1. Introduction
DNA represents the main molecule that incorporates
the genetic information of the human cells independent of the organ
tissue. It is well known that this molecule, consisting of a
specific nucleotide sequence, forms the nuclear chromatin in a
cell. The functional unit of chromatin is called a nucleosome,
whose structure consists of a DNA double helix and adjacent histone
proteins. Furthermore, all individual variations of genes form the
genotype, which is the same in all cell types within an organism.
The interactions between the genotype and the environment form
variable phenotypes, depending on the cell type (1).
As a result, different gene suppression and
activation mechanisms determine consistent phenotypic differences
between cells from different organs and even within the same
organ.
Subsequently, modifying the transcriptional
potential of DNA without changing its sequence or genetic
information, will change the chromatin tertiary structure, due to
histone-DNA interaction. However, this process will not change the
amino-acid sequence in the polypeptide chain (this represents the
unchanged genetic information). Remodeling of the chromatin
conformation, epigenetically, at the nuclear level, results in the
synthesis of the mRNA species or in suppressing this
transcriptional process (1).
Furthermore, in cytoplasm, the mRNA transcripts are controlled by
other epigenetic factors: The microRNA (miRNA or miR) species, the
transcripts of lesser dimensions (2-22 nucleotides), of the
noncoding repetitive DNA. The final product, the polypeptide chain,
may therefore be translated or not, depending on the impact of such
a post-transcriptional interference RNA network. Finally, there are
certain active proteins such as enzymes, for example, which may be
formed only following the post-translational covalent modifications
of the polypeptide chains (2).
All these processes are examples of natural,
physiological, epigenetic regulations of gene function or
expression. Epigenetics studies the variation of gene expression
that is independent of genetic information or nucleotide sequences.
It refers to gene function control through both nuclear chromatin
covalent modification and remodeling and cytoplasmic activity of
the interference RNA involving miRNAs along with post-translational
covalent modification of the newly synthetized polypeptides
(1).
Various epigenetic mechanisms could explain how a
static genome interacts with a dynamic environment. According to
Cold Spring Harbor Meeting-2009, ‘an epigenetic trait’ represents a
term designed to define ‘a stably heritable phenotype resulting
from changes in a chromosome without alterations in the DNA
sequence’ (3). Over time, different
definitions of epigenetics were suggested. Waddington was the first
to introduce the term epigenetics in 1942, as a variety of
mechanisms which promotes gene expression changes without DNA
mutation (4). In 1968, he defined
epigenesis as ‘the branch of biology which studies the causal
interactions between genes and their products which bring the
phenotype into being’ (5).
The current view of epigenetics includes the
following processes: i) Active DNA methylation, demethylation,
which imply changes in DNA tertiary structure; ii) also
post-translational histone covalent changes
(methylation/demethylation, acetylation/deacetylation,
ubiquitination), that cause broader and complex fluctuations in
DNA-histone interactions; iii) synthesis of small, non-coding RNA
molecules, called miRNAs, that modulate translational potential of
transcripts (mRNAs) at the ribosomal level; and iv)
post-translational modification of polypeptide chains (1).
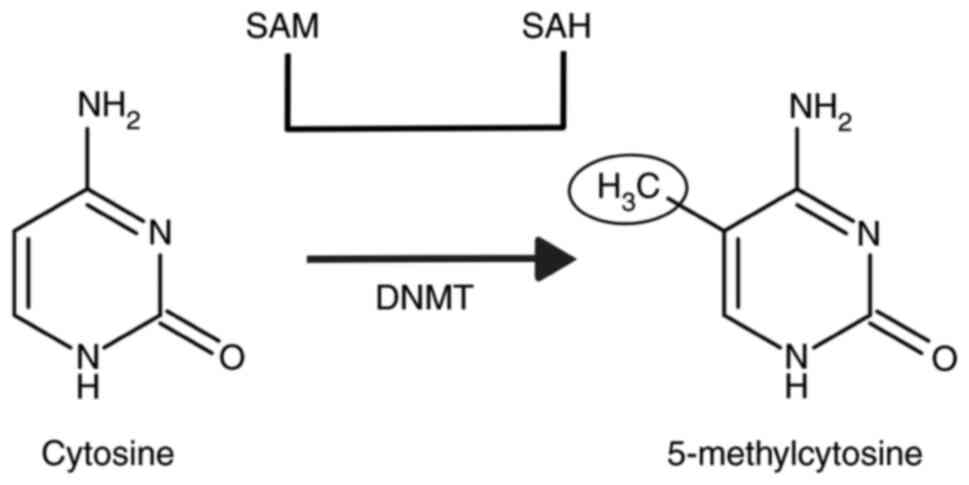
DNA methylation involves the addition of a methyl
group to a cytosine major base through a covalent interaction,
particularly at 5'-CpG-3' dinucleotide sites in DNA substrates. If
this change is symmetrical in both DNA chains, then the DNA
conformational structure will change and the replication will be
delayed (6). Usually there are
specific gene areas rich in 5'-CpG-3' nucleotides, which can be
preferentially methylated. These areas are called ‘CpG islands’, in
the case of the housekeeping genes (which have to stay active
independent of the cell types and the CpG sites should not be
methylated) (7). The extensive
methylation of the CpG repetitive regions is linked to chromatin
silencing in genes presenting tissue type expression. Over 60% of
the genes in the human genome have a high proportion of CpG
dinucleotides in the promoter region, thus being potentially
influenced by this epigenetic mechanism (8).
DNA methylation is controlled by enzymes that
transfer methyl groups from the methyl-donor, S-adenosil-methionine
(SAM), to cytosine. They are called DNA N-methyl transferases
(DNMT) (Fig. 1).
