Introduction
The coronary artery abnormal dilatation (CAAD) is an
uncommon cardiovascular disorder with an incidence ranging from 1.2
to 4.9% of patients undergoing coronary angiography (1). This pathology has been described
interchangeably using the two terms: coronary artery aneurysm (CAA)
and coronary artery ectasia (CAE) (2). However, these synonymously used terms
refer to two different phenotypes. CAA is defined as a focal
dilatation of the arteries with a diameter of 1.5 times the
adjacent normal coronary artery, whereas CAE describes similar but
more diffuse lesions (3).
According to the anatomical shape of the dilated segment, CAA is
classified into the fusiform type if the longitudinal diameter
exceeds the transverse diameter or the saccular type in the reverse
case (4). The presence of CAAD is
associated with poor long-term outcomes, regardless of concomitant
atherosclerotic coronary disease (5-7).
Clinical presentation includes asymptomatic cases
and stable angina or acute coronary syndromes (8,9).
Abnormal dilation of coronary arteries is attributed to
atherosclerosis in 50% of cases, while the origin of 20-30% are
considered inflammatory or congenital (10). Pathogenesis is multifactorial and
can be influenced by many environmental and heritable risk factors.
Understanding the underlying molecular and cellular mechanisms may
contribute to diagnosing and preventing cardiovascular events.
According to a growing body of literature, microRNAs (miRNAs) have
been implicated in regulating human physiological processes,
including gene expression in the cardiovascular system (11). MiRNAs could have a crucial role in
physiological processes and disease development, including
atherosclerosis, coronary artery disease (CAD), myocardial
infarction (MI), heart failure (HF), and cardiac arrhythmias
(12-14).
MiRNAs are short (19-25 nucleotides), single-stranded, noncoding
RNAs that function posttranscriptional regulation, attained by RNA
degradation and translation silencing. MiRNAs regulate gene
expression by binding to specific sites in the mRNA 3' untranslated
region (15). A single miRNA
downregulates numerous target genes, modulating complex
physiological processes. MiRNAs are involved in cardiovascular
remodeling, which results in cardiovascular diseases such as CAD,
abdominal aortic aneurysm (AAA), and HF (16). Our understanding the roles of serum
miRNAs in patients with CAAD is still not comprehensive; however,
essential knowledge came from the studying selected groups of
individuals with Kawasaki disease (KD) (17-19)
and AAA (20,21).
Several studies have found that the miRNA profiles
of serum exosomes or coronary artery tissues, including miR-23a,
miR-27b, miR-223, and miR-145, are associated with acute KD,
providing insight into the molecular mechanisms of the development
of cardiovascular lesions (17-19).
For example, increased levels of miR-23a contribute to
cardiomyocyte apoptosis and may promote inflammatory responses by
blocking macrophage autophagy activity (22). miR-145 is highly expressed in
vascular smooth muscle cells (VSMCs), mediating phenotype by
altering to proliferating neointimal cells (23). It is speculated that miR-145
modulates the generation of myofibroblasts from VSMC, which are
involved in arterial wall destruction in acute KD (24). Interestingly, other studies suggest
that noninflammatory vascular injury in animal models or human
diseases characterized by chronic vascular inflammation, are
associated with low levels of miR-145 (25,26).
The expression of tissue miR-145 was downregulated at the site of
experimental injury-induced lesions in animal models and human AAA
(27). Similarly, plasma or serum
levels of miR-145 in patients with stable CAD were lower than those
in healthy controls (28).
Interestingly, miR-223 is highly expressed in blood cells, such as
neutrophils, eosinophils, monocytes, and platelets, and can then
enter VSMCs to regulate their functions and atherogenesis via its
target genes (29,30).
Our study aimed to identify the circulating miRNA
signature in CAAD patients and explore its potential as a novel
biomarker for these diseases.
Materials and methods
Study design and patient
selection
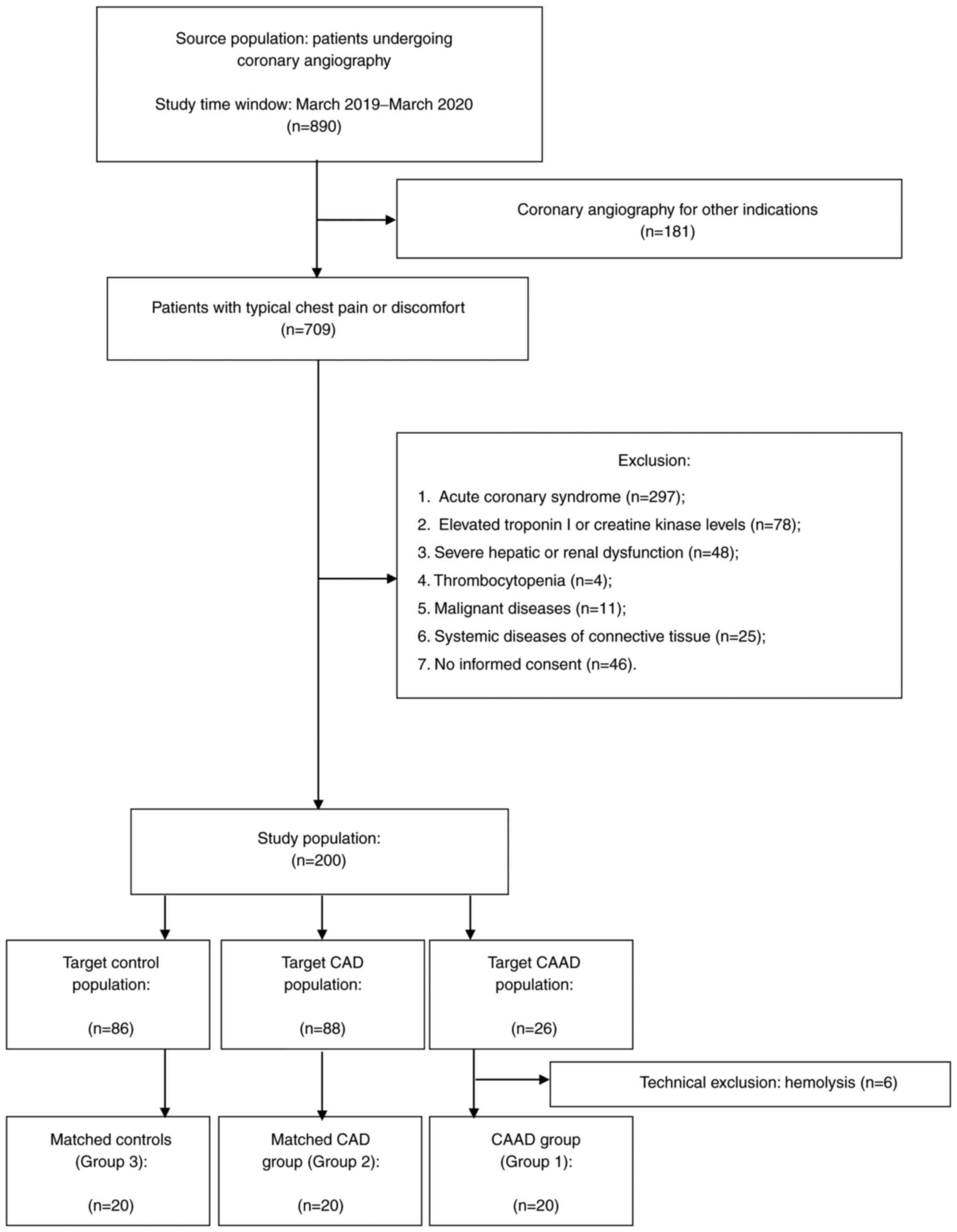
A total of two hundred patients undergoing coronary
angiography for angina symptoms were enrolled in the general cohort
between March 2019 and March 2020, including 26 consecutive
patients with CAAD. Due to technical exclusion (hemolysis), 20
patients meeting the criteria of CAE or CAA were included in the
study cohort as Group 1. Moreover, two groups of 20 patients
matched Group 1 in terms of sex and age from the overall cohort
(Group 2 and Group 3) (Fig. 1).
Patients with angiographically documented CAD were included in
Group 2 (n=20), while patients with angiographic exclusion of
coronary stenosis >50% were enrolled as control patients in
Group 3 (n=20). Group 1 and 2 included patients aged 53-77 years,
and Group 3 aged 51-78 years. The mean age was 66.1±7.2, 66.9±7.4,
65.1±7.4 for Groups 1, 2 and 3, respectively.
All patients with chest pain or discomfort were
qualified for coronary angiography, according to the European
Society of Cardiology (ESC) guidelines (31). CAE and CAA were defined as a
diffuse or focal dilatation of the coronary artery with a diameter
of 1.5 times the adjacent normal segment. The group also included
patients with associated stenosis of the coronary arteries. The
angiographic criteria for CAD were: coronary artery stenosis
>90% or intermediate stenosis (50-90%) with documented ischemia
or hemodynamically significant, defined as either fractional flow
reserve (FFR) ≤0.80 or an instantaneous wave-free ratio (iFR)
≤0.89. All patients in the control group presented with normal ECG
and echocardiography and had no evidence of ischemia during
noninvasive stress tests. The exclusion criteria were as follows:
i) Acute coronary syndrome; ii) elevated troponin I (TNI) or
creatine kinase (CK-MB) levels; iii) history of severe hepatic and
renal dysfunction; iv) leukemia, leukopenia, thrombocytopenia, or
ongoing inflammatory and malignant diseases; v) systemic diseases
of connective tissue; vi) interferon treatment; and vii) no
informed consent.
