Introduction
According to a report from the World Health
Organization (WHO), 25% of couples in developing countries
experience infertility. Infertility often causes great
psychological strain, which can eventually lead to mental illness
and ~50% of cases of infertility are caused by male factors
(1). Asthenozoospermia (AZS) is
one of the most common diseases that causes male infertility. AZS
is characterized by impaired sperm motility, which prevents sperm
from reaching the egg and leads to infertility (2). However, the pathogenesis of AZS has
remains to be elucidated. A previous study demonstrated that small
noncoding RNAs (ncRNAs), especially piwi-interacting RNAs (piRNAs),
serve key regulatory roles in the pathogenesis of AZS (3).
piRNAs are a unique class of small non-coding
(nc)RNAs that are 26-30 nucleotides in length and guide Piwi
proteins (2). These short ncRNAs
were originally discovered in mammalian testicular cells and have
been proven to be regulatory molecules involved in germ cell
maintenance in mammals (4).
Spermatogenesis is a tightly regulated process that mainly includes
spermatogonia proliferation and spermatogenesis. This process
produces abundant amounts of sperm. Abnormal expression of piRNAs
in testicular cells may lead to the failure of spermatogenesis
(4). Heyn et al (5) found that the expression of five
piRNAs (piR-DQ589977, piR-DQ591415, piR-DQ598918, piR-DQ601291 and
piR-DQ601609) was dysregulated in patients with spermatogenesis
failure and that this dysregulation was correlated with the level
of methylation of the piwi2 and Tudor domain protein 1 (tdrd1) gene
promoters. Bioinformatics analysis of these five piRNAs also
revealed some other potential target genes, such as IL-16,
kallikrein, G protein coupled receptor, histone family and Ras
oncogene family members. Kamaliyan et al (6) observed that the rs508485 mutation in
the hiwi2 gene affected the stability of mRNA or changed the
binding affinity of micro (mi)RNA and resulted in idiopathic
nonobstructive azoospermia. Hong et al (7) identified 5 piRNAs as biomarkers for
the diagnosis of AZS. However, little is known about the expression
patterns and functions of piRNAs in AZS.
Therefore, the present study aimed to determine the
expression patterns and functions of piRNAs in AZS patients.
Bioinformatics analysis was conducted to elucidate the potential
biological functions of piRNAs. These results may provide new
biomarkers for the development of novel diagnostic and therapeutic
strategies for AZS.
Materials and methods
Medical ethics statement
The protocol of the present study was approved by
the Medical Ethics Committee of Nanchang University, the Second
Affiliated Hospital and Jiangxi Maternal and Child Health Hospital
(China; approval no. 2018089). All donors provided written informed
consent for the collection and use of their samples for this
protocol.
Sample collection
The present study selected 194 patients who were
diagnosed with male sterility due to AZS and 143 normal healthy
males who visited the Maternal and Child Health Hospital in Jiangxi
Province between January 2019 and December 2020. A total of four
patient specimens and three normal specimens were randomly selected
for RNA expression analysis and 250 samples (150 AZS patient and
100 normal individuals) and 80 samples (40 AZS patient and 40
normal individuals) were randomly selected for the clinical
verification experiment by reverse transcription-quantitative
(RT-q) PCR assays. Semen samples (6-8 ml) were collected in a dry
and sealed aseptic container. Sperm morphology, concentration,
motility and quantity was used to evaluate sperm quality. According
to the fifth edition of the AZS diagnostic criteria in
2010(8), the proportions of sperm
with fast progressive motility <25% and total progressive
motility <50% in fresh ejaculates were determined. The
characteristics of normal sperm samples were as follows: Semen
concentration ≥15x106/ml, volume ≥2 ml, pH ≥7.2,
progressive motility (PR) ≥32% and PR + nonprogressive motility
(NP) ≥40%. The age range of the AZS patients and the normal
controls was 28-40 years and the groups were well matched in terms
of drinking and smoking habits. Controls affected by fertility
diseases or abnormal sperm quality were excluded from this study.
Patients with gonadal dysplasia due to congenital or inherited
disease were excluded from the study (9). The semen samples were liquefied at
37˚C for 30 min and separated by a discontinuous 45/85% Percoll
density gradient to eliminate contaminating round cells, including
germ cells and white blood cells. The spermatozoa in the lower (85%
Percoll) layer were preserved and washed 3 times with
phosphate-buffered saline (PBS) (10). The quality of the purified sperm
was examined by light microscopy (OLYMPUS, CX43,x200
magnification).
RNA isolation and RT-qPCR
Total RNA was extracted from semen samples by
TRIzol® (Thermo Fisher Scientific, Inc.; cat. no.
15596-018) reagent according to the manufacturer's specifications,
the yield was determined by a NanoDrop 2000 spectrophotometer
(Thermo Fisher Scientific, Inc.) and the integrity was assessed by
agarose gel electrophoresis with ethidium bromide staining.
Quantification was performed with a two-step
reaction process: Reverse transcription (RT) and PCR. Each RT
reaction consisted of 0.5 µg RNA, 5 µl 2XTS miRNA Reaction Mix and
0.5 µl of TransScrip miRNA RT Enzyme Mix (TransGen Biotech, AT351),
in a total volume of 10 µl. Reactions were performed in a GeneAmp
PCR System 9700 (Applied Biosystems; Thermo Fisher Scientific,
Inc.) for 60 min at 37˚C, followed by heat inactivation of RT for 5
sec at 85˚C. The 10 µl RT reaction mix was then diluted x10 in
nuclease-free water and held at -20˚C.
RT-PCR was performed with a LightCycler 480 II
Real-time PCR Instrument (Roche Diagnostics GmbH) in a 10-µl
reaction system that included 1 µl of cDNA, 5 µl of 2x PerfectStart
Green qPCR SuperMix (TransGen Biotech, AQ601), 0.2 µl of 10 µM
universal primers, 0.2 µl of 10 µM miRNA-specific primer and 3.6 µl
of nuclease-free water. The reactions were incubated in a 96-well
plate (Beijing Jiaxinheng Biotechnology Co., Ltd.) at 94˚C for 30
sec, followed by 40 cycles of 95˚C for 10 sec and 65˚C for 35 sec.
Each sample was run in triplicate. At the end of the reaction
cycle, melting curve analysis was performed to detect the product
specificity. The tailing method is used in the reverse
transcription of piRNA, so the quantitative primer only needs to
design the F primer and the R primer is the general primer sequence
of the kit (TransGen Biotech Co., Ltd.; cat. no. AT351-01),
GATCGCCCTTCTACGTCGTAT (TM=58˚C). The piRNA-specific primer
sequences were synthesized by Beijing Qingke Xinye Biotechnology
Co., Ltd. based on the piRNA sequences obtained from the piRNABase
database (regulatoryrna.org/database/piRNA; Release 20.0). 5S
rRNA was used as the reference gene and the piRNA expression levels
were calculated using the 2-ΔΔCq method (11).
Bioinformatics analysis
Briefly, small RNA sequencing and analysis were
conducted by Shanghai Oe Biotech. Co., Ltd. Data from Illumina
HiSeq sequencing are called raw reads. First, the splice sequence
was removed from the raw reads and then small RNA sequences of
different lengths were obtained. Cutadapt (version 1.14) (12), fastx Toolkit (version 0.0.13)
(13) and NGSQCToolkit (version
2.3.2) (14) software were used to
control the quality of these sequences, remove n-base sequences and
sequences with a low Q20 ratio and remove data outside the range of
15-41 NT, which were not included in the subsequent analysis. The
resulting sequences were called clean reads (15) and these reads were further
analyzed. These sequences were matched with known sequences in the
databases of piRBase (v2.0 of piRBase) miRNA (miRbase v19.0;
mirdb.org/) Rfram (version 10.0) (16) and the matched sequences were
considered to be known piRNAs, miRNAs and other small RNAs. piRNAs
were selected for subsequent analysis and functional annotation
(17-19).
