Introduction
Hepatic fibrosis (HF) is a pathological change
caused by various reasons during the repair process of chronic
liver injuries. It is a pre-pathological change of liver cirrhosis
and may even progress to liver cancer (1). Hepatic stellate cells (HSCs) are the
main effector cells involved in HF (2). Excessive production of the
extracellular matrix (ECM) secreted by HSCs in the liver is the key
event in the development of HF (3,4).
Following activation of HSCs, the deposition of ECM causes damage
to liver tissues. Eventually, abnormally activated signaling
pathways result in malignant cell growth, migration and gene
expressions; thus finally leading to HF (5). So far, clinical management of HF has
been a global challenge that lacks effective treatment and there is
an urgent need to explore the options for treatment.
MicroRNAs (miRNAs/miRs) are short non-coding RNAs
comprising only 20-24 amino acids. They are vital epigenetic
regulators in post-transcriptional translation and they are closely
linked to the development of human diseases (6). miR-122 and miR-16 are very important
during the development of HF through mediating the proliferation,
activation and apoptosis of HSCs (7,8).
Homeodomain interacting protein kinase 1 (HIPK1) is
a serine/threonine kinase located in the nucleus. It serves an
important role in regulating cell proliferation, activation and
apoptosis through mediating oligomerization of apoptosis
signal-regulating kinase 1 and activation of the mitogen-activated
protein kinase signaling pathway (9-11).
HIPK1 also participates in the DNA damage repair (12). However, the role of HIPK1 in HF has
not yet been reported. Our previous studies have shown that
overexpression of HIPK1 in the rat HSC cell line HSC-T6 can
downregulate downstream transforming growth factor β (TGF-β),
α-smooth muscle actin (α-SMA) and collagen type I α 1 chain
(COL1A1), which are closely associated with HF, suggesting that
HIPK1 may be a target for inhibiting HSC activation.
To explore the possible regulatory mechanism in the
process of HF, the present study constructed a rat model of HF and
found the abnormal expression of miR-146b-5p and HIPK1. The present
study hypothesized that HSC activation could upregulate
miR-146b-5p, thus contribute to the progression of HF.
Materials and methods
Establishment of the rat HF model
A total of 12 male Sprague-Dawley (SD) rats aged 6
weeks, weighing 180-220 g were provided by the Guangdong Medical
Laboratory Animal Center. The rats were habituated in a specific
pathogen-free environment at 22±2˚C, with a 12/12-h light/dark
cycle and free access to food and water. Rats were randomly divided
into the control group (n=6) and HF group (n=6). Rats in the HF
group were subcutaneously injected with 0.3 ml/100 g carbon
tetrachloride (CCl4) three times a week, for eight
consecutive weeks, while rats in the control group were
subcutaneously injected with the same volume of normal saline. All
rats were sacrificed at the end of the eighth week and the tissues
which were 1.5 cm from the lower edge of the left lobe of the lever
were collected for subsequent experiments. When the rats quickly
lost more than 20% of their original body weight, could not eat or
drink, exhibited mental depression with hypothermia, suffered body
organ infection or severe liver failure, the experiment was stopped
and the rats sacrificed. The rats were sacrificed by
intraperitoneal injection of sodium pentobarbital with an injection
dose of 200 mg/kg. According to AVMA Guidelines for the Euthanasia
of Animals: 2020 Edition (https://www.avma.org/resources/pet-owners/petcare/euthanasia),
mortality was confirmed by checking that the hearts had stopped
beating for 2 min and the respiratory arrest was also observed. The
animal research of this project was approved by the medical
research ethics committee of the First Affiliated Hospital of
Nanchang University, ethics approval no. 2020117.
Hematoxylin and eosin (H&E)
staining
Rat liver tissues were fixed in 4% paraformaldehyde
for 24 h at room temperature, dehydrated and transparentized with
gradient concentrations (30, 50, 75, 95 and 100%) of ethanol and
xylene, and then embedded in paraffin at 55˚C to prepare 4-µm
tissue sections. Next, they were immersed twice in xylene for 20
min, twice in anhydrous ethanol for 5 min and in 75% ethanol for 5
min and finally washed in ddH2O. The H&E staining
kit (Sangon Biotech Co., Ltd.; cat. no. E607318) was used in the
present study. Briefly, sections were stained with hematoxylin for
5 min at room temperature. Following differentiation and bluing,
they were washed in ddH2O, dehydrated in 85% and 5%
ethanol sequentially and stained with eosin for 5 min at room
temperature. After incubating three times in anhydrous ethanol and
twice in xylene, they were mounted using neutral gum for
observation under a light microscope. A total of five fields of
view were randomly selected for each tissue section and observed at
x100 magnification.
