Introduction
Pancreatic adenocarcinoma (PAAD) is one of the most
lethal types of malignancies (1).
Patients with PAAD frequently present with non-specific symptoms,
such as abdominal pain and weight loss, making it difficult to
diagnose PAAD at an early stage. Moreover, the vast majority of
patients are found to already possess tumor metastases at the time
of initial diagnosis (2,3). Despite recent advances that have been
made in terms of surgical techniques, chemotherapy, and radiation
therapy, the 5-year survival rate remains at a dismal 5% (4). Therefore, it is essential to identify
novel specific biomarkers for the application of targeted therapy
for patients with PAAD.
Agmatinase (AGMAT) belongs to the arginase family
(5), and its tertiary protein
structure comprises eight parallel β-sheets sandwiched in between
three α-helices on either side. AGMAT is a metallohydrolase whose
catalytic activity depends on Mn2+ ions (6); specifically, two Mn2+ ions
bind to the high- and low-affinity sites to activate nucleophilic
water molecules for catalysis (5-7).
AGMAT hydrolyzes agmatine, which is an endogenous polyamine
synthesized by L-arginine decarboxylase, to form urea and
putrescine. It was not confirmed until 1994 that AGMAT and agmatine
are synthesized in mammals (8). In
mammals, previous studies have shown that agmatine is involved in a
wide range of cellular biological functions; for example,
regulation of insulin release from pancreatic cells (9), control of the glomerular filtration
rate (10,11), and neuroprotection (12-14).
Therefore, a potentially important mechanistic role for AGMAT, in
terms of regulating the biological functions of agmatine in
mammalian cells was revealed (15). Subsequently, numerous studies have
been conducted to understand the physiological mechanisms through
which the levels of agmatine are regulated; however, few reports
have been published on the role of agmatine in cancer. In cancer
research, AGMAT has been shown to have an important role in
the development of tumors, including colorectal cancer (16) and lung adenocarcinoma (17). However, to the best of our
knowledge, the potential functions and molecular mechanisms of
AGMAT in PAAD have not yet been investigated. According to
the findings of the above studies on the biological functions of
AGMAT in the occurrence and development of tumors, it was
possible to speculate that AGMAT may have an important role
in the pathogenesis of PAAD, although its role remains to be
investigated.
In the present study, it was shown that AGMAT
is an oncogene that is associated with the poor prognosis of
patients with PAAD through bioinformatics analysis. Moreover, it
was demonstrated that AGMAT is able to promote the viability
of cell proliferation and metastasis of PAAD cells in vitro.
Additional mechanistic experiments demonstrated that AGMAT
is able to enhance epithelial-mesenchymal transition (EMT) via the
transforming growth factor-β (TGFβ)/Smad pathway. Taken together,
the findings of the present study suggest that AGMAT may be
a promising prognostic biomarker for PAAD, and therefore it is
potentially a novel therapeutic target.
Materials and methods
Expression dataset
RNA sequencing (RNAseq) data for AGMAT was
downloaded from the UCSC Xena database (http://xena.ucsc.edu/), and the expression of
AGMAT was analyzed in 183 PAAD tissues and 165 adjacent
normal tissues downloaded from the UCSC Xena database (http://xena.ucsc.edu/), which included PAAD tissues
from The Cancer Genome Atlas (TCGA) and normal tissues from the
Genotype Tissue Expression (GTEx) Project. In addition, the
associations between the expression level of AGMAT and
overall survival (OS) in the Kaplan-Meier plotter database
(https://kmplot.com) were also downloaded. Differences
in AGMAT expression between tumor and normal tissues, as
determined by immunohistochemical analysis in the Human Protein
Atlas (https://www.proteinatlas.org) were
subsequently compared.
Plasmids, retroviral infection, and
transfection
The full-length AGMAT gene was purchased from
Geneppl Technology Co., Ltd. and subcloned into the pCD513B vector
to generate cells that stably overexpressed AGMAT. The three
small hairpin RNA (shRNA) lentivirus plasmids were also purchased
from Geneppl Technology Co., Ltd. and cloned into a lentiviral pPLK
GFP+puro vector to generate cells with stably knocked down
AGMAT expression. The sequences of these three shRNAs were:
shRNA#1, CGATGTGAATGTCAATCTTTA; shRNA#2, CAAACCCATTTATATCAGCTT; and
shRNA#3, CGGGAAGAATCAGTGATGCTT, and the sequence of the negative
control sequence used was GTTCTCCGAACGTGTCACGTT. 293T cells were
transfected with DMEM, DNA and GM Easy Lentiviral Mix using HG
Transgene Reagent, which was purchased from Genomeditech, according
to the manufacturer's instructions. At 48 h after transfection, the
lentivirus particles were collected.
Generation of stable cell lines
The human PAAD MiaPaCa-2 (cat. no. CRM-CRL-1420),
SW1990 (cat. no. CRL-2172), Panc-1 (cat. no. CRL-1469), and BxPc3
(cat. no. CRL-1687) cells were obtained from American Type Culture
Collection (ATCC). PaTu8988s (cat. no. CL-0303) cells were obtained
from Procell Life Science & Technology Co. 293T (cat. no.
GNHu17) cells were obtained from the Cell Bank of the Chinese
Academy of Sciences (Shanghai, China). The cells were maintained at
the Central Laboratory of Shanghai Fengxian District Central
Hospital. The cell lines were cultured and maintained in
Gibco® DMEM (Thermo Fisher Scientific, Inc.)
supplemented with 10% Gibco® fetal bovine serum (FBS)
(Thermo Fisher Scientific, Inc.) in a humidified incubator supplied
with 5% CO2. The human PAAD SW1990 and BxPc3 cell lines
were infected with lentivirus plus 8 µg/ml polybrene to establish
the PAAD cell lines stably overexpressing or with stably knocked
down AGMAT, respectively. In order to obtain stably
transfected cells, the cells were cultured with puromycin
(Invitrogen®; Thermo Fisher Scientific, Inc.) for 7
days. Subsequently, western blotting and reverse
transcription-quantitative (RT-qPCR) analyses were used to detect
both the AGMAT-knockdown efficiency and
AGMAT-overexpression levels, as detailed below.
