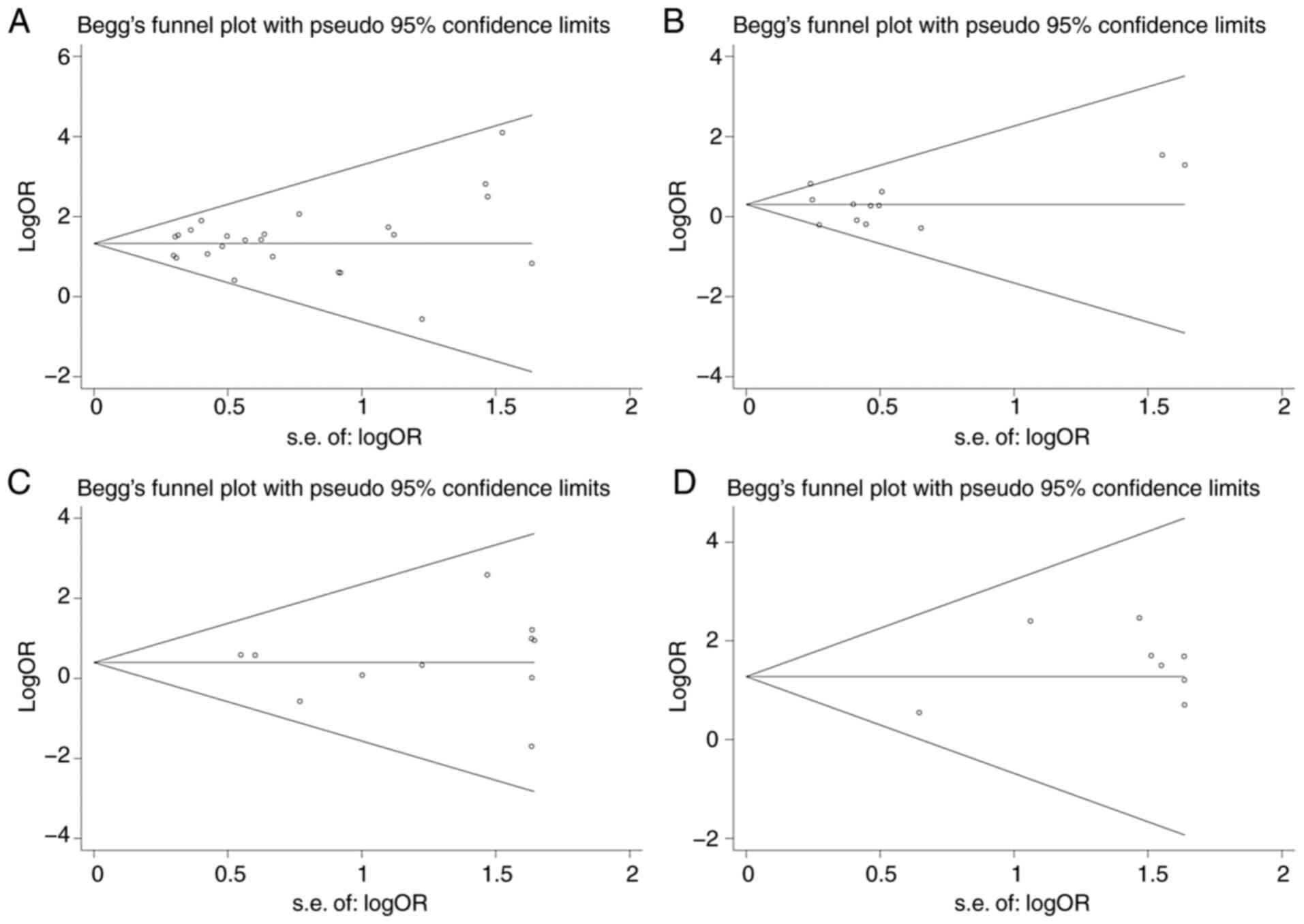
Sensitivity analysis and publication
bias
Sensitivity analysis demonstrated that none of the
included datasets significantly affected the pooled effect size
except the study of Jin et al (11) for the R62H variant as mentioned
above. The funnel plots were symmetric for each mutation (Fig. 6) and Egger's test indicated that
there was no publication bias (P>0.05).
