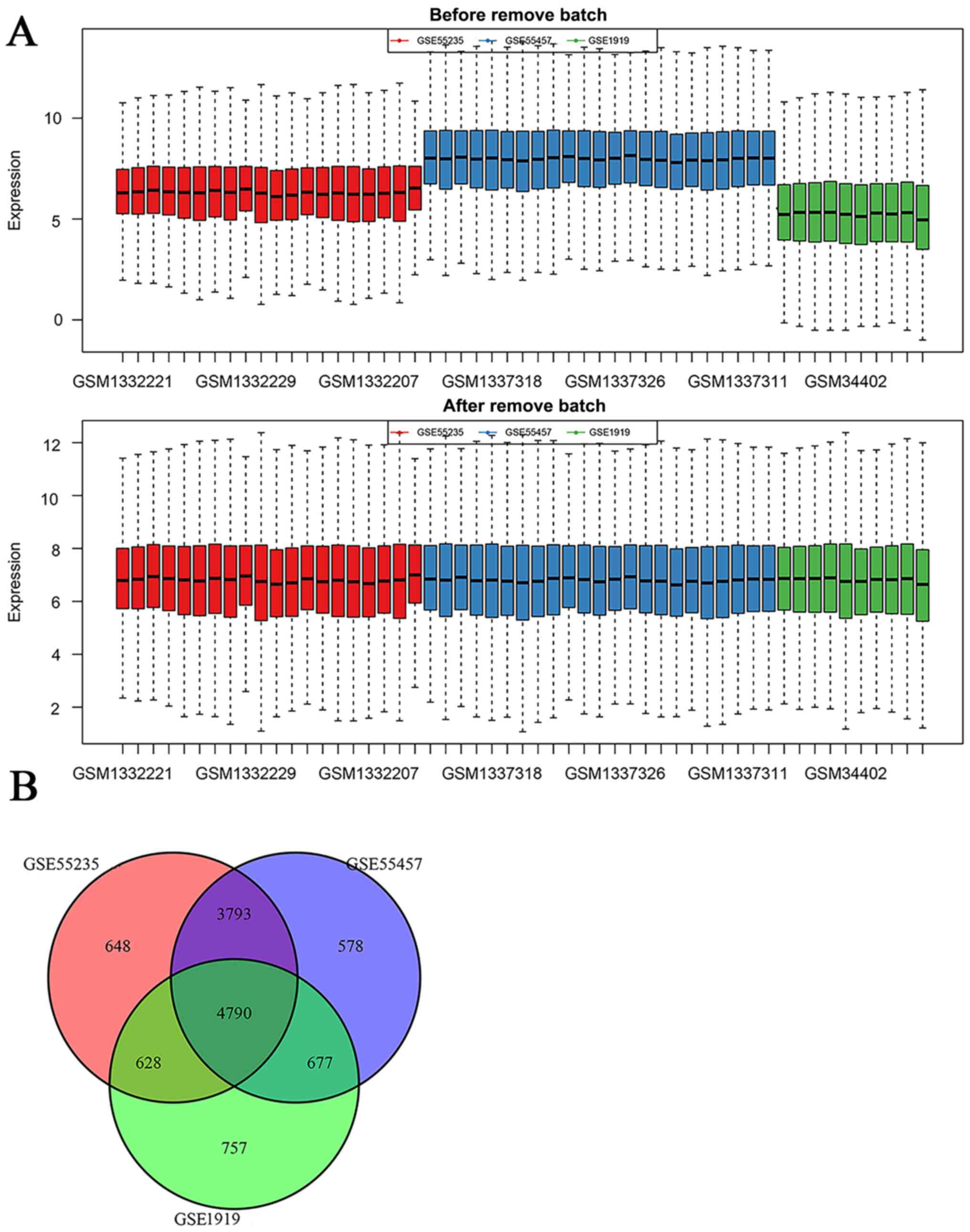
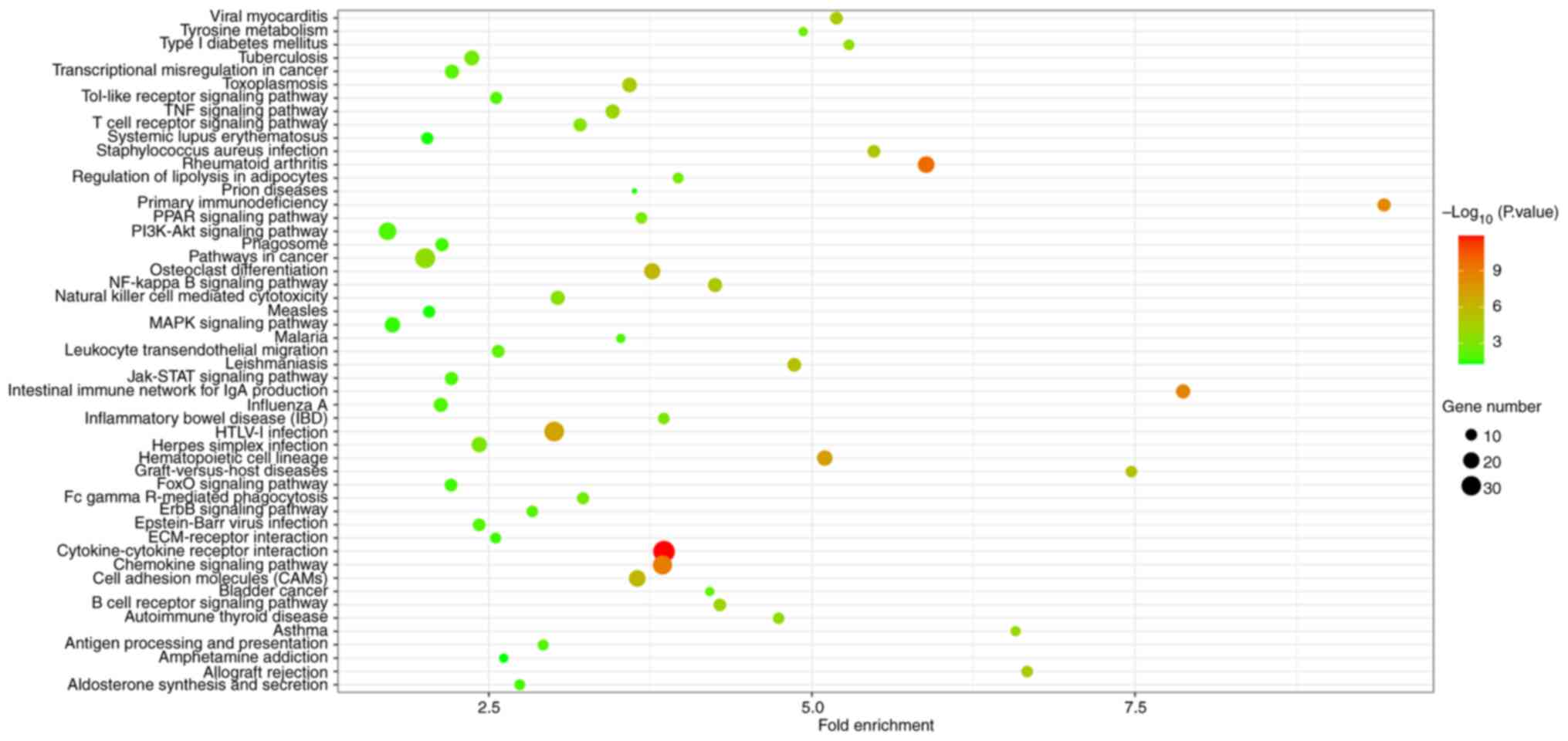
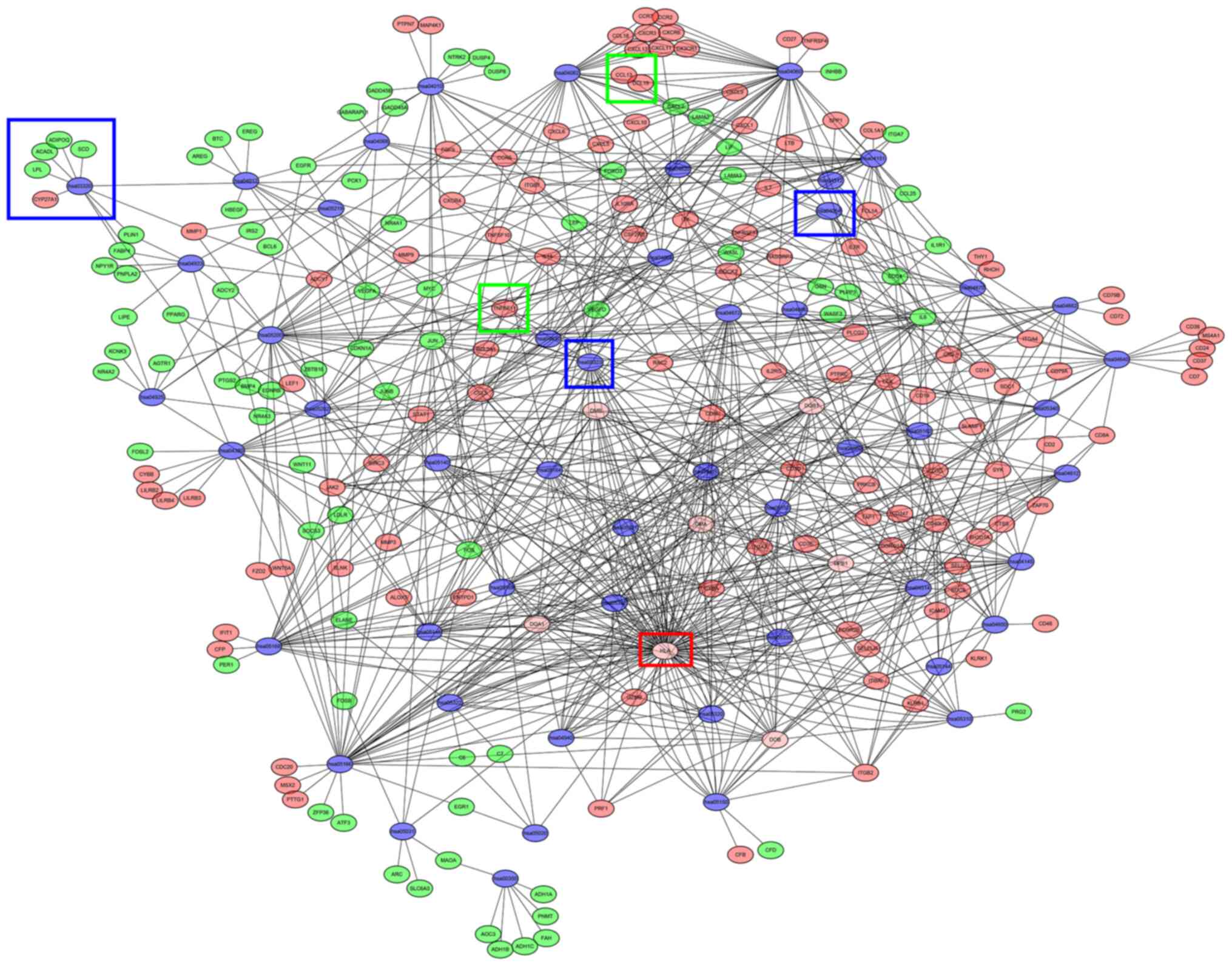
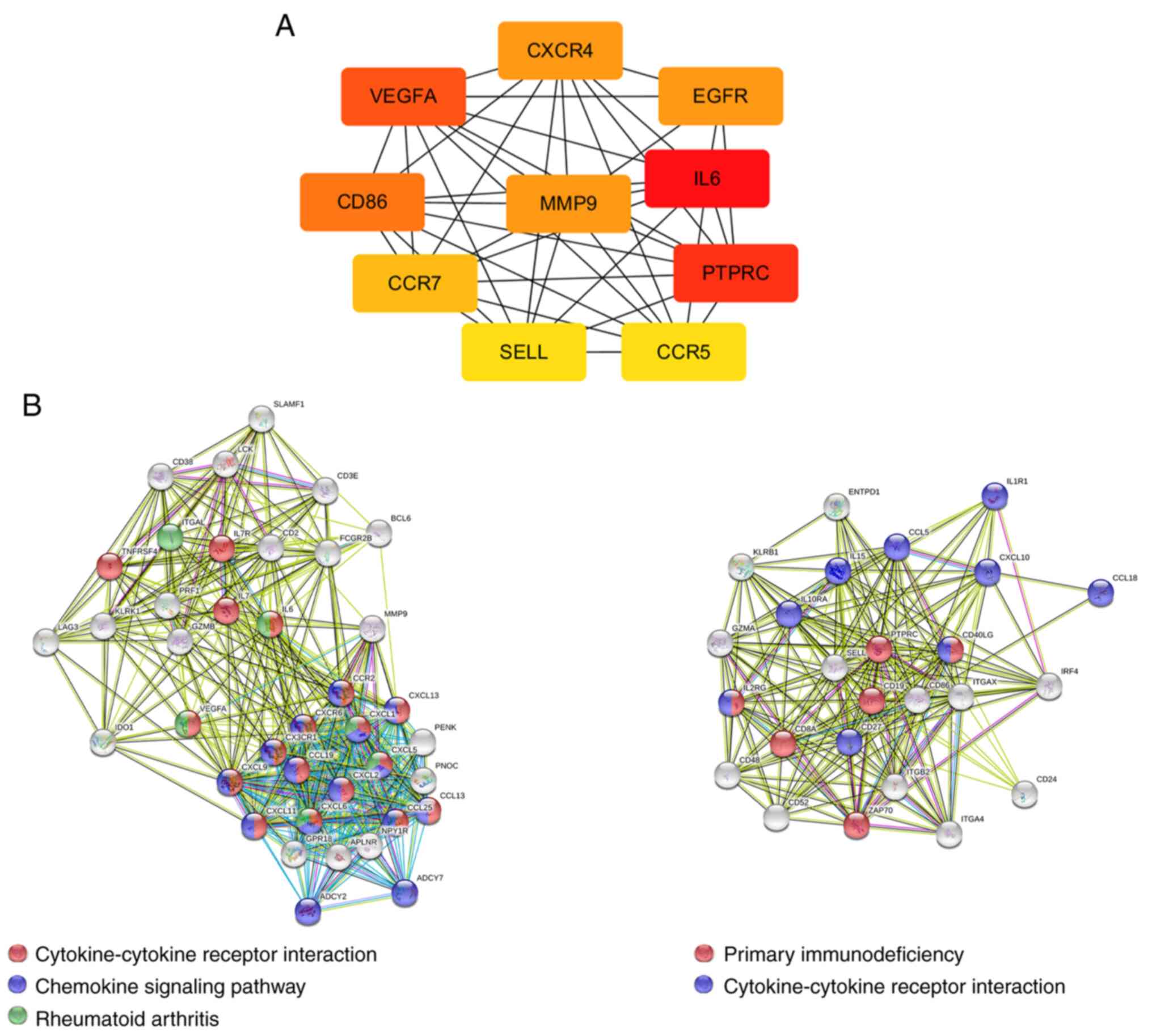
| Cytokine-cytokine
receptor interaction | hsa04060 | 38 |
1.28x10-12 | CX3CR1, CCL13,
CXCL6, CXCL9, CXCR4, CSF2RB, CXCL1, CXCR6, CXCL13, IL2RG, CXCL2,
CXCL5, CCL5, CXCR3, TNFSF10, TNFSF11, TNFRSF17, CCR7, CCL19, CCR5,
CCL18, TNFRSF4, CCR2, CCL25, IL15, IL1R1, IL10RA, LIF, INHBB,
CXCL10, CXCL11, IL6, CD40LG, IL7, LEP, CD27, LTB, IL7R |
| Rheumatoid
arthritis | hsa05323 | 21 |
1.77x10-10 | CD86, CXCL6, JUN,
IL15, MMP1, ITGB2, MMP3, FOS, ITGAL, CXCL5, VEGFA, HLA-DMA, IL6,
HLA-DMB, CCL5, HLA-DPB1, TNFSF11, LTB, HLA-DOB, HLA-DQA1,
HLA-DQB1 |
| Chemokine signaling
pathway | hsa04062 | 29 |
1.12x10-9 | CX3CR1, CCL13,
CXCL6, ITK, CXCL9, CXCR4, CXCL1, ADCY2, WASL, CXCR6, CXCL13, FOXO3,
CXCL2, ADCY7, CXCL5, CCL5, CXCR3, RAC2, CCR7, CCL19, JAK2, CCR5,
CCL18, CCR2, CCL25, STAT1, CXCL10, CXCL11, DOCK2 |
| Intestinal immune
network for IgA production | hsa04672 | 15 |
2.30x10-9 | CCL25, CD86, ITGA4,
IL15, CXCR4, HLA-DMA, IL6, HLA-DMB, CD40LG, HLA-DPB1, TNFRSF17,