RT-qPCR
Total RNA was extracted using TRIzol®
reagent (Thermo Fisher Scientific, Inc.). Subsequently, the Evo
M-MLV RT Mix Kit with gDNA Clean for qPCR (Accurate Bio Technology
Co., Ltd.) was used to reverse-transcribe total RNA to cDNA. A
SYBR® Green Premix Pro Taq HS qPCR Kit (ROX Plus)
(Accurate Bio Technology Co., Ltd.) was used to perform the qPCR
experiments. The following primer sequences were all purchased from
Sangon Biotech Co., Ltd. and were used for the RT-qPCR:
AGMAT forward primer, 5'-CTTGTCGAAGTTTCACCACCGTA-3' and
reverse primer, 5'-CTTTGGGGAGAGCACATAGCATC-3'; GAPDH forward
primer, 5'-TTGGTATCGTGGAAGGACTCA-3' and reverse primer,
5'-TGTCATCATATTTGGCAGGTT-3'. The 2-ΔΔCq method (18) was used to measure the relative mRNA
expression levels.
Western blot analysis
Total protein was extracted from the treated cells
(either stably overexpressing AGMAT or with AGMAT
stably knocked down) lysed in RIPA lysis buffer. Subsequently, the
proteins were separated using SDS-PAGE (10% gels) and then
transferred onto a nitrocellulose membrane. The membranes were
incubated with primary and secondary antibodies (see below), and
the signals were visualized using a Tanon Highly-sig ECL Western
Blotting Substrate Reagent Kit (#180-5001, Tanon Science and
Technology Co., Ltd.). The signals were visualized using an ECL kit
(Vazyme Biotech Co., Ltd.), and images were captured using a Tanon
4600 system. The following primary antibodies were used: anti-AGMAT
(1:1,000; cat. no. ab231894; Abcam), anti-E-cadherin (1:5,000; cat.
no. 20874-1-AP; Proteintech), anti-N-cadherin (1:1,000; cat.
no.22018-1-AP; Proteintech), anti-vimentin (1:1,000; cat. no.
10366-1-AP; Proteintech), anti-MMP2 (1:1,000; cat. no.10373-2-AP;
Proteintech), anti-MMP9 (1:1,000; cat. no. 10375-2-AP;
Proteintech), anti-TGFβ1 (1:1,000; cat. no. C0340; Assay Biotech),
anti-SMAD4 (1:1,000; cat. no.10231-1-AP; Proteintech), anti-Smad2/3
(1:1,000; cat. no. 8685; CST), anti-phosphorylated (p)-Smad2/3
(1:1,000; cat. no. #4511; CST), and anti-β-tubulin (1:20,000; cat.
no. 66240-1-Ig; Proteintech). The following secondary antibodies
were used: HRP-conjugated Affinipure goat anti-mouse IgG (1:5,000;
cat. no. SA00001-1; Proteintech) and HRP-conjugated Affinipure goat
anti-rabbit IgG (1:5,000; cat. no. SA00001-2; Proteintech). The
relative levels of the proteins of interest were normalized against
those of β-tubulin.
Cell proliferation and colony
formation assays
For cell proliferation assays, the treated PAAD
cells (either stably overexpressing AGMAT or with
AGMAT stably knocked down) were seeded at a density of 1,000
cells/well in 96-well plates and cultured for 24, 48, 72 and 96 h.
Aliquots of 10 µl Cell Counting Kit-8 (CCK-8) reagents (Dojindo,
Inc.) were added to each well, and the mixture was incubated at
37˚C for 1 h. Subsequently, the absorbance at 450 nm was measured.
For the colony formation assays, the treated PAAD cells (either
stably overexpressing AGMAT or with AGMAT stably
knocked down) were seeded onto a 6-well plate at a density of 1,000
cells/well and cultured for 10 or 14 days. After the incubation
period, whole colonies were treated with 4% paraformaldehyde and
stained with 0.1% crystal violet for 30 min at room temperature,
prior to capturing images with a camera (Canon, Inc.).
Cell migration and invasion
assays
Migration assays of PAAD cells overexpressing
AGMAT or with AGMAT knocked down were performed using
Transwell chambers (Corning, Inc.). Invasion assays were performed
using Corning BioCoat Matrigel Invasion Chamber (Corning, Inc.).
For the invasion assay, 5x105 cells were inoculated into
each chamber in triplicate. For the migration assay, the bottom
chamber was filled with culture medium containing 10% FBS, and
5x105 cells were suspended in serum-free medium and
plated in the upper chamber. After incubation for 24 h at 37˚C, the
cells on the lower surface of the membrane were fixed with 4%
formaldehyde in PBS and stained with 0.1% crystal violet for 30 min
at room temperature. Subsequently, the cells were counted under a
fluorescence microscope (Olympus Corporation; magnification,
x200).