This epigenetic process is carried out by different
isoforms of DNMT: DNMT 1 has a specific role in maintaining the
pre-existent methylation pattern, while DNMT 3a and DNMT 3b promote
de novo DNA methylation (6,9). In
addition, environmental factors such as nutrition, exercise and
particular chemical substances are able to modify DNMT expression
and function with consecutive changes in DNA methylation degree and
distribution, and all of these have a variable transcriptional
effect upon the gene function (10).
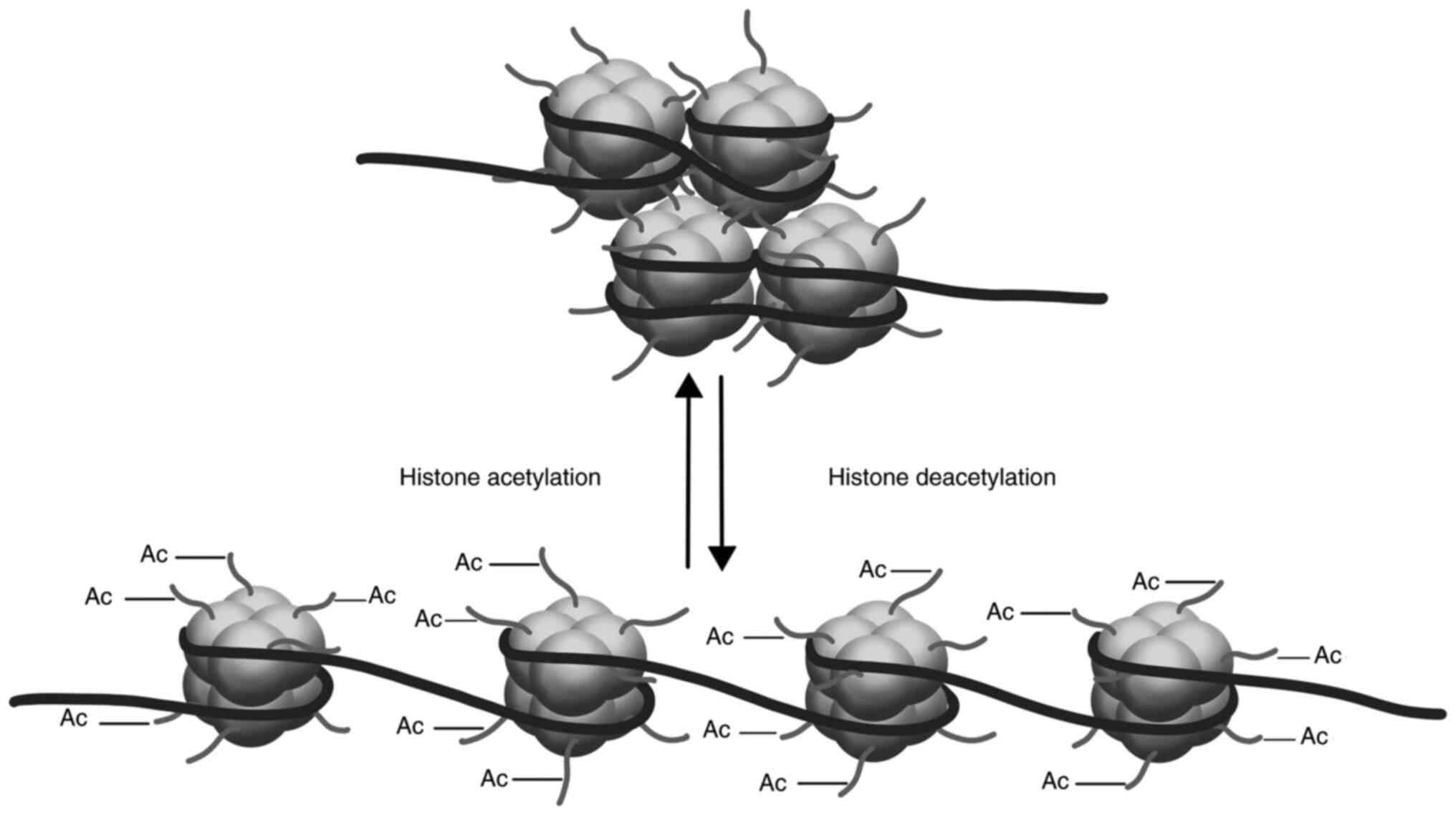
Another epigenetic nuclear mechanism involves
histone-DNA interaction and is represented by covalent binding of
acetyl groups at lysine residues within histones forming the
nucleosome core. Consequently, histone chains around which DNA
molecules are wrapped, become more relaxed, easier exposing DNA to
transcriptional factors. Conversely, if acetyl groups are removed
from the acetylated histones, the nucleosomes will appear more
compact and resistant to transcriptional factors (1) (Fig.
2).
The acetylation process is regulated by histone
acetyltransferases (HATs), while histone deacetylation is regulated
by histone deacetylases (HDACs) (11). At present, histone deacetylation is
extensively studied (12). HDACs
are classified according to their structural and functional
similarities into four classes: class I (HDACs: 1, 2, 3, 8), class
IIa (HDACs: 4, 5, 7, 9), class IIb (HDACs: 6, 10), class III
(sirtuins 1-7), and class IV (HDAC11). It is well known that
different classes of HDACs have specific intracellular locations
(HDACs from class I are predominantly located intranuclearly, while
class II HDACs shuttle between the cytoplasm and the nucleus)
(13).