ITGB7, HLA-DOB, HLA-DQA1, HLA-DQB1 |
| Primary
immunodeficiency | hsa05340 | 13 |
3.71x10-9 | TAP1, IL2RG, CD3E,
CD3D, CD79A, ZAP70, PTPRC, CD40LG, LCK, CD8A, CD19, BLNK, IL7R |
| Hematopoietic cell
lineage | hsa04640 | 18 |
4.83x10-8 | CR2, ITGA4, IL1R1,
CD3G, CD3E, CD3D, CD2, IL6, IL7, CD8A, CD19, CD7, CD38, CD37, CD14,
CD24, IL7R, MS4A1 |
| HTLV-I
infection | hsa05166 | 31 |
9.08x10-8 | CDKN1A, ITGB2,
CD3G, ADCY2, CD3E, ITGAL, IL2RG, CD3D, ADCY7, CDC20, HLA-DMA,
ZFP36, HLA-DMB, WNT11, PTTG1, MYC, HLA-DOB, HLA-DQA1, EGR1, JUN,
FZD2, MSX2, IL15, IL1R1, WNT5A, FOS, IL6, LCK, HLA-DPB1, ATF3,
HLA-DQB1 |
| Cell adhesion
molecules (CAMs) | hsa04514 | 21 |
8.92x10-7 | CD86, SELPLG, SDC4,
ITGA4, SDC3, ITGB2, ICAM3, ITGAL, CD2, HLA-DMA, HLA-DMB, PTPRC,
CD40LG, SELL, CD8A, HLA-DPB1, SDC1, ITGB7, HLA-DOB, HLA-DQA1,
HLA-DQB1 |
| Osteoclast
differentiation | hsa04380 | 20 |
1.07x10-6 | JUN, SYK, IL1R1,
STAT1, CYBB, CYBA, LILRB2, FOS, LILRB3, LILRB4, FOSL2, SOCS3, LCK,
BLNK, PLCG2, FOSB, TNFSF11, PPARG, FCGR2B, JUNB |
| Leishmaniasis | hsa05140 | 14 |
4.10x10-6 | JUN, ITGA4, STAT1,