The Institutional Review Board (or Ethics Committee)
of Poznan University of Medical Sciences (protocol code, 985/18;
date of approval, October 11, 2018) approved the protocols, and the
study was conducted following the Declaration of Helsinki. We
obtained written informed consent from each individual.
Sample collection and miRNA
isolation
EDTA-blood samples (10 ml) were collected from all
patients on the first day after the cardiac catheterization
procedure and were processed within 30 min's of collection. Samples
were centrifuged at 1300 g for 15 min at room temperature. The
supernatant was transferred to RNase-free tubes and then stored at
-80˚C.
The miRNAs were isolated from individual 200 µl
frozen plasmas using the miRNeasy Serum/Plasma Advanced Kit (cat.
no. 217204, Qiagen, Dusseldorf, Germany) according to the
manufacturer's instructions. To correct sample-to-sample variation,
a synthetic set of spike-ins UniSp2, UniSp4 and UniSp5 was applied
[RNA Spike-In Kit, for reverse transcription (RT), cat. no. 339390,
Qiagen]. Three of the RNA spike-in templates (UniSp2, UniSp4, and
UniSp5) were premixed in one vial, each at a different
concentration in 100-fold increments of UniSp2 (2 fmol/µl), UniSp4
(0.02 fmol/µl) and UniSp5 (0.00002 fmol/µl). Before starting the
RNA isolation procedure, 1 µl of this RNA spike-in mix per RNA prep
was aliquoted into 60 µl RPL lysis buffer. Approximate RNA quantity
and quality were estimated using a NanoDrop 2000 spectrophotometer
(Thermo Scientific, Waltham MA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Four microliters of each total RNA was reverse
transcribed with a miRCURY LNA RT Kit (cat. no. 339340, Qiagen).
The UniSp6 RNA spike-in from the miRCURY LNA RT Kit and
cel-miR-39-3p RNA from the miRCURY LNA RT Kit were used as
controls. RT temperature protocol: reverse transcription step 60
min at 42˚C, inactivation of reaction 5 min at 95˚C. All reverse
transcribed samples were verified using the miRCURY LNA miRNA QC
PCR Panel (cat. no. 339331, Qiagen). This panel contains eight
samples per 96 well plate, controls for RNA isolation quality, and
monitors cDNA synthesis in the reverse-transcription experiment.
The QC panel contains primers for hsa-miR-103a-3p, mmu-miR-191-5p,
hsa-miR-451a, hsa-miR-23a-3p, UniSp6, UniSp2, UniSp4, UniSp5,
miR-39-3p, UniSp3, hsa-miR-124-3p and hsa-miR-30c-5p. Quantitative
qPCR using the miRCURY LNA miRNA QC PCR Panel (cat. no. 339331,
Qiagen), 0.8 µl of RT reaction product per sample, and the miRCURY
LNA SYBR Green PCR Kit (cat. no. 339345, 339346, 339347, Qiagen)
was used to control samples on a LightCycler 480 (Roche, Basel,
Switzerland). According to manufacturer instruction we applied
following cycling conditions: denaturation 2 min 95˚C, cycling: 50
cycles as follows 10 sec at 95˚C, 69 sec at 56˚C. Also according to
the manufacturer instructions ‘miRCURY LNA miRNA QC PCR Panel
Handbook,’ we used samples only when they passed the following
conditions: i) UniSp2, 4, and 5 showed consistent values across the
sample (<2-3 Cq) and ΔCq=5-7 between spike-Ins. cDNA synthesis
was evaluated using UniSp6 and cel-39-3p. Samples passed
quantification when their Cq values were consistent across the
sample set with low variance (<1-2 Cq). ii) Samples passed the
qPCR reaction after evaluation by UniSp3, which showed consistent
values across the dataset (<2 Cq across the dataset). Evaluation
of hemolysis miR-451 and 2. iii) Hemolysis was evaluated by
measuring miR-23a and miR-451 expression. The plasma sample quality
was sufficient if ΔCq (miR-23a-miR-451) was lower than 7.
A total of 20 µl cDNA product of the miRCURY LNA RT
Kit was applied for each qPCR. qPCR was performed using a miRCURY
LNA SYBR Green PCR Kit (cat. no. 339345, 339346, 339347, Qiagen) on
miRCURY LNA Serum/Plasma Focus PCR Panels (cat. no. YAHS-106YF-8,
Qiagen).
Statistical analysis
All continuous variables were presented as means
with standard deviation for normal distribution or medians (upper
and lower quartile) for non-normal distribution. The normality of
the distribution of variables was tested using the
Kolmogorov-Smirnov test. Categorical variables were presented as
counts and percentages or frequencies. The significance of
differences between the mean values of the continuous data
consistent with the normal distribution was assessed using one-way
ANOVA and Tukey's Test. Mann-Whitney and Kruskal-Wallis tests with
Benjamini-Hochberg post-hoc analysis were used to compare the
continuous data inconsistent with the normal distribution.
Categorical variables were compared using the χ2
test.
Analysis of array data was performed with GENEGLOBE
online software (https://geneglobe.qiagen.com/pl/analyze). The global
mean was used as a reference for the PCR-array analysis. The p
values were calculated based on Student's t-test of the replicate
2(-ΔCq) values for each miRNA in the control and treatment groups
(32). The P-value calculation
used in this analysis is based on a parametric, two-sample equal
variance, unpaired, two-tailed analysis. The alpha level was set a
priori at 0.05.
We performed an additional multivariate analysis to
evaluate the impact of selected comorbidities and cardiovascular
risk factors on the obtained miRNA results. Due to the limited size
of the group, it was possible to include up to three variables in
one analysis. Two logistic regression models were built to consider
the essential variables. All miRNAs, which expression was
significantly different (P<0.05) in CAAD group than in the other
groups, were included in the multivariate analysis. The results
were obtained separately to compare Group 1 vs. Group 2 and Group 1
vs. Group 3. We used PQStat Software (PQStat v.1.8.0.476, Poland)
for statistical analysis.
Results
Clinical characteristics of the study
population
The clinical characteristics of the study
populations are summarized in Table
I. Patients with CAAD had a significantly higher BMI than the
patients with CAD and the control group. There were no significant
differences in other cardiovascular risk factors between Group 1
and Group 2. The control patients had no history of cardiovascular
events and were characterized by a significantly lower risk of
hypertension and diabetes. Despite a comparable frequency of HF in
all groups, left ventricular ejection fraction (LVEF) was
significantly higher in the control patients than in those with
CAD. Moreover, a significantly lower percentage of patients in
Group 3 were treated with aspirin, and none of them had indications
for dual antiplatelet therapy (DAPT). Clopidogrel has been
significantly more often used in patients with CAD.
 | Table IBaseline clinical
characteristics. |
Table I
Baseline clinical
characteristics.
| | P-value |
|---|
| Baseline data | Group 1 (n=20) | Group 2 (n=20) | Group 3 (n=20) | Group 1 vs. Group
2 | Group 1 vs. Group
3 | Group 2 vs. Group
3 |
|---|
| Sex, male, n
(%) | 15 (75.0) | 15 (75.0) | 15 (75.0) | 0.5 | 0.7 | 0.3 |
| Age, years, mean ±
SD | 66.1±7.2 | 66.9±7.4 | 65.1±7.4 | 0.8 | 0.8 | 0.6 |
| BMI,
kg/m2, mean ± SD | 31.9±4.7 | 28.6±5.1 | 28.0±4.1 | 0.04 | 0.002 | 0.4 |
| Previous MI, n
(%) | 5 (25.0) | 7 (35.0) | 0 | 0.3 | 0.01 | <0.001 |
| Previous PCI, n
(%) | 8 (40.0) | 8 (40.0) | 0 | 0.9 | 0.001 | <0.001 |
| Previous CABG, n
(%) | 2 (10.0) | 3 (15.0) | 0 | 0.3 | 0.3 | 0.06 |
| Hypertension, n
(%) | 18 (90.0) | 18 (90.0) | 13(65) | 0.3 | 0.04 | 0.01 |
| Heart failure, n
(%) | 11 (55.0) | 12 (60.0) | 9 (45.0) | 0.2 | 0.9 | 0.2 |
| Median LVEF, %
(Q1-Q3) | 60.0
(42.5-60.0) | 52.5
(45.0-55.0) | 60.0
(53.7-61.2) | 0.5 | 0.1 | 0.01 |
| Diabetes mellitus,
n (%) | 8 (40.0) | 6 (30.0) | 1 (5.0) | 0.2 | 0.02 | 0.02 |
| Hiperlipidemia, n
(%) | 17 (85.0) | 16 (80.0) | 13 (65.0) | 0.3 | 0.1 | 0.6 |
| Cigarette smoking,
n (%) | 8 (40.0) | 8 (40.0) | 8 (40.0) | 0.8 | 0.9 | 0.9 |
| Aortic aneurysm, n
(%) | 1 (5.0) | 2 (10.0) | 1 (5.0) | 0.7 | 0.7 | 0.5 |
| CKD, n (%) | 12 (60.0) | 13 (65.0) | 13 (65.0) | 0.2 | 0.2 | 1 |
| Drug
administration | | | | | | |
|
Statin, n
(%) | 16 (80.0) | 17 (85.0) | 17 (85.0) | 0.3 | 0.9 | 0.3 |
|
CCB, n
(%) | 7 (35.0) | 5 (25.0) | 7 (35.0) | 0.5 | 0.9 | 0.6 |
|
Beta-blocker,
n (%) | 18 (90.0) | 18 (90.0) | 15 (75.0) | 0.3 | 0.08 | 0.02 |
|
Aspirin, n
(%) | 16 (80.0) | 17 (85.0) | 10 (50.0) | 0.3 | 0.02 | 0.003 |
|
Clopidogrel,
n (%) | 8 (40.0) | 14 (70.0) | 0 | 0.03 | 0.001 | <0.001 |
|
ACEI/ARB, n
(%) | 16 (80.0) | 17 (85.0) | 14 (70.0) | 0.3 | 0.3 | 0.052 |
| Laboratory
tests | | | | | | |
|
LDL
cholesterol, mmol/l | 2.3±1.3 | 2.4±1.2 | 2.6±1.2 | 0.7 | 0.4 | 0.6 |
|
Creatinine,
mmol/l | 90.7±25.8 | 91.9±26.1 | 93.6±26.7 | 0.3 | 0.2 | 0.5 |
|
GFR,
ml/min | 74.0±14.5 | 73.2±14.2 | 73.2±14.5 | 0.4 | 0.6 | 0.6 |
The angiographic characteristics of Group 1 are
presented in Table II. CAE was
diagnosed in the majority of patients (75%). CAAD, both diffuse and
focal, often involves only one vessel in the right coronary artery
(RCA). CAD was angiographically documented in 55% of patients.