Differentially expressed genes (DEGs) between the
AZS patients and the fertile controls were identified using DEG-seq
software (version 1.18.0). Genes with q-values (FDRs) <0.05 and
fold-changes >2.0 were considered differentially expressed
(20,21). The miRanda algorithm (version 3.3a)
was used to predict piRNA target genes and the threshold parameters
were s ≥150, ΔG ≤-30 kcal/mol and strict 5' seed pairing (18,22,23).
Gene Ontology (GO; geneontology.org) functional analysis and the Kyoto
Encyclopedia of Genes and Genomes (KEGG; genome.jp/kegg/) database were used to annotate and
analyze the pathways of the target genes of the different piRNAs.
Significant enrichment items were screened according to a P-value
≤0.05.
Statistical analysis
Differences in demographic variables and expression
levels of piRNA in the AZS group and the fertile control group were
evaluated via the χ2 test. Fisher's exact test and
hypergeometric distribution test were performed to assess the
enrichment significance of GO items or KEGG pathways in the
differentially expressed genes.
To evaluate the specificity and sensitivity of each
piRNA in the diagnostic value of AZS, the ROC curves and AUC were
analyzed. A high Youden's index (sensitivity and specificity-1) was
used to calculate the cut-off value of the optimal diagnostic point
of the ROC curve. Binary logistic regression and ROC curves were
used to improve the diagnostic efficiency. All the statistical
analyses were performed in SPSS (Version 23, SPSS Inc.) and R
software (v 3.5.0; r-project.org/). P<0.05 was considered to indicate
a statistically significant difference.
Results
Patient data analysis and seminal
plasma piRNA profiling
A total of seven semen samples were collected from
AZS patients and male healthy controls according to the WHO
guidelines. The basic information and sperm parameters of the AZS
patients and healthy controls are presented in Table I. There were no distinct
differences in age, BMI and smoking or drinking habits between the
AZS and control groups (P>0.05). However, there were obvious
differences in motility and sperm morphology (P<0.001) between
the AZS and control groups. The semen samples were observed under a
light microscope equipped with a 100x oil objective and were found
to be free of seminiferous epithelium cells, somatic cells and
white blood cells.
 | Table IBasic information and parameters of
AZS and fertile controls. |
Table I
Basic information and parameters of
AZS and fertile controls.
| Characteristic | AZS (194.0) | Fertile controls
(143) | P-value |
|---|
| Age (years) | 34.00±5.3 | 33.5±5.32 | 0.927 |
| BMI | 23.50±3.69 | 24.6±2.79 | 0.923 |
| Smoking | | | 0.87 |
|
Yes | 146 (75.00%) | 107 (75%) | |
|
No | 48 (25%) | 36 (25%) | |
| Drinking | | | 0.96 |
|
Yes | 139 (71%) | 1033 (71%) | |
|
No | 55 (29%) | 40.00 (29%) | |
| Sperm volume
(ml) | 4±0.68 | 3±0.63 | 0.87 |
| pH | 7.6±0.15 | 7.7±0.18 | 0.56 |
| Sperm density
(x106/ml) | 58.59±6.34 | 34.12±5.43 |
<0.001a |
| PR | 16.18±3.2 | 62.55±3.8 |
<0.001a |
| Sperm activity
rate | 22.12±5.64 | 77.34±3.2 |
<0.001a |
Small RNAs were extracted from the semen specimens
of healthy controls and AZS patients and analyzed by Illumina
high-throughput sequencing. First, the splice sequences were
eliminated from the raw reads and small RNA sequences of different
lengths were obtained. After quality control steps, small RNAs in
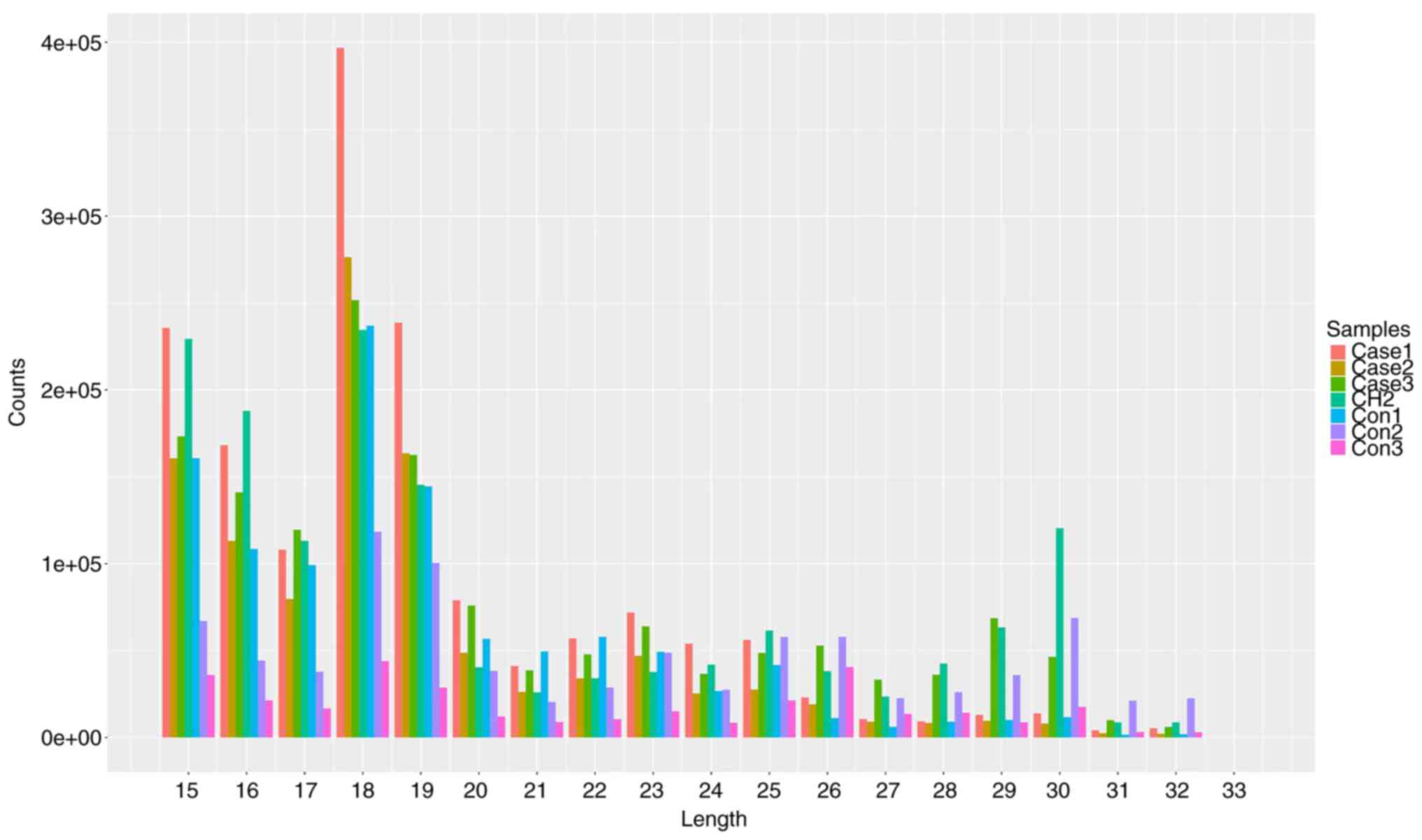
the range of 10 to 45 nucleotides (nt) were obtained. Length
distribution analysis showed that all the samples contained many
RNAs shorter than 45 nt, which was consistent with the lengths of
miRNAs (18-224 nt) and piRNAs (26-330 nt; Fig. 1). In general, piRNAs account for a
part of the total RNA content in seminal plasma. Among the 77
million piRNAs annotated in the piRBase database, 1,585,622 piRNAs
were detected in the healthy control and AZS samples.