ITGB2, CYBA, FOS, PTGS2, HLA-DMA, HLA-DMB, HLA-DPB1, JAK2, HLA-DOB,
HLA-DQA1, HLA-DQB1 |
| Graft-vs. -host
disease | hsa05332 | 10 |
4.14x10-6 | CD86, IL6, HLA-DMA,
HLA-DMB, HLA-DPB1, PRF1, GZMB, HLA-DOB, HLA-DQA1, HLA-DQB1 |
| Staphylococcus
aureus infection | hsa05150 | 12 |
7.83x10-6 | CFD, HLA-DMA,
SELPLG, HLA-DMB, ITGB2, HLA-DPB1, ITGAL, FCGR2B, HLA-DOB, CFB,
HLA-DQA1, HLA-DQB1 |
| NF-κB signaling
pathway | hsa04064 | 15 |
8.49x10-6 | CCL13, SYK, BCL2A1,
IL1R1, PTGS2, ZAP70, CD40LG, LCK, BLNK, PLCG2, TNFSF11, CD14, LTB,
CCL19, BIRC3 |
| Allograft
rejection | hsa05330 | 10 |
1.16x10-5 | CD86, HLA-DMA,
HLA-DMB, CD40LG, HLA-DPB1, PRF1, GZMB, HLA-DOB, HLA-DQA1,
HLA-DQB1 |
| Viral
myocarditis | hsa05416 | 12 |
1.35x10-5 | CD86, HLA-DMA,
HLA-DMB, CD40LG, ITGB2, RAC2, HLA-DPB1, PRF1, ITGAL, HLA-DOB,
HLA-DQA1, HLA-DQB1 |
| Toxoplasmosis | hsa05145 | 16 |
3.17x10-5 | LAMA2, STAT1,
IL10RA, LAMA3, HLA-DMA, HLA-DMB, CD40LG, ALOX5, HLA-DPB1, JAK2,
CCR5, HLA-DOB, LDLR, HLA-DQA1, BIRC3, HLA-DQB1 |
| B cell receptor
signaling pathway | hsa04662 | 12 |
8.69x10-5 | CD79B, CD79A, CR2,
JUN, CD72, SYK, CD19, BLNK, PLCG2, RAC2, FOS, FCGR2B |