 | Table IIAngiographic characteristics of Group
1 (CAAD group; n=20). |
Table II
Angiographic characteristics of Group
1 (CAAD group; n=20).
| Baseline data | Group 1, n (%) |
|---|
| CAE | 15 (75.0) |
| CAA | 4 (20.0) |
| Both | 1 (5.0) |
| Number of vessels
involved | |
|
1 | 14 (70.0) |
|
2 | 5 (25.0) |
|
3 | 1 (5.0) |
| Vessel
localization | |
|
LM | 0 |
|
RCA | 10 (50.0) |
|
LAD | 9 (45.0) |
|
LCx | 8 (40.0) |
| Concomitant
CAD | 11 (55.0) |
Expression profile of miRNAs in the
plasma of Group 1 vs. Group 2 and Group 3
We analyzed the miRNA expression profiles in the
plasma of all patients from the study cohort a using miRNA PCR
array and compared the results among the three studied groups. Due
to the presence of hemolysis in the 3 tested samples, 57 patients
were finally included in molecular analysis, 19 from Group 1, 18
from Group 2 and 20 from Group 3.
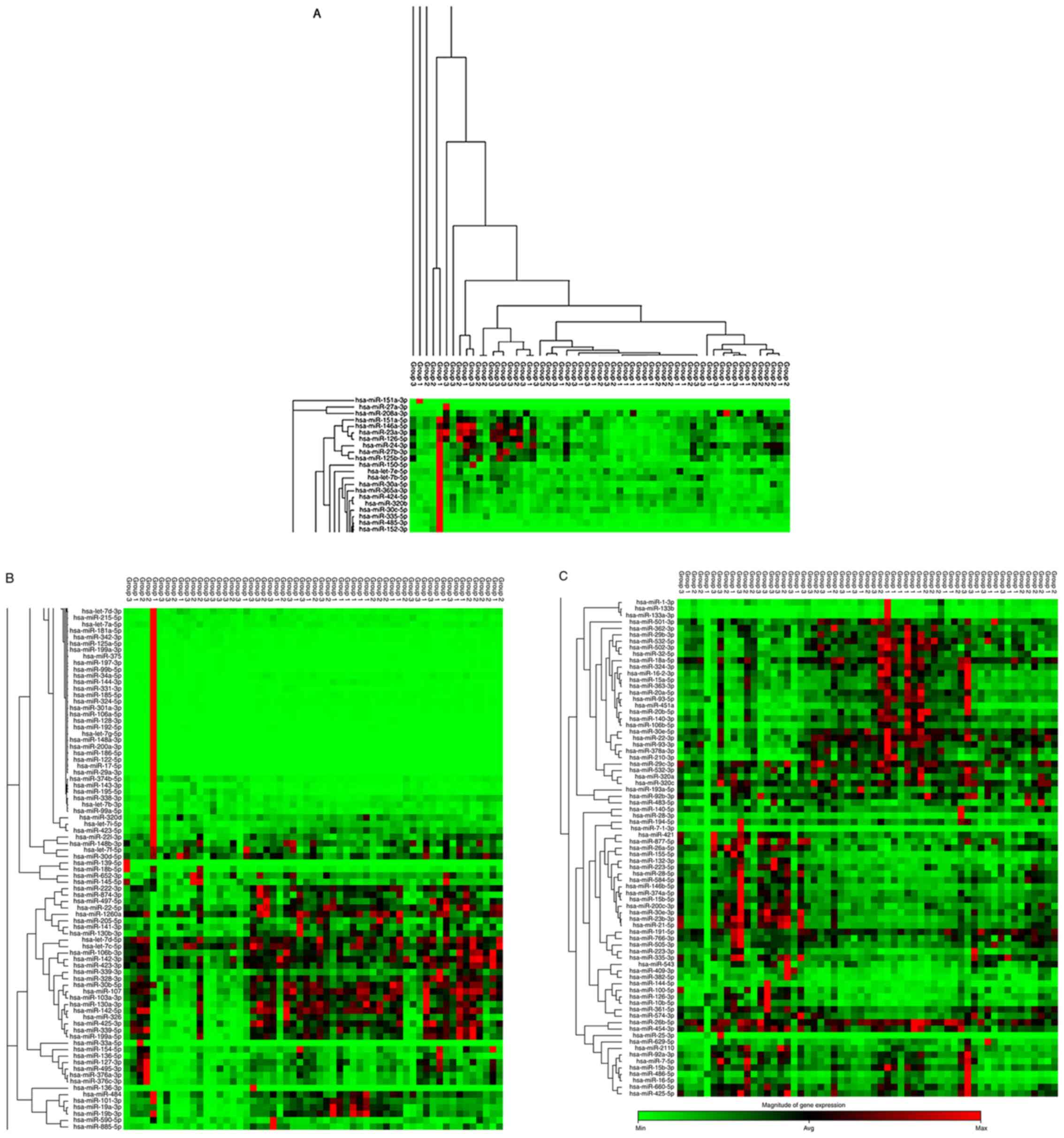
The levels of circulating miRNAs differed profoundly
between Group 1 and Group 3, as illustrated in the heat map diagram
shown in Fig. 2. We identified
twenty-three significantly deregulated miRNAs (fold change >2
and P<0.05, see P-values in Table
III). Twenty-one were upregulated, and two were downregulated
in patients with aneurysmal coronary artery dilatation compared to
the control group (Table
III).
 | Table IIIDeregulated miRNAs in the circulation
of Group 1 compared with Group 3. |
Table III
Deregulated miRNAs in the circulation
of Group 1 compared with Group 3.