Discussion
The present meta-analysis systematically
investigated the genetic associations of four frequently reported
rare variants of TREM2 with AD susceptibility. The results
indicated that carriers of the R47H, R62H or H157Y variants were
more vulnerable to AD.
TREM2 encodes a transmembrane immune receptor on
microglial cells and exerts its effect by regulating the number of
myeloid cells, enhancing phagocytosis and modulating the
inflammation response in the CNS (6,41). Its
pivotal role in neuroimmunology and neuroinflammation may indicate
an implication of TREM2 in the pathogenesis of neurodegenerative
disease (41). Association studies
have identified rare variants in the coding sequence of TREM2 as
susceptibility markers for amyotrophic lateral sclerosis (42), Parkinson's disease and
frontotemporal dementia (43), as
well as AD. The involvement of TREM2 in the etiology of
neurodegenerative diseases, particularly AD, is complex. Numerous
studies have revealed that disease-associated variants (mainly
R47H) of TREM2 impact amyloid pathology (44), modulate neuritic dystrophy (34), influence tau hyperphosphorylation
and aggregation (45) and affect
synaptic and neuronal loss (46).
Although the pathogenic mechanism of TREM2 in AD may not have been
conclusive, TREM2 variants are conclusively susceptibility markers
for AD, as demonstrated by the present meta-analysis.
R47H is the most frequently investigated variant in
TREM2. The substitution of Arg by His leads to a marked reduction
in soluble TREM2 levels (47) and
the binding ability to cells, APOE and various lipids (45,48,49).
However, whether it reduces the mRNA and protein expression in
humans remains controversial (50).
Furthermore, experiments have revealed that the heterozygous R47H
variant may confer a loss of TREM2 function and enhance neuritic
dystrophy around plaques, thus increasing the risk of AD (51), which was ascertained by
epidemiological investigations in various populations (4,8,40). The
R47H variant is enriched in AD cases (1.39%) but less prevalent in
cognitively normal controls (0.35%). Most of the identified
variants are heterozygous (98%, 514/524) and homozygous variants
are detected in AD cases only. The present meta-analysis indicated
an approximately 4-fold risk of AD in R47H carriers compared to
non-carriers, indicating an effect size similar to that of the APOE
Ɛ4 allele (4). However, APOE Ɛ4 is
much more prevalent in populations and should still be the main
genetic determinant of AD susceptibility.
In addition to risk, the R47H variant may also
contribute to an earlier age of onset in AD. Slattery et al
(32) reported a markedly younger
age at onset in patients with the R47H variant than in those
without the variant (55.2 vs. 61.7 years on average). Similar
results were obtained in Icelandic and Dutch populations (4), implying an association between R47H
and susceptibility to EOAD. In the present study, pooled analysis
of 3 studies (27,28,37)
suggested a higher prevalence (2.22%) of R47H in EOAD cases.
Carriers had 4.86-fold risk of developing EOAD, which was even
higher than the effect size observed in the total AD samples.
The R62H variant disrupts TREM2 recognition of cells
and APOE (52) but does not alter
the binding to lipid ligands (53).
Jin et al (11) determined
that it is associated with an increased risk of AD in a population
of Americans of European descent (11). The present study revealed a mildly
increased risk of AD in individuals harboring the R62H variant
(OR=1.38, 95% CI: 1.11-1.70, P=0.004). However, the effect size may
be largely attributed to the aforementioned study (11), as indicated by the sensitivity
analysis. The association was no longer significant after excluding
this study (P=0.192). A significant association was only observed
in the study by Jin et al (11), but not in any other study. It should
be noted that the study by Jin et al (11) had a large sample size (>3,700
subjects), and thus, it may have more statistical power than the
other studies to identify the association of a rare variant with AD
susceptibility. Therefore, more studies with a large sample size
are required to confirm the association.