Statistical analysis
GraphPad Prism 8 (GraphPad Software, Inc.) was used
for the statistical analysis. Data are presented as the means ±
standard error of the mean (SEM) or the means ± standard deviation
(SD) of three independent experiments. The Student's t-test or
one-way ANOVA followed by post hoc Dunnett's test was used to
estimate the significant differences between different groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
AGMAT is upregulated in PAAD and
positively associated with a poor prognosis
In order to explore the mechanism through which
AGMAT is dysregulated in PAAD, the expression levels of
AGMAT in PAAD tumor tissues and adjacent normal pancreatic
tissues were first investigated. The AGMAT mRNA expression
data of 183 PAAD tumors and 165 adjacent pancreatic tissues were
analyzed using the online bioinformatics tool UCSC Xena. The
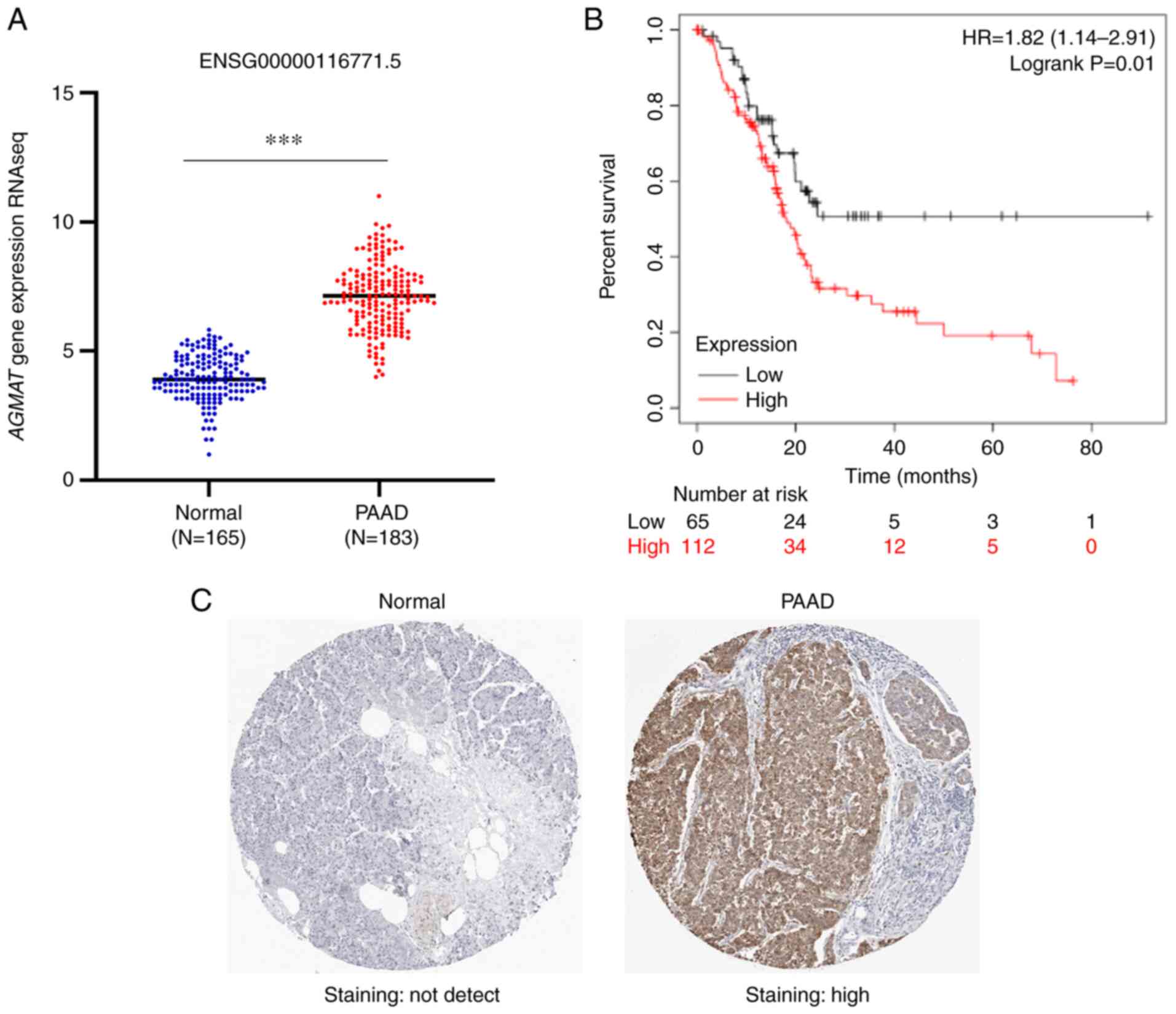
results from this analysis showed that AGMAT was
significantly upregulated in the PAAD tumor tissues (Fig. 1A). Accordingly, the expression
levels of AGMAT were found to be inversely correlated with
the OS rate. The survival analysis revealed that the PAAD patients
with AGMAT overexpression had higher mortality rates and
shorter OS rates compared with patients with lower expression
levels of AGMAT (Fig. 1B).
Furthermore, the immunohistochemical results showed that there were
no appreciable levels of AGMAT expression in normal
pancreatic tissues, although there was high expression of
AGMAT in PAAD (Fig. 1C).