| miRNA ID | Fold change | P-value |
|---|
| hsa-miR-210-3p | 6.44 | 0.000314 |
| hsa-miR-326 | 5.22 | 0.020272 |
| hsa-miR-142-5p | 4.79 | 0.020595 |
| hsa-miR-19a-3p | 4.76 | 0.002408 |
| hsa-miR-19b-3p | 4.35 | 0.012555 |
| hsa-miR-339-5p | 4.03 | 0.019595 |
| hsa-miR-874-3p | 3.79 | 0.034911 |
| hsa-miR-497-5p | 3.30 | 0.005334 |
| hsa-miR-425-3p | 3.30 | 0.023547 |
| hsa-miR-20a-5p | 2.85 | 0.017399 |
|
hsa-miR-106b-5p | 2.85 | 0.003684 |
|
hsa-miR-378a-3p | 2.74 | 0.007929 |
| hsa-miR-532-5p | 2.72 | 0.008666 |
| hsa-miR-502-3p | 2.61 | 0.001956 |
| hsa-miR-20b-5p | 2.54 | 0.028569 |
| hsa-miR-328-3p | 2.47 | 0.019208 |
| hsa-miR-107 | 2.37 | 0.005988 |
|
hsa-miR-130b-3p | 2.31 | 0.026696 |
|
hsa-miR-103a-3p | 2.21 | 0.013745 |
| hsa-miR-93-3p | 2.10 | 0.021729 |
| hsa-miR-339-3p | 2.09 | 0.024420 |
| hsa-miR-23a-3p | -2.36 | 0.001293 |
|
hsa-miR-125b-5p | -2.55 | 0.001279 |
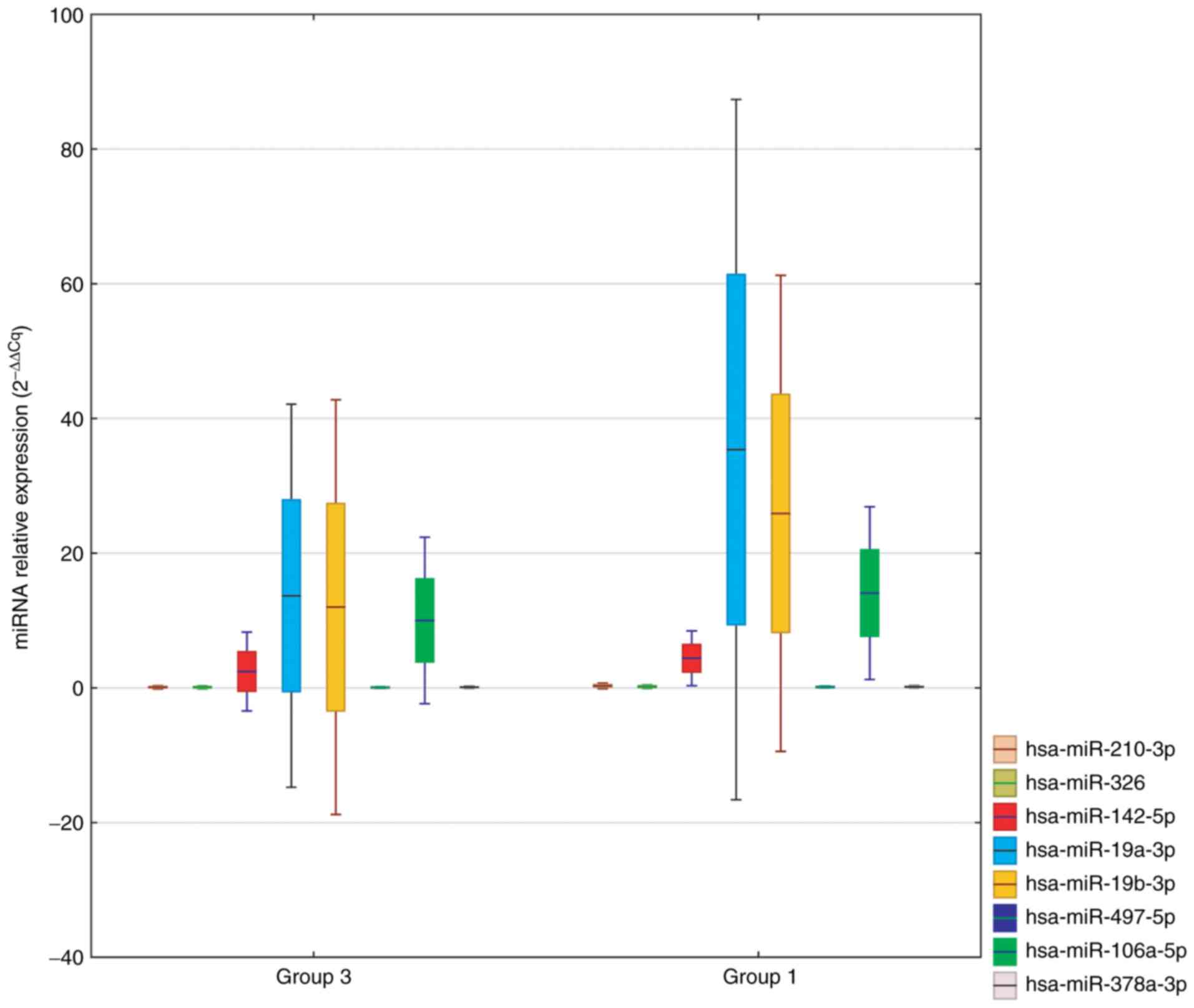
A comparison of circulating miRNAs, characterized by
the greatest fold change in Group 1 vs. Group 3, is shown in
Fig. 3. We selected:
hsa-miR-210-3p, hsa-miR-326, hsa-miR-142-5p, hsa-miR-19a-3p,
hsa-miR-19b-3p, hsa-miR-339-5p, hsa-miR-874-3p and hsa-miR-425-3p
as potential markers.
Moreover, we compared the miRNA expression profiles
in the plasma of Group 2 with Group 3 and identified eight
significantly deregulated miRNAs (fold change >2 and P<0.05
see P-values in Table IV). Six
were upregulated, and two were downregulated in patients with CAD
compared to controls (Table
IV).
 | Table IVDeregulated miRNAs in the circulation
of Group 2 compared with Group 3. |
Table IV
Deregulated miRNAs in the circulation
of Group 2 compared with Group 3.
| miRNA | Fold change | P-value |
|---|
| hsa-miR-885-5p | 3.22 | 0.024577 |
|
hsa-miR-133a-3p | 3.07 | 0.024260 |
| hsa-miR-483-5p | 2.77 | 0.002122 |
| hsa-miR-425-3p | 2.61 | 0.042032 |
| hsa-miR-328-3p | 2.42 | 0.012738 |
| hsa-miR-191-5p | 2.21 | 0.049314 |
| hsa-miR-145-5p | -2.15 | 0.013063 |
|
hsa-miR-125b-5p | -2.32 | 0.007553 |
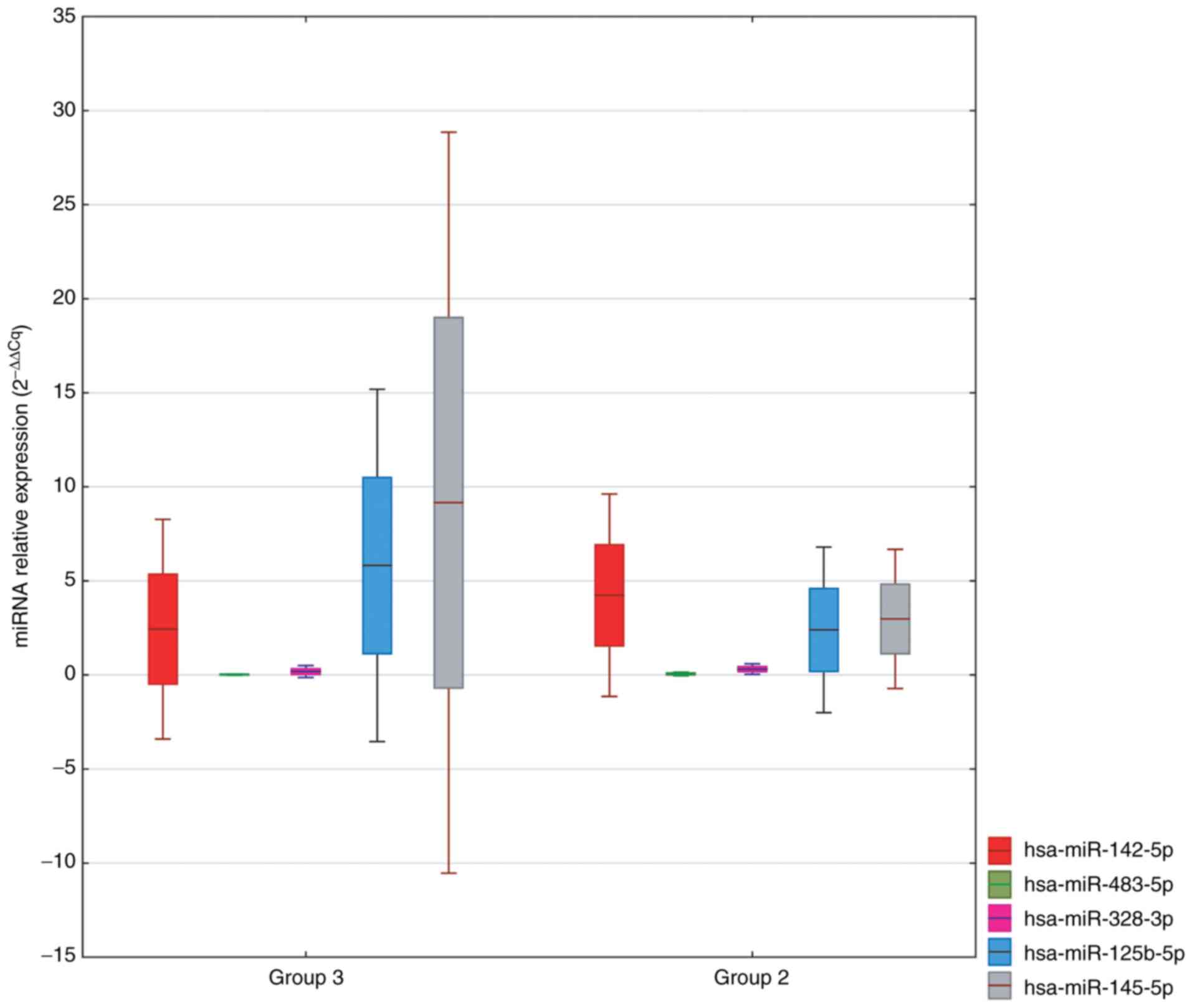
A comparison of circulating miRNAs characterized by
the greatest fold change in Group 3 vs. Group 2 is shown in
Fig. 4. We selected:
hsa-miR-885-5p, hsa-miR-133a-3p, hsa-miR-483-5p, hsa-miR-425-3p,
hsa-miR-328-3p, hsa-miR-191-5p, hsa-miR-145-5p and hsa-miR-125b-5p
as potential markers.