| TNF signaling
pathway | hsa04668 | 15 |
9.23x10-5 | JUN, IL15, MMP3,
LIF, CXCL1, FOS, PTGS2, CXCL2, MMP9, CXCL10, SOCS3, IL6, CCL5,
JUNB, BIRC3 |
| Asthma | hsa05310 | 8 |
1.49x10-4 | HLA-DMA, HLA-DMB,
CD40LG, PRG2, HLA-DPB1, HLA-DOB, HLA-DQA1, HLA-DQB1 |
| Autoimmune thyroid
disease | hsa05320 | 10 |
2.01x10-4 | CD86, HLA-DMA,
HLA-DMB, CD40LG, HLA-DPB1, PRF1, GZMB, HLA-DOB, HLA-DQA1,
HLA-DQB1 |
| Type I diabetes
mellitus | hsa04940 | 9 |
2.29x10-4 | CD86, HLA-DMA,
HLA-DMB, HLA-DPB1, PRF1, GZMB, HLA-DOB, HLA-DQA1, HLA-DQB1 |
| Pathways in
cancer | hsa05200 | 32 |
2.37x10-4 | CDKN1A, LAMA2,
LAMA3, LEF1, CXCR4, ADCY2, PTGS2, RASGRP1, ADCY7, EGFR, EDNRB,
WNT11, FGF9, MYC, PLCG2, RAC2, JUN, FZD2, PRKCB, MMP1, STAT1,
ZBTB16, WNT5A, VEGFD, FOS, MMP9, VEGFA, BMP4, IL6, AGTR1, PPARG,
BIRC3 |
| Natural killer cell
mediated cytotoxicity | hsa04650 | 15 |
3.77x10-4 | SYK, PRKCB, SH2D1A,
ITGB2, PRF1, GZMB, ITGAL, ZAP70, KLRK1, LCK, TNFSF10, PLCG2, RAC2,
CD48, CD247 |
| T cell receptor
signaling pathway | hsa04660 | 13 |
6.53x10-4 | ITK, JUN, CD3G,
FOS, CD3E, RASGRP1, CD3D, ZAP70, PTPRC, CD40LG, LCK, CD8A,