Masson staining
Rat liver tissues were dewaxed and dehydrated in
xylene and ethanol with gradient concentrations (100, 85, 75 and
30%). After nuclei staining using the Weigert's iron hematoxylin
solution for 5-10 min and washing in flowing water, sections were
later counterstained with Ponceau S and acid fuchsin for 5-10 min.
Following washing in 2% acetic acid aqueous solution,
differentiation in 1% phosphomolybdic acid solution for 3-5 min,
staining in aniline blue WS for 5 min and washing in 0.2% acetic
acid aqueous solution, sections were washed in 95% ethanol and
anhydrous ethanol. All the aforementioned steps were performed at
room temperature. Sections were permeabilized using xylene, mounted
using neutral gum and observed under a light microscope. A total of
five fields of view were randomly selected for each tissue section
and observed at x100 magnification.
Immunohistochemistry (IHC)
Rat liver sections were incubated three times in
xylene for 15 min and they were dehydrated twice in anhydrous
ethanol for 5 min, 85% ethanol for 5 min and 75% ethanol for 5 min.
After washing in ddH2O, sections were immersed in
citrate buffer (pH 6.0, Sangon Biotech Co., Ltd.; cat. no. E673000)
for 15 min antigen retrieval at room temperature. They were
incubated in 3% H2O2 in the dark for 25 min
and blocked in 3% BSA (Sangon Biotech Co., Ltd.; cat. no. E661003)
at 37˚C for 30 min. Incubation of primary antibodies was performed
at 4˚C overnight, and on the next day, sections were washed three
times in PBS (pH 7.4) for 5 min and incubated with HRP-labeled
secondary antibodies (ABclonal Biotech Co., Ltd.; cat. no. AS029;
1:500 dilution) at room temperature for 2 h. Later, sections were
counterstained with DAB and hematoxylin for 3 min to distinguish
between cytoplasm and nucleus. After dehydration in
ddH2O, 75% ethanol for 5 min, 85% ethanol for 5 min,
twice in anhydrous ethanol for 5 min and n-butanol for 5 min and
permeabilization in xylene for 5 min, sections were mounted using
neutral gum. Positive staining of cells was observed under a
microscope (XSP-8CA; Shanghai Optical Instrument Co. Ltd.). A total
of five fields of view were randomly selected for each tissue
section and observed at x100 magnification. The following primary
antibodies (1:200 dilution) were purchased from ABclonal Biotech
Co., Ltd.: α-SMA (cat. no. A17910) and HIPK1 (cat. no. A7414).
Cell culture
The rat HSC cell line HSC-T6 (Procell Life Science
& Technology Co., Ltd.; cat. no. CL-0116) was cultivated in
Dulbecco's Modified Eagle Medium (DMEM) (Procell Life Science &
Technology Co., Ltd.; cat. no. 164210-500) containing 10% FBS
(Procell Life Science & Technology Co., Ltd.; cat. no.
PM150210) and 1% penicillin and streptomycin in a humidified
incubator containing 5% CO2 and 95% air at 37˚C.
Cryopreservation using DMEM containing 40% FBS, 10% DMSO and 55%
glucose was performed until the cells reached a confluence of more
than 80% by trypsin digestion.
293T cells were purchased from the ATCC and used for
lentiviral packaging. The cell culture protocol was the same as
that of HSC-T6 cells. When the cells were >80% confluent, they
were digested with trypsin and inoculated into a new culture dish.
When the cells were adherent, they were incubated with serum-free
Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.) for 4 h, after
which transfection and lentiviral packaging were performed.
HSC-T6 cell activation
HSC-T6 cell activation was performed by
lipopolysaccharide (LPS) induction at 37˚C for 48 h. Briefly, cells
were cultured in DMEM containing 10% FBS, 1% penicillin and
streptomycin and 0.1 µg/ml LPS at 37˚C. At 48 h, the expression
level of the HF marker α-SMA in HSC-T6 cells was determined using
RT-qPCR and western blotting. Upregulated α-SMA suggested
activation of HSC-T6 cells.