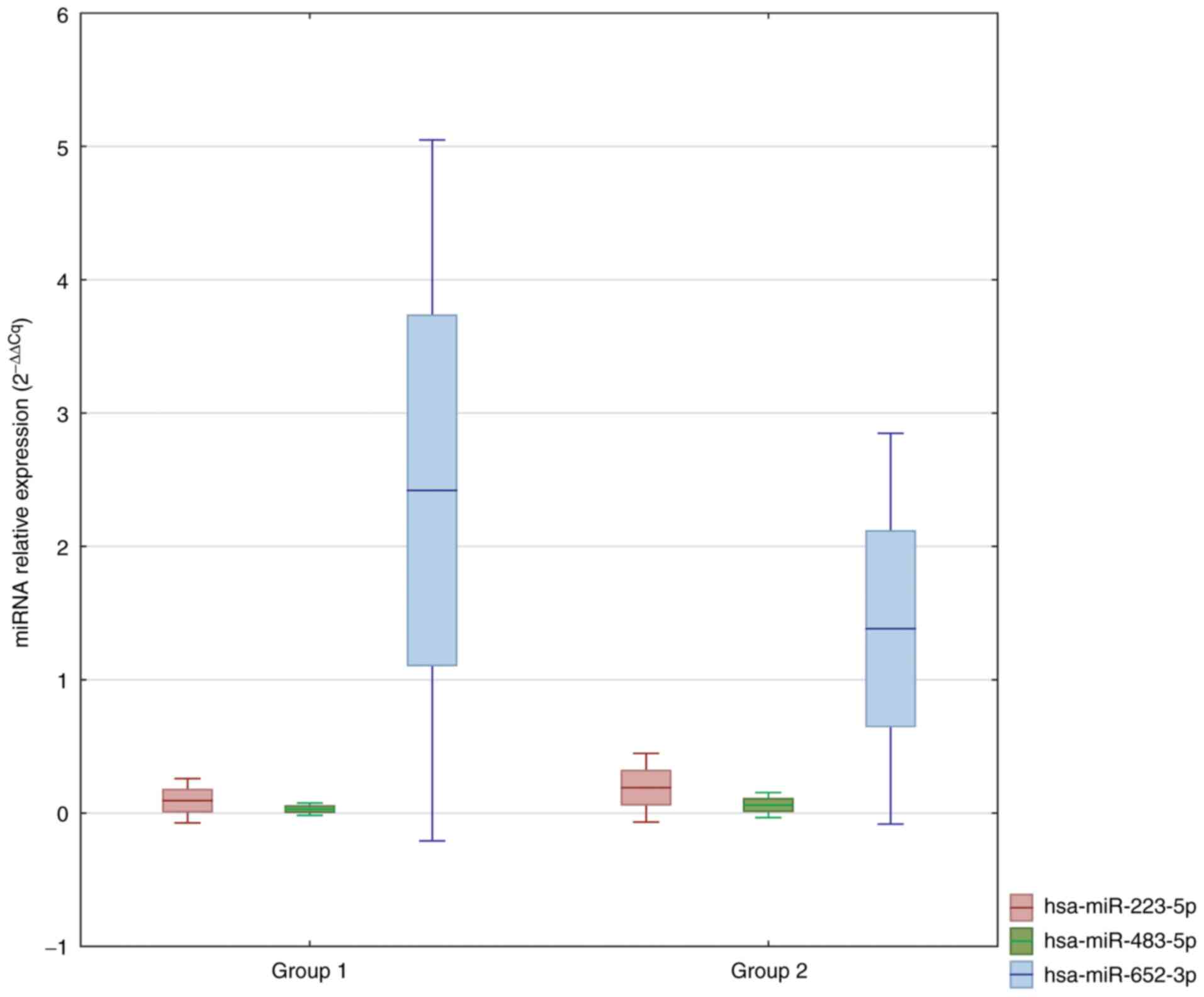
Comparing the miRNA expression profiles in the
plasma of Group 2 and Group 1 identified seven significantly
deregulated miRNAs (fold change >2 and P<0.05 see P-values in
Table V). Three of these miRNAs
were upregulated, and four were downregulated in Group 2 compared
to Group 1 (Table V).
 | Table VDeregulated miRNAs in the circulation
of Group 2 compared with Group 1. |
Table V
Deregulated miRNAs in the circulation
of Group 2 compared with Group 1.
| miRNA ID | Fold change | P-value |
|---|
| hsa-miR-223-5p | 2.35 | 0.009035 |
| hsa-miR-483-5p | 2.06 | 0.015292 |
| hsa-miR-375 | 2.04 | 0.026221 |
|
hsa-miR-16-2-3p | -2.08 | 0.017763 |
| hsa-miR-210-3p | -2.22 | 0.036750 |
| hsa-miR-652-3p | -2.25 | 0.005766 |
| hsa-miR-18b-5p | -2.50 | 0.034395 |
A comparison of circulating miRNAs characterized by
the greatest fold change in Group 3 vs. Group 2 is shown in
Fig. 5. We selected:
hsa-miR-885-5p, hsa-miR-133a-3p, hsa-miR-483-5p, hsa-miR-425-3p,
hsa-miR-328-3p, hsa-miR-191-5p, hsa-miR-145-5p and hsa-miR-125b-5p
as potential markers.
Multivariate analysis of miRNA
expression profile in Group 1 (n=19) compared to Group 3
(n=20)
In the first logistic regression model, the obtained
results of miRNA expression were divided according to the presence
of arterial hypertension, diabetes, and HF, which allowed the
selection of eight miRNAs that were independent risk factors for
coronary aneurysms compared to the control group (Table VI).
 | Table VIMultivariate analysis of miRNA
expression profile in Group 1 compared to Group 3, adjusted for
heart failure, diabetes and hypertension. |
Table VI
Multivariate analysis of miRNA
expression profile in Group 1 compared to Group 3, adjusted for
heart failure, diabetes and hypertension.
| miRNA | ORa [95% CI] | P-value |
|---|
|
hsa-miR-103a-3p | 2.6 [1.3, 3.1] | 0.033 |
| hsa-miR-20a-5p | 3.2 [1.1, 5.2] | 0.049 |
| hsa-miR-107 | 1.5 [1.1, 2.6] | 0.036 |
| hsa-miR-19a-3p | 2.7 [1.6, 3.8] | 0.009 |
| hsa-miR-19b-3p | 1.4 [1.03,
3.8] | 0.011 |
|
hsa-miR-106b-5p | 1.6 [1.1, 2.4] | 0.032 |
| hsa-miR-142-5p | 2.8 [1.3, 3.9] | 0.031 |
|
hsa-miR-16-2-3p | 9.0 [1.1, 2.9] | 0.048 |
Similarly, in the second regression model, which
included smoking, renal failure, and dyslipidemia, a profile of
eight miRNAs were identified. The upregulation of aforementioned
miRNAs was significantly correlated with the risk of CAAD (Table VII). In addition, in a separate
regression model, miRNA expression results were randomized on the
basis of the use of antiplatelet drugs. The results are presented
in Table VIII. In multivariate
analysis, miRNA expression did not differ between Groups 1 and
2.
 | Table VIIMultivariate analysis of miRNA
expression profile in Group 1 compared to Group 3, adjusted for
smoking, renal failure, and dyslipidemia. |
Table VII
Multivariate analysis of miRNA
expression profile in Group 1 compared to Group 3, adjusted for
smoking, renal failure, and dyslipidemia.
| miRNA | ORa [95% CI] | P-value |
|---|
|
hsa-miR-103a-3p | 9.1 [1.1,
14.1] | 0.022 |
| hsa-miR-20a-3p | 5.5 [2.8, 7.2] | 0.042 |
| hsa-miR-107 | 3.2 [2.1, 4.3] | 0.024 |
| hsa-miR-19a-3p | 6.8 [4.2, 8.2] | 0.004 |
| hsa-miR-19b-3p | 1.2 [1.0, 1.9] | 0.004 |
|
hsa-miR-106b-5p | 4.2 [2.5, 7.2] | 0.022 |
| hsa-miR-425-3p | 2.6 [1.1, 4.2] | 0.048 |
| hsa-miR-339-5p | 1.2 [1.0, 2.1] | 0.038 |
 | Table VIIIMultivariate analysis of miRNA
expression profile in Group 1 compared to Group 3, adjusted for the
use of antiplatelet drugs. |
Table VIII
Multivariate analysis of miRNA
expression profile in Group 1 compared to Group 3, adjusted for the
use of antiplatelet drugs.
| miRNA | ORa [95% CI] | P-value |
|---|
|
hsa-miR-103a-3p | 2.7 [1.2, 3.8] | 0.036 |
| hsa-miR-107 | 1.05 [1.0,
2.18] | 0.047 |
| hsa-miR-19a-3p | 2.02 [1.8,
3.6] | 0.012 |
| hsa-miR-19b-3p | 8.4 [6.2,
10.1] | 0.014 |
|
hsa-miR-106b-5p | 1.5 [1.1, 2.2] | 0.042 |
| hsa-miR-142-5p | 7.4 [3.2, 8.6] | 0.029 |
Discussion
The present study demonstrates that the plasma
miRNAs detected in the blood were different in patients with CAAD
than in other groups. We identified a specific signature of plasma
miRNAs that were upregulated and downregulated in Group 1 compared
to Group 2 and Group 3. In addition, we selected potential miRNA
markers for further validation in a larger independent patient
cohort.
