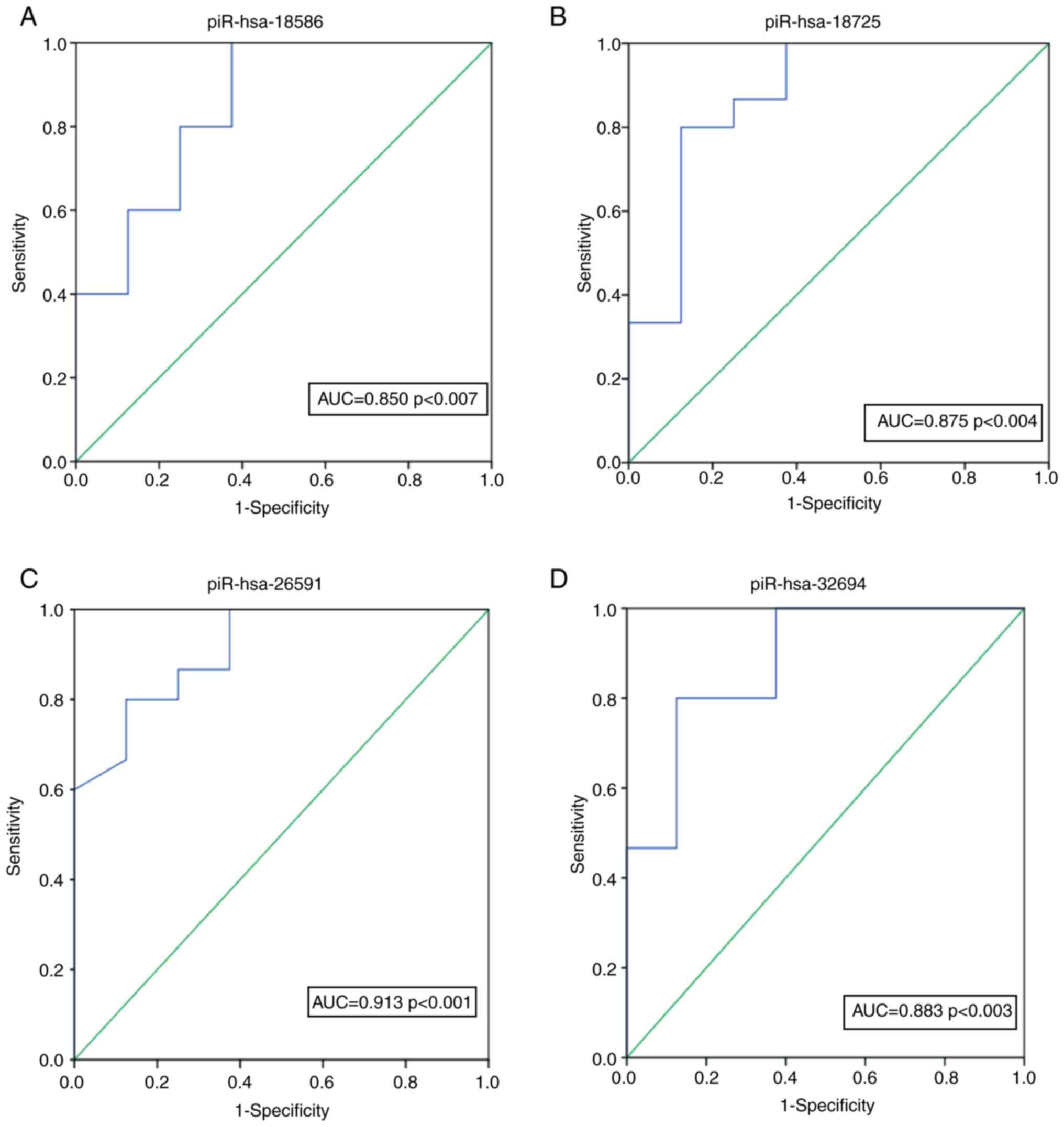
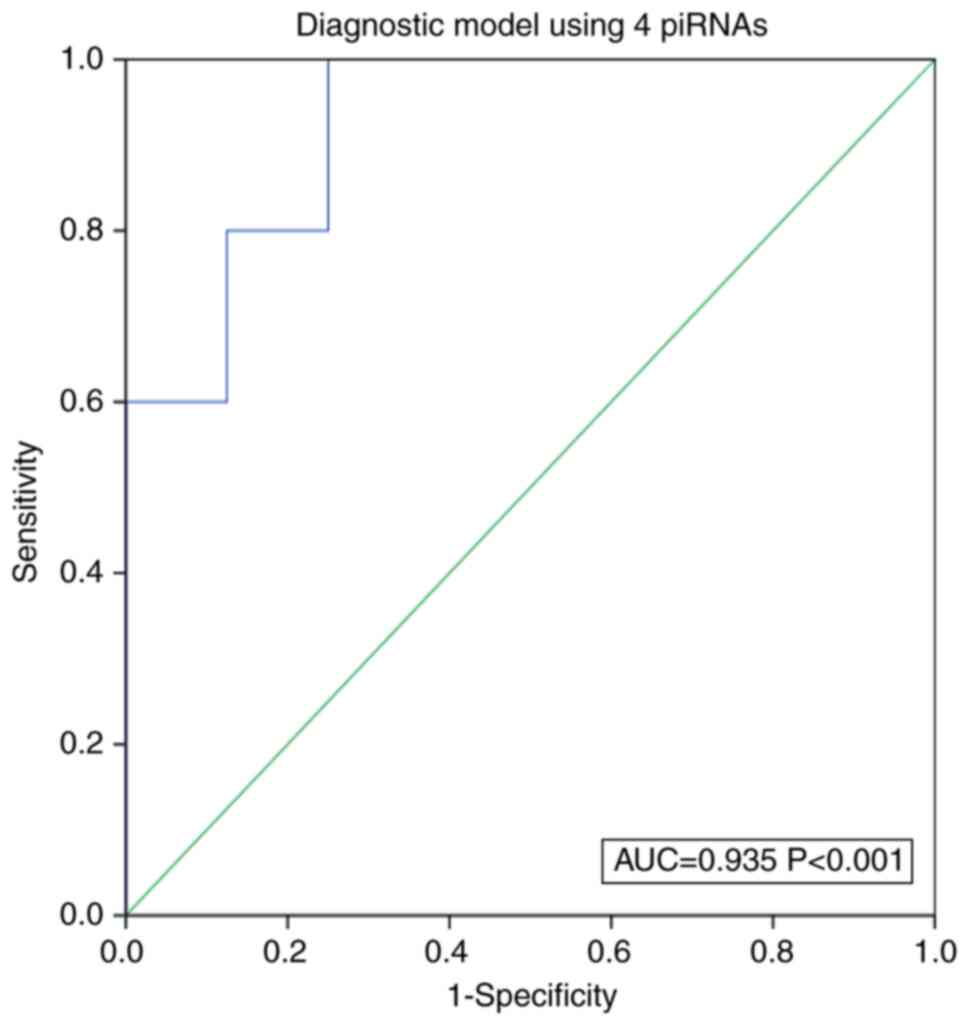
|
1
|
Vander Borght M and Wyns C: Fertility and
infertility: Definition and epidemiology. Clin Biochem. 62:2–10.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wu X, Pan Y, Fang Y, Zhang J, Xie M, Yang
F, Yu T, Ma P, Li W and Shu Y: The biogenesis and functions of
piRNAs in human diseases. Mol Ther Nucleic Acids. 21:108–20.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ozata DM, Gainetdinov I, Zoch A, O'Carroll
D and Zamore PD: PIWI-interacting RNAs: Small RNAs with big
functions. Nat Rev Genet. 20:89–108. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Charlesworth AG, Nitschko V, Renaud MS and
Claycomb JM: PIWI puts spermatogenesis in its place. Dev Cell.
57:149–151. 2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Heyn H, Ferreira HJ, Bassas L, Bonache S,
Sayols S, Sandoval J, Esteller M and Larriba S: Epigenetic
disruption of the PIWI pathway in human spermatogenic disorders.
PLoS One. 7(e47892)2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kamaliyan Z, Pouriamanesh S, Soosanabadi
M, Gholami M and Mirfakhraie R: Investigation of piwi-interacting
RNA pathway genes role in idiopathic non-obstructive azoospermia.