The D87N variant resides within the ectodomain
proximal to the stalk region of TREM2. The mutation leads to
enhanced interaction of TREM2 with certain ligands (53). However, the present study did not
indicate any association of the variant with AD susceptibility when
11 datasets were pooled together. Of note, D87N was identified in 4
out of 5 patients in a family affected by AD (54). It may be enriched in familial cases
in contrast to sporadic cases. Another variant, H157Y, is located
on the stalk domain of TREM2 but has no impact on binding activity
(53). Specifically, it is located
at the TREM2 cleavage site by metalloprotease ADAM metallopeptidase
domain 10 and results in increased shedding of TREM2 and reduced
cell surface expression of TREM2(55). In addition, in silico
programs [SIFT (http://sift.jcvi.org/) and Polyphen2
(http://genetics.bwh.harvard.edu/pph2/)] predicted that
the variant may have a deleterious/possibly damaging impact on
functions of TREM2. The present analysis, comprising 10,096 cases
and 10,099 controls, revealed a significantly increased AD
susceptibility in carriers of H157Y, although the variant is rare
in AD (0.28%).
In the present study, a subgroup analysis was
performed with regard to ethnicity, indicating an
ethnicity-specific pattern of TREM2 variants. R47H and R62H were
mostly identified in Caucasians but rare in Asians. Only one study
from Asian (10) reported a low
frequency of R47H (0.06%), whereas the other studies did not
(18-22).
Therefore, R47H and R62H mutations were only associated with an
increased risk of AD in Caucasians. Contrary to R47H and R62H,
H157Y was more frequent in Asians. Miyashita et al (10) reported 10 carriers out of 4,631
participants, while Jiang et al (12) identified 9 carriers among 2,352
participants. However, the H157Y variant was not associated with AD
risk in Japanese (10) but
increased the risk in a Han Chinese population (12). Of note, in Caucasians, the H157Y
variant was only harbored by AD cases but by none of the controls,
indicating that H157Y was more likely to be a causative variant of
AD in Caucasians.
In summary, the present meta-analysis, involving
>73,000 individuals, supported that rare coding variants of
TREM2 are associated with AD susceptibility and may be used as
predictive markers for neurodegenerative disease.
Supplementary Material
Forest plot for the association of
triggering receptors expressed on myeloid cells 2 variant R47H with
susceptibility to Alzheimer's disease in Caucasians. OR, odds
ratio; M-H, Mantel-Haentzel method; d.f., degree of freedom.
Forest plot for the association of
triggering receptors expressed on myeloid cells 2 variant R47H with
susceptibility to early-onset Alzheimer's disease. OR, odds ratio;
M-H, Mantel-Haentzel method; d.f., degree of freedom.
Forest plot for the association of
triggering receptors expressed on myeloid cells 2 variant R62H with
susceptibility to Alzheimer's disease in Caucasians. OR, odds
ratio; M-H, Mantel-Haentzel method; d.f., degree of freedom.
Forest plot for the association of
triggering receptors expressed on myeloid cells 2 variant R62H with
susceptibility to Alzheimer's disease after excluding the study by
Jin et al (11). OR, odds ratio;
M-H, Mantel-Haentzel method; d.f., degree of freedom.
Forest plot for the association of
triggering receptors expressed on myeloid cells 2 variant R62H with
susceptibility to Alzheimer's disease in Caucasians after excluding
the study by Jin et al (11). OR,
odds ratio; M-H, Mantel-Haentzel method; d.f., degree of
freedom.
Forest plot for the association of
triggering receptors expressed on myeloid cells 2 variant D87N with
susceptibility to Alzheimer's disease in Caucasians. OR, odds
ratio; M-H, Mantel-Haentzel method; d.f., degree of freedom.
Forest plot for the association of
triggering receptors expressed on myeloid cells 2 variant H157Y
with susceptibility to Alzheimer's disease in Caucasians. OR, odds
ratio; M-H, Mantel-Haentzel method; d.f., degree of freedom.
Forest plot for the association of
triggering receptors expressed on myeloid cells 2 variant H157Y
with susceptibility to Alzheimer's disease in Asians. OR, odds
ratio; D-L, DerSimonian-Laird method; d.f., degree of freedom.
Acknowledgements
Not applicable.
