Introduction
Breast cancer is one of the most common malignant
tumors in women. According to the latest global cancer data in
2020, there were 2.26 million new cases of breast cancer worldwide
in 2020, accounting for 11.7% of the total cancer cases, and
~860,000 deaths, accounting for 6.9% of the total cancer-related
deaths (1). There are five
different types of breast cancer: Luminal A breast cancer, Luminal
B breast cancer, triple negative breast cancer (TNBC), human
epidermal growth factor 2 receptor (HER2) negative breast cancer
and HER2 positive breast cancer. TNBC accounts for ~15% of breast
cancers and is characterized by loss of expression of estrogen
receptor, progesterone receptor and HER2 (2,3).
Local invasion and distant spread are the main causes of tumor
progression (4). Although surgery
and chemotherapy are effective, they still fail to meet
expectations (5). Therefore, the
development of effective and safe treatment strategies for TNBC
should not be underestimated.
Previous studies have found that p53 mutations are
diagnostic and prognostic indicators of breast cancer, and breast
cancer with p53 mutations is more aggressive, such as TNBC
(6,7). Glutamate-rich WD-repeat-containing
protein 1 (GRWD1) is a multifunctional protein rich in glutamic
acid and is involved in numerous cellular regulatory pathways,
particularly ribosome metabolism and cell growth (8). Recently, GRWD1 was found to bind to
the tumor suppressor gene p53 and negatively regulate the
expression of p53(9), which leads
to the attention of GRWD1 as a potential oncogene in an increasing
number of tumors. For example, GRWD1 promotes tumor cell growth and
drug resistance by regulating p53 in colorectal cancer (10). Concurrently, highly expressed GRWD1
can promote the growth and metastasis of non-small cell lung cancer
(NSCLC) through the Notch pathway (11). However, the role of GRWD1 in TNBC
has not been studied.
The present study aimed to detect the expression
levels of GRWD1 in breast cancer cells and to explore the
regulatory effect of GRWD1 on the proliferation, apoptosis,
invasion and migration of breast cancer cells through the Notch
pathway.
Materials and methods
Cell culture
Normal human breast epithelial cells (MCF-10A) and
human breast cancer cells (MDA-MB-231, HCC1937, MCF-7, SK-BR-3 and
BT474) were provided from Procell Life Science & Technology
Co., Ltd. The MCF-10A cells were cultured in Dulbecco's modified
Eagle's medium (DMEM)/F12 (Procell Life Science & Technology
Co., Ltd.) containing 5% horse serum (Procell Life Science &
Technology Co., Ltd.), 20 ng/ml EGF, 0.5 µg/ml hydrocortisone, 10
µg/ml insulin, 1% non-essential amino acids and 1%
penicillin/streptomycin. The breast cancer cells were cultured in
DMEM (Hyclone; Cytiva) containing 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific; Inc.) and 1%
penicillin/streptomycin. All cells were cultured at 37˚C in a
humidified atmosphere of 95% air and 5% CO2.
Cell transfection
Small interfering RNA (siRNA)-negative control (NC)
(siB06525141922-1-5), siRNA-GRWD1 (siG000083743A-1-5),
overexpression (Ov)-NC and Ov-Notch1 were synthesized and purchased
from Guangzhou RiboBio Co., Ltd. A total of 50 nM siRNA-NC,
siRNA-GRWD1, Ov-NC and Ov-Notch1 were transfected into MDA-MB-231
and HCC1937 cells when their confluence reached 80% using
Lipofectamine® 3000 kit (cat. no. L3000015; Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol.
Then, cells were cultured at 37˚C for 48 h before subsequent
experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from MDA-MB-231 and HCC1937
cells (1x106 cells) using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. Then, the PrimeScript™ First Strand cDNA
Synthesis kit (Takara Bio, Inc.) was used to reverse transcribe the
RNA to first-stranded cDNA at 30˚C for 10 min. qPCR was performed
with SYBR Premix Ex Taq (Takara Bio, Inc.) on a CFX96 System
(Bio-Rad Laboratories, Inc.). The thermocycling conditions were as
follows: 10 min initial denaturation at 94˚C, 15 sec denaturation
at 94˚C and 30 sec of annealing at 55˚C (40 cycles) and final
extension for 1 min at 72˚C. GAPDH were used as internal control.
The relative expression of genes was calculated using the
2-ΔΔCq method (12).
The primer sequences used for qPCR (designed by Thermo Fisher
Scientific, Inc.) were as follows: GRWD1 forward,
5'-ATCACACAGTGGGACCTGGCA-3' and reverse,
5'-TCAGACGCTGATGGTGCGGAA-3'; Notch1 forward,
5'-CTGGTCAGGGAAATCGTG-3' and reverse, 5'-TGGGCAGTGGCAGATGTAG-3';
Notch4 forward, 5'-ACACACACATGAGGATCTCTGGCA-3' and reverse,
5'-AGTTGGCCTTGTCTTTCTGGTCCT-3'; and GAPDH forward,
5'-GACAGTCAGCCGCATCTTCT-3' and reverse,
5'-TTAAAAGCAGCCCTGGTGAC-3'.
Western blot analysis
Proteins were extracted from MDA-MB-231 and HCC1937
cells using RIPA lysis buffer (Thermo Fisher Scientific, Inc.). The
protein concentration was detected by the BCA Protein Assay kit
(Thermo Fisher Scientific, Inc.). Proteins (15 µg) were separated
by 12% sodium dodecyl sulfate-polyacrylamide gel and transferred
onto polyvinylidene difluoride membranes. The membranes were
blocked with 5% non-fat milk for 2 h at room temperature and
subsequently incubated with primary antibodies against GRWD1 (cat.
no. ab176815; 1:2,000), Ki67 (cat. no. ab92742; 1:5,000),
proliferating cell nuclear antigen (PCNA; cat. no. ab92552;
1:1,000), Bcl-2 (cat. no. ab32124; 1:1,000), cleaved caspase-3
(cat. no. ab32042; 1:500), Bax (cat. no. ab32503; 1:1,000),
caspase-3 (cat. no. ab32351; 1:5,000), MMP9 (cat. no. ab76003;
1:1,000), MMP2 (cat. no. ab92536; 1:1,000), E-cadherin (cat. no.
ab40772; 1:10,000), N-cadherin (cat. no. ab76011; 1:5,000),
Vimentin (cat. no. ab92547; 1:1,000), Notch1 (cat. no. ab52627;
1:1,000), Notch4 (cat. no. ab184742; 1:1,000), Hes1 (cat. no.
ab108937; 1:1,000), Hes5 (cat. no. ab194111; 1:1,000), Hey1 (cat.
no. ab154077; 1:1,000), Hey2 (cat. no. ab167280; 1:1,000), p21
(cat. no. ab109520; 1:1,000), c-Myc (cat. no. ab32072; 1:1,000),
cyclin D1 (CCND1; cat. no. ab16663; 1:200), HER2 (cat. no.
ab134182; 1:1,000), NF-κB (cat. no. ab32536; 1:1,000) and GAPDH
(cat. no. ab9485; 1:2,500; all from Abcam) overnight at 4˚C.
Following 0.1% Tween TBST washing, membranes were incubated with
the HRP-conjugated goat anti-rabbit IgG secondary antibody (ab6721;
1:2,000; Abcam) for 1 h at room temperature. The blots were
developed with enhanced chemiluminescence Western Blotting
Substrate (Pierce; Thermo Fisher Scientific, Inc.), visualized
using the Gel Imager System (Bio-Rad Laboratories, Inc.) and
quantified using ImageJ version 1.8.0 (National Institutes of
Health).
Cell Counting Kit-8 (CCK-8) assay
The MDA-MB-231 and HCC1937 cells were inoculated in
96-well plates with each well including 2,000 cells. Following
transfection, 10 µl of CCK-8 solution (Dojindo Molecular
Technologies, Inc.) was added to each well; then, MDA-MB-231 and
HCC1937 cells were incubated for 24, 48 and 72 h at 37˚C.