HATs are represented by a vast family of proteins
such as cAMP-response element binding (protein) (CREB) binding
protein (CBP), that acts in a phosphorylation dependent manner:
Once phosphorylated, a HAT molecule will be activated, while
dephosphorylating will lead to HAT inactivation. The equilibrium
between HATs and HDACs is termed ‘acetylation homeostasis’ and will
finally dictate the degree of DNA exposure to transcriptional
factors inside the nucleosome (14). Histone acetylation and deacetylation
regulate cellular processes such as aging and oncogenesis.
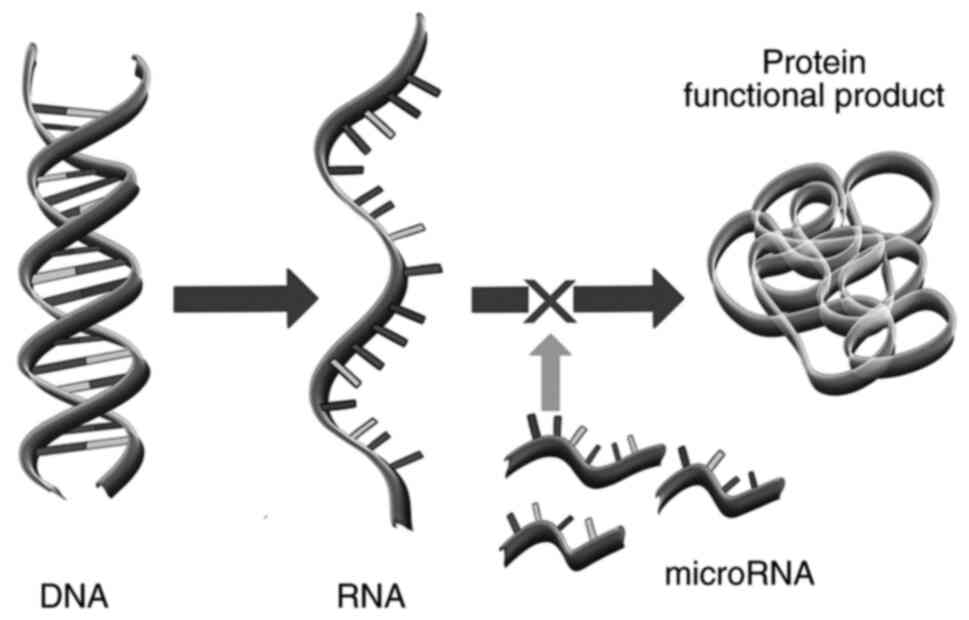
Conversely, miRNAs are small non-coding
single-stranded RNA species, composed of 15-30 bases, that are
involved in the post-transcriptional control of gene function or
expression in the cytoplasm (15).
Its effect of silencing is achieved by altering mRNA stability and
blocking the mRNA elongation, thus terminating protein synthesis
(Fig. 3).
Briefly, following maturation of pre-miRNA (formed
in the nucleus and exported and processed in the cytoplasm) into
miRNA by enzymatic cleavage of the hairpin structures, the leading
strands of the miRNA are integrated into the protein RNA-induced
silencing complex (RISC). The leading strand is thermodynamically
unstable relative to the passenger strand and directs the RISC to
the complementary strand of mRNA. miRNAs usually recognize binding
sites in the 3' untranslated region (UTR) of mRNA transcripts.
Perfect base complementarity between miRNA and mRNA cleaves the
mRNA by the slicing activity of Argonaute-2 (AGO2), whereas
imperfect binding leads to translational suppression and
slicer-independent mRNA damage (16).
Since miRNA represents epigenetic markers that
modulate terminal cell differentiation and developmental changes,
there has been great interest in achieving new miRNA-targeted
epigenetic therapies and identifying new epigenetic markers for
various clinical paradigms (17).
At present, there are numerous miRNA species recognized, being
independently and organ-specifically coded which can modulate the
protein synthesis and function in health and disease.
Although the screening tests and therapeutic
algorithms are well-established in various hepatic diseases,
including liver transplantation as etiologic therapy in end-stage
liver disease, strategies to identify reversible lesions in early
stages and to develop new epigenetic therapies should be
prioritized (18-20).
2. Methods: Selection of studies
A systematic review was performed on articles in
English published in databases including PubMed, Elsevier or Scopus
until August 2021, using the following key word: ‘epigenetics in
liver diseases’. Articles referring to any of the following hepatic
diseases were included: Non-alcoholic fatty liver disease (NAFLD),
alcoholic fatty liver disease (AFLD), autoimmune hepatitis (AIH),
viral hepatitis (VH) and Wilson disease (WD). Studies focusing on
end-stage liver disease were excluded. The most relevant articles
published in the last 15 years were added.
Some relevant articles from the domain of
epigenetics, in general, were further included, independent of the
year of publication, for a better description of fundamental
epigenetic mechanisms. After duplication removal and screening for
eligibility, 65 articles consisting of 19 clinical studies, 22
experimental studies and 24 review articles were included.
3. Epigenetics in various liver
diseases
Epigenetics in NAFLD
NAFLD represents a serious condition linked with an
inappropriate high-fat diet, which is more obvious in developed
countries where obesity and unhealthy dietary habits represent
public health issues. These liver changes are in correlation with
other multisystemic changes such as type II diabetes, various
cardiovascular or renal diseases, as consequences of the
interaction between the environment (due to various dietary habits)
and the organism.
In NAFLD, epigenetic DNA changes have been observed,
altering the insulin metabolism or producing dysregulations, at the
cellular level of the various metabolic pathways. Ahrens et
al observed that genes encoding insulin-like growth-factor 1
(IGF-1) and insulin-like factor binding protein 2 (IGFBP-2) could
be hypermethylated in NAFLD, inducing gene silencing and consequent
impairments in glucose metabolism. Other genes including pyruvate
carboxylases and ATP citrate lyase involved in the glucose cycle
could also be epigenetically silenced (21).