CD247 |
| Inflammatory bowel
disease | hsa05321 | 10 |
9.86x10-4 | IL6, HLA-DMA, JUN,
HLA-DMB, STAT1, HLA-DPB1, IL2RG, HLA-DOB, HLA-DQA1, HLA-DQB1 |
| Herpes simplex
infection | hsa05168 | 18 | 0.001109261 | JUN, IL15, STAT1,
TAP1, FOS, IFIT1, CFP, PER1, SOCS3, HLA-DMA, IL6, HLA-DMB, CCL5,
HLA-DPB1, JAK2, HLA-DOB, HLA-DQA1, HLA-DQB1 |
| PPAR signaling
pathway | hsa03320 | 10 | 0.001376807 | CYP27A1, FABP4,
ACADL, MMP1, SCD, ADIPOQ, LPL, PPARG, PLIN1, PCK1 |
| Regulation of
lipolysis in adipocytes | hsa04923 | 9 | 0.001688948 | LIPE, FABP4, NPY1R,
IRS2, ADCY2, PLIN1, PTGS2, ADCY7, PNPLA2 |
| Fc gamma R-mediated
phagocytosis | hsa04666 | 11 | 0.001936067 | GSN, PTPRC, SYK,
PRKCB, PLCG2, RAC2, WASL, DOCK2, FCGR2B, PLPP3, WASF3 |
| Tuberculosis | hsa05152 | 17 | 0.002050272 | SYK, STAT1, IL10RA,
ITGB2, CORO1A, CTSS, HLA-DMA, IL6, HLA-DMB, ITGAX, HLA-DPB1, CD14,
FCGR2B, JAK2, HLA-DOB, HLA-DQA1, HLA-DQB1 |
| Tyrosine
metabolism | hsa00350 | 7 | 0.002490246 | PNMT, AOC3, ADH1C,
MAOA, ADH1B, ADH1A, FAH |
| Bladder cancer | hsa05219 | 7 | 0.005633865 | CDKN1A, MMP1, MYC,
MMP9, EGFR, VEGFA, HBEGF |