Identification of differential piRNA
expression
DEG-seq software was used to analyze the differences
in piRNA expression levels between the AZS and normal groups. A
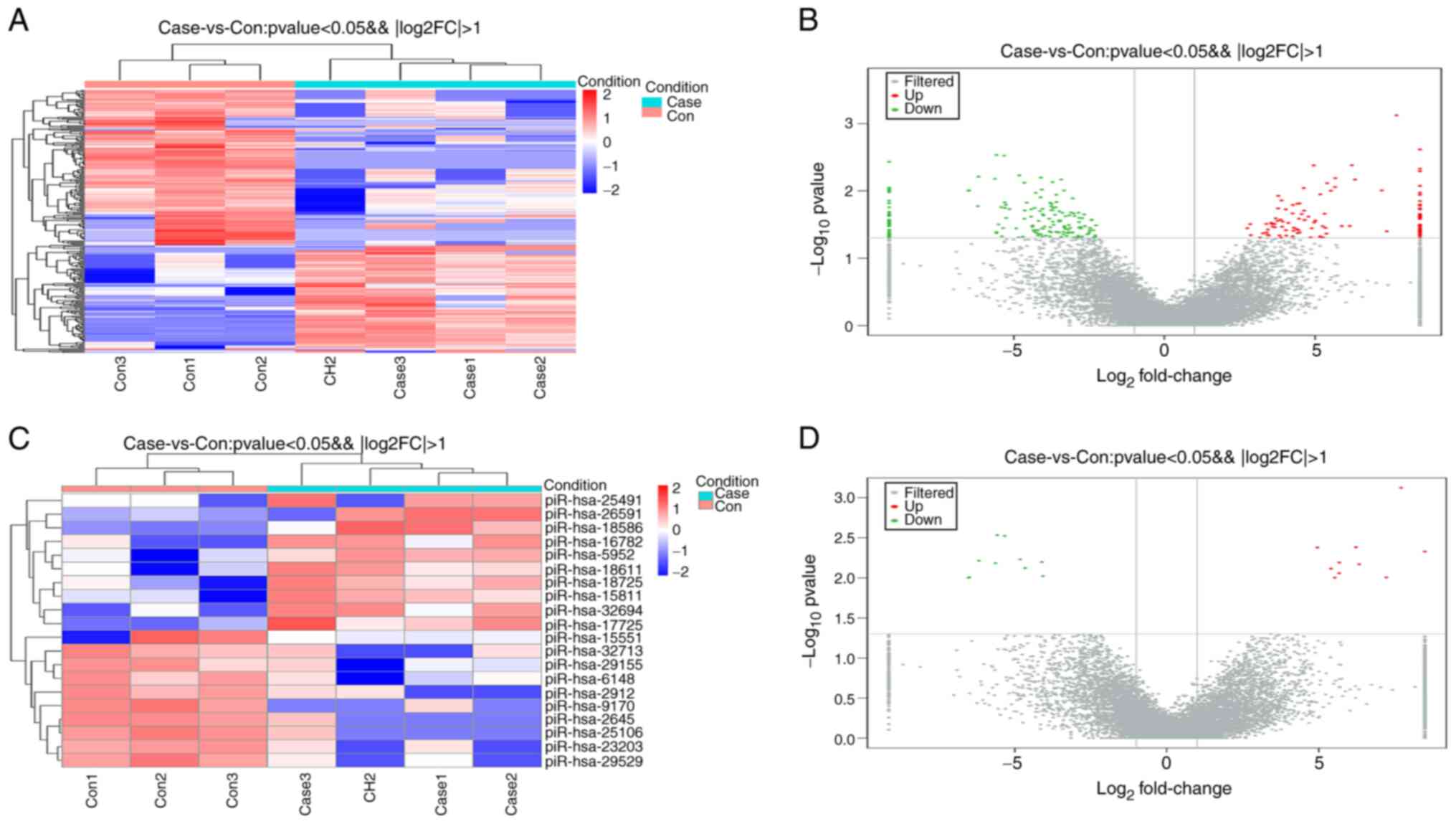
total of 283 differentially expressed piRNAs were obtained with a
significance threshold of |log2FC|≥1 and P-value ≤0.05, including
114 upregulated DEGs and 169 downregulated DEGs. Volcano plots and
heatmaps of the DEGs are shown in Fig.
2A and B. Fig. 2C and D and Table
II present 20 DEpiRNAs, including the top 10 most strongly
upregulated DEpiRNAs and the top 10 most strongly downregulated
DEpiRNAs.
 | Table IIDifferentially expressed genes
between AZS and controls (top 10). |
Table II
Differentially expressed genes
between AZS and controls (top 10).
| Upregulated | Downregulated |
|---|
| Gene | logFC | P-value | Gene | logFC | P-value |
|---|
| piR-26591 | 8.490985 | 0.004693 | piR-15551 | -6.52024 | 0.009999 |
| piR-32694 | 7.712583 | 0.000754 | piR-29529 | -6.49304 | 0.009822 |
| piR-18725 | 7.221809 | 0.009867 | piR-25106 | -6.17803 | 0.006124 |
| piR-16782 | 6.333706 | 0.006759 | piR-2645 | -5.63236 | 0.006583 |
| piR-25491 | 6.224344 | 0.004153 | piR-9170 | -5.57597 | 0.002927 |
| piR-5952 | 5.67597 | 0.006451 | piR-2912 | -5.32921 | 0.003 |
| piR-18611 | 5.669888 | 0.008762 | piR-23203 | -4.82064 | 0.005891 |
| piR-18586 | 5.527972 | 0.009949 | piR-6148 | -4.6561 | 0.007541 |
| piR-15811 | 5.406727 | 0.007682 | piR-32713 | -4.10666 | 0.006346 |
| piR-17725 | 4.956704 | 0.004188 | piR-29155 | -4.07376 | 0.009545 |
| 114 | | | 169 | | |
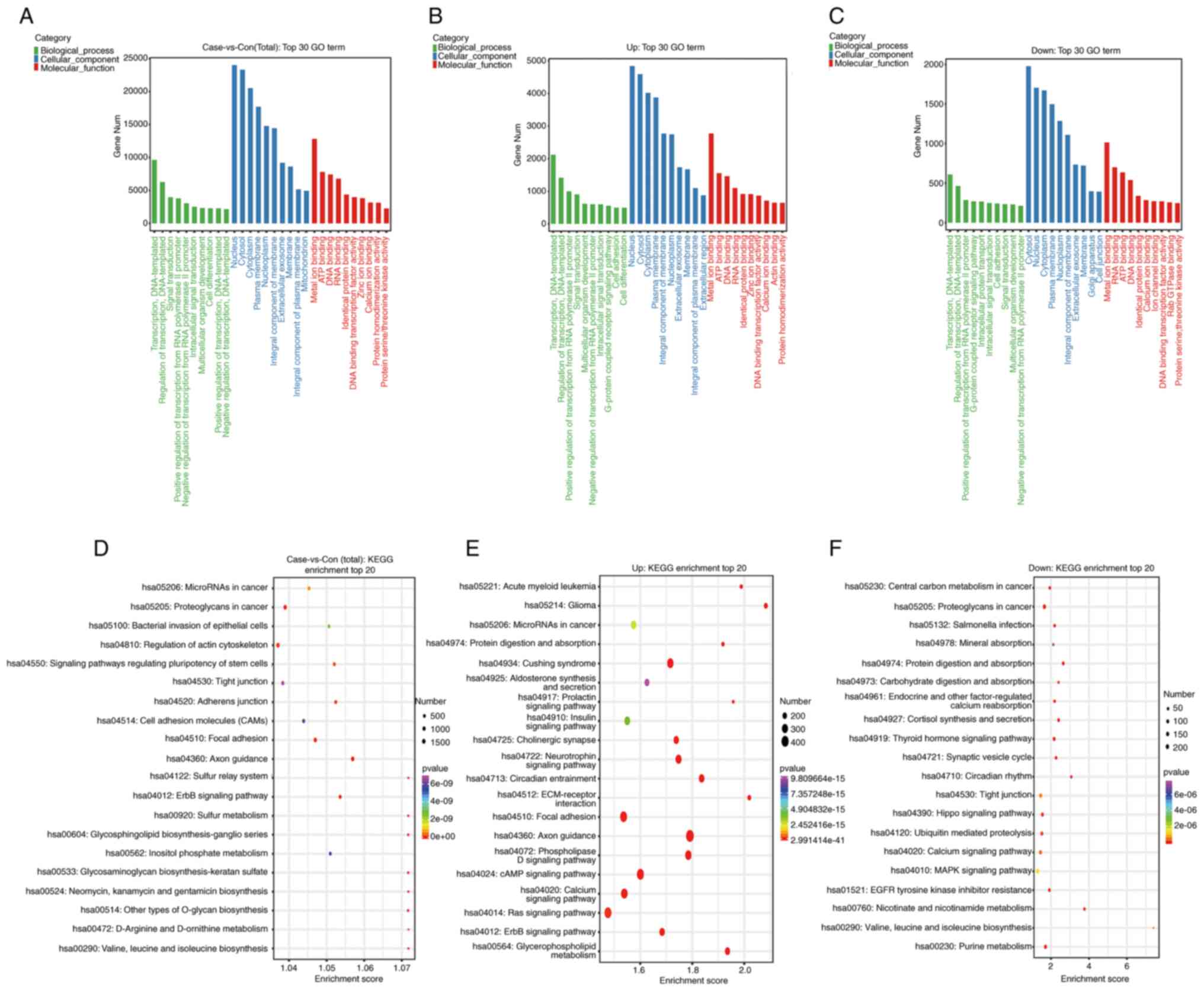
GO and KEGG pathway analysis
GO enrichment analysis was used to explore the
potential functions of these DEpiRNA-target genes. The top 30 most
frequent GO terms are shown in Fig.
3 and Table III. As shown in
Fig. 3A-C, the top 3 cellular
component (CC) terms were nucleus, cytosol and cytoplasm; the top 3
biological process (BP) terms were transcription, regulation of
transcription and signal transduction; and the top 3 molecular
function terms (MF) were metal ion binding, ATP and DNA
binding.
 | Table IIITop 30 GO terms of DEpiRNA-target
genes. |
Table III
Top 30 GO terms of DEpiRNA-target
genes.
| | Biological
process | Cellular
component | Molecular
function |
|---|
| Top 30 GO terms of
total DEpiRNA-target genes | Transcription,
DNA-templated | Nucleus | Metal ion
binding |
| Regulation of
transcription, DNA-templated | Cytosol | ATP binding |
| Signal
transduction | Cytoplasm | DNA binding |
| Positive regulation
of transcription from RNA polymerase II promoter | Plasma
membrane | RNA binding |
| Negative regulation
of transcription from RNA polymerase II promoter | Nucleoplasm | Identical protein
binding |
| Intracellular
signal transduction | Integral component