Histone deacetylation was also observed to interfere
with lipid metabolism. Silent information regulator factor
2-related enzyme 1 (SIRT-1) improves hepatic steatosis and
circadian rhythm (22,23). Other histone deacetylases such as
HDAC-3 and HDAC-8 promote triglyceride metabolism and insulin
sensitivity (24). De novo
liver lipogenesis could also be epigenetically controlled due to
certain histone changes: The interaction between host cell factor 1
(HCF-1) and carbohydrate response element binding protein (ChREBP)
regulates hepatic lipogenic genes (25).
In NAFLD, miRNAs species have been identified as
epigenetic markers for liver injury. miR-122 is one of the most
abundant small non-coding RNAs expressed in the liver. In NAFLD,
miR-122 is downregulated. Experimental study models have revealed
that the downregulation of this marker promotes lipogenesis and
liver inflammation (26).
Conversely, upregulation of miR-21 has deleterious effects on the
degree of hepatic steatosis and glucose metabolism (27). Other overexpressed liver miRNAs in
NAFLD are miR-24, miR-34a and miR-124, which could interfere with
lipid metabolism and insulin sensitivity (26,28).
miR-155 modulates the crosstalk between adipose tissue and the
liver in NAFLD induced by a high-fat diet (29).
In conclusion, NAFLD is a highly epigenetically
controlled liver pathology, whose complex mechanisms involve
factors associated with dietary habits, alterations in tertiary DNA
and histone structure and specific miRNA expression.
Epigenetics in (AIH)
AIH exhibits various phenotypes, reflecting the
complexity of underlying immune mechanisms. There are two forms:
AIH type I, present particularly in middle-aged women and AIH type
2, which is more common in children. AIH type 1 is characterized by
an increased titer of antinuclear antibodies, soluble liver
antigen/liver pancreas antibodies, and smooth muscle antibodies,
while AIH type 2 exhibits large amounts of the liver kidney
microsomal 1 antibodies (30). As
the main immune mechanism of the disease, the imbalance between pro
and anti-inflammatory promoting T-cells is extensively studied.
Treg cells are T-cells with anti-inflammatory properties, whose
presence is linked with the disease activity and liver inflammation
(31).
The data in the literature is rather focused on the
miRNA-mediated epigenetic changes in AIH than on DNA or histone
changes. Similar to other inflammatory hepatic diseases, miR-122,
the most abundant liver miRNA species, is upregulated in AIH as
well, serving as a marker of the disease activity (32). Consequently, this epigenetic marker
could serve as biomarker for therapy response or disease control.
miR-21 is also upregulated and is inversely correlated with the
degree of fibrosis (33). miR-223
suppresses proinflammatory liver activity via the NF-κB pathway,
inhibiting the macrophage function. In an AIH experimental model,
the overexpression of miR-223 was revealed to have a
liver-protective effect (34).
miR-155 could affect AIH progression as well. The literature
offers, however, contradictory data. miR-155 regulates the
inflammatory response by influencing the Th17 cells, with no effect
on IL-10-mediated Treg response (35).
Other epigenetic markers with an undefined role in
AIH are: miR-218, miR-363, miR-518f, miR-628-5p, miR-888, miR-523,
miR-141, miR-302b, miR-643 and miR-573(36).
Although there is insufficient data to characterize
epigenetic histone changes in AIH, certain studies have revealed
anti-histone auto-antibodies in AIH, whose interaction with the
histones could be reduced due to the epigenetic aforementioned
potential structural change (37).
This paradigm should be, however, further explored.
A previous clinical study reported a correlation
between the DNA methylation status in certain immune cells such as
CD4+ and CD19+ lymphocytes and disease
activity. Altered expression of enzymes involved in DNA
methylation, TET1 and DNMT3A, characterizes lymphocytes in AIH
(38). Further studies are required
in order to confirm this association.
From an epigenetic perspective, AIH still represents
an open field for research: To date, miRNAs represent the only area
which have offered a perspective regarding epigenetic modulating
mechanisms in this disease.
Epigenetics in VH
The interplay between a hepatic virus and the liver
leaves, in the majority of cases, an epigenetic signature. Due to
complex modulatory mechanisms, these signatures act either as a
prognostic tool or as a therapeutic response (39). Subsequently, epigenetic therapies
are evoked as novel therapies against these viruses. For example
DNMT inhibitors could be useful in VHC-associated HCC, while HDAC
inhibitors could reduce the VHB replication (39).
Hepatitis B virus (HBV) is a DNA virus, whose
genetic structure consists of covalently closed circular (ccc)DNA
incorporated into hepatocytes. The translational process is
realized using the host nuclear enzymes. The cccDNA methylation in
the region of GpG islands could, however, reduce the translational
potential of the viral DNA. There are three CpG regions defined in
the viral DNA: i) The start site of the S gene; ii) the region
surrounding enhancer I, the HBx gene promoter (Xp), and the core
promoter (Cp); and iii) the region harboring the Sp1 promoter and
the start codon of the Pol gene (40). According to Zhang et al, the
start site of the S gene is variably methylated among the different
HBV genotypes, while the other two regions are more stably
methylated (41). The methylation
of the second island is linked with a decreased viremia, while the
methylation of the third island influences carcinogenesis (41). This epigenetic change is observed
mainly in the nuclear cccDNA, integrated in the host cells and not
in the histone-free cytoplasmic DNA or circulating virions. The
role of DNMTs in HBV infection is not fully understood. According
to the literature, DNMT1, DNMT2 and DNMT3, are upregulated in HBV
infection leading to hypermethylation in host cells and,
consequently, to a reduction in virus replication (42). cccDNA replication is also modulated
by epigenetic changes of the histones. Hypomethylation of H3 and H4
histones and the recruitment of HDAC 1 nearby cccDNA, could reduce
the replication potential of the virus (43). Furthermore, epigenetic therapies
targeting upregulation of DNMTs and histone hypomethylation linked
with immunomodulatory therapy could represent the future in chronic
HBV infection.