Taken together, these results showed that the expression levels of
the AGMAT gene was markedly upregulated in PAAD, suggesting
that AGMAT may exert a positive effect in PAAD and could be
of use as a prognostic biomarker.
AGMAT promotes PAAD cell proliferation
and colony formation in PAAD
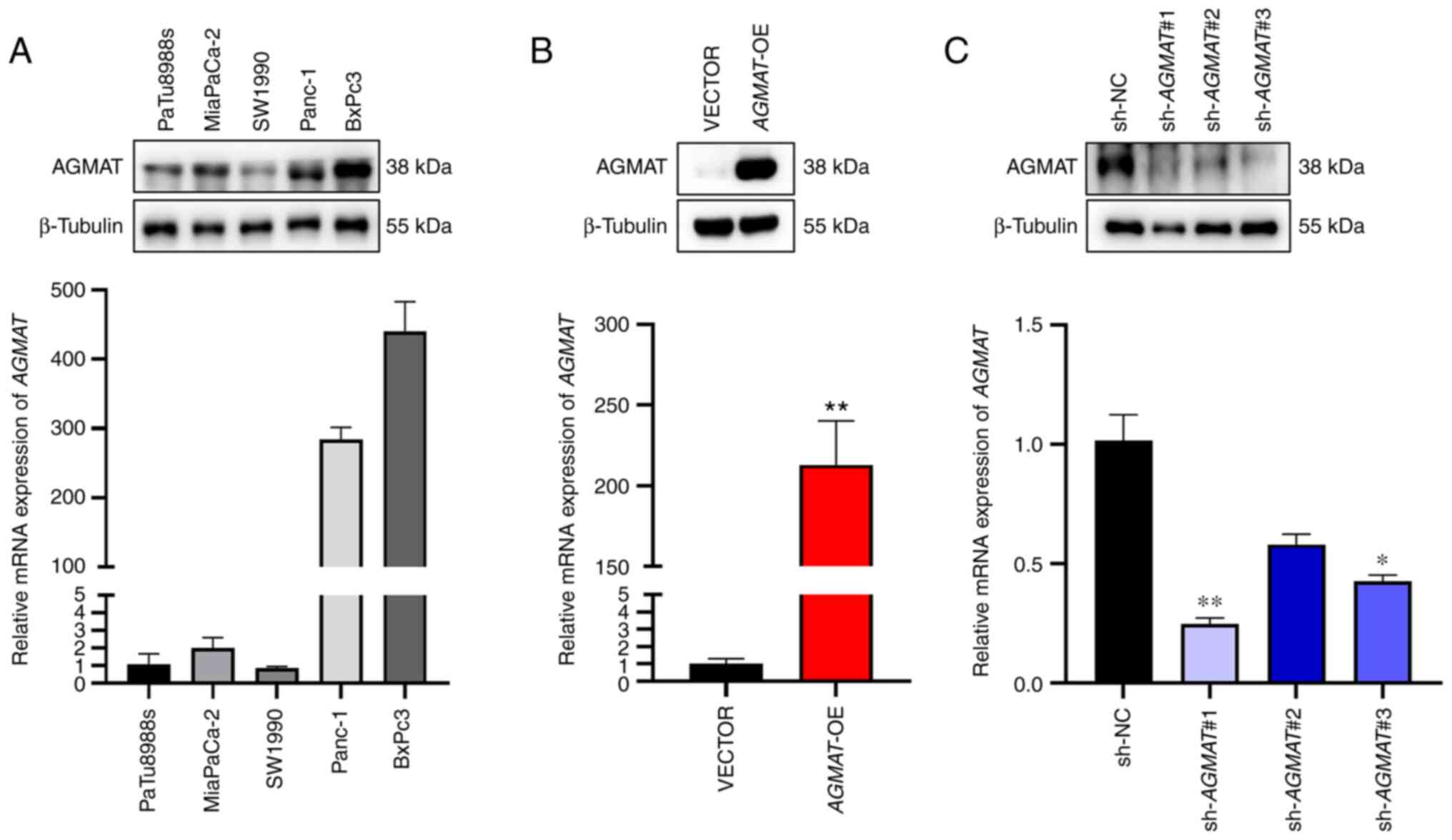
The protein and mRNA expression levels of
AGMAT in various PAAD cell lines, including PaTu8988s,
MiaPaCa-2, SW1990, Panc-1 and BxPc3 cells, were first examined by
western blot and RT-qPCR analyses (Fig. 2A). As a result of these
experiments, SW1990 cells were selected to construct the stably
overexpressing AGMAT cells, and BxPc3 cells were used to
construct cells that had stably knocked down AGMAT using a
lentiviral vector. The efficiency of stable overexpression, and
knockdown efficiency, of AGMAT was analyzed by both western
blot and RT-qPCR analyses (Fig. 2B
and C).
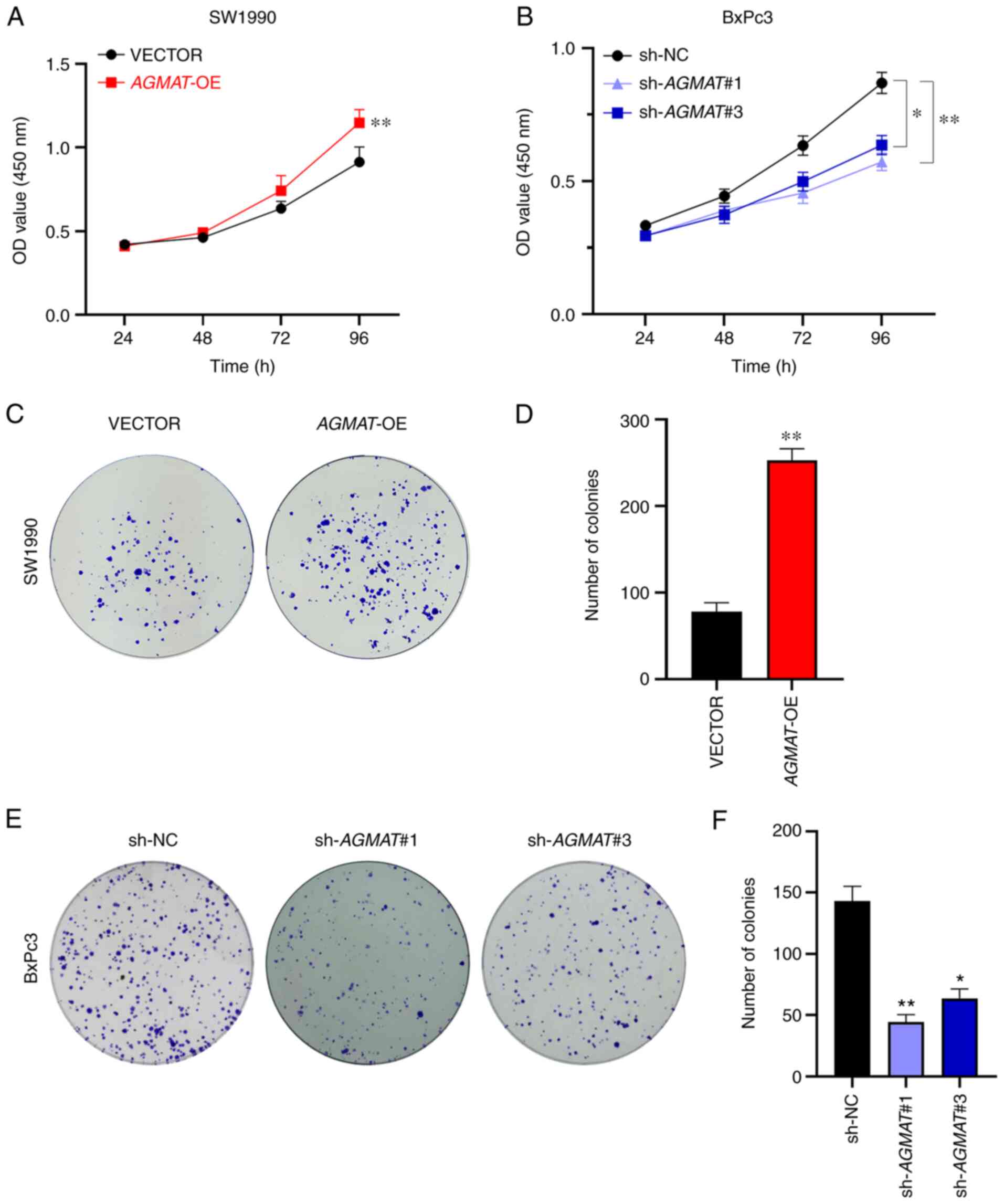
To explore the biological function of AGMAT
in PAAD, the proliferation of AGMAT-overexpressing SW1990
cells and AGMAT-knockdown BxPc3 cells were examined via
CCK-8 assays and colony formation assays, respectively. The results
of the CCK-8 assays showed that cell proliferation was
significantly enhanced after stably overexpressing AGMAT
(Fig. 3A). The same results were
obtained in the colony formation assays, AGMAT could promote
the formation of pancreatic cancer cell colonies (Fig. 3C and D). Conversely, the CCK-8 assays revealed
that cell proliferation was significantly inhibited after the
stable knockdown of AGMAT (Fig.