of membrane | DNA binding
transcription factor activity |
| Multicellular
organism development | Extracellular
exosome | Zinc ion
binding |
| Cell
differentiation | Membrane | Calcium ion
binding |
| Positive regulation
of transcription, DNA-templated | Integral component
of plasma membrane | |
| Protein
homodimerization activity | | |
| Negative regulation
of transcription, DNA-templated | Mitochondrion | Protein
serine/threonine kinase activity |
| Top 30 GO terms of
upregulated | Transcription,
DNA-templated | Nucleus | Metal ion
binding |
| Regulation of
transcription, DNA-templated | Cytosol | |
| DEpiRNA-target
genes | ATP binding | | |
| Positive regulation
of transcription from RNA polymerase II promoter | Cytoplasm | |
| DNA binding | | |
| Signal
transduction | Plasma
membrane | RNA binding |
| Multicellular
organism development | Integral component
of membrane | Identical protein
binding |
| Negative regulation
of transcription from RNA polymerase II promoter | Nucleoplasm | Zinc ion
binding |
| Intracellular
signal transduction | Extracellular
exosome | DNA binding
transcription factor activity |
| G-protein coupled
receptor signaling pathway | Membrane | Calcium ion
binding |
| Cell adhesion | Integral component
of plasma membrane | Actin binding |
| Cell
differentiation | Extracellular
region | Protein
homodimerization activity |
| The top 30 GO terms
of downregulated | Transcription,
DNA-templated | Cytosol | Metal ion
binding |
| Regulation of
transcription, DNA-templated | Nucleus | RNA binding |
| DEpiRNA-target
gene | Positive regulation
of transcription from RNA polymerase II promoter | Cytoplasm | ATP binding |
| G-protein coupled
receptor signaling pathway | Plasma
membrane | DNA binding |
| Intracellular
protein transport | Nucleoplasm | Identical protein
binding |
| Intracellular
signal transduction | Integral component
of membrane | |
| Calcium ion
binding | | |
| Cell adhesion | Extracellular
exosome | Ion channel
binding |
| Signal
transduction | Membrane | DNA binding
transcription factor activity |
| Multicellular
organism development | Golgi
apparatus | Rab GTPase
binding |
| Negative regulation
of transcription from RNA polymerase II promoter | Cell junction | Protein
serine;threonine kinase activity |
KEGG pathway analysis was used to identify all the
pathways and the top 20 most highly enriched pathways related to
the DEpiRNA-target genes (Fig.
3D-F, Table IV). The target
genes enriched among the AZS DEGs were mainly involved in focal
adhesion, actin cytoskeleton, axon guidance, ErbB signaling pathway
and other signal transduction pathways.
 | Table IVTop 20 KEGG pathways of
DEpiRNA-target genes (upregulated and downregulated). |
Table IV
Top 20 KEGG pathways of
DEpiRNA-target genes (upregulated and downregulated).