Considering the miRNA species as potential
epigenetic biomarkers in HBV, miR-146 predicts the evolution to
fibrosis in HBV-infected patients (44).
Conversely, hepatitis C virus (HCV) virus is an RNA
virus, whose particular epigenetic signature reveals the risk of
carcinogenesis even in the presence of the sustained viral
response. Specific histone changes, H3K4Me3 and H3K9Ac, promote the
persistence of the virus following successful direct antiviral
therapy, acting as an epigenetic signature (45).
DNA methylation is another epigenetic change,
influencing carcinogenesis in HCV-infected patients. The two most
common repetitive elements in humans, long interspersed nuclear
element-1 (LINE-1) and Alu element (Alu), have been linked with
carcinogenesis. HCV may cause hepatocellular carcinoma (HCC) by
suppressing host defenses through DNA methylation that controls the
mobilization of repetitive elements (46).
Certain miRNA species are used as prognostic tools,
being particularly associated with the risk for HCV patients to
develop various complications such as HCC, fibrosis or cirrhosis.
miR-122-5p, miR-486-5p and miRNA-142-3p could predict the
development of HCC in HCV-infected patients (47).
According to Shrivastava et al, miR-20a and
miR-92a are epigenetic biomarkers, which promote the evolution from
an acute to a chronic state (48).
Let-7c is another epigenetic biomarker, which could predict the
evolution to fibrosis (49).
miR-494 is associated to the therapeutic response,
while miR-34a is upregulated in fatty liver compared with chronic
HCV (50,51).
Epigenetics in alcoholic fatty liver
disease (ALFD)
Diet-induced epigenetic changes are common and one
of the first described modulatory factors which could lead to
epigenetic changes according to the target tissue. The most exposed
tissues developing epigenetic modulatory mechanisms secondary to
various dietary factors are the brain, hematopoietic system or
liver (52,53). Alcoholic liver disease (ALD) has a
significant influence on the life quality, having a very important
impact on various health systems.
Alcohol-induced oxidative stress in hepatocytes
interferes with all main mechanisms of chromosomal epigenetic
control including DNA hypomethylation, histone acetylation and
phosphorylation and miRNA alteration.
Histone H3 acetylation at Lys 9 (H3AcK9) in
alcohol-exposed hepatocytes was observed in experimental studies
(54,55). HDAC inhibition, particularly SIRT1,
and HAT enzyme activation are responsible for this change (54). Phosphorylation of the histone H3 at
serine residues could synergistically influence the epigenetic
signature in hepatocytes exposed to alcohol (55). Another experimental study revealed a
differential methylation pattern of the histone H3 and H4 in
alcohol-exposed hepatocytes (56).
Whether chronic or acute exposure to alcohol could induce these
histone alterations is, however, controversial.
S-adenosil-methionine (SAM) is an important methyl
donor involved in CpG island DNA methylation. According to
Dannenberg et al, SAM concentration is reduced in ALD, which
consequently interferes with the DNA methylation process (57). This hypomethylation state promotes
DNA damage and strand breaks in ALD (57). DNA hypomethylation also disturbs
alcohol-metabolizing enzymes, such as alcohol dehydrogenase 1B
(ADH1) (58). An interplay between
histone acetylation and DNA methylation is also possible in ALD:
DNA hypomethylation in promoter regions triggers histone
deacetylation. The exact effect of this epigenetic crosstalk is,
however, unknown.
miRNA species represent other epigenetic markers in
ALD. miR-155 is increased in liver macrophages secondary to alcohol
intake (59). miR-212 is
upregulated secondary to alcohol intake, affecting the intestinal
permeability by downregulating tight junction protein zonula
occludens 1(60). Further studies
are required, in order to understand the exact prognostic and
therapeutic potential of these epigenetic markers in ALD.
Epigenetics in WD
WD is an autosomal recessive disease characterized
by accumulation of copper in the liver and brain. The key gene
causing this disease is the copper-transporting gene ATP7B, which
blocks copper extraction through the biliary tract. The knowledge
of the disease physiopathology is continuously evolving. At
present, it is widely accepted that alteration of this gene
interferes with at least eight transmembrane active transporters of
copper from the hepatocytes (61).
There is a large variability in WD considering the
onset, sex, severity, response to treatment and target organ (the
liver vs. the brain). This disease inconsistency is partially
explained through certain epigenetic modulatory mechanisms. WD
impairs the methionine metabolism, which is the main methyl
supplier for DNA and histone methylation. The aberrant
SAM/S-adenosil-homocysteine (SAH) ratio impairs the methylation
process (62).
Furthermore, traces of heavy metals impair the
mitochondrial metabolism, thus increasing the amount of reactive
oxygen species (ROS). This metabolic change dysregulates the
activity of TET enzymes, inhibiting the DNA demethylation (63).
In animal models, the hepatic accumulation of copper
is inversely correlated with DNMT3a and DNMT3b levels, impairing
the DNA methylation process (64).
This alteration is particularly important in utero.
Consequently, choline and penicillamine treatment, differentially
modify the methylation status of the DNA in mice according to the
sex (65).
4. Final considerations
Liver pathology is a vast field with incomplete
knowledge, which requires profound expertise. The outcome in
advanced and irreversible chronic hepatic injury in the absence of
a liver transplant (cirrhosis and end-stage liver disease) is poor,
with short and long-term socio-economic consequences. Therefore,
new strategies to identify, stratify and treat these hepatic
diseases while still being in a reversible state are highly
required.