Funding
Funding: No funding received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RL designed and supervised the study; RL and XW
performed literature search, study selection, data curation and
formal analysis; RL and PH performed quality control of studies; RL
and XW prepared the original draft; RL, XW and PH critically
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Castellani RJ, Rolston RK and Smith MA:
Alzheimer disase. Dis Mon. 56:484–546. 2010.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gatz M, Reynolds CA, Fratiglioni L,
Johansson B, Mortimer JA, Berg S, Fiske A and Pedersen NL: Role of
genes and environments for explaining Alzheimer disease. Arch Gen
Psychiat. 63:168–174. 2006.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Naj AC and Schellenberg GD: Alzheimer's
Disease Genetics Consortium (ADGC). Genomic variants, genes, and
pathways of Alzheimer's disease: An overview. Am J Med Genet B
Neuropsychiatr Genet. 174:5–26. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jonsson T, Stefansson H, Steinberg S,
Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J,
Levey AI, Lah JJ, et al: Variant of TREM2 associated with the risk
of Alzheimer's disease. New Engl J Med. 368:107–116.
2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sims R, van der Lee SJ, Naj AC, Bellenguez
C, Badarinarayan N, Jakobsdottir J, Kunkle BW, Boland A, Raybould
R, Bis JC, et al: Rare coding variants in PLCG2, ABI3, and TREM2
implicate microglial-mediated innate immunity in Alzheimer's
disease. Nat Genet. 49:1373–1384. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Rohn TT: The triggering receptor expressed
on myeloid cells 2: ‘TREM-ming’ the inflammatory component
associated with Alzheimer's disease. Oxid Med Cell Longev.
2013(860959)2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Paloneva J, Manninen T, Christman G,
Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R,
Salmaggi A, et al: Mutations in two genes encoding different
subunits of a receptor signaling complex result in an identical
disease phenotype. Am J Hum Genet. 71:656–662. 2002.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Guerreiro R, Wojtas A, Bras J,
Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe
JS, Younkin S, et al: TREM2 variants in Alzheimer's disease. N Engl
J Med. 368:117–127. 2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Jin SC, Carrasquillo MM, Benitez BA,
Skorupa T, Carrell D, Patel D, Lincoln S, Krishnan S, Kachadoorian
M, Reitz C, et al: TREM2 is associated with increased risk for
Alzheimer's disease in African Americans. Mol Neurodegener.
10(19)2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Miyashita A, Wen Y, Kitamura N, Matsubara
E, Kawarabayashi T, Shoji M, Tomita N, Furukawa K, Arai H, Asada T,
et al: Lack of genetic association between TREM2 and late-onset
Alzheimer's disease in a Japanese population. J Alzheimers Dis.
41:1031–1038. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jin SC, Benitez BA, Karch CM, Cooper B,
Skorupa T, Carrell D, Norton JB, Hsu S, Harari O, Cai Y, et al:
Coding variants in TREM2 increase risk for Alzheimer's disease. Hum
Mol Genet. 23:5838–5846. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Jiang T, Tan L, Chen Q, Tan MS, Zhou JS,
Zhu XC, Lu H, Wang HF, Zhang YD and Yu JT: A rare coding variant in
TREM2 increases risk for Alzheimer's disease in Han Chinese.
Neurobiol Aging. 42:217.e1–e3. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Dardiotis E, Siokas V, Pantazi E, Dardioti
M, Rikos D, Xiromerisiou G, Markou A, Papadimitriou D, Speletas M
and Hadjigeorgiou GM: A novel mutation in TREM2 gene causing
Nasu-Hakola disease and review of the literature. Neurobiol Aging.
53:194.e13–194.e22. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mehrjoo Z, Najmabadi A, Abedini SS,
Mohseni M, Kamali K, Najmabadi H and Khorram Khorshid HR:
Association study of the TREM2 gene and identification of a novel
variant in exon 2 in iranian patients with late-onset Alzheimer's
disease. Med Prin Pract. 24:351–354. 2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Peplonska B, Berdynski M, Mandecka M,
Barczak A, Kuzma-Kozakiewicz M, Barcikowska M and Zekanowski C:
TREM2 variants in neurodegenerative disorders in the Polish
population. Homozygosity and compound heterozygosity in FTD
patients. Amyotroph Lateral Scler Frontotemporal Degener.