Subsequently, the cells were incubated with CCK-8 solution for
another 2 h and the absorbance at 450 nm was detected by a
SpectraMax Absorbance Reader (Molecular Devices, LLC).
5-Ethynyl-2'-deoxyuridine (EdU)
staining assay
The MDA-MB-231 and HCC1937 cells were seeded into
96-well plates with each well including 1x104 cells for
overnight culture at 37˚C. Each well was supplemented with EdU (10
µM) and plates were incubated for 2 h at 37˚C. After that, cells
were fixed with 4% polyoxymethylene at room temperature for 20 min
and decolorized with glycine (2 mg/ml) at room temperature for 10
min; Hoechst 33342 was added to stain the nucleus at room
temperature for 25 min. The fluorescence of cells was observed by a
fluorescent microscope (magnification, x200; BX41; Olympus
Corporation). A total of 3 fields per group were counted. The cell
proliferation was quantified using ImageJ version 1.8.0 (National
Institutes of Health).
TUNEL assay
The MDA-MB-231 and HCC1937 cells (1x105
cells) were seeded in 24-well plate and transfected for 48 h. Then,
cells were fixed with 4% paraformaldehyde for 0.5 h at room
temperature and incubated with 0.3% Triton X-100 for 5 min at 4˚C.
Subsequently, 50 µl TUNEL reaction buffer was added and the plate
was incubated for 1 h at 37˚C; 1 mg/ml DAPI was added to
counterstain the nucleus for 1 min at room temperature in dark; the
slides were mounted with neutral balsam mounting media (Sangon
Biotech Co., Ltd.). Finally, the stained apoptotic cells were
visualized under a fluorescent microscope (BX41; Olympus
Corporation) and quantified using ImageJ version 1.8.0 (National
Institutes of Health). A total of three randomly selected fields
per group were counted.
Transwell invasion assay
MDA-MB-231 and HCC1937 cells were seeded at a
density of 5x104 cells/well in the upper chamber with
8-µm pore filters (MilliporeSigma) of Transwell plates precoated
with Matrigel at 37˚C for 30 min. The upper chamber contained the
serum-free medium and the lower chamber contained DMEM supplemented
with 10% FBS. After 48 h of incubation at 37˚C, cells that passed
through the membrane were fixed with 4% paraformaldehyde for 15 min
at room temperature and stained with 0.1% crystal violet at room
temperature for 20 min. Invasive cells were then observed under a
light microscope (magnification, x100) and quantified using ImageJ
version 1.8.0 (National Institutes of Health). A total of 3 fields
per group were counted.
Wound healing assay
MDA-MB-231 and HCC1937 cells were seeded in a
six-well plate with each well containing 5x105 cells and
incubated overnight at 37˚C. The next day, a sterile 200 µl pipette
tip was used to create a 0.5~1 cm horizontal line, followed by PBS
washing. Then, cells were incubated with the serum-free medium at
37˚C with 5% CO2 for 24 h. Cells migrating into the
scratch were observed under a light microscope (magnification,
x100) at 0 and 24 h. The relative mobility of the cells was then
calculated using ImageJ version 1.8.0 (National Institutes of
Health).
Statistical analysis
Data analysis was conducted by GraphPad Prism
version 7 (GraphPad Software, Inc.) and data were expressed as the
means ± standard deviation (SD) of at least three independent
experiments. ANOVA followed by Tukey's post hoc test was used to
compare differences between ≥3 groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
GRWD1 is highly expressed in breast
cancer cells
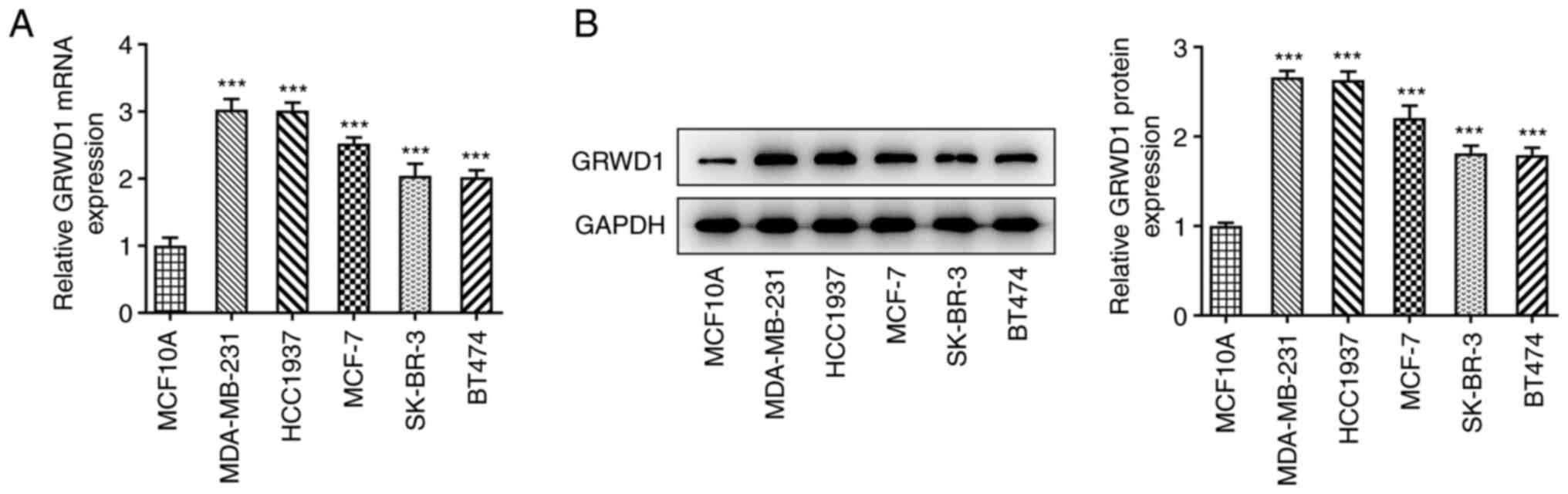
GRWD1 mRNA expression in breast cancer cells was
significantly higher than that in MCF10A cells (Fig. 1A). In addition, the GRWD1 protein
expression was higher in breast cancer cells compared with MCF10A
cells (Fig. 1B). The
aforementioned results indicated that the expression of GRWD1 in
MDA-MB-231 and HCC1937 cells was higher than that in other breast
cancer cells. Therefore, MDA-MB-231 and HCC1937 cells were chosen
for the subsequent experiments.