Sci Rep. 8(142)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hong Y, Wang C, Fu Z, Liang H, Zhang S, Lu
M, Sun W, Ye C, Zhang CY, Zen K, et al: Systematic characterization
of seminal plasma piRNAs as molecular biomarkers for male
infertility. Sci Rep. 6(24229)2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lu JC, Huang YF and Lü N: WHO laboratory
manual for the examination and processing of human semen: Its
applicability to andrology laboratories in China. Zhonghua Nan Ke
Xue. 16:867–871. 2010.PubMed/NCBI(In Chinese).
|
|
9
|
None. WHO laboratory manual for the
examination of human semen and sperm-cervical mucus interaction. J
Androl. 17(442)1996.
|
|
10
|
Satoru K, Shigeru O, Kiyoshi K, Toshifumi
K, Hideo M and Rihachi I: Purification of human sperm by a
discontinuous Percoll density gradient with an innercolumn. Biol
Reprod. 35:1059–1063. 1986.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Martin M: Cutadapt removes adapter
sequences from high-throughput sequencing reads. EMBnet J.
17:10–12. 2011.
|
|
13
|
Gordon A and Hannon GJ: Fastx-toolkit.
FASTQ/A short-reads pre-processing tools, 2010.
http://hannonlabcshl.edu/fastx_toolkit.
|
|
14
|
Patel RK and Jain M: NGS QC toolkit: A
toolkit for quality control of next generation sequencing data.