Upon detailed analysis, we noticed that miRNA-210-3p
was significantly upregulated in Group 1 vs. Group 2 and Group 3.
In contrast, no differences in the expression of this miRNA were
found between Groups 2 and 3. miR-210 represents prominent
hypoxia-inducible miRs, also known as hypoxemic, which are
expressed in a wide range of primary and transformed cells
(33). Moreover, Fasanaro et
al first reported that hypoxia-driven miR-210 supporting the
angiogenic response in endothelial cells (34). The effect mainly resulted from the
downregulation of EFNA3, an ephrin family member involved in
vascular development (34). The
proangiogenic effect of miR-210 was evaluated in MI, which was
proved by the improvement of endothelial cell survival after the
delivery of miR-210 in the heart (35). The involvement of miR-210 in the
regulation of pathophysiological angiogenesis has also been
demonstrated in renal ischemia/reperfusion (I/R) injury, indicating
that miR-210 induction is necessary to drive the expression of VEGF
and VEGFR2 in endothelial cells (36). The higher expression of miR-210-3p
in Group 1 compared to Groups 2 and 3 remains challenging to
explain. It would seem that the most severe ischemia occurs among
CAD patients in Group 2, and thus the level of miR-210-3p
expression should be the highest in this group. However, it is
important to note that as many as 55% of patients with CAAD have
been diagnosed with CAD. Furthermore, coronary dilatation can cause
a turbulent and stagnant flow, possibly increasing the risk of
thrombus formation and peripheral microembolization, which can also
be a source of ischemia. The influence of miR-210-3p on the
angiogenesis process also seems to be important. Ischemia, which
arises from significant coronary stenosis, activates angiogenesis
processes that lead to vessel remodeling and branching of new blood
vessels. Angiogenesis requires many molecular mechanisms regulated
by pro- and antiangiogenic factors (37,38).
The imbalance between them can lead to thinning of the vessel walls
and subsequent coronary aneurysm formation.
In addition, hsa-miR-328-3p and hsa-miR-425-3p were
upregulated, and hsa-miR-125b-5p was downregulated in both Group 1
and Group 2 compared to Group 3. Data on the involvement of the
above miRNAs in the pathogenesis of CAAD and CAD are minimal. Qin
et al showed that miR-328-3p protects vascular endothelial
cells against ox-LDL-induced injury by targeting FOXO4(39). Its expression was downregulated in
oxidized low-density lipoprotein (ox-LDL) treated human umbilical
vein endothelial cells (HUVECs). Overexpression of miR-328-3p
attenuated ox-LDL-induced inhibition of cell viability, migration,
and invasion and stimulation of apoptosis, autophagy and
inflammation in HUVECs. Moreover, numerous studies have shown that
miR-328-3p and miR-425-3p may play a crucial role in malignant
tumor progression in osteosarcoma, glioblastoma, and bladder cancer
(40-44).
miR-125b-5p was downregulated in the aneurysmal tissue of AAA,
specifically in the FDG-uptake site, compared with a negative zone
in the same aneurysm and with no FDG uptake aneurysms (45). Furthermore, it was inversely
correlated with the expression of some of their potential gene
targets at the positive uptake site, most notably matrix
metalloproteinase 13. Increase of miR-133 in Group 2 has been found
to regulate endothelial function and angiogenesis, vascular smooth
muscle cell differentiation, apoptosis, and cardiac myocyte
differentiation, and repress cardiac hypertrophy. Its role in these
processes may be explained by the regulation of targets such as
FSCN1, LASP1, purine nucleoside phosphorylase (PNP), and transgelin
2 (TAGLN2) (46). High expression
of miR-483-5p in Group 2, has been found to inhibit angiogenesis by
targeting serum response factor (SRF), providing a clue for
combating angiogenesis in CAD patients (47). In conclusion, our findings
correlated with the observed upregulation of plasma miR-483-5p and
may be favorable for CAD patients protection against adverse
effects caused by plaque rupture (48). Our results also revealed
downregulated miR-145-3p in the patients with CAD compared to the
control group. These findings are in line with previous reports,
which showed that chronic vascular inflammation is associated with
the level of miR-145. The authors postulated that miR-145 is a
negative regulator of TGF-b signaling and thus may contribute to
the downregulation of inflammation in the arterial wall (22,29,48).
Nevertheless, the mechanism of miR-145 reduction in diseases
associated with chronic inflammation is still unclear and requires
further studies on animal models. Due to the varied etiology of
CAAD, the level of miR-145-3p in Group 1 did not differ
significantly from that in the other groups.
miR-145 expression levels strongly correlate with
miR-223, suggesting that a shared mechanism may regulate these
miRNAs (29,30). In our study, miR-223 expression was
downregulated in Group 1 compared to Group 2. We know from previous
studies that the level of miR-223 in the blood serum of KD was
higher in patients with vascular injury than in patients without
vascular complications, and it was reduced after immunoglobulin
treatment (17). One of the target
genes for miR-223 is STAT1 mRNA at the 3-UTR. JAK/STAT signaling is
one of the most important regulatory pathways in many inflammatory
processes. It initiates innate and acquired immunity and mediates
various cytokine signaling pathways (49,50).
Dysfunction in the regulation of miRNA-223 and related target genes
in immune and myocardial cells is believed to contribute to the
development of heart disease, including CAD and HF. These data may
indicate the most advanced inflammatory process and subsequent
vessel damage in patients with atherosclerosis.
Reduced expression of miR-23a-3p in CAAD group
compared to control is also worth discussing. This miRNA belongs to
the miR-23/27/24 cluster members on chromosome 19 (19p13.13) and
plays a role in cell cycle control, proliferation, differentiation,
and apoptosis (51,52). It is involved in the angiogenesis
process by regulating the growth of cardiomyocytes, inducing the
proliferation of VSMCs, and inhibiting VSMC apoptosis by targeting
the BCL2L11 (BIM) gene (51).
Several lines of evidence revealed that miR-23a-3p was
downregulated in the walls of large AAA, supporting the hypothesis
that the downregulation of miR-23a-3p can contribute to AAA
development and progression (53).
The above data may suggest common pathogenesis of AAA and CAAD.
Multivariate analysis revealed that the several
miRNAs with higher expression in Group 1 than in Group 3. One of
these miRNAs is hsa-miR-103a-3p, whose expression is altered in the
state of inflammation, immune disorders, and cancer (54-57).
Li et al reported that miR-103a-3p is involved in septic
injury. Its overexpression reduced lipopolysaccharide (LPS)-induced
inflammation (23). Moreover,
studies revealed that miR-103a-3p is proinflammatory also through
the renal pathway. Increased miR-103a-3β expression enhances
angiotensin II-induced renal inflammation and injury. Researchers
have also found the increased levels of type I and IV collagen
protein and mRNA in the kidneys (58). In addition, Jiao et al
presented in vitro and in a murine model that have showed
that miR-103a-3p could also be involved in AAA by targeting
ADAM10(59). Plana et al
revealed the overexpression of miR-103a-3p in AAA tissue compared
to healthy tissues (25).
Moreover, the studies showed that the overexpression of miR-103a-3p
inhibited high mobility group Box 1 (HMGB1) expression. HMGB1 is a
universal, nonhistone DNA-binding protein and a common inflammatory
regulator that sensitizes many inflammation-related signaling
pathways, leading to the production of proinflammatory cytokines
(60).
Similar results were obtained for hsa-miR-107,
hsa-miR-19a-3p and hsa-miR-19b-3p. Interestingly, abnormal
expression of miR-107-5p, member 3 of the solute carrier 24 family
(SLC24A3), an integral membrane protein 2C (ITM2C), was identified
in acute aortic dissection (AD) (61). miR-107-5p expression was higher in
AD samples in comparison with normal aortic samples. In turn, the
expression of ITM2C in AD tissue was lower than that in normal
aortic samples. Moreover, miR-107-5p inhibited the progression of
acute AD by targeting ITM2C.
The miR-19a-3p family is one of the most important
factors regulating heart disease and cancer development, including
the extensive invasion of malignancies (62). The expression level of miR-19a-3p
is also upregulated after MI. For example, miR-19 inhibited
apoptosis and promoted cardiomyocyte proliferation. In addition,
the upregulation of miR-19 reduces the formation of endothelial
cells and regulates the expression level of cyclin D1 and
fibroblast growth factor receptor 2, blocking the cell cycle
(62). Moreover, an essential role
of miR-19a-3p in the regulation of angiogenesis has been
demonstrated. It has also been shown that the inhibition of
miR-19a-3p promotes angiogenesis in mice with MI (63).
Circulating miRNAs are influenced by many factors,
most of them uncontrolled (12,13).