GRWD1 knockdown suppresses TNBC cell
proliferation
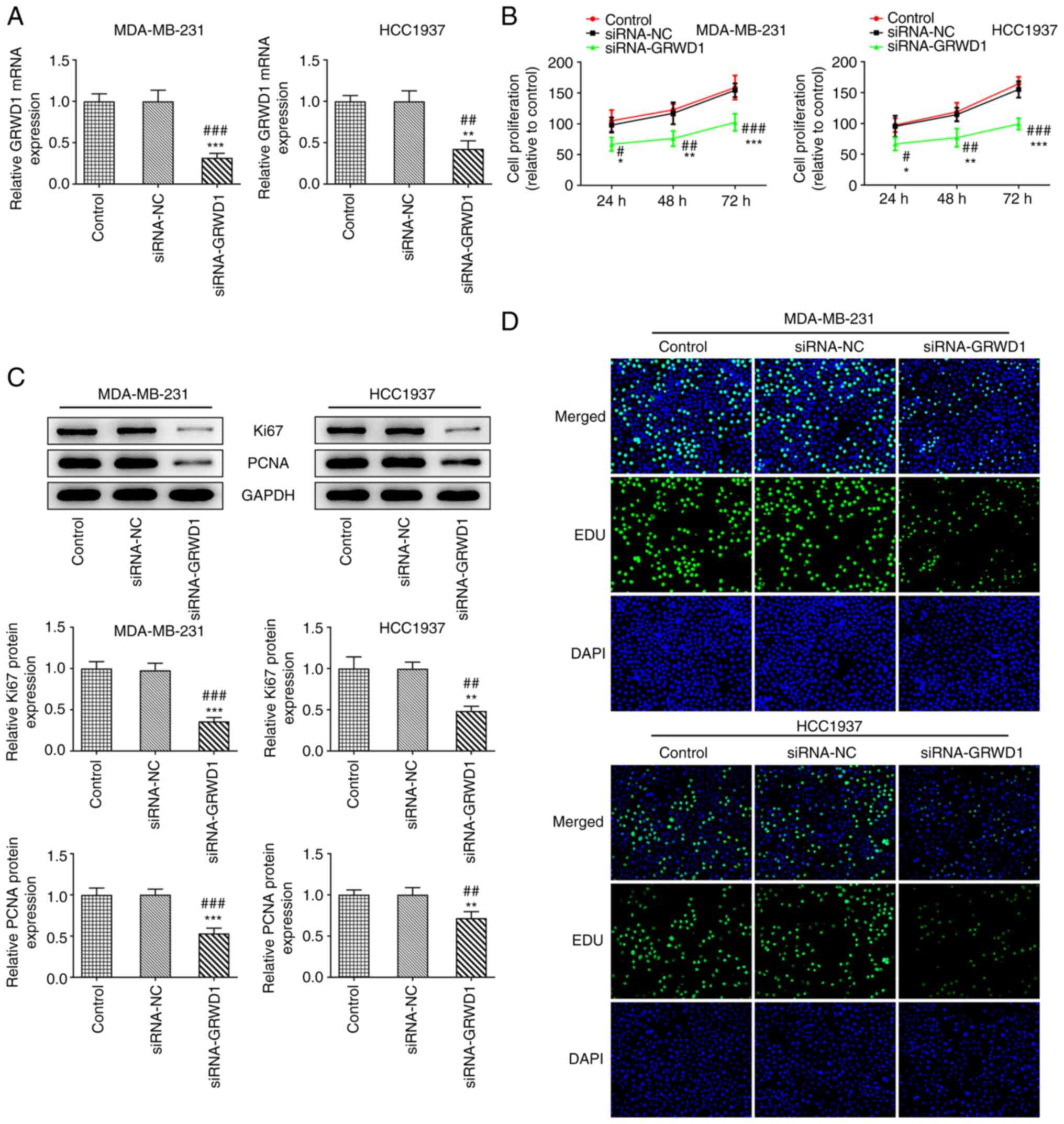
When MDA-MB-231 and HCC1937 cells were transfected
with siRNA-GRWD1, the GRWD1 mRNA expression in both cell lines was
decreased compared with the siRNA-NC group (Fig. 2A). Downregulation of GRWD1 resulted
in the decreased proliferation of MDA-MB-231 and HCC1937 cells
(Fig. 2B), accompanied with the
decreased expression of Ki67 and PCNA (Fig. 2C). The Edu staining also indicated
that the proliferation of MDA-MB-231 and HCC1937 cells was
inhibited in the siRNA-GRWD1 group (Fig. 2D).
GRWD1 knockdown promotes TNBC cell
apoptosis
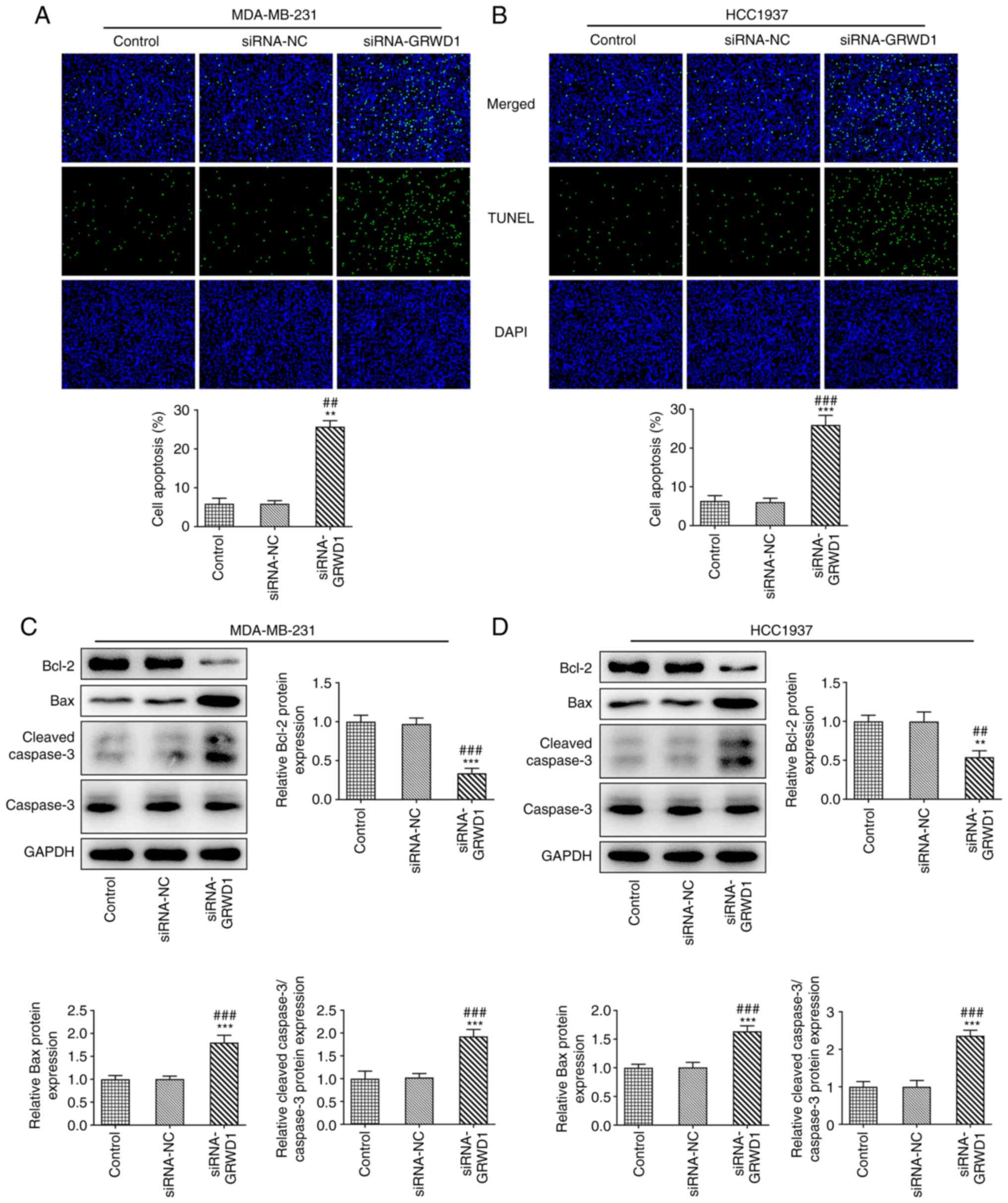
There were no obvious changes in apoptosis of
MDA-MB-231 and HCC1937 cells between the control group and the
siRNA-NC group. The apoptosis of MDA-MB-231 and HCC1937 cells was
significantly increased after siRNA-GRWD1 transfection (Fig. 3A and B). GRWD1 knockdown suppressed the
expression of Bcl-2 and promoted the expression of Bax and cleaved
caspase-3 in both cell lines (Fig.
3C and D).