Cell transfection
miR-146b-5p mimics, miR-146b-5p inhibitor and their
negative controls mimic-NC and inhibitor-NC were provided by
RiboBio Co., Ltd. (Guangzhou, China) (mimics:
5'-UGAGAACUGAAUUCCAUAGGCUGU-3'; mimic-NC:
5'-UUCUCCGAACGUGUCACGUTT-3'; inhibitor:
5'-ACAGCCUAUGGAAUUCAGUUCUCA-3'; inhibitor-NC:
5'-CAGUACUUUUGUGUAGUACAA-3'). Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used for
cell transfection. Briefly, cells were seeded in a 12-well plate
(1x105 cells per well) and cultured to 80% confluence.
After 4-h starvation in serum-free DMEM, a 100 nM transfection
mixture containing the mimics, inhibitor or the mimic-NC or
inihibitor-NC (1.6 µg/well) and Lipofectamine® 2000
reagent (4 µl/well) was added for 40 min incubation at 37˚C. Fresh
medium was replaced at 24 h and transfection efficacy was examined
by reverse transcription-quantitative (RT-q)PCR. Subsequent
experiments were performed 2 weeks after the transfection
experiment.
RT-qPCR
Total RNAs in HSC-T6 cells or liver tissues were
isolated using TRIzol® (Thermo Fisher Scientific, Inc.),
and 1 µg total RNA was reverse transcribed to cDNA using the Takara
PrimeScript RT reagent kit. Subsequently, qPCR was performed using
a SYBR Premix Ex Taq II with Tli RNaseH (Takara Bio, Inc.) on an
ABI Prism 7500 system (Thermo Fisher Scientific, Inc.). All
experimental steps were performed in accordance with the
manufacturer's protocols. In brief, total RNA induced with gDNA
eraser and reverse transcribed to cDNA was subjected to
thermocycling at 95˚C for 15 min, followed by 40 cycles at 95˚C for
5 sec, 60˚C for 30 sec and 72˚C for 40 sec and finally annealing at
72˚C for 10 min. The relative level was measured using an ABI Prism
7500 system (Thermo Fisher Scientific, Inc.) and calculated using
the 2-ΔΔCq method with GAPDH or U6 as the internal
reference (13). Each experiment
was repeated three times. Primer sequences are listed in Table I.
 | Table IPrimers used in the present
study. |
Table I
Primers used in the present
study.
| Gene | Primers
(5'-3') |
|---|
|
Rno-miR-146b-5p-F |
AACACGCTGAGAACTGAATTCC |
|
Rno-miR-146b-5p-R |
GTCGTATCCAGTGCAGGGTCCG
AGGTATTCGCACTGGATACGAC ACAGCC |
| HIPK1-F |
CAGCATCAGCCAATCATC |
| HIPK1-R |
ATTAGACCTCGCCTTCAG |
| α-SMA-F |
CCGAGATCTCACCGACTACC |
| α-SMA-R |
TCCAGAGCTACATAGCACAG |
| COL1A1-F |
CCGAGGTATGCTTGATCT |
| COL1A1-R |
GACAGTCCAGTTCTTCATTG |
| TGF-β-F |
CACCATCCATGACATGAACC |
| TGF-β-R |
TCATGTTGGACAACTGCTCC |
| GAPDH-F |
AAGCTCACTGGCATGGCCTT |
| GAPDH-R |
CGGCATGTCAGATCCACAAC |
| U6-F |
CTCGCTTCGGCAGCACA |
| U6-R |
AACGCTTCACGAATTTGCGT |
Western blotting
Total proteins in HSC-T6 cells or liver tissues were
isolated using RIPA buffer (Beyotime Institute of Biotechnology)
containing phenylmethylsulfonyl fluoride and protease inhibitor.
Protein concentrations were measured using the BCA protein assay
kit (Beyotime Institute of Biotechnology). Protein samples (50 µg)
were loaded onto 10% gels for sodium dodecyl sulfate-polyacrylamide
gel electrophoresis (SDS-PAGE) and transferred on polyvinylidene
fluoride (PVDF) membranes at 300 mA. Non-specific antigens on PVDF
membranes were blocked by immersing in 1X TBST (0.1% Tween)
containing 5% skimmed milk. After immunoblotting with primary
antibodies (1:1,000) at 4˚C overnight and secondary antibodies
(1:1,000) at room temperature for 1 h, band exposure was performed
using the Bio-Rad Universal Hood II Gel Doc Imaging system and the
gray value was analyzed using ImageJ v. 1.8.0.112 (National
Institutes of Health,). All the antibodies were purchased from
Abclonal as listed: α-SMA (cat. no. A17910), COL1A1 (cat. no.