19:407–412. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Arboleda-Bustos CE, Ortega-Rojas J,
Mahecha MF, Arboleda G, Vásquez R, Pardo R and Arboleda H: The
p.R47H variant of TREM2 gene is associated with late-onset
Alzheimer disease in Colombian population. Alzheimer Dis Assoc
Disord. 32:305–308. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Stang A: Critical evaluation of the
Newcastle-Ottawa scale for the assessment of the quality of
nonrandomized studies in meta-analyses. Eur J Epidemiol.
25:603–605. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yu JT, Jiang T, Wang YL, Wang HF, Zhang W,
Hu N and Tan L, Sun L, Tan MS, Zhu XC and Tan L: Triggering
receptor expressed on myeloid cells 2 variant is rare in late-onset
Alzheimer's disease in Han Chinese individuals. Neurobiol Aging.
35:937.e1–e3. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chung SJ, Kim MJ, Kim J, Kim YJ, You S,
Koh J, Kim SY and Lee JH: Exome array study did not identify novel
variants in Alzheimer's disease. Neurobiol Aging. 35:1958.e13–e14.
2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ma J, Zhou Y, Xu J, Liu X, Wang Y, Deng Y,
Wang G, Xu W, Ren R, Liu X, et al: Association study of TREM2
polymorphism rs75932628 with late-onset Alzheimer's disease in
Chinese Han population. Neurol Res. 36:894–896. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jiao B, Liu X, Tang B, Hou L, Zhou L,
Zhang F, Zhou Y, Guo J, Yan X and Shen L: Investigation of TREM2,
PLD3, and UNC5C variants in patients with Alzheimer's disease from
mainland China. Neurobiol Aging. 35:2422.e9–2422.e11.
2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang P, Guo Q, Zhou Y, Chen K, Xu Y, Ding
D, Hong Z and Zhao Q: Lack of association between triggering
receptor expressed on myeloid cells 2 polymorphism rs75932628 and
late-onset Alzheimer's disease in a Chinese Han population.
Psychiat Genet. 28:16–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Giraldo M, Lopera F, Siniard AL,
Corneveaux JJ, Schrauwen I, Carvajal J, Muñoz C, Ramirez-Restrepo
M, Gaiteri C, Myers AJ, et al: Variants in triggering receptor
expressed on myeloid cells 2 are associated with both behavioral
variant frontotemporal lobar degeneration and Alzheimer's disease.
Neurobiol Aging. 34:2077.e11–e18. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bertram L, Parrado AR and Tanzi RE: TREM2
and neurodegenerative disease. N Engl J Med.
369(1565)2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tosto G, Vardarajan B, Sariya S, Brickman
AM, Andrews H, Manly JJ, Schupf N, Reyes-Dumeyer D, Lantigua R,
Bennett DA, et al: Association of variants in PINX1 and TREM2 with
late-onset Alzheimer disease. JAMA Neurol. 76:942–948.
2019.PubMed/NCBI View Article : Google Scholar : (Online ahead of
print).
|
|
26
|
Sirkis DW, Bonham LW, Aparicio RE, Geier
EG, Ramos EM, Wang Q, Karydas A, Miller ZA, Miller BL, Coppola G
and Yokoyama JS: Rare TREM2 variants associated with Alzheimer's
disease display reduced cell surface expression. Acta Neuropathol
Commun. 4(98)2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Pottier C, Wallon D, Rousseau S,
Rovelet-Lecrux A, Richard AC, Rollin-Sillaire A, Frebourg T,
Campion D and Hannequin D: TREM2 R47H variant as a risk factor for
early-onset Alzheimer's disease. J Alzheimers Dis. 35:45–49.
2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Benitez BA, Cooper B, Pastor P, Jin SC,
Lorenzo E, Cervantes S and Cruchaga C: TREM2 is associated with the
risk of Alzheimer's disease in Spanish population. Neurobiol Aging.