| Gene item | Top 20 KEGG
pathways |
|---|
| Total
DEpiRNA-target genes | MicroRNAs in
cancer |
| | Proteoglycans in
cancer |
| | Bacterial invasion
of epithelial cells |
| | Regulation of actin
cytoskeleton |
| | Signaling pathways
regulating pluripotency of stem cells |
| | Tight junction |
| | Adherens
junction |
| | Cell adhesion
molecules (CAMs) |
| | Focal adhesion |
| | Axon guidance |
| | Sulfur relay
system |
| | ErbB signaling
pathway |
| | Sulfur
metabolism |
| | Glycosphingolipid
biosynthesis-ganglio series |
| | Inositol phosphate
metabolism |
| | Glycosaminoglycan
biosynthesis-keratan sulfate |
| | Neomycin, kanamycin
and gentamicin biosynthesis |
| | Other types of
O-glycan biosynthesis |
| | D-Arginine and
D-ornithine metabolism |
| | Valine, leucine and
isoleucine biosynthesis |
| Upregulated
DEpiRNA-target genes | MicroRNAs in
cancer |
| | Protein digestion
and absorption |
| | Cushing
syndrome |
| | Aldosterone
synthesis and secretion |
| | Protein digestion
and absorption |
| | Insulin signaling
pathway |
| | Cholinergic
synapse |
| | Neurotrophin
signaling pathway |
| | Circadian
entrainment |
| | ECM-receptor
interaction |
| | Focal adhesion |
| | Axon guidance |
| | Phospholipase D
signaling pathway |
| | cAMP signaling
pathway |
| | Calcium signaling
pathway |
| | Ras signaling
pathway |
| | ErbB signaling
pathway |
| | Glycerophospholipid
metabolism |
| | Acute myeloid
leukemia |
| | Glioma |
| Downregulated
DEpiRNA-target genes | Central carbon
metabolism in cancer |
| | Proteoglycans in
cancer |
| | Salmonella
infection |
| | Mineral
absorption |
| | Protein digestion
and absorption |
| | Carbohydrate
digestion and absorption |
| | Endocrine and other
factor-regulated calcium reabsorption |
| | Cortisol synthesis
and secretion |
| | Thyroid hormone
signaling pathway |
| | Synaptic vesicle
cycle |
| | Circadian
rhythm |
| | Tight junction |
| | Hippo signaling
pathway |
| | Ubiquitin mediated
proteolysis |
| | Calcium signaling
pathway |
| | MAPK signaling
pathway |
| | EGFR tyrosine
kinase inhibitor resistance |
| | Nicotinate and
nicotinamide metabolism |
| | Valine, leucine and
isoleucine biosynthesis |
| | Purine
metabolism |
Validation of piRNAs expression
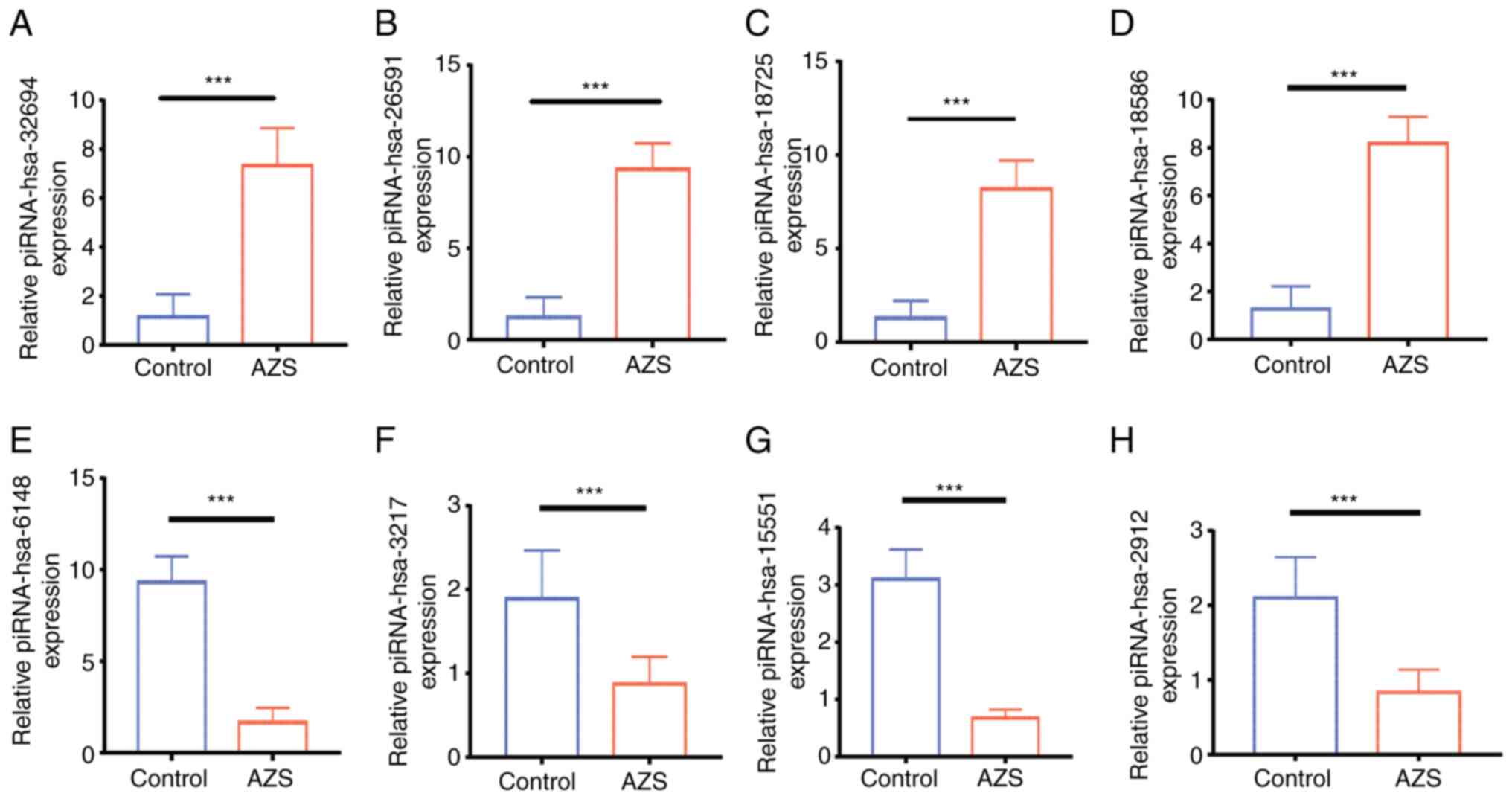
To validate the small RNA-seq results, RT-qPCR
analysis was performed on 8 piRNAs among the DEGs. The present
study selected four upregulated piRNAs (hsa-32694, hsa-26591,
hsa-18725 and hsa-18586) and four downregulated piRNAs (hsa-6148,
hsa-32713, hsa-2912 and hsa-15551) to quantify their expression in
the AZS and normal group semen samples. The primer sequences used
for investigating piRNA expression are listed in Table V. As shown in Fig. 4, the expression of piR-hsa-32694,
piR-hsa-26591, piR-hsa-18725 and piR-hsa-18586 was upregulated in
the AZS group compared with the normal group and the downregulated
expression patterns were also consistent with the small RNA-seq
results.
 | Table VPrimers sequences of the piRNA. |
Table V
Primers sequences of the piRNA.