| Leukocyte
transendothelial migration | hsa04670 | 12 | 0.006560639 | ITK, ITGA4, PRKCB,
ITGB2, PLCG2, RAC2, CXCR4, RHOH, CYBA, THY1, ITGAL, MMP9 |
| Transcriptional
misregulation in cancer | hsa05202 | 15 | 0.007463353 | CD86, CDKN1A,
BCL2A1, ZBTB16, MMP3, GZMB, MMP9, IL6, NR4A3, BCL6, MYC, ITGB7,
PPARG, CD14, ELANE |
| ErbB signaling
pathway | hsa04012 | 10 | 0.008189004 | BTC, CDKN1A, JUN,
PRKCB, MYC, PLCG2, AREG, EGFR, EREG, HBEGF |
| Epstein-Barr virus
infection | hsa05169 | 12 | 0.010097479 | CDKN1A, CR2,
ENTPD1, JUN, SYK, MYC, CD19, PLCG2, HLA-DPB1, ITGAL, HLA-DQA1,
HLA-DQB1 |
| Toll-like receptor
signaling pathway | hsa04620 | 11 | 0.010274377 | CD86, CXCL10, IL6,
CXCL11, CXCL9, JUN, STAT1, CCL5, SPP1, FOS, CD14 |
| Influenza A | hsa05164 | 15 | 0.010556604 | JUN, PRKCB, STAT1,
CXCL10, SOCS3, HLA-DMA, IL6, HLA-DMB, CCL5, TNFSF10, HLA-DPB1,
JAK2, HLA-DOB, HLA-DQA1, HLA-DQB1 |
| Antigen processing
and presentation | hsa04612 | 9 | 0.011202712 | HLA-DMA, HLA-DMB,
CD8A, HLA-DPB1, TAP1, HLA-DOB, CTSS, HLA-DQA1, HLA-DQB1 |
| PI3K-Akt signaling
pathway | hsa04151 | 24 | 0.012003195 | CDKN1A, SYK, LAMA2,