3B), and according to the cell formation assays, AGMAT
knockdown resulted in a significant suppression of the numbers of
cell colonies formed (Fig. 3E and
F). Therefore, these results
suggest that AGMAT exerts an important role in the growth of
cells in vitro.
Effects of AGMAT on migration and
invasion of pancreatic cancer cells
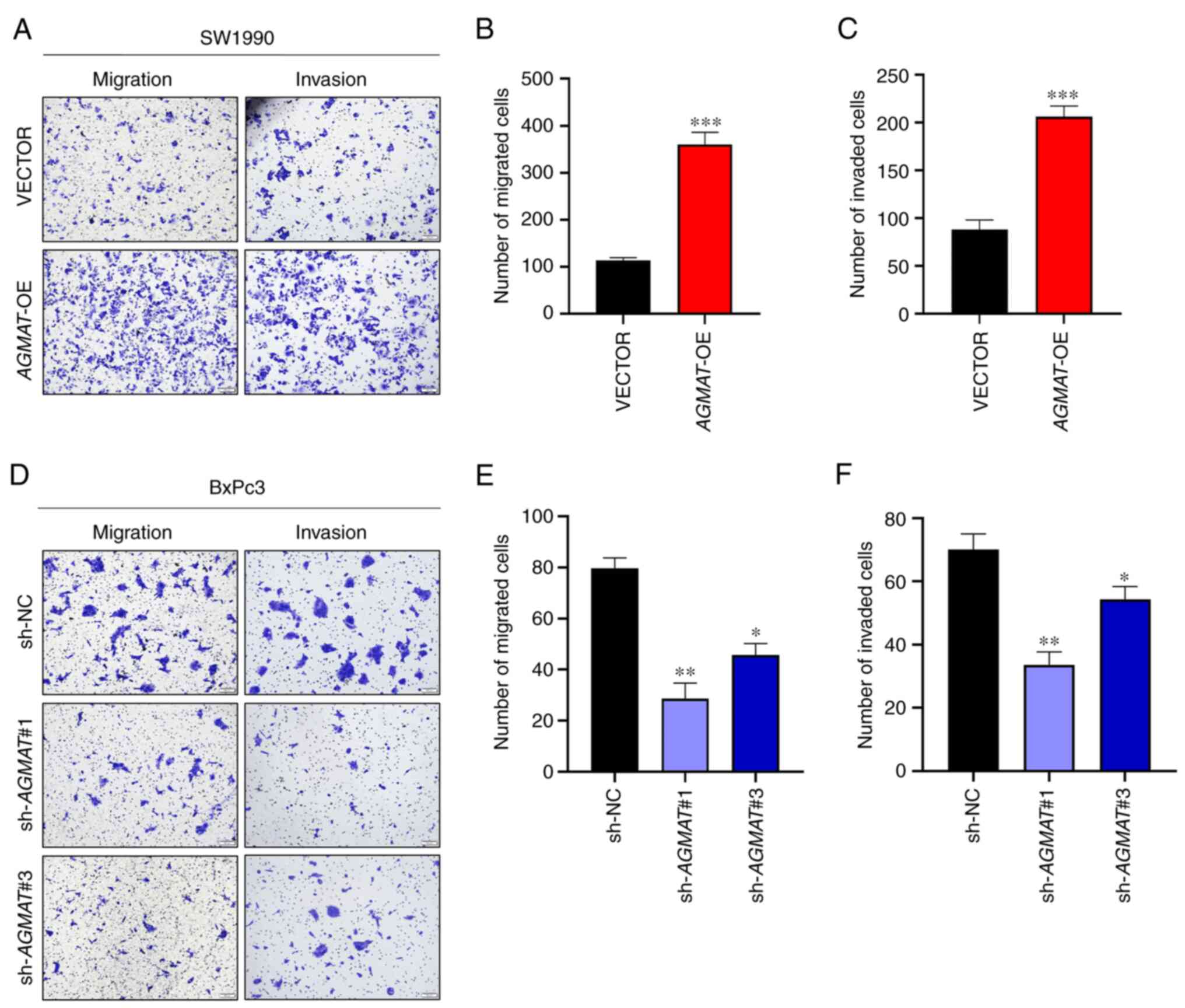
Subsequently, the function of AGMAT in PAAD
metastasis was examined. As shown in Fig. 4A, the results revealed that both
cell migration and invasion were enhanced after stably
overexpressing AGMAT in SW1990 cells. In contrast, cell
migration and invasion were inhibited in the AGMAT-knockdown
BxPc3 cells, as shown in Fig. 4D.
Then, the numbers of cells that had migrated and invaded were
counted and analyzed, as shown in Fig.
4B, C, E and F.
These results demonstrated that AGMAT could promote the
migration and invasion capabilities of the PAAD cells.