GRWD1 knockdown suppresses TNBC cell
invasion and migration
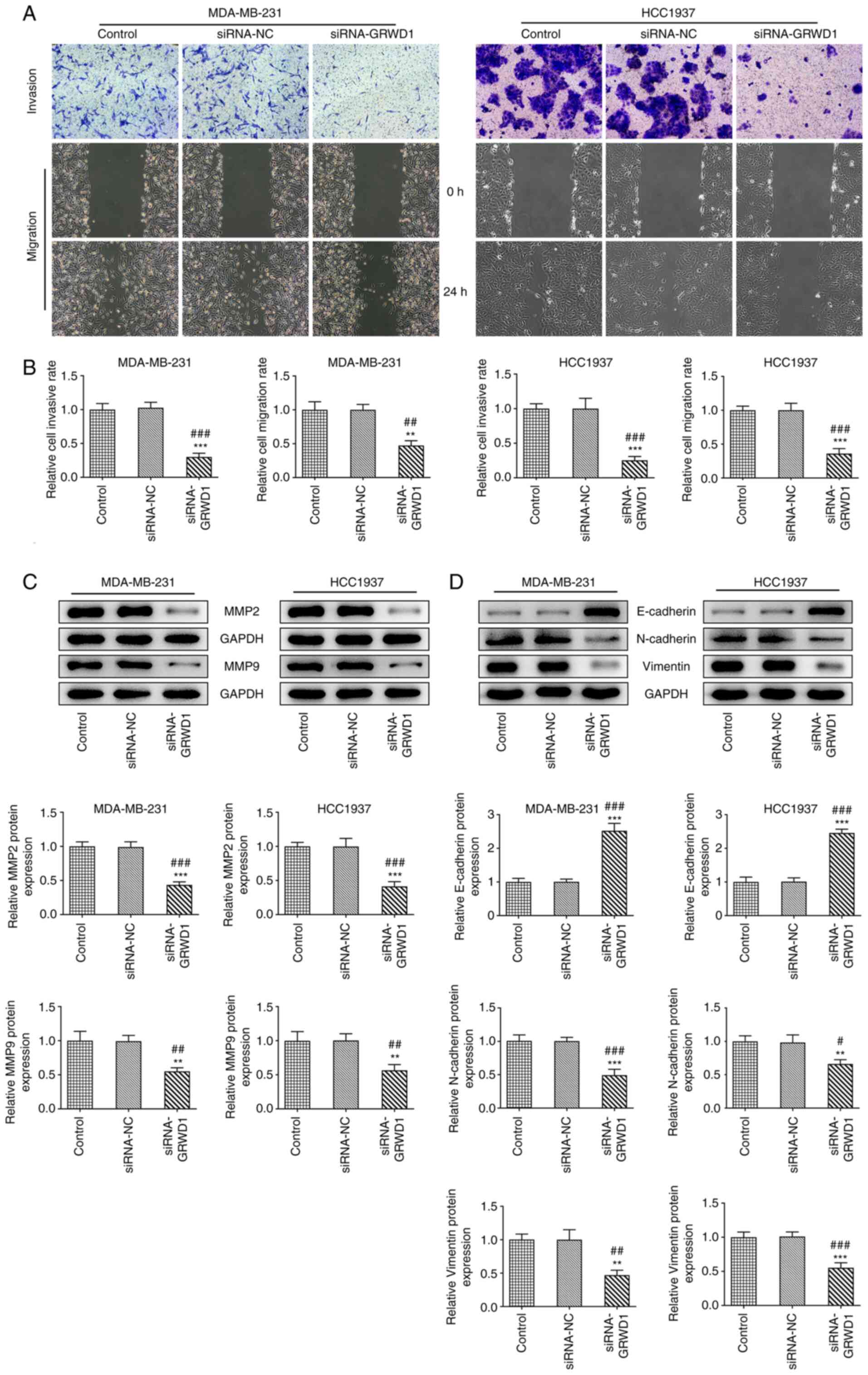
The invasion and migration of MDA-MB-231 and HCC1937
cells was not changed in the siRNA-NC group compared with the
control group and was inhibited in siRNA-GRWD1 group (Fig. 4A and B). The expression of MMP9 and MMP2 was
also decreased in both MDA-MB-231 and HCC1937 cells transfected
with siRNA-GRWD1 (Fig. 4C).
siRNA-GRWD1 also enhanced the expression of E-cadherin and
suppressed the expression of N-cadherin and Vimentin in both
MDA-MB-231 and HCC1937 cells (Fig.
4D).
GRWD1 knockdown inactivates the Notch
signaling pathway
The mRNA and protein expression levels of Notch1 and
Notch4 in MDA-MB-231 and HCC1937 cells were increased compared with
MCF10A cells (Fig. 5A and B). Following transfection of both cell
lines with siRNA-GRWD1, the protein expression of Notch1 and Notch4
was decreased (Fig. 5C). GRWD1
knockdown reduced the expression of Hes1, Hes5, Hey1, Hey2, p21,
c-Myc, CCND1, HER2 and NF-κB (Fig.
5D and E) in both cell
lines.
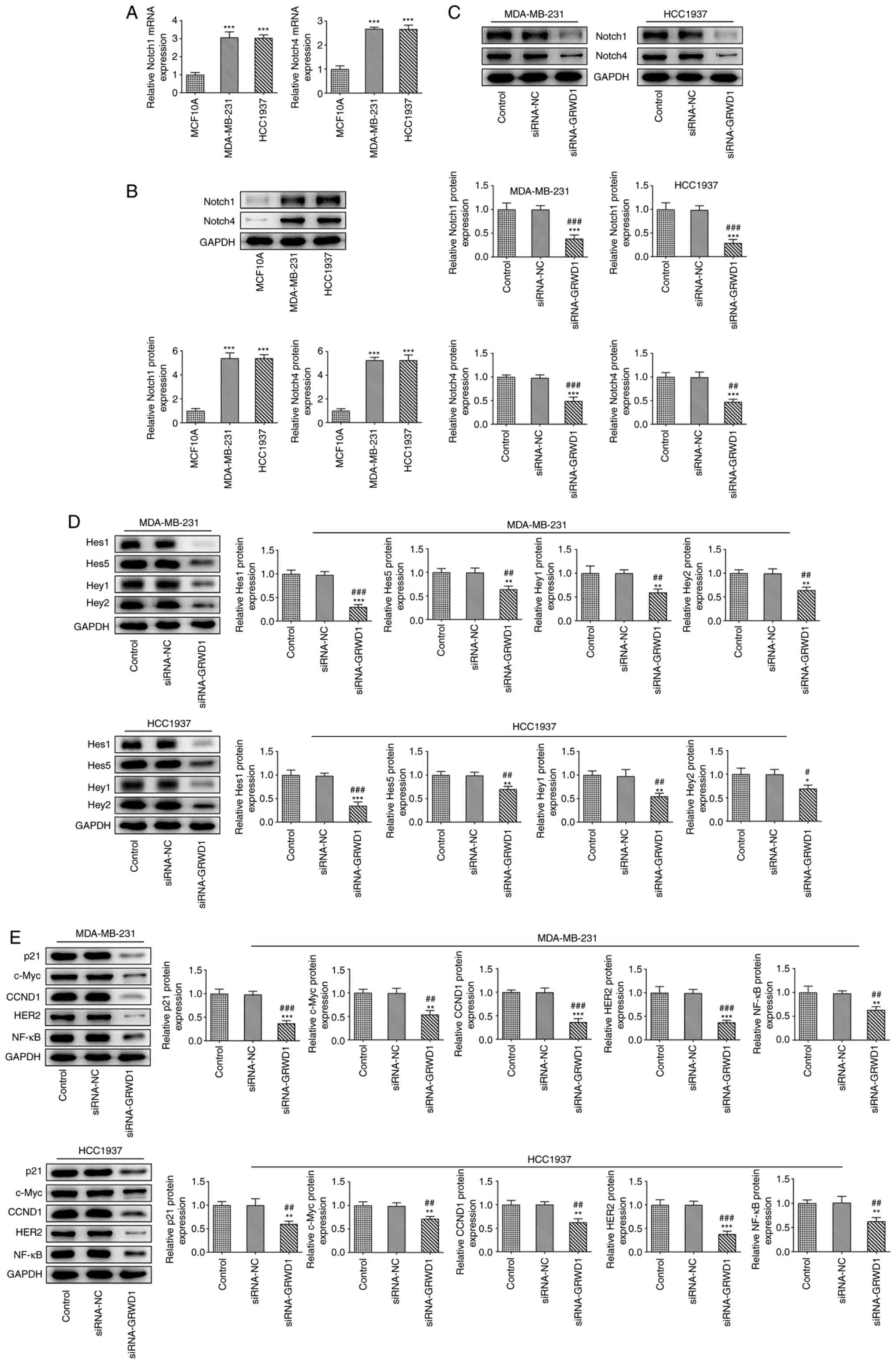 | Figure 5GRWD1 knockdown inactivates the Notch
signaling pathway. (A) The mRNA and (B) protein expression of
Notch1 and Notch4 in breast cancer cells and normal human breast
epithelial cells was detected by reverse transcription-quantitative
PCR and western blotting, respectively. ***P<0.001
vs. MCF-10A group. (C) The expression of Notch1 and Notch4 in