The idea is to look for the most stable markers and signature of
the patient's condition. In our study, the prevalence of almost all
risk factors for ischemic disease and comorbidities was comparable
in Groups 1 and 2. Antiplatelet therapy affects the expression of
some miRNAs, such as miR-19b, miR-191, miR-223. However, due to the
disease profile, the frequency of its use was significantly lower
in the control group.
The current outcomes of miRNA profile in serum are
uncertain in clinical diagnosis of CAAD, but we believe there is
nonetheless a regular distribution of miRNA expression in a large
number of patients' serum. Statistic randomness of miRNA in a serum
profile is a kind of order that emerges only in a large number of
repeats. On the other hand, our results, like many other earlier
studies of miRNA serum profile, suggest that miRNA profiles are not
statistically ‘haphazard’ (15).
The precision of CAAD diagnostics upon miRNA profiles will probably
increase in the future by applying new models for meta-analysis
incorporating several studies on CAAD patients. The number of
sufficient samples, cannot be predicted to estimate probability
since the number of factors determining miRNA profiles is
unknown.
Our study identified miRNAs as potential biomarkers
of CAAD. However, the relationship of miRNAs with the process of
atherosclerosis and other aneurysms has not been demonstrated.
Regardless, the relationship of miRNAs with the angiogenesis
process and cancer pathogenesis remains interesting. Undoubtedly,
the obtained results require further validation.
Acknowledgements
Authors thank Ms Agnieszka Hertel (Poznan University
of Medical Sciences) for her helpful assistance in every step of
miRNA profiling in blood. We also thank Dr Agata
Maciejak-Jastrzębska (Medical University of Warsaw) for her helpful
assistance in PCR-array analysis.
Funding
Funding: This research was funded by Poznan University of
Medical Sciences, Poland, Young Scientists 2018.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the NCBI Gene Expression Omnibus
repository and are accessible through GEO Series accession number
GSE198885 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE198885).
Authors' contributions
SI, TL and AA designed the current study and wrote
the manuscript. AC, AR, KM and MW collected clinical samples and
prepared them for further analyses. TL and SI conducted the
experiments and statistical analyses. PJ, MG and ML made
substantial contribiutions to conception and design, and revised
this manuscript. SI and TL confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The study was conducted according to the guidelines
of the Declaration of Helsinki, and approved by the Institutional
Ethics Committee of Poznan University of Medical Sciences (protocol
code, 985/18; date of approval, October 11, 2018). Written informed
consent was obtained from all subjects involved in the study.
Patient clinical data remains anonymous and does not identify the
patient. The consent form has been approved by the Institutional
Ethics Committee of Poznan University of Medical Sciences.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Swaye PS, Fisher LD, Litwin P, Vignola PA,
Judkins MP, Kemp HG, Mudd JG and Gosselin AJ: Aneurysmal coronary
artery disease. Circulation. 67:134–138. 1983.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Baman TS, Cole JH, Devireddy CM and
Sperling LS: Risk factors and outcomes in patients with coronary
artery aneurysms. Am J Cardiol. 93:1549–1551. 2004.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Warisawa T, Naganuma T, Tomizawa N, Fujino
Y, Ishiguro H, Tahara S, Kurita N, Nojo T and Nakamura S and
Nakamura S: High prevalence of coronary artery events and
non-coronary events in patients with coronary artery an-eurysm in
the observational group. Int J Cardiol Heart Vasc. 10:29–31.
2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Markis JE, Joffe CD, Cohn PF, Feen DJ,
Herman MV and Gorlin R: Clinical significance of coronary arterial
ectasia. Am J Cardiol. 37:217–222. 1976.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Aboeata AS, Sontineni SP, Alla VM and
Esterbrooks DJ: Coronary artery ectasia: Current concepts and
interventions. Front Biosci (Elite Ed). 4:300–310. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Doi T, Kataoka Y, Noguchi T, Shibata T,
Nakashima T, Kawakami S, Nakao K, Fujino M, Nagai T, Kanaya T, et
al: Coronary artery ectasia predicts future cardiac events in
patients with acute myocardial infarction. Arterioscler Thromb Vasc
Biol. 37:2350–2355. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Saglam M, Karakaya O, Barutcu I, Esen AM,
Turkmen M, Kargin R, Esen O, Ozdemir N and Kaymaz C: Identifying
cardiovascular risk factors in a patient population with coronary
artery ectasia. Angiology. 58:698–703. 2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kühl M and Varma C: A case of acute
coronary thrombosis in diffuse coronary artery ectasia. J Invasive
Cardiol. 20:E23–E25. 2008.PubMed/NCBI
|
|
9
|
Luo Y, Tang J and Liu X, Qiu J, Ye Z, Lai
Y, Yao Y, Li J, Wang X and Liu X: Coronary artery aneurysm differs
from coronary artery ectasia: Angiographic characteristics and
cardiovascular risk factor analysis in patients referred for
coronary angiography. Angiology. 68:823–830. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Araszkiewicz A, Grygier M, Lesiak M and
Grajek S: From positive remodelling to coronary artery ectasia. Is
coronary artery aneurysm a benign form of coronary disease? Kardiol
Pol. 67:1390–1395. 2009.PubMed/NCBI(In Polish).
|
|
11
|
Wojciechowska A, Braniewska A and
Kozar-Kamińska K: MicroRNA in cardiovascular biology and disease.
Adv Clin Exp Med. 26:865–874. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Barwari T, Joshi A and Mayr M: MicroRNAs
in cardiovascular disease. J Am Coll Cardiol. 68:2577–2584.
2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Montgomery RL and van Rooij E: MicroRNA
regulation as a therapeutic strategy for cardiovascular disease.
Curr Drug Targets. 11:936–942. 2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Viereck J and Thum T: Circulating
noncoding RNAs as biomarkers of cardiovascular disease and injury.
Circ Res. 120:381–399. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Mohr AM and Mott JL: Overview of microRNA
biology. Semin Liver Dis. 35:3–11. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kumar S, Boon RA, Maegdefessel L, Dimmeler
S and Jo H: Role of noncoding RNAs in the pathogenesis of abdominal
aortic aneurysm. Circ Res. 124:619–630. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Rowley AH, Pink AJ, Reindel R, Innocentini
N, Baker SC, Shulman ST and Kim KY: A study of cardiovascular miRNA
biomarkers for Kawasaki disease. Pediatr Infect Dis J.
33:1296–1299. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chu M, Wu R, Qin S, Hua W, Shan Z, Rong X,
Zeng J, Hong L, Sun Y, Liu Y, et al: Bone marrow-derived
Mi-croRNA-223 works as an endocrine genetic signal in vascular
endothelial cells and participates in vascular injury from kawasaki
disease. J Am Heart Assoc. 6(e004878)2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bang C, Fiedler J and Thum T:
Cardiovascular importance of the microRNA-23/27/24 family.
Microcirculation. 19:208–214. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang H, Bian C, Tu S, Yin F, Guo P, Zhang
J, Wu Y, Yin Y, Gou J and Han Y: Construction of the
circRNA-miRNA-mRNA regulatory network of an abdominal aortic
aneurysm to explore its potential pathogenesis. Dis Markers.
2021(9916881)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li T, Wang T, Yan L and Ma C:
Identification of potential novel biomarkers for abdominal aortic
aneurysm based on comprehensive analysis of circRNA-miRNA-mRNA
networks. Exp Ther Med. 22(1468)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shimizu C, Kim J, Stepanowsky P, Trinh C,
Lau HD, Akers JC, Chen C, Kanegaye JT, Tremoulet A, Ohno-Machado L
and Burns JC: Differential expression of miR-145 in children with
Kawasaki disease. PLoS One. 8(e58159)2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li R, Liang P, Yuan J and He F: Exosomal
miR-103a-3p ameliorates lipopolysaccharide-induced immune response
in BEAS-2B cells via NF-κB pathway by targeting transducin β-like
1X related protein 1. Clin Exp Pharmacol Physiol. 47:620–627.
2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Allantaz F, Cheng DT, Bergauer T,
Ravindran P, Rossier MF, Ebeling M, Badi L, Reis B, Bitter H,
D'Asaro M, et al: Expression profiling of human immune cell subsets
identifies miRNA-mRNA regulatory relationships correlated with cell
type specific expression. PLoS One. 7(e29979)2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Plana E, Gálvez L, Medina P, Navarro S,
Fornés-Ferrer V, Panadero J and Miralles M: Identification of Novel
microRNA Profiles Dysregulated in Plasma and Tissue of Abdominal
Aortic Aneurysm Patients. Int J Mol Sci. 21(4600)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gou L, Xue C, Tang X and Fang Z:
Inhibition of Exo-miR-19a-3p derived from cardiomyocytes promotes
angiogenesis and improves heart function in mice with myocardial
infarction via targeting HIF-α. Aging (Albany NY). 12:23609–23618.
2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Chen C, Ponnusamy M, Liu C, Gao J, Wang K
and Li P: MicroRNA as a therapeutic target in cardiac remodeling.