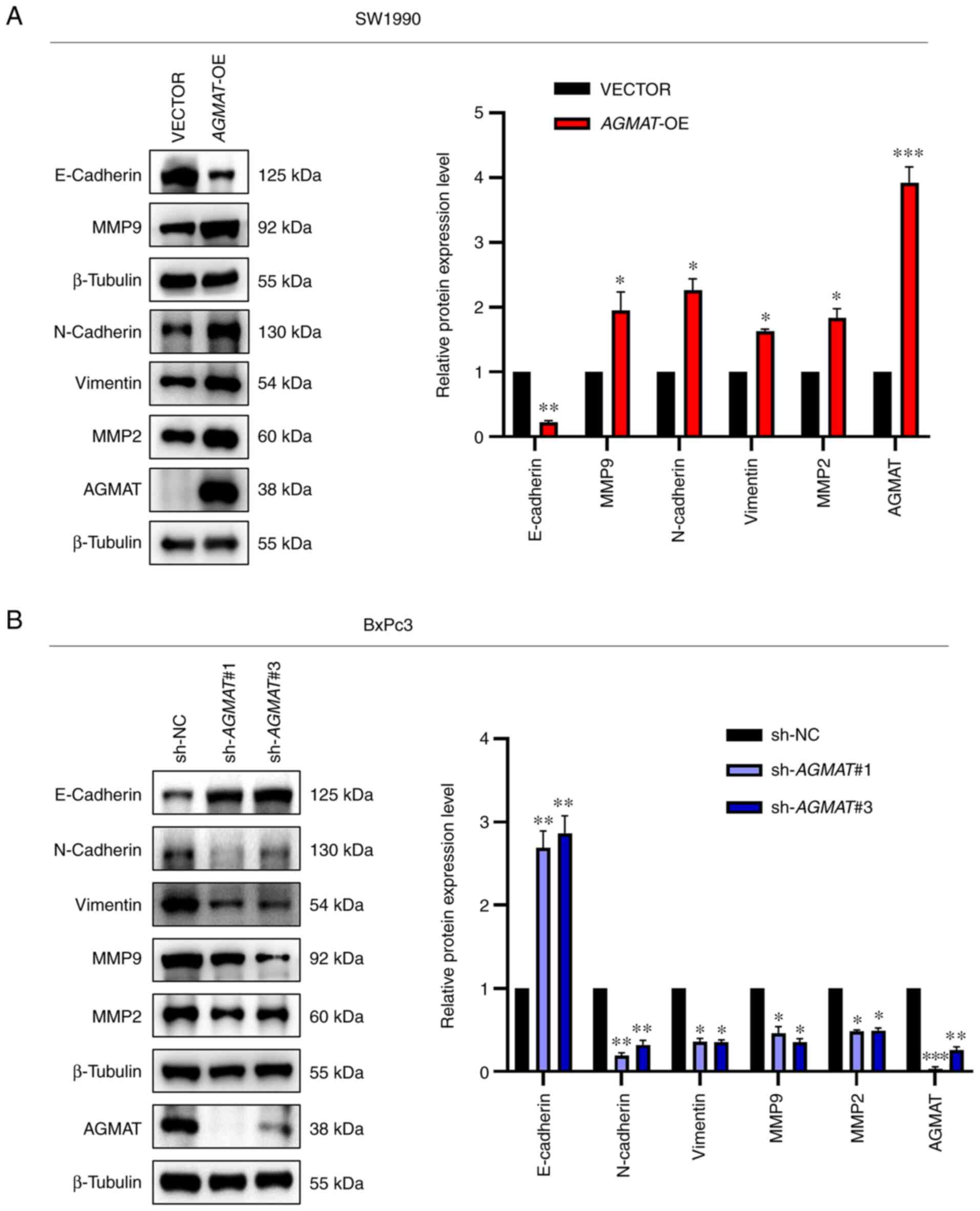
AGMAT induces EMT in PAAD cells and
accelerates the process induced by the TGFβ/Smad signaling
pathway
Epithelial-mesenchymal transition (EMT) refers to
the transition of epithelial to mesenchymal cells. During this
process, the cells lose their epithelial characteristics, including
their polarity, which confers on them a migratory behavior
(19,20). At the same time, the protein
expression levels of three key epithelial marker proteins
(E-cadherin, vimentin and N-cadherin) are characteristically
altered, which leads to a decrease in the adhesion of cells, and a
loss of polarity and tight junctions (19). The matrix metalloproteinases (MMPs)
are both a large family and an important class of proteolytic
enzymes, which are able to degrade various protein components in
the extracellular matrix, destroy the histological barrier of tumor
cell invasion, and serve a key role in tumor invasion and
metastasis (21). A previous study
revealed that the overexpression of MMP2 and MMP9 is associated
with tumors (22). These findings
motivated us to examine the effect of AGMAT on the
EMT-associated proteins, MMP2 and MMP9, in PAAD cells. To meet this
end, western blot analysis was performed. The results obtained
showed that the SW1990-constructed cell line that stably
overexpressed AGMAT exhibited a significantly suppressed
expression levels of E-cadherin and significantly upregulated
levels of N-cadherin, vimentin, MMP2 and MMP9 (Fig. 5A). Conversely, the opposite trends
were showed in BxPc3 cells with AGMAT-knockdown (Fig. 5B). Taken together, all the above
results demonstrated that overexpression of AGMAT could
promote cell migration and invasion in PAAD cells through promoting
EMT.
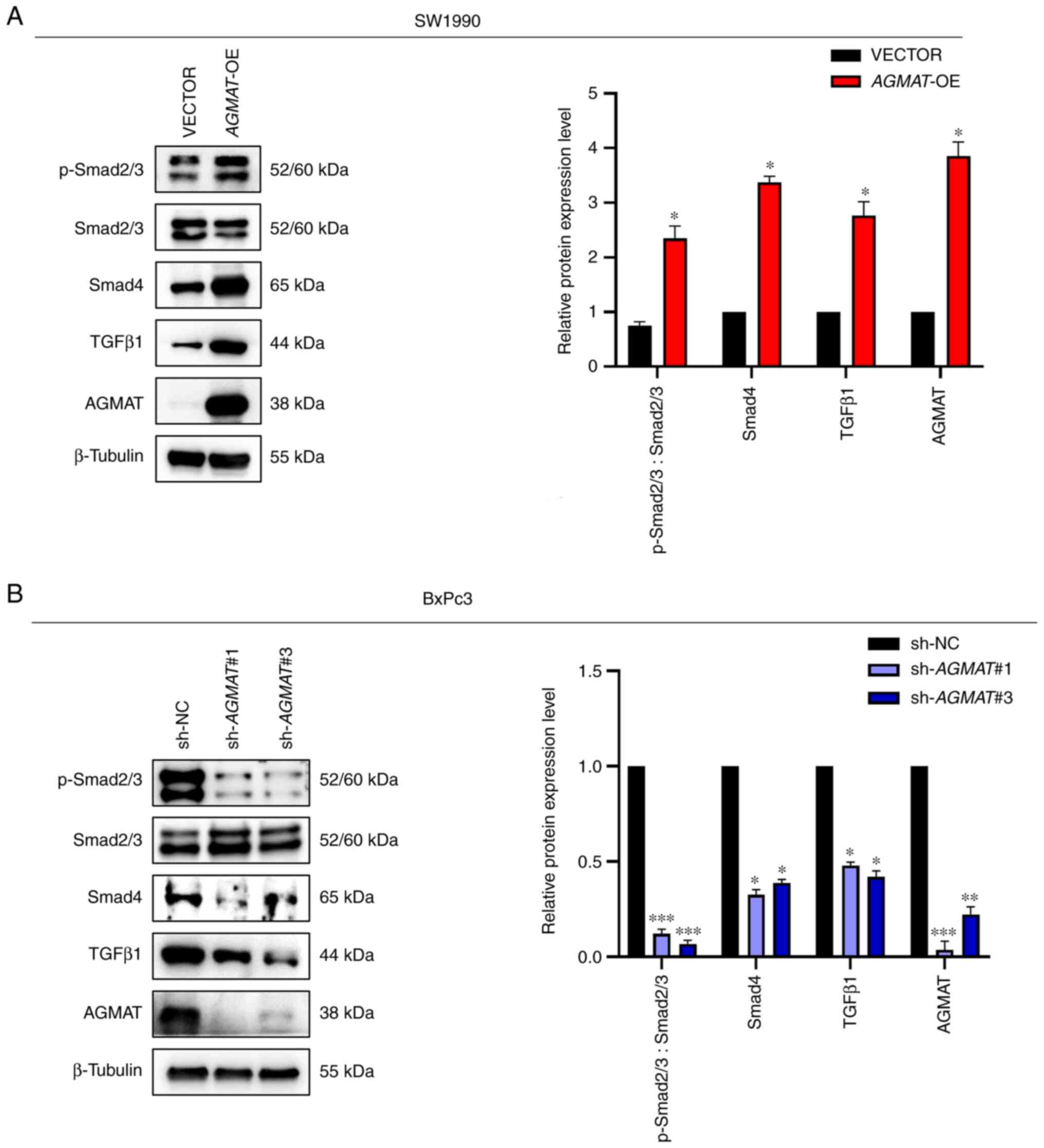
Subsequently, the mechanism of AGMAT in PAAD
cells was explored in more precise detail. A previous study
demonstrated that TGFβ is an important factor in the tumor
microenvironment; in particular, it was shown that TGFβ is able to
promote tumor metastasis via inducing the so-called EMT (23). Therefore, we focused on whether
AGMAT could accelerate the process of EMT by inducing the
TGFβ/Smad signaling pathway in PAAD. To meet this aim, western
blotting was used to detect the protein expression levels of TGFβ1,
Smad4 and p-Smad2/3, which serve important roles in the pathway. As
anticipated, AGMAT positively regulated the TGFβ/Smad
signaling pathway. The protein expression levels of p-Smad2/3,
Smad4 and TGFβ1 were significantly upregulated in SW1990 cells with
AGMAT overexpression, whereas the expression levels of these
proteins were significantly decreased in BxPc3 cells with
AGMAT expression knocked down (Fig. 6A and B). Collectively, these results indicated
that AGMAT could induce EMT in PAAD cells, and that this
process of induction was accelerated via the TGFβ/Smad signaling
pathway.
Discussion
In the present study, a novel function of
AGMAT in PAAD was uncovered. First, it was shown that the
AGMAT mRNA levels were overexpressed in PAAD tissues and
cells. Functionally, AGMAT was found to promote the growth
and aggressiveness of PAAD cells in vitro. During tumor
development, epithelial-mesenchymal transition (EMT) has been
demonstrated to fulfill a role in the progression of PAAD (24), and the present study revealed that
AGMAT could promote cell migration and invasion in PAAD
cells through promoting EMT. In addition, several studies have
shown that the TGFβ/Smad signaling pathway is a critical regulator
of EMT (24,25). Therefore, it was crucial to