MDA-MB-231 and HCC1937 cells transfected with siRNA-GRWD1 was
detected by western blot analysis. (D) The expression of Hes1,
Hes5, Hey1 and Hey2 in MDA-MB-231 and HCC1937 cells transfected
with siRNA-GRWD1 was detected by western blot analysis. (E) The
expression of p21, c-Myc, CCND1, HER2 and NF-κB in MDA-MB-231 and
HCC1937 cells transfected with siRNA-GRWD1 was detected by western
blotting. *P<0.05, **P<0.01 and
***P<0.001 vs. Control group. #P<0.05,
##P<0.01 and ###P<0.001 vs. siRNA-NC
group. GRWD1, glutamate-rich WD-repeat-containing protein 1; siRNA,
small interfering RNA; NC, negative control. |
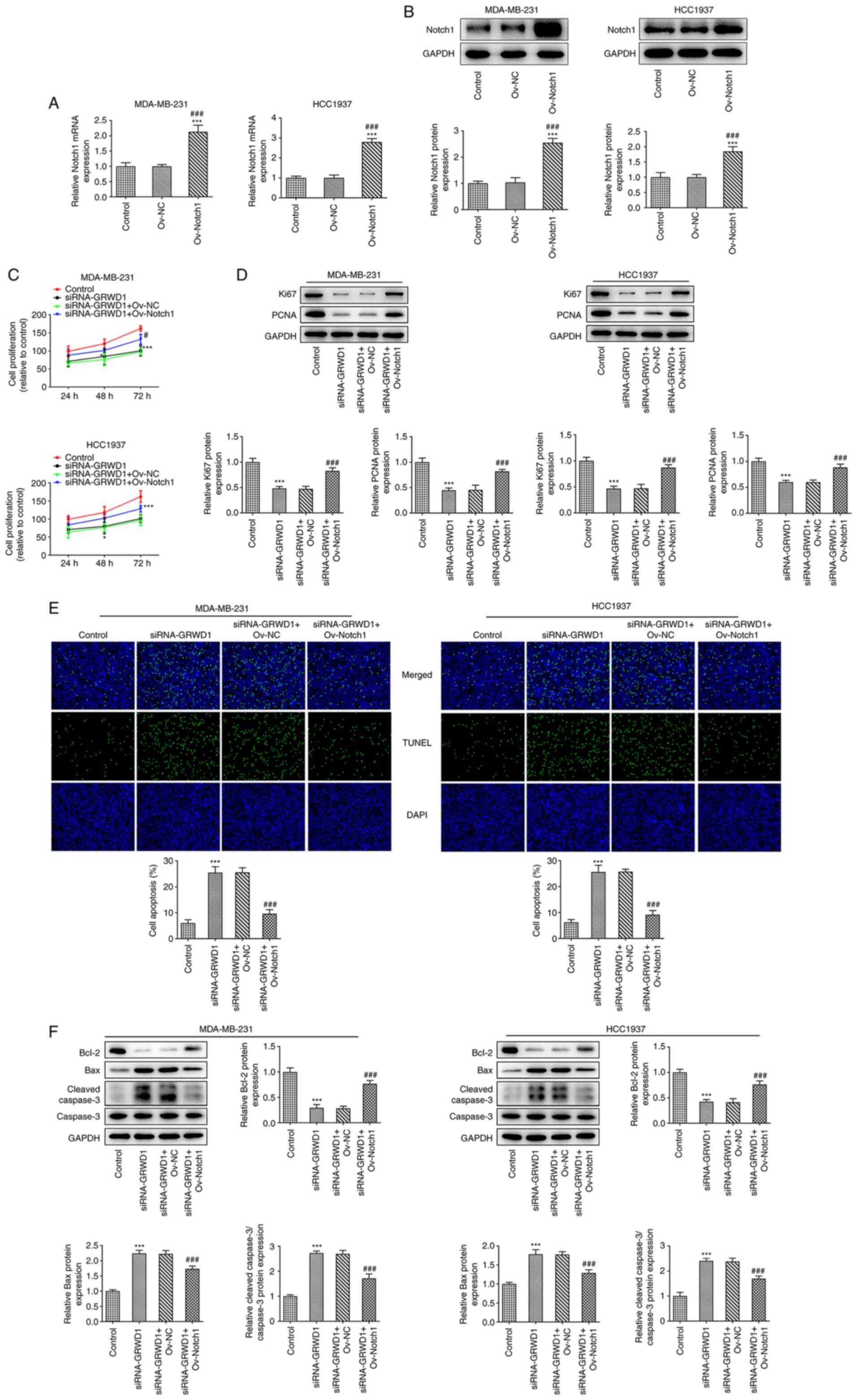
Notch1 overexpression reverses the
effect of GRWD1 knockdown on TNBC cell proliferation and
apoptosis
The mRNA and protein expression of Notch1 was
increased in MDA-MB-231 and HCC1937 cells transfected with
Ov-Notch1 (Fig. 6A and B). Notch1 overexpression improved the
decreased proliferation of MDA-MB-231 and HCC1937 cells induced by
GRWD1 knockdown, as well as increased the expression of Ki67 and
PCNA (Fig. 6C and D). Notch1 overexpression reduced the
apoptosis of MDA-MB-231 and HCC1937 cells which were transfected
with siRNA-GRWD1 (Fig. 6E). Notch1
overexpression upregulated the expression of Bcl-2 and
downregulated the expression of Bax and cleaved caspase-3 in
siRNA-GRWD1- transfected MDA-MB-231 and HCC1937 cells (Fig. 6F).