34:1711.e15–e17. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gonzalez Murcia JD, Schmutz C, Munger C,
Perkes A, Gustin A, Peterson M, Ebbert MT, Norton MC, Tschanz JT,
Munger RG, et al: Assessment of TREM2 rs75932628 association with
Alzheimer's disease in a population-based sample: The cache county
study. Neurobiol Aging. 34:2889.e11–e13. 2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ruiz A, Dols-Icardo O, Bullido MJ, Pastor
P, Rodríguez-Rodríguez E, López de Munain A, de Pancorbo MM,
Pérez-Tur J, Alvarez V, Antonell A, et al: Assessing the role of
the TREM2 p.R47H variant as a risk factor for Alzheimer's disease
and frontotemporal dementia. Neurobiol Aging. 35:444.e1–e4.
2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Cuyvers E, Bettens K, Philtjens S, Van
Langenhove T, Gijselinck I, van der Zee J, Engelborghs S,
Vandenbulcke M, Van Dongen J, Geerts N, et al: Investigating the
role of rare heterozygous TREM2 variants in Alzheimer's disease and
frontotemporal dementia. Neurobiol Aging. 35:726.e11–e19.
2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Slattery CF, Beck JA, Harper L, Adamson G,
Abdi Z, Uphill J, Campbell T, Druyeh R, Mahoney CJ, Rohrer JD, et
al: R47H TREM2 variant increases risk of typical early-onset
Alzheimer's disease but not of prion or frontotemporal dementia.
Alzheimers Dement. 10:602–608.e4. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Finelli D, Rollinson S, Harris J, Jones M,
Richardson A, Gerhard A, Snowden J, Mann D and Pickering-Brown S:
TREM2 analysis and increased risk of Alzheimer's disease. Neurobiol
Aging. 36:546.e9–e13. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Roussos P, Katsel P, Fam P, Tan W, Purohit
DP and Haroutunian V: The triggering receptor expressed on myeloid
cells 2 (TREM2) is associated with enhanced inflammation,
neuropathological lesions and increased risk for Alzheimer's
dementia. Alzheimers Dement. 11:1163–1170. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Rosenthal SL, Bamne MN, Wang X, Berman S,
Snitz BE, Klunk WE, Sweet RA, Demirci FY, Lopez OL and Kamboh MI:
More evidence for association of a rare TREM2 mutation (R47H) with
Alzheimer's disease risk. Neurobiol Aging. 36:2443.e21–e26.
2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ghani M, Sato C, Kakhki EG, Gibbs JR,
Traynor B, St George-Hyslop P and Rogaeva E: Mutation analysis of
the MS4A and TREM gene clusters in a case-control Alzheimer's
disease data set. Neurobiol Aging. 42:217.e7–217.e13.
2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Bellenguez C, Charbonnier C, Grenier-Boley
B, Quenez O, Le Guennec K, Nicolas G, Chauhan G, Wallon D, Rousseau
S, Richard AC, et al: Contribution to Alzheimer's disease risk of
rare variants in TREM2, SORL1, and ABCA7 in 1,779 cases and 1,273
controls. Neurobiol Aging. 59:220.e1–220.e9. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Landoulsi Z, Ben Djebara M, Kacem I,
Sidhom Y, Kefi R, Abdelhak S, Gargouri-Berrechid A and Gouider R:
Genetic Analysis of TREM2 variants in tunisian patients with
Alzheimer's disease. Med Prin Pract. 27:317–322. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Dalmasso MC, Brusco LI, Olivar N, Muchnik
C, Hanses C, Milz E, Becker J, Heilmann-Heimbach S, Hoffmann P,
Prestia FA, et al: Transethnic meta-analysis of rare coding
variants in PLCG2, ABI3, and TREM2 supports their general
contribution to Alzheimer's disease. Transl Psychiatry.
9(55)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ayer AH, Wojta K, Ramos EM, Dokuru D, Chen
JA, Karydas AM, Papatriantafyllou JD, Agiomyrgiannakis D,
Kamtsadeli V, Tsinia N, et al: Frequency of the TREM2 R47H variant
in various neurodegenerative disorders. Alzheimer Dis Assoc Disord.
33:327–330. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Jay TR, von Saucken VE and Landreth GE:
TREM2 in neurodegenerative diseases. Mol Neurodegener.