Biomed Res Int. 2017(1278436)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kim H, Yang JM, Jin Y, Jheon S, Kim K, Lee
CT, Chung JH and Paik JH: MicroRNA expression profiles and
clinicopathological implications in lung adenocarcinoma according
to EGFR, KRAS, and ALK status. Oncotarget. 8:8484–8498.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu
Q, Deitch EA, Huo Y, Delphin ES and Zhang C: MicroRNA-145, a novel
smooth muscle cell phenotypic marker and modulator, controls
vascular neointimal lesion formation. Circ Res. 105:158–166.
2009.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gatsiou A, Boeckel JN, Randriamboavonjy V
and Stellos K: MicroRNAs in platelet biogenesis and function:
Implications in vascular homeostasis and inflammation. Curr Vasc
Pharmacol. 10:524–531. 2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Neumann FJ, Sousa-Uva M, Ahlsson A,
Alfonso F, Banning AP, Benedetto U, Byrne RA, Collet JP, Falk V,
Head SJ, et al: 2018 ESC/EACTS guidelines on myocardial
revascularization. Eur Heart J. 40:87–165. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Camps C, Buffa FM, Colella S, Moore J,
Sotiriou C, Sheldon H, Harris AL, Gleadle JM and Ragoussis J:
hsa-miR-210 Is induced by hypoxia and is an independent prognostic
factor in breast cancer. Clin Cancer Res. 14:1340–1348.
2008.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Fasanaro P, D'Alessandra Y, Di Stefano V,
Melchionna R, Romani S, Pompilio G, Capogrossi MC and Martelli F:
MicroRNA-210 modulates endothelial cell response to hypoxia and
inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol
Chem. 283:15878–15883. 2008.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hu S, Huang M, Li Z, Jia F, Ghosh Z,
Lijkwan MA, Fasanaro P, Sun N, Wang X, Martelli F, et al:
MicroRNA-210 as a novel therapy for treatment of ischemic heart
disease. Circulation. 122 (11 Suppl):S124–S131. 2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Liu F, Lou YL, Wu J, Ruan QF, Xie A, Guo
F, Cui SP, Deng ZF and Wang Y: Upregulation of microRNA-210
regulates renal angiogenesis mediated by activation of VEGF
signaling pathway under ischemia/perfusion injury in vivo and in
vitro. Kidney Blood Press Res. 35:182–191. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cai W and Schaper W: Mechanisms of
arteriogenesis. Acta Biochim Biophys Sin (Shanghai). 40:681–692.
2008.PubMed/NCBI
|
|
38
|
Paulus P, Jennewein C and Zacharowski K:
Biomarkers of endothelial dysfunction: Can they help us deciphering
systemic inflammation and sepsis? Biomarkers. 16 (Suppl 1):S11–S21.
2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Qin X and Guo J: MicroRNA-328-3p protects
vascular endothelial cells against oxidized low-density lipoprotein
induced injury via targeting forkhead box protein O4 (FOXO4) in
atherosclerosis. Med Sci Monit. 26(e921877)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ma W, Ma CN, Zhou NN, Li XD and Zhang YJ:
Upregulation of miR-328-3p sensitizes non-small cell lung cancer to
radiotherapy. Sci Rep. 6(31651)2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ma Y, Yuwen D, Chen J, Zheng B, Gao J, Fan
M, Xue W, Wang Y, Li W, Shu Y, et al: Exosomal transfer of
cisplatin-induced miR-425-3p confers cisplatin resistance in NSCLC
through activating autophagy. Int J Nanomedicine. 14:8121–8132.
2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Shi J, An G, Guan Y, Wei T, Peng Z, Liang
M and Wang Y: miR-328-3p mediates the anti-tumor effect in
osteosarcoma via directly targeting MMP-16. Cancer Cell Int.
19(104)2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yan T and Ye XX: MicroRNA-328-3p inhibits
the tumorigenesis of bladder cancer through targeting ITGA5 and
inactivating PI3K/AKT pathway. Eur Rev Med Pharmacol Sci.
23:5139–5148. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Yuwen D, Ma Y, Wang D, Gao J, Li X, Xue W,
Fan M, Xu Q, Shen Y and Shu Y: prognostic role of circulating
exosomal miR-425-3p for the response of NSCLC to platinum-based
chemotherapy. Cancer Epidemiol Biomarkers Prev. 28:163–173.
2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Courtois A, Nusgens B, Garbacki N, Hustinx
R, Gomez P, Defraigne JO, Colige AC and Sakalihasan N: Circulating
microRNAs signature correlates with positive
[18F]fluorodeoxyglucose-positron emission tomography in
patients with abdominal aortic aneurysm. J Vasc Surg. 67:585–595
e3. 2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Xiao Y, Zhao J, Tuazon JP, Borlongan CV
and Yu G: MicroRNA-133a and myocardial infarction. Cell Transplant.
28:831–838. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Qiao Y, Ma N, Wang X, Hui Y, Li F, Xiang
Y, Zhou J, Zou C, Jin J, Lv G, et al: MiR-483-5p controls
angiogenesis in vitro and targets serum response factor. FEBS Lett.
585:3095–3100. 2011.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Li S, Lee C, Song J, Lu C, Liu J, Cui Y,
Liang H, Cao C, Zhang F and Chen H: Circulating microRNAs as
potential biomarkers for coronary plaque rupture. Oncotarget.
8:48145–48156. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Chmielewski S, Olejnik A, Sikorski K,
Pelisek J, Błaszczyk K, Aoqui C, Nowicka H, Zernecke A, Heemann U,
Wesoly J, et al: STAT1-dependent signal integration between IFNү
and TLR4 in vascular cells reflect pro-atherogenic responses in
human atherosclerosis. PLoS One. 9(e113318)2014.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Chmielewski S, Piaszyk-Borychowska A,
Wesoly J and Bluyssen HA: STAT1 and IRF8 in vascular inflammation
and cardiovascular disease: Diagnostic and therapeutic potential.
Int Rev Immunol. 35:434–454. 2016.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Liu L, Cheng Z and Yang J: miR-23
regulates cell proliferation and apoptosis of vascular smooth
muscle cells in coronary heart disease. Pathol Res Pract.
214:1873–1878. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Li X, Teng C, Ma J, Fu N, Wang L, Wen J
and Wang TY: miR-19 family: A promising biomarker and therapeutic
target in heart, vessels and neurons. Life Sci.
232(116651)2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Chen L, Lu Q, Deng F, Peng S, Yuan J, Liu
C and Du X: miR-103a-3p Could attenuate sepsis-induced liver injury
by targeting HMGB1. Inflammation. 43:2075–2086. 2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Rangrez AY, Massy ZA, Metzinger-Le Meuth V
and Metzinger L: miR-143 and miR-145: Molecular keys to switch the
phenotype of vascular smooth muscle cells. Circ Cardiovasc Genet.
4:197–205. 2011.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Cordes KR, Sheehy NT, White MP, Berry EC,
Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN and Srivastava D:
miR-145 and miR-143 regulate smooth muscle cell fate and
plasticity. Nature. 460:705–710. 2009.PubMed/NCBI View Article : Google Scholar
|
|
56
|
He M, Chen Z, Martin M, Zhang J, Sangwung
P, Woo B, Tremoulet AH, Shimizu C, Jain MK, Burns JC and Shyy JY:
miR-483 Targeting of CTGF suppresses endothelial-to-mesenchymal
transition: Therapeutic implications in kawasaki disease. Circ Res.
120:354–365. 2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Serafin A, Foco L, Zanigni S, Blankenburg
H, Picard A, Zanon A, Giannini G, Pichler I, Facheris MF, Cortelli
P, et al: Overexpression of blood microRNAs 103a, 30b, and 29a in
L-dopa-treated patients with PD. Neurology. 84:645–653.
2015.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Lu Q, Ma Z, Ding Y, Bedarida T, Chen L,
Xie Z, Song P and Zou MH: Circulating miR-103a-3p contributes to
angiotensin II-induced renal inflammation and fibrosis via a
SNRK/NF-κB/p65 regulatory axis. Nat Commun. 10(2145)2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Jiao T, Yao Y, Zhang B, Hao DC, Sun QF, Li
JB, Yuan C, Jing B, Wang YP and Wang HY: Role of Mi-croRNA-103a
Targeting ADAM10 in abdominal aortic aneurysm. Biomed Res Int.
2017(9645874)2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Cerna V, Ostasov P, Pitule P, Mollacek J,
Treska V and Pesta M: The expression profile of MicroRNAs in small
and large abdominal aortic aneurysms. Cardiol Res Pract.
2019(8645840)2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Elia L, Quintavalle M, Zhang J, Contu R,
Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J,
et al: The knockout of miR-143 and -145 alters smooth muscle cell
maintenance and vascular homeostasis in mice: Correlates with human
disease. Cell Death Differ. 16:1590–1598. 2009.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Wang Z, Zhuang X, Chen B, Feng D, Li G and
Wei M: The role of miR-107 as a potential biomarker and cellular
factor for acute aortic dissection. DNA Cell Biol. 39:1895–1906.
2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Shimizu C, Oharaseki T, Takahashi K,
Kottek A, Franco A and Burns JC: The role of TGF-β and
myofibroblasts in the arteritis of Kawasaki disease. Hum Pathol.
44:189–198. 2013.PubMed/NCBI View Article : Google Scholar
|