| Gene symbol
(piRNA) | Primer 5'→3' |
|---|
| piR-hsa-26591 |
GACGAGGTGGCCGAGTGGTT |
| piR-hsa-32694 |
GAACAAGAAGACACTCGTGG |
| piR-hsa-18725 |
GACACTCGTGGAGGCGTCA |
| piR-hsa-18586 |
AGACACTCGTGGAGACGTC |
| piR-hsa-32713 |
CCTAGAAGACTGACTCTTTGC |
| piR-hsa-6148 |
TAGTGGTTATCACTTTCGCCT |
| piR-hsa-2912 |
TCCAACTGCTCTACGACACA |
| piR-hsa-15551 |
TGCGACGAGATGCTGCTTC |
| hsa-5S |
GGAGACCGCCTGGGAATA |
Evaluation of piRNAs as a diagnostic
marker for AZS
Illumina high-throughput sequencing analysis
indicated that the piR-hsa-32694, piR-hsa-26591, piR-hsa-18725 and
piR-hsa-18586 expression levels were obviously different between
AZS patients and controls. Therefore, the diagnostic value of these
four piRNAs as potential biomarkers for AZS was assessed and
evaluated. ROC curve analysis was used to calculate whether
piR-hsa-32694, piR-hsa-26591, piR-hsa-18725 and piR-hsa-18586 could
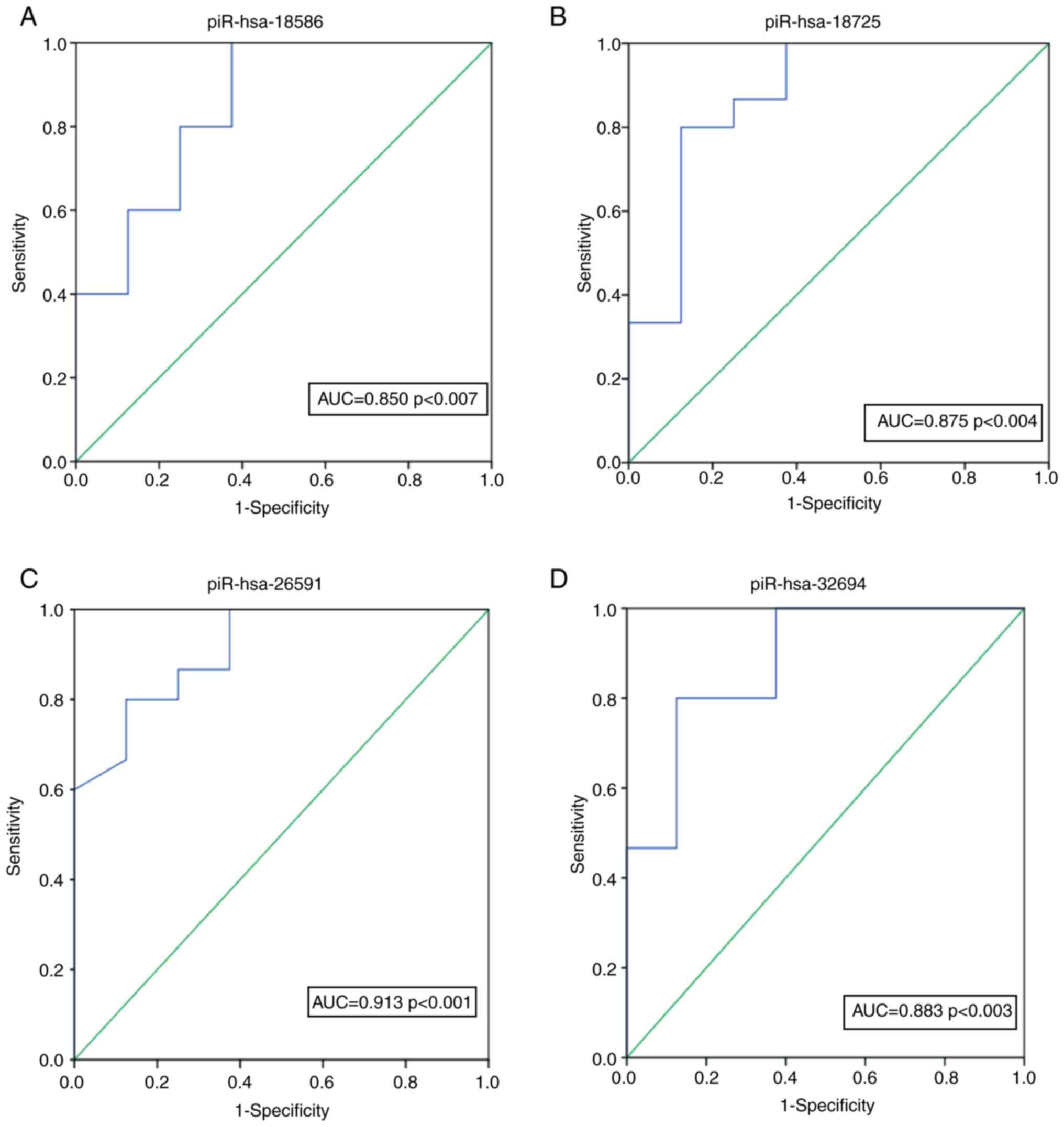
distinguish AZS patients from controls. As shown in Fig. 5, 150 patients with AZS and 100
normal controls were used as a training cohort to build a
diagnostic model using binary logistic regression. The AUCs of
piR-hsa-18586, piR-hsa-18725, piR-hsa-26591 and piR-hsa-32694 were
0.850 (95% CI: 0.861-0.9644 P<0.007), 0.875 (95% CI:
0.761-0.9244 P<0.004), 0.913 (95% CI: 0.751-0.9134 P<0.001)
and 0.883 (95% CI: 0.721-0.9744 P<0.003), respectively. To
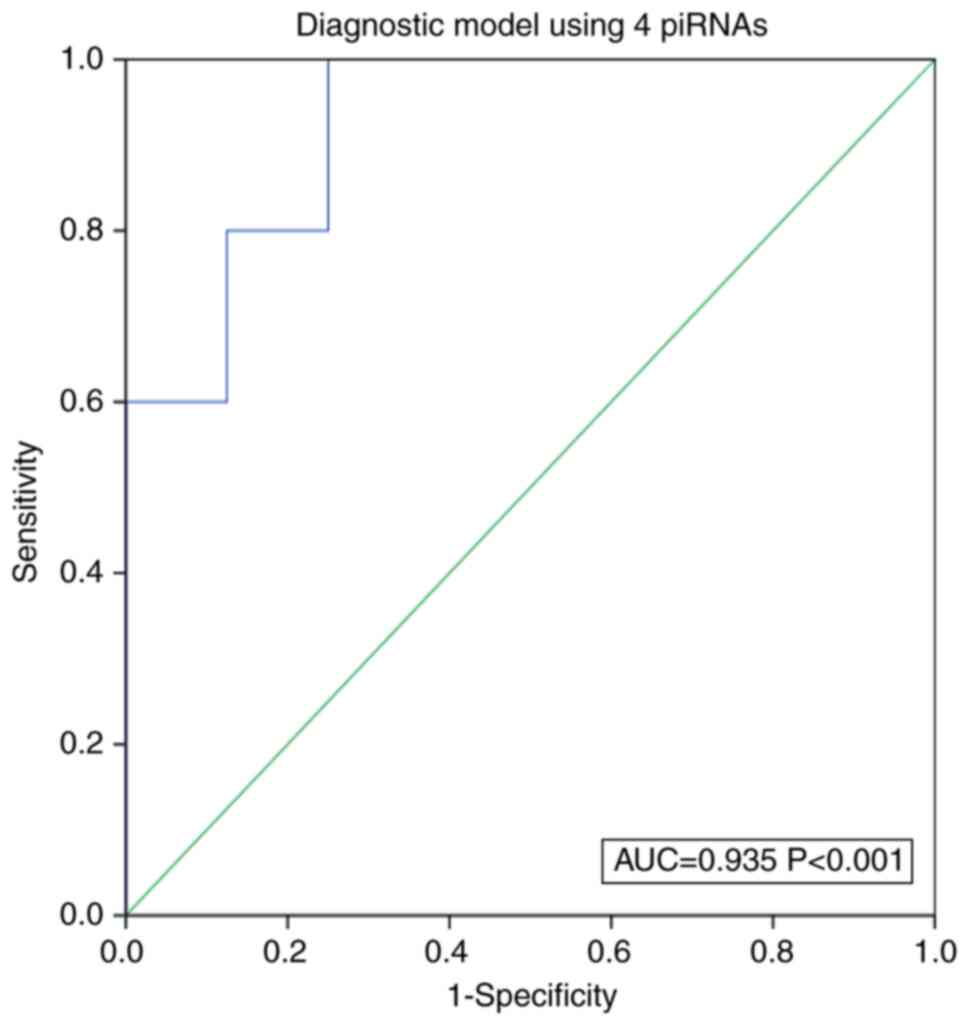
improve the diagnostic power, these four piRNAs were combined as
diagnostic model. An AUC of 0.935 (95% CI: 0.741-0.978; P<0.001)
was observed when the model was applied to a testing cohort (250
specimens mentioned above and the recollected samples of 40
controls and 40 AZS patients) to estimate the diagnostic accuracy
of the joint diagnosis model (Fig.