ITGA4, LAMA3, VEGFD, FOXO3, IL2RG, EGFR, VEGFA, COL1A1, NR4A1, IL6,
TCL1A, FGF9, IL7, MYC, CD19, SPP1, ITGA7, ITGB7, JAK2, PCK1,
IL7R |
| Malaria | hsa05144 | 7 | 0.01339692 | IL6, KLRK1, CD40LG,
KLRB1, ITGB2, SDC1, ITGAL |
| Jak-STAT signaling
pathway | hsa04630 | 13 | 0.014076056 | IL15, STAT1,
IL10RA, LIF, CSF2RB, IL2RG, SOCS3, IL6, IL7, MYC, LEP, JAK2,
IL7R |
| Aldosterone
synthesis and secretion | hsa04925 | 9 | 0.016086366 | NR4A2, LIPE, NR4A1,
PRKCB, AGTR1, ADCY2, ADCY7, LDLR, KCNK3 |
| Phagosome | hsa04145 | 13 | 0.01799255 | ITGB2, TAP1, CYBA,
CORO1A, CTSS, HLA-DMA, HLA-DMB, HLA-DPB1, CD14, FCGR2B, HLA-DOB,
HLA-DQA1, HLA-DQB1 |
| FoxO signaling
pathway | hsa04068 | 12 | 0.019364257 | IL6, CDKN1A,
GABARAPL1, GADD45B, BCL6, GADD45A, TNFSF10, IRS2, PCK1, FOXO3,
IL7R, EGFR |
| ECM-receptor
interaction | hsa04512 | 9 | 0.023784926 | COL1A1, SDC4,
LAMA2, ITGA4, LAMA3, SPP1, SDC1, ITGA7, ITGB7 |
| MAPK signaling
pathway | hsa04010 | 18 | 0.02689659 | MAP4K1, DUSP4,
NTRK2, JUN, GADD45B, IL1R1, PRKCB, GADD45A, FOS, DUSP8, RASGRP1,
EGFR, NR4A1, FGF9, MYC, RAC2, CD14, PTPN7 |