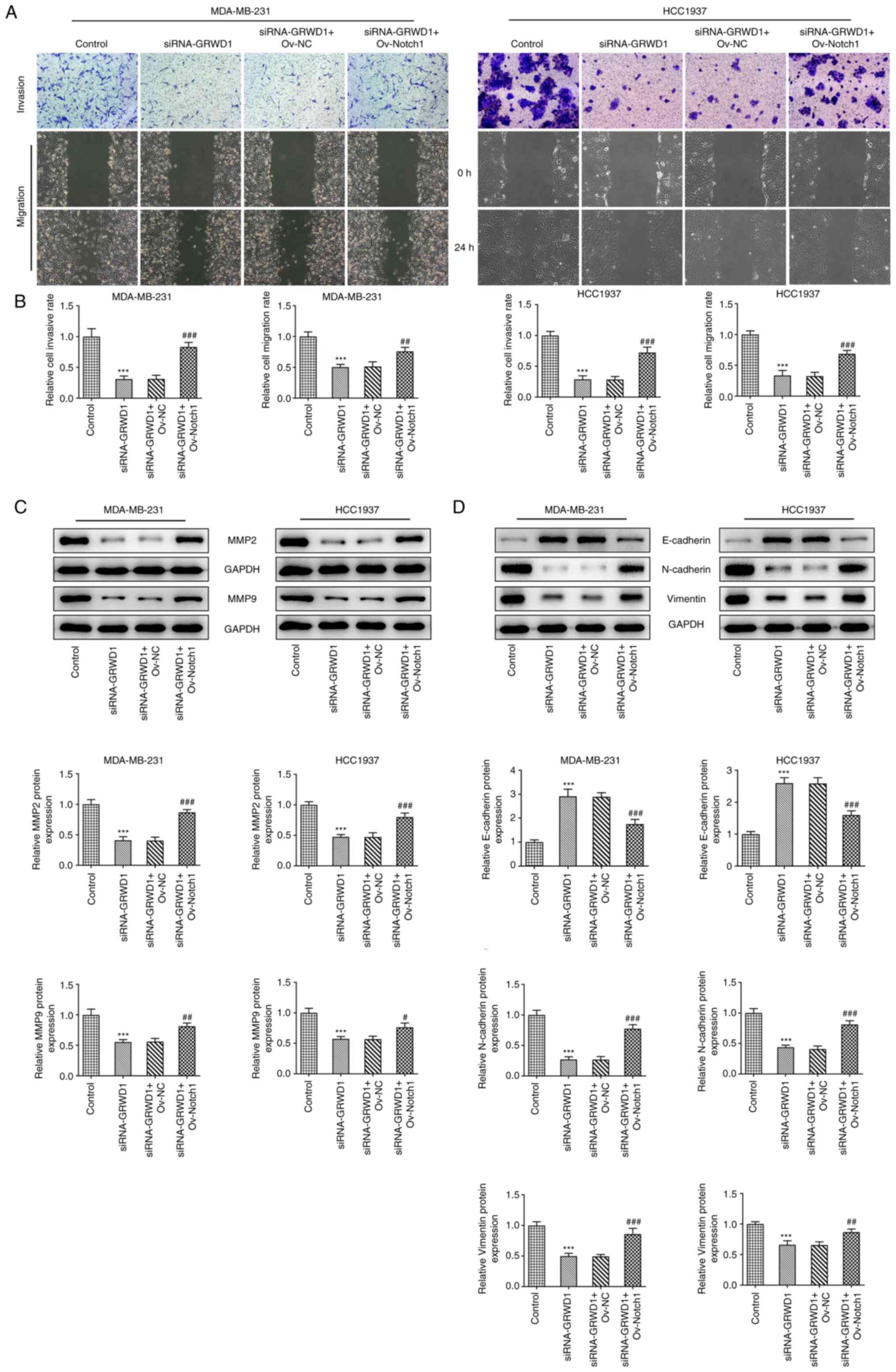
Notch1 overexpression reverses the
effect of GRWD1 knockdown on TNBC cell invasion and migration
The invasion and migration of MDA-MB-231 and HCC1937
cells were suppressed by GRWD1 knockdown, which was reversed by
Notch1 overexpression (Fig. 7A and
B). Notch1 overexpression
upregulated the expression of MMP2 and MMP9 which were inhibited by
GRWD1 knockdown in MDA-MB-231 and HCC1937 cells (Fig. 7C). Both cell lines co-transfected
with siRNA-GRWD1 and Ov-Notch1 presented decreased expression of
E-cadherin and increased expression of N-cadherin and Vimentin
compared with the siRNA-GRWD1 group (Fig. 7D).
Notch1 overexpression reverses the
effect of GRWD1 knockdown on the Notch signaling pathway in TNBC
cells
The expression of p21, c-Myc, CCND1, HER2 and NF-κB
in MDA-MB-231 and HCC1937 cells was reduced in siRNA-GRWD1 group,
which was reversed by Ov-Notch1 transfection (Fig. 8).
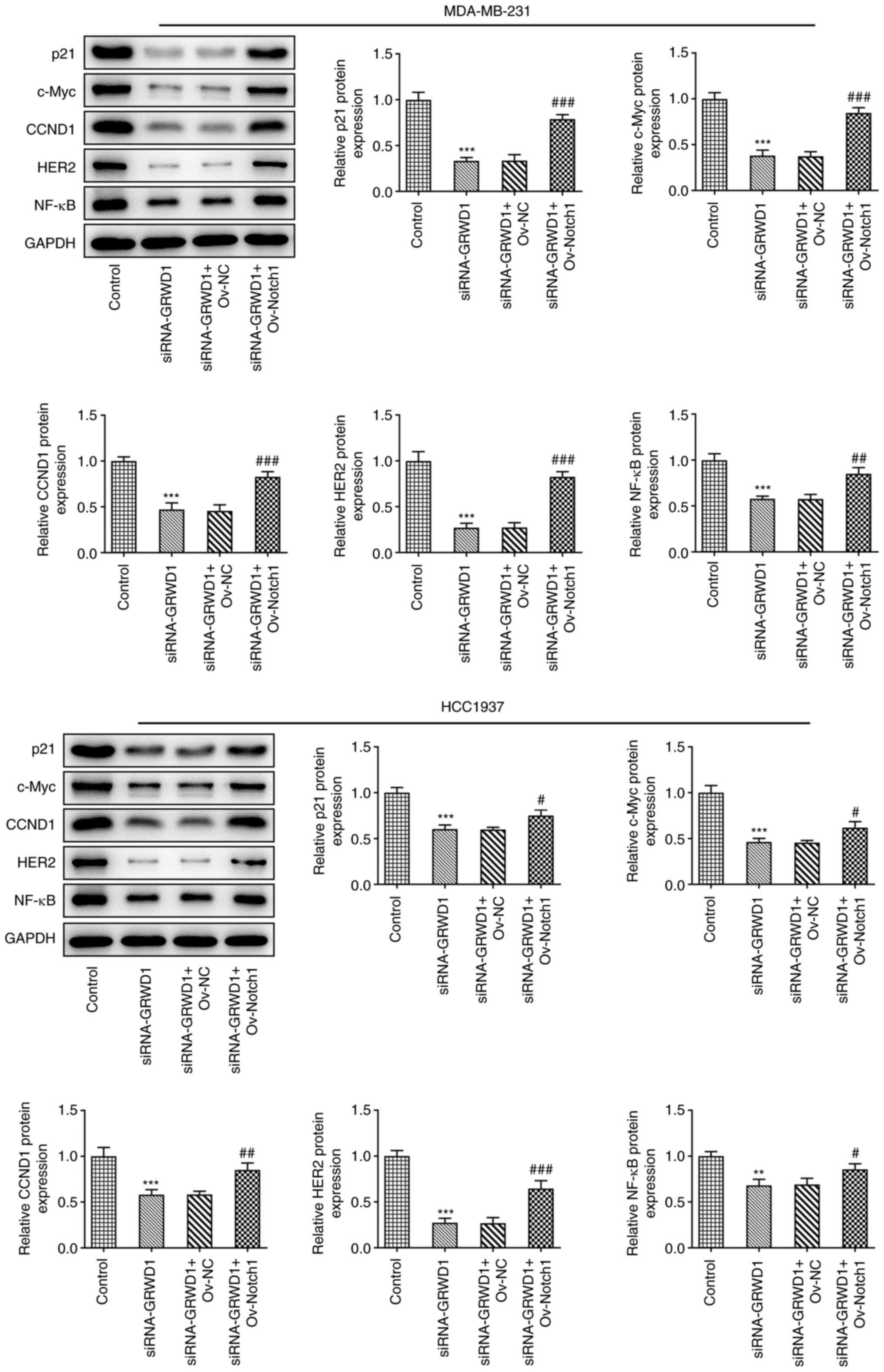 | Figure 8Notch1 overexpression reverses the
effect of GRWD1 knockdown on the Notch signaling pathway in triple
negative breast cancer cells. The expression of p21, c-Myc, CCND1,
HER2 and NF-κB in MDA-MB-231 and HCC1937 cells co-transfected with
siRNA-GRWD1 and Ov-Notch1 was detected by western blotting.
**P<0.01 and ***P<0.001 vs. Control
group. #P<0.05, ##P<0.01 and
###P<0.001 vs. siRNA-GRWD1 + Ov-NC group. GRWD1,
glutamate-rich WD-repeat-containing protein 1; CCND1, cyclin D1;
HER2, human epidermal growth factor 2 receptor; siRNA, small
interfering RNA; NC, negative control; Ov, overexpression. |
Discussion
Breast cancer is a malignant tumor occurring in the
glandular epithelial tissue of the breast, and the cancer cells are
easy to fall off and dissociate, leading to distal metastasis. The
metastasis of vital organs such as lung, bone and brain can
directly threaten the lives of patients, adding great difficulties
to the clinical treatment of breast cancer (13). Therefore, effectively inhibiting
the malignant biology of breast cancer cells has become an
important measure for the treatment of breast cancer (14).