6). These results showed that the combined diagnostic model
could distinguish AZS patients from controls (specificity=95%,
sensitivity=90%).
Discussion
The present study identified the comprehensive piRNA
expression pattern in the semen samples of AZS patients. The small
RNA sequencing data revealed that a total of 283 significant
DEpiRNAs (114 upregulated and 169 downregulated) were obtained at a
threshold of |log2FC|≥1 and P≤0.05. Further analyses, including
DEpiRNA analysis and GO and KEGG pathway analyses, were performed
to predict the functions of DEpiRNAs, which revealed the critical
roles of piRNAs in regulating spermatogenesis. These sequencing
results were validated by RT-qPCR analysis of eight piRNAs,
specifically, four upregulated piRNAs (piR-hsa-32694,
piR-hsa-26591, piR-hsa-18725 and piR-hsa-18586) and four
downregulated piRNAs (piR-hsa-6148, piR-hsa-32713, piR-hsa-2912 and
piR-hsa-15551). Of these, piR-hsa-32694, piR-hsa-26591,
piR-hsa-18725, piR-hsa-18586 and piR-hsa-2912 produced RT-qPCR
results consistent with the sequencing results, whereas
piR-hsa-6148, piR-hsa-32713 and piR-hsa-15551 did not show
significant differences in expression between the AZS and control
samples. The expression of piR-hsa-32694, piR-hsa-26591,
piR-hsa-18725 and piR-hsa-18586 was significantly upregulated in
patients with AZS. The diagnostic power of these four piRNAs was
further analyzed using ROC analysis and piR-hsa-26591 was found to
be the piRNA with the most potential for diagnosing AZS, with an
AUC of 0.913 (95% CI: 0.795-0.994). A logistic regression model
including all four piRNAs was constructed and ROC analysis revealed
that the combination model achieved good diagnostic efficacy (AUC:
0.935). Therefore, these piRNAs were identified as hub genes that
participate in AZS pathogenesis and could serve as biomarkers for
this disease.
Human spermatogenesis is a carefully coordinated and
precisely regulated biological process that includes rigorous
regulation of gene expression at specific phases (24). ncRNAs, such as circular RNAs, long
non-coding RNAs and miRNAs, are critical regulators of gene
transcription and translation in different phases of
spermatogenesis (25-27).
piRNAs are small ncRNAs that are mainly expressed in human germ
cells. piRNAs can maintain cell genome integrity, inhibit
transposon transcription and translation and participate in cell
heterochromatin formation, epigenetic regulation and mammalian germ
cell genesis (28). To date, there
have been few reports describing the changes in the piRNA profile
in human AZS patients. In light of the role of piRNAs in male
reproduction, the changes in the expression profile of piRNAs in
sperm from patients with AZS was studied and the the differential
expression of piRNAs in AZS patients and controls analyzed to
explore the unknown pathological mechanism of AZS.
Studies have shown that piRNAs affect gene silencing
mainly by binding to Piwi subfamily proteins of the Ago/Piwi
protein family. Human piwi family proteins, PIWIL1, PIWIL2, PIWIL3
and PIWIL4 are expressed in testis tissues (1,3,4). Dai
et al (29) showed that a
piwi mutation is pathogenic, causing male infertility and
established a clear relationship between the human piwi gene and
male infertility. In addition to its main functions in transposon
silencing, the piwi/piRNA mechanism affects the degradation of
generous mRNA transcripts in various germ cells through miRNA-
or/and short interfering RNA-like mechanisms (30,31).
The present study predicted the target genes of piRNAs that were
differentially expressed between the control group and the AZS
group to explore how the DEpiRNAs might alter sperm function. In
addition, enrichment analysis based on these predicted target genes
showed that cell macromolecule biosynthesis, spermatogenesis, male
gametogenesis, cytoplasmic transport and RNA metabolism were
altered in the AZS group. Enrichment pathway analysis showed that
the DEpiRNA target genes were involved in focal adhesion kinase
(FAK), actin cytoskeleton, axon guidance, the ErbB signaling
pathway and other signal transduction pathways. A notable study
showed that FAK coordinates the transport of spermatozoa and
prealbumin spermatids through the epithelium along the functional
axis of the acroplasmic thinning-blood testicular barrier basement
membrane (32). A study of pig
sperm found that altered expression of many mRNAs is associated
with infertility and involved in metabolic processes. Specifically,
these altered genes ensure the production of fertile sperm by
promoting 26S proteasome exchange, nucleoprotein digestion and
cytoplasmic residue absorption and supporting cell phagocytosis to
eliminate damaged RNA (33,34).
Through the analysis of animal models and human patient data,
López-Lemus et al (35)
found that testis (germinal epithelium) is one of the organs
damaged by the multisystem effect of Nonalcoholic fatty liver
disease (NAFLD). Previous studies on animal models reported that
NAFLD does not affect sperm morphology by reducing testosterone
synthesis (35,36). A study of the European eel
(Anguilla anguilla) shows that the liver seems to serve a
role in determining the number of sperm by producing some fatty
acids (37). The results of the
present study also supported the hypothesis that the metabolism of
FAK, fatty acids and other macromolecules affects sperm development
and maturation.
Previous reports identified mRNAs that are related
to sperm motility, particularly transcripts encoding
flagella-associated protein 45 (CFAP45) and bromodomain-containing
2 (BRD2). The content of CFAP45 in low motile power sperm is
significantly higher compared with that in high motile power sperm,
while the content of BRD2 mRNA in sperm from AZS patients is
significantly lower (38,39). This indicates that low sperm
motility could be attributed to the abnormal expression of CFAP45
and BRD2. The present study found that the expression of four
piRNAs, piR-hsa-32694, piR-hsa-26591, piR-hsa-18725 and
piR-hsa-18586, was significantly upregulated in AZS patients.
Furthermore, the ROC curve analysis revealed that the AUC of the
combination of the four piRNAs was equal to 0.935 and demonstrated
93.8% sensitivity and 89.3% specificity. These results suggested
that these four piRNAs were significantly and independently
associated with sperm motility and male infertility. Bioinformatics
analysis showed that CAFP45 and BRD2 were the potential target
genes of piR-hsa-26591. Whether piR-hsa-26591 silences the
expression of CAFP45 and BRD2 and leads to decreased sperm motility
and infertility in AZS patients requires further study. Overall,
the present study may provide insights into the use of human sperm
cell-specific piRNAs as biomarkers for AZS screening. The present
study has some limitations. First, the small number of samples
limited the statistical power of the microarray analysis. Second,
it did not use a luciferase reporter assay to further verify the
relationship between piR-hsa-26591 and CAFP45.
In summary, the present study identified a piRNA
expression profile and defined the potential function of DEpiRNAs
in AZS. Moreover, the findings also provide some information
regarding the pathological mechanisms of male infertility.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the Provincial Natural
Science Foundation of Jiangxi (grant nos. 20181BAB205024 and
20181BAB205013), Project of Jiangxi Provincial Health Department
(grant nos. 20181075, 20181084 and 2017A100), Science and
Technology Research Project of Jiangxi Provincial Department of
Education (grant nos. 20160165, 171353, 170096 and 170014) and
Graduate Innovation Fund Project of Jiangxi Province (grant no.