investigate whether AGMAT could induce EMT via the TGFβ/Smad
signaling pathway. The mechanistic analysis suggested that
AGMAT could indeed induce EMT via the TGFβ/Smad signaling
pathway. However, the associations between AGMAT, EMT and
the TGFβ/Smad signaling pathway need to be further explored
utilizing TGFβ receptor inhibitors or agonists, among other
approaches. In pursuing these avenues, the mechanisms of
AGMAT in PAAD will be delineated more precisely.
AGMAT significantly affects the polyamine
biosynthetic pathway, functioning as the key enzyme of the
polyamine metabolism alternative pathway (26). Previous studies have demonstrated
that abnormalities in enzymes may be linked with a number of
diseases, especially cancer (17).
In a previous study, colon cancer samples were found to have low
levels of agmatine (27).
Furthermore, agmatine was found to suppress the proliferation of
tumor cells, which were derived from colon, liver, neuronal,
leukemia, among other types of cancer (28,29).
Based on the aforementioned studies, we hypothesized that AGMAT may
hydrolyze agmatine, thereby suppressing the antitumor function of
agmatine and facilitating the progression of PAAD. For almost a
decade, studies that have targeted polyamine metabolism have
intensely focused on cancer treatment and prevention, as regulating
polyamine levels is clearly one of the most potentially effective
strategies for treating cancer. Therefore, examining more closely
the molecular regulatory mechanisms of polyamine metabolism will be
of great significance for the treatment of cancer. In the future,
our studies will focus on whether and how AGMAT may hydrolyze
agmatine to inhibit the antitumor properties in PAAD.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the Shanghai Municipal
Health Commission Foundation (grant no. 20204Y0181).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XZ and YZ conceived and designed the experiments and
wrote the paper. YZ and XZ participated in all experiments. LC, YZ
and YX contributed to the plasmid construction and cell
transfection and interpretation of the experimental results. CW and
XL performed and analyzed the WB experiments. YX and YZ performed
and analyzed the IF experiments. JC and XZ revised the manuscript
and analyzed all data. XZ, YZ and JC confirm the authenticity of
all the raw data. All authors have read and approved the final
manuscript for publication.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhang L, Sanagapalli S and Stoita A:
Challenges in diagnosis of pancreatic cancer. World J
Gastroenterol. 24:2047–2060. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chu LC, Goggins MG and Fishman EK:
Diagnosis and detection of pancreatic cancer. Cancer J. 23:333–342.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Dowling DP, Di Costanzo L, Gennadios HA
and Christianson DW: Evolution of the arginase fold and functional
diversity. Cell Mol Life Sci. 65:2039–2055. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Carvajal N, López V, Salas M, Uribe E,
Herrera P and Cerpa J: Manganese is essential for catalytic
activity of Escherichia coli agmatinase. Biochem Biophys Res
Commun. 258:808–811. 1999.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Uribe E, Reyes MB, Martinez I, Mella K,
Salas M, Tarifeño-Saldivia E, López V, García-Robles M,
Martínez-Oyanedel J, Figueroa M, et al: Functional analysis of the
Mn2+ requirement in the catalysis of ureohydrolases
arginase and agmatinase-a historical perspective. J Inorg Biochem.