12(56)2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cady J, Koval ED, Benitez BA, Zaidman C,
Jockel-Balsarotti J, Allred P, Baloh RH, Ravits J, Simpson E, Appel
SH, et al: TREM2 variant p.R47H as a risk factor for sporadic
amyotrophic lateral sclerosis. JAMA Neurol. 71:449–453.
2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Rayaprolu S, Mullen B, Baker M, Lynch T,
Finger E, Seeley WW, Hatanpaa KJ, Lomen-Hoerth C, Kertesz A, Bigio
EH, et al: TREM2 in neurodegeneration: Evidence for association of
the p.R47H variant with frontotemporal dementia and Parkinson's
disease. Mol Neurodegener. 8(19)2013.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Yuan P, Condello C, C Keene D, Wang Y,
Bird TD, Paul SM, Luo W, Colonna M, Baddeley D and Grutzendler J:
TREM2 haplodeficiency in mice and humans impairs the microglia
barrier function leading to decreased amyloid compaction and severe
axonal dystrophy. Neuron. 92:252–264. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang Y, Cella M, Mallinson K, Ulrich JD,
Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S,
Zinselmeyer BH, et al: TREM2 lipid sensing sustains the microglial
response in an Alzheimer's disease model. Cell. 160:1061–1071.
2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Lue LF, Schmitz CT, Serrano G, Sue LI,
Beach TG and Walker DG: TREM2 Protein expression changes correlate
with Alzheimer's disease neurodegenerative pathologies in
post-mortem temporal cortices. Brain Pathol. 25:469–480.
2015.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Cheng Q, Danao J, Talreja S, Wen P, Yin J,
Sun N, Li CM, Chui D, Tran D, Koirala S, et al: TREM2-activating
antibodies abrogate the negative pleiotropic effects of the
Alzheimer's disease variant Trem2R47H on murine myeloid
cell function. J Biol Chem. 293:12620–12633. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Kober DL, Alexander-Brett JM, Karch CM,
Cruchaga C, Colonna M, Holtzman MJ and Brett TJ: Neurodegenerative
disease mutations in TREM2 reveal a functional surface and distinct
loss-of-function mechanisms. Elife. 5(e20391)2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Bailey CC, DeVaux LB and Farzan M: The
triggering receptor expressed on myeloid cells 2 binds
apolipoprotein E. J Biol Chem. 290:26033–26042. 2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Xiang X, Piers TM, Wefers B, Zhu K,
Mallach A, Brunner B, Kleinberger G, Song W, Colonna M, Herms J, et
al: The Trem2 R47H Alzheimer's risk variant impairs splicing and
reduces Trem2 mRNA and protein in mice but not in humans. Mol
Neurodegener. 13(49)2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Cheng-Hathaway PJ, Reed-Geaghan EG, Jay
TR, Casali BT, Bemiller SM, Puntambekar SS, von Saucken VE,
Williams RY, Karlo JC, Moutinho M, et al: The Trem2 R47H variant
confers loss-of-function-like phenotypes in Alzheimer's disease.
Mol Neurodegener. 13(29)2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Yeh FL, Wang Y, Tom I, Gonzalez LC and
Sheng M: TREM2 binds to apolipoproteins, including APOE and
CLU/APOJ, and thereby facilitates uptake of amyloid-beta by
microglia. Neuron. 91:328–340. 2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Song W, Hooli B, Mullin K, Jin SC, Cella
M, Ulland TK, Wang Y, Tanzi RE and Colonna M: Alzheimer's
disease-associated TREM2 variants exhibit either decreased or
increased ligand-dependent activation. Alzheimer Dement.
13:381–387. 2017.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Ghani M, Lang AE, Zinman L, Nacmias B,
Sorbi S, Bessi V, Tedde A, Tartaglia MC, Surace EI, Sato C, et al:
Mutation analysis of patients with neurodegenerative disorders
using NeuroX array. Neurobiol Aging. 36:545.e9–e14. 2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Schlepckow K, Kleinberger G, Fukumori A,
Feederle R, Lichtenthaler SF, Steiner H and Haass C: An
Alzheimer-associated TREM2 variant occurs at the ADAM cleavage site
and affects shedding and phagocytic function. EMBO Mol Med.
9:1356–1365. 2017.PubMed/NCBI View Article : Google Scholar
|