PLoS One. 7(e30619)2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Fu Y, He W, Zhou C, Fu X, Wan Q, He L and
Wei B: Bioinformatics analysis of circRNA expression and
construction of ‘circRNA-miRNA-mRNA’ competing endogenous RNAs
networks in bipolar disorder patients. Front Genet.
12(718976)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang J, Zhang P, Lu Y, Li Y, Zheng Y, Kan
Y, Chen R and He S: piRBase: A comprehensive database of piRNA
sequences. Nucleic Acids Res. 47:D175–D180. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34 (Database
Issue):D140–D144. 2006.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Griffiths-Jones S, Bateman A, Marshall M,
Khanna A and Eddy SR: Rfam: An RNA family database. Nucleic Acids
Res. 31:439–441. 2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tino P: Basic properties and information
theory of Audic-Claverie statistic for analyzing cDNA arrays. BMC
Bioinformatics. 10(310)2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2(e363)2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5(R1)2003.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Alexa A and Rahnenfuhrer J: topGO:
Enrichment analysis for gene ontology. R package version.
28:2013.
|
|
25
|
Manfrevola F, Chioccarelli T, Cobellis G,
Fasano S, Ferraro B, Sellitto C, Marella G, Pierantoni R and
Chianese R: CircRNA role and circRNA-dependent network (ceRNET) in
asthenozoospermia. Front Endocrinol (Lausanne).
11(395)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lu H, Xu D, Wang P, Sun W, Xue X, Hu Y,
Xie C and Ma Y: RNA-sequencing and bioinformatics analysis of long
noncoding RNAs and mRNAs in the asthenozoospermia. Biosci Rep.
40(BSR20194041)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Heidary Z, Zaki-Dizaji M, Saliminejad K
and Khorram Khorshid HR: MicroRNA profiling in spermatozoa of men
with unexplained asthenozoospermia. Andrologia.
51(e13284)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Larriba E and Del Mazo J: An integrative
piRNA analysis of mouse gametes and zygotes reveals new potential
origins and gene regulatory roles. Sci Rep. 8(12832)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Dai P, Wang X and Liu MF: A dual role of
the PIWI/piRNA machinery in regulating mRNAs during mouse
spermiogenesis. Sci China Life Sci. 63:447–449. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gou LT, Kang JY, Dai P, Wang X, Li F, Zhao
S, Zhang M, Hua MM, Lu Y, Zhu Y, et al: Ubiquitination-deficient
mutations in human piwi cause male infertility by impairing
histone-to-protamine exchange during spermiogenesis. Cell.
169:1090–1104.e13. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hasuwa H, Ishino K and Siomi H: Human PIWI
(HIWI) is an azoospermia factor. Sci China Life Sci. 61:348–350.
2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Xiao X, Mruk DD, Wong CK and Cheng CY:
Germ cell transport across the seminiferous epithelium during
spermatogenesis. Physiology (Bethesda). 29:286–298. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yang CC, Lin YS, Hsu CC, Tsai MH, Wu SC
and Cheng WT: Seasonal effect on sperm messenger RNA profile of
domestic swine (Sus Scrofa). Anim Reprod Sci. 119:76–84.
2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Percipalle P: New insights into
co-transcriptional sorting of mRNA for cytoplasmic transport during
development. Semin Cell Dev Biol. 32:55–62. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
López-Lemus UA, Garza-Guajardo R,
Barboza-Quintana O, Rodríguez-Hernandez A, García-Rivera A,
Madrigal-Pérez VM, Guzmán-Esquivel J, García-Labastida LE,
Soriano-Hernández AD, Martínez-Fierro ML, et al: Association
between nonalcoholic fatty liver disease and severe male
reproductive organ impairment (germinal epithelial loss): Study on
a mouse model and on human patients. Am J Mens Health. 12:639–648.
2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Li Y, Liu L, Wang B, Xiong J, Li Q, Wang J
and Chen D: Impairment of reproductive function in a male rat model
of non-alcoholic fatty liver disease and beneficial effect of N-3
fatty acid supplementation. Toxicol Lett. 222:224–232.
2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Baeza R, Mazzeo I, Vílchez MC, Gallego V,
Peñaranda DS, Pérez L and Asturiano JF: Relationship between sperm
quality parameters and the fatty acid composition of the muscle,
liver and testis of European eel. Comp Biochem Physiol A Mol Integr
Physiol. 181:79–86. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Dougherty GW, Mizuno K, Nöthe-Menchen T,
Ikawa Y, Boldt K, Ta-Shma A, Aprea I, Minegishi K, Pang YP,
Pennekamp P, et al: CFAP45 deficiency causes situs abnormalities
and asthenospermia by disrupting an axonemal adenine nucleotide
homeostasis module. Nat Commun. 11(5520)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Jodar M, Kalko S, Castillo J, Ballescà JL
and Oliva R: Differential RNAs in the sperm cells of
asthenozoospermic patients. Hum Reprod. 27:1431–1438.
2012.PubMed/NCBI View Article : Google Scholar
|