GRWD1 was confirmed to be a novel negative regulator
of p53 induced by nucleolar stress and a latent oncogene (15). GRWD1 can competitively bind
ribosomal protein L11, thus promoting the recovery of MDM2
ubiquitin activity and negatively regulating p53(16). GRWD1 overexpression could promote
tumor cell growth by inhibiting p53 (9,16).
In a previous study by Pan et al (17), 156 patients with TNBC were include
and it was found that the mutation rate of p53 reached 71.3%, and
the mutation of p53 was closely related to TNBC histological
grading. In a recent study by Xu et al (18), the expression and prognosis of p53
in TNBC tissues was investigated and it was found that p53 was
highly expressed in TNBC tissues and was associated with tumor node
metastasis staging, histological grade, lymph node metastasis and
poor prognosis in patients with TNBC. Considering that GRWD1
overexpression could inhibit the highly expressed p53 in TNBC
tissues, it was found in the present study that GRWD1 expression
was also upregulated in MDA-MB-231 and HCC1937 cells, and
downregulation of GRWD1 could suppress the proliferation and
promoted the apoptosis of these two cell lines.
In tumor cells, in addition to the downregulation of
E-cadherin expression, the expression of N-cadherin protein is also
upregulated, and the downregulation of E-cadherin expression is a
marker for the occurrence of epithelial-mesenchymal transition
(EMT) (19). In the present study,
it was observed that downregulation of GRWD1 increased the
expression of E-cadherin while decreased the expression of
N-cadherin and Vimentin, suggesting that GRWD1 could promote the
EMT process of TNBC cells. MMP2 and MMP9 belong to the MMP family,
whose main function is to maintain the dynamic balance of
remodeling and degradation of extracellular matrix, and participate
in the process of tumor cell migration and invasion (20,21).
In the present study, the expression of MMP2 and MMP9 was increased
in TNBC cells, which was decreased by GRWD1 knockdown.
The Notch signaling pathway is a common regulated
signaling pathway mainly involved in organ development,
differentiation, cell proliferation and apoptosis. Previous studies
have shown that it can be involved in the regulation of malignant
tumor diseases such as NSCLC, breast cancer and T-cell leukemia
(22-24).
Activated Notch protein is overexpressed in alveolar epithelial
cells and can interact with Myc Pathway co-promoting the
development of NSCLC (25). Notch1
inhibits p53-mediated apoptosis by regulating the stability of p53,
and is necessary for the growth of lung adenocarcinoma (26). In addition, the Notch pathway plays
an important role in the proliferation, metastasis and treatment of
breast cancer (23,27). Notch1 was demonstrated to be
related to the invasion and migration steps which characterize the
EMT process in TNBC (28). Notch1
was highly expressed in Cisplatin-resistant MDA-MB-231 TNBC cells,
and this helped to induce chemoresistance via activating the AKT
pathway and promoting EMT (29).
It was indicated in the present study that Notch1 expression was
also higher in MDA-MB-231 and HCC1937 cells than MCF10A cells.
Furthermore, Notch1 overexpression could enhance the proliferation,
invasion and migration and suppress the apoptosis of MDA-MB-231 and
HCC1937 cells which were transfected with siRNA-GRWD1. Despite
being epithelial and lymphoblast cells, respectively, MDA-MB-231
and HCC1937 cells are both TNBC cells and yielded similar results,
which is consistent with previous studies (30-32).
The KEGG PATHWAY database (https://www.genome.jp/kegg/pathway.html) indicates
that Notch1/4 affects downstream expression of p21, c-Myc, CCND1,
HER2 and NF-κB through Hes1/5 and HEY in TNBC. The most well-known
Notch target genes are transcription factors of the Hes and Hey
families (33). Hes and Hey
members are helix-loop-helix proteins, forming homo-or heterodimers
that regulate transcription of genes relating to cell fate
determination (33-35).
Other Notch pathway targets include cell cycle regulators CCND1 and
p21, NF-κB family members, c-Myc and Deltex (36-39).
Hes1 protein is downstream of the Notch1 signaling pathway and
affects cell proliferation and differentiation (40-42).
Salidroside inhibited the hepatocellular carcinoma cell metastasis
by suppressing the Notch1 signaling pathway, which was manifested
by the downregulation of Hey1, Hes1 and Hes5(43). In the present study, it was
revealed that GRWD1 knockdown suppressed the expression of
Notch1/4, Hes1/5, Hey1/2, p21, c-Myc, CCND1, HER2 and NF-κB and
Notch1 overexpression promoted the expression of p21, c-Myc, CCND1,
HER2 and NF-κB in both TNBC cell lines used. In addition,
regulation of the Notch signaling pathway could indeed affect the
proliferation, invasion and migration and apoptosis of TNBC
cells.
In conclusion, GRWD1 knockdown suppressed the
proliferation, invasion and migration and promoted the apoptosis of
TNBC cells through the activation of the Notch signaling pathway.
The present study may provide a potential biomarker for the
diagnosis and treatment of TNBC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LY wrote the manuscript and analyzed the data. FT
carried out the experiments, supervised the present study, searched
the literature and revised the manuscript. LY and FT confirm the
authenticity of all the raw data. Both authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Costa RLB and Gradishar WJ:
Triple-negative breast cancer: Current practice and future
directions. J Oncol Pract. 13:301–303. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bauer KR, Brown M, Cress RD, Parise CA and
Caggiano V: Descriptive analysis of estrogen receptor
(ER)-negative, progesterone receptor (PR)-negative, and
HER2-negative invasive breast cancer, the so-called triple-negative
phenotype: A population-based study from the California cancer
registry. Cancer. 109:1721–1728. 2007.PubMed/NCBI View Article : Google Scholar
|
|
4
|
de Foucher T, Roussel H, Hivelin M, Rossi
L, Cornou C, Bats AS, Deloménie M, Lécuru F and Ngô C: Atypical
distant metastasis of breast malignant phyllodes tumors: A case
report and literature review. Case Rep Obstet Gynecol.
2017(8963013)2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sutter SA, Slinker A, Balumuka DD and
Mitchell KB: Surgical management of breast cancer in africa: A
continent-wide review of intervention practices, barriers to care,
and adjuvant therapy. J Glob Oncol. 3:162–168. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Huszno J and Grzybowska E: TP53 mutations
and SNPs as prognostic and predictive factors in patients with
breast cancer. Oncol Lett. 16:34–40. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Langerød A, Zhao H, Borgan Ø, Nesland JM,
Bukholm IR, Ikdahl T, Kåresen R, Børresen-Dale AL and Jeffrey SS:
TP53 mutation status and gene expression profiles are powerful
prognostic markers of breast cancer. Breast Cancer Res.
9(R30)2007.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Gratenstein K, Heggestad AD, Fortun J,
Notterpek L, Pestov DG and Fletcher BS: The WD-repeat protein
GRWD1: Potential roles in myeloid differentiation and ribosome
biogenesis. Genomics. 85:762–773. 2005.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Fujiyama H, Tsuji T, Hironaka K, Yoshida
K, Sugimoto N and Fujita M: GRWD1 directly interacts with p53 and
negatively regulates p53 transcriptional activity. J Biochem.
167:15–24. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Deng X, Li S, Kong F, Ruan H, Xu X, Zhang
X, Wu Z, Zhang L, Xu Y, Yuan H, et al: Long noncoding RNA PiHL
regulates p53 protein stability through GRWD1/RPL11/MDM2 axis in
colorectal cancer. Theranostics. l10:265–280. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang Q, Ren H, Xu Y, Jiang J, Wudu M, Liu
Z, Su H, Jiang X, Zhang Y, Zhang B and Qiu X: GRWD1 promotes cell
proliferation and migration in non-small cell lung cancer by
activating the notch pathway. Exp Cell Res.