YC2012-B011).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
All the authors were involved in the conception and
design of the study and data interpretation. LH and HC performed
the experiments, XW and RW analyzed the data and PG, WH and WS
helped perform the experiments. LH prepared the manuscript. HC
oversaw the project and proofread the manuscript. LH and HC confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The protocol of the present study was approved by
the Medical Ethics Committee of Nanchang University, the Second
Affiliated Hospital and Jiangxi Maternal and Child Health Hospital
(China; approval no. 2018089). All donors provided informed consent
for the collection and use of their samples for this protocol.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vander Borght M and Wyns C: Fertility and
infertility: Definition and epidemiology. Clin Biochem. 62:2–10.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wu X, Pan Y, Fang Y, Zhang J, Xie M, Yang
F, Yu T, Ma P, Li W and Shu Y: The biogenesis and functions of
piRNAs in human diseases. Mol Ther Nucleic Acids. 21:108–20.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ozata DM, Gainetdinov I, Zoch A, O'Carroll
D and Zamore PD: PIWI-interacting RNAs: Small RNAs with big
functions. Nat Rev Genet. 20:89–108. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Charlesworth AG, Nitschko V, Renaud MS and
Claycomb JM: PIWI puts spermatogenesis in its place. Dev Cell.
57:149–151. 2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Heyn H, Ferreira HJ, Bassas L, Bonache S,
Sayols S, Sandoval J, Esteller M and Larriba S: Epigenetic
disruption of the PIWI pathway in human spermatogenic disorders.
PLoS One. 7(e47892)2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kamaliyan Z, Pouriamanesh S, Soosanabadi
M, Gholami M and Mirfakhraie R: Investigation of piwi-interacting
RNA pathway genes role in idiopathic non-obstructive azoospermia.
Sci Rep. 8(142)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hong Y, Wang C, Fu Z, Liang H, Zhang S, Lu
M, Sun W, Ye C, Zhang CY, Zen K, et al: Systematic characterization
of seminal plasma piRNAs as molecular biomarkers for male
infertility. Sci Rep. 6(24229)2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lu JC, Huang YF and Lü N: WHO laboratory
manual for the examination and processing of human semen: Its
applicability to andrology laboratories in China. Zhonghua Nan Ke
Xue. 16:867–871. 2010.PubMed/NCBI(In Chinese).
|
|
9
|
None. WHO laboratory manual for the
examination of human semen and sperm-cervical mucus interaction. J
Androl. 17(442)1996.
|
|
10
|
Satoru K, Shigeru O, Kiyoshi K, Toshifumi
K, Hideo M and Rihachi I: Purification of human sperm by a
discontinuous Percoll density gradient with an innercolumn. Biol
Reprod. 35:1059–1063. 1986.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Martin M: Cutadapt removes adapter
sequences from high-throughput sequencing reads. EMBnet J.
17:10–12. 2011.
|
|
13
|
Gordon A and Hannon GJ: Fastx-toolkit.
FASTQ/A short-reads pre-processing tools, 2010.
http://hannonlabcshl.edu/fastx_toolkit.
|
|
14
|
Patel RK and Jain M: NGS QC toolkit: A
toolkit for quality control of next generation sequencing data.
PLoS One. 7(e30619)2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Fu Y, He W, Zhou C, Fu X, Wan Q, He L and
Wei B: Bioinformatics analysis of circRNA expression and
construction of ‘circRNA-miRNA-mRNA’ competing endogenous RNAs
networks in bipolar disorder patients. Front Genet.
12(718976)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang J, Zhang P, Lu Y, Li Y, Zheng Y, Kan
Y, Chen R and He S: piRBase: A comprehensive database of piRNA
sequences. Nucleic Acids Res. 47:D175–D180. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34 (Database
Issue):D140–D144. 2006.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Griffiths-Jones S, Bateman A, Marshall M,
Khanna A and Eddy SR: Rfam: An RNA family database. Nucleic Acids
Res. 31:439–441. 2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tino P: Basic properties and information
theory of Audic-Claverie statistic for analyzing cDNA arrays. BMC
Bioinformatics. 10(310)2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2(e363)2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5(R1)2003.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Alexa A and Rahnenfuhrer J: topGO:
Enrichment analysis for gene ontology. R package version.
28:2013.
|
|
25
|
Manfrevola F, Chioccarelli T, Cobellis G,
Fasano S, Ferraro B, Sellitto C, Marella G, Pierantoni R and
Chianese R: CircRNA role and circRNA-dependent network (ceRNET) in
asthenozoospermia. Front Endocrinol (Lausanne).
11(395)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lu H, Xu D, Wang P, Sun W, Xue X, Hu Y,
Xie C and Ma Y: RNA-sequencing and bioinformatics analysis of long
noncoding RNAs and mRNAs in the asthenozoospermia. Biosci Rep.
40(BSR20194041)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Heidary Z, Zaki-Dizaji M, Saliminejad K
and Khorram Khorshid HR: MicroRNA profiling in spermatozoa of men
with unexplained asthenozoospermia. Andrologia.
51(e13284)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Larriba E and Del Mazo J: An integrative
piRNA analysis of mouse gametes and zygotes reveals new potential
origins and gene regulatory roles. Sci Rep. 8(12832)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Dai P, Wang X and Liu MF: A dual role of
the PIWI/piRNA machinery in regulating mRNAs during mouse
spermiogenesis. Sci China Life Sci. 63:447–449. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gou LT, Kang JY, Dai P, Wang X, Li F, Zhao
S, Zhang M, Hua MM, Lu Y, Zhu Y, et al: Ubiquitination-deficient
mutations in human piwi cause male infertility by impairing
histone-to-protamine exchange during spermiogenesis. Cell.
169:1090–1104.e13. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hasuwa H, Ishino K and Siomi H: Human PIWI
(HIWI) is an azoospermia factor. Sci China Life Sci. 61:348–350.
2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Xiao X, Mruk DD, Wong CK and Cheng CY:
Germ cell transport across the seminiferous epithelium during
spermatogenesis. Physiology (Bethesda). 29:286–298. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yang CC, Lin YS, Hsu CC, Tsai MH, Wu SC
and Cheng WT: Seasonal effect on sperm messenger RNA profile of
domestic swine (Sus Scrofa). Anim Reprod Sci. 119:76–84.
2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Percipalle P: New insights into
co-transcriptional sorting of mRNA for cytoplasmic transport during
development. Semin Cell Dev Biol. 32:55–62. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
López-Lemus UA, Garza-Guajardo R,
Barboza-Quintana O, Rodríguez-Hernandez A, García-Rivera A,
Madrigal-Pérez VM, Guzmán-Esquivel J, García-Labastida LE,
Soriano-Hernández AD, Martínez-Fierro ML, et al: Association
between nonalcoholic fatty liver disease and severe male
reproductive organ impairment (germinal epithelial loss): Study on
a mouse model and on human patients. Am J Mens Health. 12:639–648.
2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Li Y, Liu L, Wang B, Xiong J, Li Q, Wang J
and Chen D: Impairment of reproductive function in a male rat model
of non-alcoholic fatty liver disease and beneficial effect of N-3
fatty acid supplementation. Toxicol Lett. 222:224–232.
2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Baeza R, Mazzeo I, Vílchez MC, Gallego V,
Peñaranda DS, Pérez L and Asturiano JF: Relationship between sperm
quality parameters and the fatty acid composition of the muscle,
liver and testis of European eel. Comp Biochem Physiol A Mol Integr
Physiol. 181:79–86. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Dougherty GW, Mizuno K, Nöthe-Menchen T,
Ikawa Y, Boldt K, Ta-Shma A, Aprea I, Minegishi K, Pang YP,
Pennekamp P, et al: CFAP45 deficiency causes situs abnormalities
and asthenospermia by disrupting an axonemal adenine nucleotide
homeostasis module. Nat Commun. 11(5520)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Jodar M, Kalko S, Castillo J, Ballescà JL
and Oliva R: Differential RNAs in the sperm cells of
asthenozoospermic patients. Hum Reprod. 27:1431–1438.
2012.PubMed/NCBI View Article : Google Scholar
|