The epigenetic approach of all these diseases is
more than welcomed, taking into consideration that conventional
therapeutic strategies are almost exhausted. This is generally
valid for ALD and NAFLD, viral hepatitis, AIH as well as other
metabolic conditions.
Analysis of gene function and expression independent
on the ‘heritable’ character of the genome, consisting in analysis
of histone-DNA interactions and small non-coding RNA synthesis, is
an extremely valuable tool for future diagnostic and therapeutic
strategies of hepatic diseases, whose molecular etiologies are far
from being completely elucidated.
Acknowledgements
The authors would like to thank Ms Irina Matache,
researcher at the Department of Physiology II and Neurosciences of
‘Carol Davila’ University of Medicine and Pharmacy (Bucharest,
Romania), for her help with manuscript reviewing.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
TI contributed to literature research, manuscript
writing and reviewing. SI contributed to manuscript writing and
reviewing. RR contributed to manuscript reviewing and design of
figures. MC contributed to manuscript reviewing. LI coordinated and
reviewed the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Margueron R and Reinberg D: Chromatin
structure and the inheritance of epigenetic information. Nat Rev
Genet. 11:285–296. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Ryšlavá H, Doubnerová V, Kavan D and Vaněk
O: Effect of posttranslational modifications on enzyme function and
assembly. J Proteomics. 92:80–109. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Berger SL, Kouzarides T, Shiekhattar R and
Shilatifard A: An operational definition of epigenetics. Genes Dev.
23:781–783. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Waddington CH: Towards a Theoretical
Biology. Edinburgh University Press, Edinburgh, 1968.
|
|
5
|
Jeltsch A: On the enzymatic properties of
Dnmt1: Specificity, processivity, mechanism of linear diffusion and
allosteric regulation of the enzyme. Epigenetics. 1:63–66.
2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Fischer A, Sananbenesi F, Mungenast A and
Tsai LH: Targeting the correct HDAC(s) to treat cognitive
disorders. Trends Pharmacol Sci. 31:605–617. 2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhu J, He F, Hu S and Yu J: On the nature
of human housekeeping genes. Trends Genet. 24:481–484.
2008.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bibikova M: Chapter 2. DNA Methylation.
Microarrays. In: Epigenomics in Health and Disease. Fraga MF and
Fernández AF (eds). Academic Press, Boston, MA, pp19-46, 2016.
|
|
9
|
Goll MG and Bestor TH: Eukaryotic cytosine
methyltransferases. Annu Rev Biochem. 74:481–514. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Keil KP and Lein PJ: DNA methylation: A
mechanism linking environmental chemical exposures to risk of
autism spectrum disorders? Environ Epigenet.
2(dvv012)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mielcarek M, Zielonka D, Carnemolla A,
Marcinkowski JT and Guidez F: HDAC4 as a potential therapeutic
target in neurodegenerative diseases: A summary of recent
achievements. Front Cell Neurosci. 9(42)2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Saha RN and Pahan K: HATs and HDACs in
neurodegeneration: A tale of disconcerted acetylation homeostasis.
Cell Death Differ. 13:539–550. 2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Peserico A and Simone C: Physical and
functional HAT/HDAC interplay regulates protein acetylation
balance. J Biomed Biotechnol. 2011(371832)2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Varela MA, Roberts TC and Wood MJ:
Epigenetics and ncRNAs in brain function and disease: Mechanisms
and prospects for therapy. Neurotherapeutics. 10:621–631.
2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kim VN: MicroRNA biogenesis: Coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385.
2005.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Isac S, Panaitescu AM, Iesanu MI, Zeca V,
Cucu N, Zagrean L, Peltecu G and Zagrean AM: Maternal
citicoline-supplemented diet improves the response of the immature
hippocampus to perinatal asphyxia in rats. Neonatology.
117:729–735. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Isac T, Isac S, Ioanitescu S, Mihaly E,
Tanasescu MD, Balan DG, Tulin A and Iliescu L: Dynamics of serum
α-fetoprotein in viral hepatitis C without hepatocellular
carcinoma. Exp Ther Med. 22(749)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wu K, Ye C, Lin L, Chu Y, Ji M, Dai W,
Zeng X and Lin Y: Inhibiting miR-21 attenuates experimental hepatic
fibrosis by suppressing both the ERK1 pathway in HSC and hepatocyte
EMT. Clin Sci (Lond). 130:1469–1480. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Popescu I, Ionescu M, Braşoveanu V,
Hrehoreţ D, Copca N, Lupaşcu C, Botea F, Dorobanţu B, Alexandrescu
S, Grigorie M, et al: The Romanian National program for liver
transplantation -852 procedures in 815 patients over 17 years
(2000-2017): A continuous evolution to success. Chirurgia (Bucur).
112:229–243. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ahrens M, Ammerpohl O, von Schönfels W,
Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M,
Hinrichsen H, et al: DNA methylation analysis in nonalcoholic fatty
liver disease suggests distinct disease-specific and remodeling
signatures after bariatric surgery. Cell Metab. 18:296–302.
2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Cao Y, Xue Y, Xue L, Jiang X, Wang X,
Zhang Z, Yang J, Lu J, Zhang C, Wang W and Ning G: Hepatic menin
recruits SIRT1 to control liver steatosis through histone
deacetylation. J Hepatol. 59:1299–1306. 2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Nakahata Y, Kaluzova M, Grimaldi B, Sahar
S, Hirayama J, Chen D, Guarente LP and Sassone-Corsi P: The
NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin
remodeling and circadian control. Cell. 134:329–340.