202(110812)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Li G, Regunathan S, Barrow CJ, Eshraghi J,
Cooper R and Reis DJ: Agmatine: An endogenous clonidine-displacing
substance in the brain. Science. 263:966–969. 1994.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Su CH, Liu IM, Chung HH and Cheng JT:
Activation of I2-imidazoline receptors by agmatine improved insulin
sensitivity through two mechanisms in type-2 diabetic rats.
Neurosci Lett. 457:125–128. 2009.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Satriano J, Cunard R, Peterson OW, Dousa
T, Gabbai FB and Blantz RC: Effects on kidney filtration rate by
agmatine requires activation of ryanodine channels for nitric oxide
generation. Am J Physiol Renal Physiol. 294:F795–F800.
2008.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Penner SB and Smyth DD: Natriuresis
following central and peripheral administration of agmatine in the
rat. Pharmacology. 53:160–169. 1996.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Cunha AS, Matheus FC, Moretti M, Sampaio
TB, Poli A, Santos DB, Colle D, Cunha MP, Blum-Silva CH, Sandjo LP,
et al: Agmatine attenuates reserpine-induced oral dyskinesia in
mice: Role of oxidative stress, nitric oxide and glutamate NMDA
receptors. Behav Brain Res. 312:64–76. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lee WT, Hong S, Yoon SH, Kim JH, Park KA,
Seong GJ and Lee JE: Neuroprotective effects of agmatine on
oxygen-glucose deprived primary-cultured astrocytes and nuclear
translocation of nuclear factor-kappa B. Brain Res. 1281:64–70.
2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Moretti M, Matheus FC, de Oliveira PA,
Neis VB, Ben J, Walz R, Rodrigues AL and Prediger RD: Role of
agmatine in neurodegenerative diseases and epilepsy. Front Biosci
(Elite Ed). 6:341–359. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Mistry SK, Burwell TJ, Chambers RM,
Rudolph-Owen L, Spaltmann F, Cook WJ and Morris SM Jr: Cloning of
human agmatinase. An alternate path for polyamine synthesis induced
in liver by hepatitis B virus. Am J Physiol Gastrointest Liver
Physiol. 282:G375–G381. 2002.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Snezhkina AV, Krasnov GS, Lipatova AV,
Sadritdinova AF, Kardymon OL, Fedorova MS, Melnikova NV, Stepanov
OA, Zaretsky AR, Kaprin AD, et al: The dysregulation of polyamine
metabolism in colorectal cancer is associated with overexpression
of c-Myc and C/EBPβ rather than enterotoxigenic bacteroides
fragilis infection. Oxid Med Cell Longev.
2016(2353560)2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhu HE, Yin JY, Chen DX, He S and Chen H:
Agmatinase promotes the lung adenocarcinoma tumorigenesis by
activating the NO-MAPKs-PI3K/Akt pathway. Cell Death Dis.
10(854)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tsai JH and Yang J: Epithelial-mesenchymal
plasticity in carcinoma metastasis. Genes Dev. 27:2192–2206.
2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Xu J, Lamouille S and Derynck R:
TGF-beta-induced epithelial to mesenchymal transition. Cell Res.
19:156–172. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lengyel E, Schmalfeldt B, Konik E, Späthe
K, Härting K, Fenn A, Berger U, Fridman R, Schmitt M, Prechtel D
and Kuhn W: Expression of latent matrix metalloproteinase 9 (MMP-9)
predicts survival in advanced ovarian cancer. Gynecol Oncol.
82:291–298. 2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Roomi MW, Monterrey JC, Kalinovsky T, Rath
M and Niedzwiecki A: In vitro modulation of MMP-2 and MMP-9
in human cervical and ovarian cancer cell lines by cytokines,
inducers and inhibitors. Oncol Rep. 23:605–614. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Peinado H, Quintanilla M and Cano A:
Transforming growth factor beta-1 induces snail transcription
factor in epithelial cell lines: Mechanisms for epithelial
mesenchymal transitions. J Biol Chem. 278:21113–21123.
2003.PubMed/NCBI View Article : Google Scholar
|
|
24
|
van Staalduinen J, Baker D, ten Dijke P
and van Dam H: Epithelial-mesenchymal-transition-inducing
transcription factors: New targets for tackling chemoresistance in
cancer? Oncogene. 37:6195–6211. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhao L, Liu S, Che X, Hou K, Ma Y, Li C,
Wen T, Fan Y, Hu X, Liu Y and Qu X: Bufalin inhibits TGF-β-induced
epithelial-to-mesenchymal transition and migration in human lung
cancer A549 cells by downregulating TGF-β receptors. Int J Mol Med.
36:645–652. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Laube G and Bernstein HG: Agmatine:
Multifunctional arginine metabolite and magic bullet in clinical
neuroscience? Biochem J. 474:2619–2640. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Molderings GJ, Kribben B, Heinen A,
Schroder D, Brüss M and Göthert M: Intestinal tumor and agmatine
(decarboxylated arginine): Low content in colon carcinoma tissue
specimens and inhibitory effect on tumor cell proliferation in
vitro. Cancer. 101:858–868. 2004.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wolf C, Brüss M, Hänisch B, Göthert M, von
Kügelgen I and Molderings GJ: Molecular basis for the
antiproliferative effect of agmatine in tumor cells of colonic,
hepatic, and neuronal origin. Mol Pharmacol. 71:276–283.
2007.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Haenisch B, Bönisch H, Cichon S, Allam JP,
Novak N and Molderings GJ: Effects of exogenous agmatine in human
leukemia HMC-1 and HL-60 cells on proliferation, polyamine
metabolism and cell cycle. Leuk Res. 35:1248–1253. 2011.PubMed/NCBI View Article : Google Scholar
|