| Measles | hsa05162 | 11 | 0.042383892 | IL6, STAT1, SH2D1A,
TNFSF10, CD3G, CD3E, FCGR2B, JAK2, IL2RG, CD3D, SLAMF1 |
| Systemic lupus
erythematosus | hsa05322 | 11 | 0.044240314 | CD86, HLA-DMA,
HLA-DMB, C6, CD40LG, C7, HLA-DPB1, HLA-DOB, ELANE, HLA-DQA1,
HLA-DQB1 |
| Prion diseases | hsa05020 | 5 | 0.046727411 | EGR1, IL6, C6, C7,
CCL5 |
| Amphetamine
addiction | hsa05031 | 7 | 0.049531294 | ARC, JUN, MAOA,
PRKCB, FOSB, FOS, SLC6A3 |
| Complement and
coagulation cascades | hsa04610 | 7 | 0.059219897 | CFD, CR2, SERPINA1,
C6, C7, CFB, SERPINA5 |
| AMPK signaling
pathway | hsa04152 | 10 | 0.060492183 | LIPE, SCD, LEP,
FASN, ADIPOQ, IRS2, PPARG, PCK1, FOXO3, ACACB |
| Adipocytokine
signaling pathway | hsa04920 | 7 | 0.062683741 | SOCS3, LEP, ADIPOQ,
IRS2, PCK1, JAK2, ACACB |
| Focal adhesion | hsa04510 | 14 | 0.073426753 | JUN, LAMA2, ITGA4,
PRKCB, LAMA3, VEGFD, EGFR, VEGFA, COL1A1, SPP1, RAC2, ITGA7, ITGB7,
BIRC3 |
| Pertussis | hsa05133 | 7 | 0.081761396 | CXCL6, IL6, JUN,
ITGB2, FOS, CD14, CXCL5 |
| HIF-1 signaling
pathway | hsa04066 | 8 | 0.092748632 | HK3, IL6, CDKN1A,
PRKCB, PLCG2, EGFR, PDK1, VEGFA |