2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Xu F and Guo W: The progress of
epigenetics in the development and progression of non-alcoholic
fatty liver disease. Liver Res. 4:118–123. 2020.
|
|
25
|
Lane EA, Choi DW, Garcia-Haro L, Levine
ZG, Tedoldi M, Walker S and Danial NN: HCF-1 regulates de novo
lipogenesis through a nutrient-sensitive complex with ChREBP. Mol
Cell 25:. 75:357–371.e7. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hsu SH, Wang B, Kota J, Yu J, Costinean S,
Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, et al: Essential
metabolic, anti-inflammatory, and anti-tumorigenic functions of
miR-122 in liver. J Clin Invest. 122:2871–2883. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Calo N, Ramadori P, Sobolewski C, Romero
Y, Maeder C, Fournier M, Rantakari P, Zhang FP, Poutanen M, Dufour
JF, et al: Stress-activated miR-21/miR-21* in hepatocytes promotes
lipid and glucose metabolic disorders associated with high-fat diet
consumption. Gut. 65:1871–1881. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liu X, Zhao J, Liu Q, Xiong X, Zhang Z,
Jiao Y, Li X, Liu B, Li Y and Lu Y: MicroRNA-124 promotes hepatic
triglyceride accumulation through targeting tribbles homolog 3. Sci
Rep. 6(37170)2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ying W, Riopel M, Bandyopadhyay G, Dong Y,
Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez-Carretero A,
Fu W, et al: Adipose tissue macrophage-derived exosomal miRNAs can
modulate in vivo and in vitro insulin sensitivity. Cell.
171:372–384.e12. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kerkar N and Vergani D: De novo autoimmune
hepatitis -is this different in adults compared to children? J
Autoimmun. 95:26–33. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Behairy BE, El-Araby HA, Abd El Kader HH,
Ehsan NA, Salem ME, Zakaria HM and Khedr MA: Assessment of
intrahepatic regulatory T cells in children with autoimmune
hepatitis. Ann Hepatol. 15:682–690. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bandiera S, Pfeffer S, Baumert TF and
Zeisel MB: MiR-122-a key factor and therapeutic target in liver
disease. J Hepatol. 62:448–457. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Migita K, Komori A, Kozuru H, Jiuchi Y,
Nakamura M, Yasunami M, Furukawa H, Abiru S, Yamasaki K, Nagaoka S,
et al: Circulating microRNA profiles in patients with type-1
autoimmune hepatitis. PLoS One. 10(e0136908)2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen L, Lu FB, Chen DZ, Wu JL, Hu ED, Xu
LM, Zheng MH, Li H, Huang Y, Jin XY, et al: BMSCs-derived
miR-223-containing exosomes contribute to liver protection in
experimental autoimmune hepatitis. Mol Immunol. 93:38–46.
2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Xia G, Wu S, Wang X and Fu M: Inhibition
of microRNA-155 attenuates concanavalin-A-induced autoimmune
hepatitis by regulating Treg/Th17 cell differentiation. Can J
Physiol Pharmacol. 96:1293–1300. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Liu Q, Li Y, Xiong M and Tang R:
Epigenetics of autoimmune liver diseases: Current progress and
future directions. J Bio-X Res. 2:46–55. 2019.
|
|
37
|
Chen M, Shirai M, Czaja AJ, Kurokohchi K,
Arichi T, Arima K, Kodama T and Nishioka M: Characterization of
anti-histone antibodies in patients with type 1 autoimmune
hepatitis. J Gastroenterol Hepatol. 13:483–489. 1998.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zachou K, Arvaniti P, Lyberopoulou A,
Sevdali E, Speletas M, Ioannou M, Koukoulis GK, Renaudineau Y and
Dalekos GN: Altered DNA methylation pattern characterizes the
peripheral immune cells of patients with autoimmune hepatitis.
Liver Int: Feb 2, 2022 (Epub ahead of print).
|
|
39
|
Nehme Z, Pasquereau S and Herbein G:
Control of viral infections by epigenetic-targeted therapy. Clin
Epigenetics. 11(55)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Jain S, Chang TT, Chen S, Boldbaatar B,
Clemens A, Lin SY, Yan R, Hu CT, Guo H, Block TM, et al:
Comprehensive DNA methylation analysis of hepatitis B virus genome
in infected liver tissues. Sci Rep. 5(10478)2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhang Y, Li C, Zhang Y, Zhu H, Kang Y, Liu
H, Wang J, Qin Y, Mao R, Xie Y, et al: Comparative analysis of CpG
islands among HBV genotypes. PLoS One. 8(e56711)2013.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hong X, Kim ES and Guo H: Epigenetic
regulation of hepatitis B virus covalently closed circular DNA:
Implications for epigenetic therapy against chronic hepatitis B.
Hepatology. 66:2066–2077. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Pollicino T, Belloni L, Raffa G, Pediconi
N, Squadrito G, Raimondo G and Levrero M: Hepatitis B virus
replication is regulated by the acetylation status of hepatitis B
virus cccDNA-bound H3 and H4 histones. Gastroenterology.
130:823–837. 2006.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wang TZ, Lin DD, Jin BX, Sun XY and Li N:
Plasma microRNA: A novel non-invasive biomarker for HBV-associated
liver fibrosis staging. Exp Ther Med. 17:1919–1929. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Perez S, Kaspi A, Domovitz T, Davidovich
A, Lavi-Itzkovitz A, Meirson T, Alison Holmes J, Dai CY, Huang CF,
Chung RT, et al: Hepatitis C virus leaves an epigenetic signature
post cure of infection by direct-acting antivirals. PLoS Genet.