387(111806)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Atwal D, Ramos JM and Makhoul I: Late
breast cancer recurrence with bone marrow metastases and acute
pulmonary hypertension. Proc (Bayl Univ Med Cent). 31:213–215.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
McAllister SD, Murase R, Christian RT, Lau
D, Zielinski AJ, Allison J, Almanza C, Pakdel A, Lee J, Limbad C,
et al: Pathways mediating the effects of cannabidiol on the
reduction of breast cancer cell proliferation, invasion, and
metastasis. Breast Cancer Res Treat. 129:37–47. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Takafuji T, Kayama K, Sugimoto N and
Fujita M: GRWD1, a new player among oncogenesis-related
ribosomal/nucleolar proteins. Cell Cycle. 16:1397–1403.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kayama K, Watanabe S, Takafuji T, Tsuji T,
Hironaka K, Matsumoto M, Nakayama KI, Enari M, Kohno T, Shiraishi
K, et al: GRWD1 negatively regulates p53 via the RPL11-MDM2 pathway
and promotes tumorigenesis. EMBO Rep. 18:123–137. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Pan Y, Yuan Y, Liu G and Wei Y: P53 and
Ki-67 as prognostic markers in triple-negative breast cancer
patients. PLoS One. 12(e0172324)2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xu L, Wang S, Zhao Z and Wang K:
Expression and prognosis of VEGF and P53 in tissues and serum of
patients with triple negative breast cancer. Journal of Nanjing
Medical University (Natural Sciences). 41:118–121. 2021.(In
Chinese).
|
|
19
|
Aigner K, Dampier B, Descovich L, Mikula
M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist
P, et al: The transcription factor ZEB1 (deltaEF1) promotes tumour
cell dedifferentiation by repressing master regulators of
epithelial polarity. Oncogene. 26:6979–6988. 2007.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Choi BD, Jeong SJ, Wang G, Park JJ, Lim
DS, Kim BH, Cho YI, Kim CS and Jeong MJ: Secretory leukocyte
protease inhibitor is associated with MMP-2 and MMP-9 to promote
migration and invasion in SNU638 gastric cancer cells. Int J Mol
Med. 28:527–534. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Burlaka AP, Ganusevich II, Gafurov MR,
Lukin SM and Sidorik EP: Stomach cancer: Interconnection between
the redox state, activity of MMP-2, MMP-9 and stage of tumor
growth. Cancer Microenviron. 9:27–32. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yuan X, Wu H, Han N, Xu H, Chu Q, Yu S,
Chen Y and Wu K: Notch signaling and EMT in non-small cell lung
cancer: Biological significance and therapeutic application. J
Hematol Oncol. 7(87)2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Giuli MV, Giuliani E, Screpanti I,
Bellavia D and Checquolo S: Notch signaling activation as a
hallmark for triple-negative breast cancer subtype. J Oncol.
2019(8707053)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chiang MY, Radojcic V and Maillard I:
Oncogenic notch signaling in t-cell and B-cell lymphoproliferative
disorders. Curr Opin Hematol. 23:362–370. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Allen TD, Rodriguez EM, Jones KD and
Bishop JM: Activated notch1 induces lung adenomas in mice and
cooperates with myc in the generation of lung adenocarcinoma.
Cancer Res. 71:6010–6018. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Licciulli S, Avila JL, Hanlon L, Troutman
S, Cesaroni M, Kota S, Keith B, Simon MC, Puré E, Radtke F, et al:
Notch1 is required for Kras-induced lung adenocarcinoma and
controls tumor cell survival via p53. Cancer Res. 73:5974–5984.
2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kar R, Jha NK, Jha SK, Sharma A, Dholpuria
S, Asthana N, Chaurasiya K, Singh VK, Burgee S and Nand P: A
‘NOTCH’ deeper into the epithelial-to-mesenchymal transition (EMT)
program in breast cancer. Genes. 10(961)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kong P, Chen L, Yu M, Tao J, Liu J, Wang
Y, Pan H, Zhou W and Wang S: miR-3178 inhibits cell proliferation
and metastasis by targeting Notch1 in triple-negative breast
cancer. Cell Death Dis. 9(1059)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Xiao YS, Zeng D, Liang YK, Wu Y, Li MF, Qi
YZ, Wei XL, Huang WH, Chen M and Zhang GJ: Major vault protein is a
direct target of notch1 signaling and contributes to
chemoresistance in triple-negative breast cancer cells. Cancer
Lett. 440-441:156–167. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sun D, Lei W, Hou X, Li H and Ni W: PUF60
accelerates the progression of breast cancer through downregulation
of PTEN expression. Cancer Manag Res. 11:821–830. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lin Y, Zhang J, Li Y, Guo W, Chen L, Chen
M, Chen X, Zhang W, Jin X, Jiang M, et al: CTPS1 promotes malignant
progression of triple-negative breast cancer with transcriptional
activation by YBX1. J Transl Med. 20(17)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Eskiler GG, Sahin E, Ozkan AD, Kaya OT and
Kaleli S: Curcumin induces DNA damage by mediating homologous
recombination mechanism in triple negative breast cancer. Nutr
Cancer. 72:1057–1066. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Iso T, Kedes L and Hamamori Y: HES and
HERP families: Multiple effectors of the notch signaling pathway. J
Cell Physiol. 194:237–255. 2003.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Stifani S, Blaumueller CM, Redhead NJ,
Hill RE and Artavanis-Tsakonas S: Human homologs of a drosophila
enhancer of split gene product define a novel family of nuclear
proteins. Nat Genet. 2:119–127. 1992.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Maier MM and Gessler M: Comparative
analysis of the human and mouse Hey1 promoter: Hey genes are new
notch target genes. Biochem Biophys Res Commun. 275:652–660.
2000.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ronchini C and Capobianco AJ: Induction of
cyclin D1 transcription and CDK2 activity by Notch(ic): Implication
for cell cycle disruption in transformation by Notch(ic). Mol Cell
Biol. 21:5925–5934. 2001.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Oswald F, Liptay S, Adler G and Schmid RM:
NF-kappaB2 is a putative target gene of activated Notch-1 via
RBP-Jkappa. Mol Cell Biol. 18:2077–2088. 1998.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Weng AP, Millholland JM, Yashiro-Ohtani Y,
Arcangeli ML, Lau A, Wai C, Bianco CD, Rodriguez CG, Sai H, Tobias
J, et al: c-Myc is an important direct target of Notch1 in T-cell
acute lymphoblastic leukemia/lymphoma. Genes Dev. 20:2096–2109.
2006.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Choi JW, Pampeno C, Vukmanovic S and
Meruelo D: Characterization of the transcriptional expression of
notch-1 signaling pathway members, Deltex and HES-1, in developing
mouse thymocytes. Dev Comp Immunol. 26:575–588. 2002.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kobayashi T, Mizuno H, Imayoshi I,
Furusawa C, Shirahige K and Kageyama R: The cyclic gene Hes1
contributes to diverse differentiation responses of embryonic stem
cells. Genes Dev. 23:1870–1875. 2009.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Suh JH, Lee HW, Lee JW and Kim JB: Hes1
stimulates transcriptional activity of Runx2 by increasing protein
stabilization during osteoblast differentiation. Biochem Biophys
Res Commun. 367:97–102. 2008.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Himes AD and Raetzman LT: Premature
differentiation and aberrant movement of pituitary cells lacking
both Hes1 and Prop1. Dev Biol. 325:151–161. 2009.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Lu L, Liu S, Dong Q and Xin Y: Salidroside
suppresses the metastasis of hepatocellular carcinoma cells by
inhibiting the activation of the notch1 signaling pathway. Mol Med
Rep. 19:4964–4972. 2019.PubMed/NCBI View Article : Google Scholar
|