15(e1008181)2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zheng Y, Hlady RA, Joyce BT, Robertson KD,
He C, Nannini DR, Kibbe WA, Achenbach CJ, Murphy RL, Roberts LR and
Hou L: DNA methylation of individual repetitive elements in
hepatitis C virus infection-induced hepatocellular carcinoma. Clin
Epigenetics. 11(145)2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Weis A, Marquart L, Calvopina DA, Genz B,
Ramm GA and Skoien R: Serum MicroRNAs as biomarkers in hepatitis C:
Preliminary evidence of a MicroRNA panel for the diagnosis of
hepatocellular carcinoma. Int J Mol Sci. 20(864)2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Shrivastava S, Petrone J, Steele R, Lauer
GM, Di Bisceglie AM and Ray RB: Up-regulation of circulating
miR-20a is correlated with hepatitis C virus-mediated liver disease
progression. Hepatology. 58:863–871. 2013.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Matsuura K, De Giorgi V, Schechterly C,
Wang RY, Farci P, Tanaka Y and Alter HJ: Circulating let-7 levels
in plasma and extracellular vesicles correlate with hepatic
fibrosis progression in chronic hepatitis C. Hepatology.
64:732–745. 2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
El-Diwany R, Wasilewski LN, Witwer KW,
Bailey JR, Page K, Ray SC, Cox AL, Thomas DL and Balagopal A: Acute
hepatitis C virus infection induces consistent changes in
circulating MicroRNAs that are associated with nonlytic hepatocyte
release. J Virol. 89:9454–9464. 2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Liu XL, Pan Q, Zhang RN, Shen F, Yan SY,
Sun C, Xu ZJ, Chen YW and Fan JG: Disease-specific miR-34a as
diagnostic marker of non-alcoholic steatohepatitis in a Chinese
population. World J Gastroenterol. 22:9844–9852. 2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Isac S, Panaitescu AM, Iesanu M, Grigoras
IF, Totan A, Udriste A, Cucu N, Peltecu G, Zagrean L and Zagrean
AM: Maternal high-fat diet modifies the immature hippocampus
vulnerability to perinatal asphyxia in rats. Neonatology.
114:355–361. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Lieber CS, Leo MA, Wang X and Decarli LM:
Effect of chronic alcohol consumption on Hepatic SIRT1 and
PGC-1alpha in rats. Biochem Biophys Res Commun. 370:44–48.
2008.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lee YJ and Shukla SD: Histone H3
phosphorylation at serine 10 and serine 28 is mediated by p38 MAPK
in rat hepatocytes exposed to ethanol and acetaldehyde. Eur J
Pharmacol. 573:29–38. 2007.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Pal-Bhadra M, Bhadra U, Jackson DE,
Mamatha L, Park PH and Shukla SD: Distinct methylation patterns in
histone H3 at Lys-4 and Lys-9 correlate with up- &
down-regulation of genes by ethanol in hepatocytes. Life Sci.
81:979–987. 2007.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lu SC, Huang ZZ, Yang H, Mato JM, Avila MA
and Tsukamoto H: Changes in methionine adenosyltransferase and
S-adenosylmethionine homeostasis in alcoholic rat liver. Am J
Physiol Gastrointest Liver Physiol. 279:G178–G185. 2000.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Dannenberg LO, Chen HJ, Tian H and
Edenberg HJ: Differential regulation of the alcohol dehydrogenase
1B (ADH1B) and ADH1C genes by DNA methylation and histone
deacetylation. Alcohol Clin Exp Res. 30:928–937. 2006.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Mandrekar P: Epigenetic regulation in
alcoholic liver disease. World J Gastroenterol. 17:2456–2464.
2011.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Bala S, Marcos M, Kodys K, Csak T,
Catalano D, Mandrekar P and Szabo G: Up-regulation of microRNA-155
in macrophages contributes to increased tumor necrosis factor
{alpha} (TNF{alpha}) production via increased mRNA half-life in
alcoholic liver disease. J Biol Chem. 286:1436–1444.
2011.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Tang Y, Banan A, Forsyth CB, Fields JZ,
Lau CK, Zhang LJ and Keshavarzian A: Effect of alcohol on miR-212
expression in intestinal epithelial cells and its potential role in
alcoholic liver disease. Alcohol Clin Exp Res. 32:355–364.
2008.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Medici V and LaSalle JM: Genetics and
epigenetic factors of Wilson disease. Ann Transl Med. 7 (Suppl
2)(S58)2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Li M, Li Y, Chen J, Wei W, Pan X, Liu J,
Liu Q, Leu W, Zhang L, Yang X, et al: Copper ions inhibit
S-adenosylhomocysteine hydrolase by causing dissociation of NAD+
cofactor. Biochemistry. 46:11451–11458. 2007.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Yin R, Mo J, Dai J and Wang H: Nickel(II)
inhibits tet-mediated 5-methylcytosine oxidation by high affinity
displacement of the cofactor iron(II). ACS Chem Biol. 12:1494–1498.
2017.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Le A, Shibata NM, French SW, Kim K,
Kharbanda KK, Islam MS, LaSalle JM, Halsted CH, Keen CL and Medici
V: Characterization of timed changes in hepatic copper
concentrations, methionine metabolism, gene expression, and global
DNA methylation in the Jackson toxic milk mouse model of Wilson
disease. Int J Mol Sci. 15:8004–8023. 2014.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Medici V, Kieffer DA, Shibata NM, Chima H,
Kim K, Canovas A, Medrano JF, Islas-Trejo AD, Kharbanda KK, Olson
K, et al: Wilson disease: Epigenetic effects of choline
supplementation on phenotype and clinical course in a mouse model.
Epigenetics. 11:804–818. 2016.PubMed/NCBI View Article : Google Scholar
|