A1352), TGF-β (cat. no. A18692), HIPK1 (cat. no. A19580), GAPDH
(cat. no. A19056) and the secondary antibody (cat. no. AS014).
MTT assay
HSC-T6 cells were seeded in a 96-well plate
(5x103 cells per well) and 200 µl of DMEM containing 10%
FBS and 1% penicillin and streptomycin was added per well. Briefly,
15 µl of MTT solution (15 mg/ml, Sangon Biotech Co., Ltd.; cat. no.
A600799) was added at the indicated time points at 37˚C. After cell
culture for 4 h, cells per well were lysed in 150 µl of DMSO for 10
min at room temperature and optical density at 490 nm was measured
using the HBS-1101 microplate reader (Nanjing Detie Experimental
Equipment Co., Ltd.).
Dual-luciferase reporter assay
The wild-type plasmid pGL3-HIPK1-WT was constructed
by amplifying complementary sequences in the HIPK1 3'UTR and the
promoter region of miR-146b-5p and cloning them into the pGL3 3'UTR
(Shanghai GeneChem Co., Ltd.). The mutant-type plasmid
pGL3-HIPK1-Mut was constructed using the site-directed mutagenesis
kit (Sangon Biotech Co., Ltd.; cat. no. B639281). Wild-type and
mutant-type plasmids were co-transfected into cells with
miR-146b-5p mimics or mimics-NC and Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) for 48 h. Relative Firefly and
Renilla luciferase activities were measured using the
Dual-Luciferase Reporter Gene Assay kit (Promega Corporation).
Lentivirus infection
Overexpressing lentivirus plasmid GV367-HIPK1
(Shanghai GeneChem Co., Ltd.) was generated by cleavage of the
GV367 plasmid using AgeI/NheI and GV367 was used as
blank control. Lentivirus packaging was performed using the
second-generation lentivirus packaging kit (Shanghai GeneChem Co.,
Ltd.). The lentiviral plasmid, packaging vector and envelope vector
were mixed at a 4:3:2 ratio for a total DNA mass of 20 µg and
incubated with 1 ml of Lenti-Easy Packaging Mix (Shanghai GeneChem
Co., Ltd.) for 15 min. The mixture was then incubated for another
20 min in Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) and it was applied into 293T cell culture medium for 6 h at
37˚C. 293T cells were seeded in a 12-well plate at a density of
2.5x105 cells/well and cultured to 80% confluence. After
incubation in serum-free DMEM for 4 h, cells were transfected as
mentioned above with lentiviruses for 3 days. Transfected cells
were filtered using a 0.45 µM mesh, which were concentrated at
70,000 g at 4˚C for 2 h. The supernatant was collected for
detecting viral titers. HSC-T6 cells cultured to more than 80%
confluence were cultured with diluted lentiviruses and GFP-labeled
cells with lentivirus transfection rate >80% at 72 h were
screened out. Transfection efficacy of HIPK1 was finally verified
by RT-qPCR.
Statistical analysis
Data were expressed as mean ± standard deviation
from three replicates and processed by GraphPad Prism 8.0 (GraphPad
Software, Inc.). Differences between groups were compared by the
unpaired Student's t test. One-way ANOVA was used to assess
differences among the groups and Tukey's and Bonferroni's tests
were used for post hoc testing following ANOVA. Pearson's analysis
was used in correlation analysis between miR-146b-5p and HIPK1.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-146b-5p is upregulated in rats
with HF
An in vivo HF model in rats was established
by administering a subcutaneous injection of CCl4 for 8
weeks. After sacrifice, rat liver tissues were collected and
prepared for H&E staining, Masson staining, IHC, RT-qPCR and
western blotting. Compared with rat liver tissues from the control
group, tissues from the HF group presented mixed nodules of varying
sizes and a widened fibrosis interval (Fig. 1A). In addition, upregulated mRNA
levels of miR-146b-5p, α-SMA, COL1A1 and TGF-β, as well as
upregulated protein levels of α-SMA, COL1A1 and TGF-β, were
observed in HF liver tissues (Fig.
1B and C). A negative
correlation was identified between the relative levels of
miR-146b-5p and HIPK1 (Fig. 1D).
As shown in IHC staining images, the HIPK1 was significantly
observed both in cytoplasmic and nuclear regions and the expression
level of HIPK1 was significantly downregulated in liver sections
from the HF group, while the level of α-SMA was upregulated
compared with those in the control group (Fig. 1E). This suggested that upregulated
miR-146b-5p in liver tissues of rats with HF could be involved in
the progression of HF.
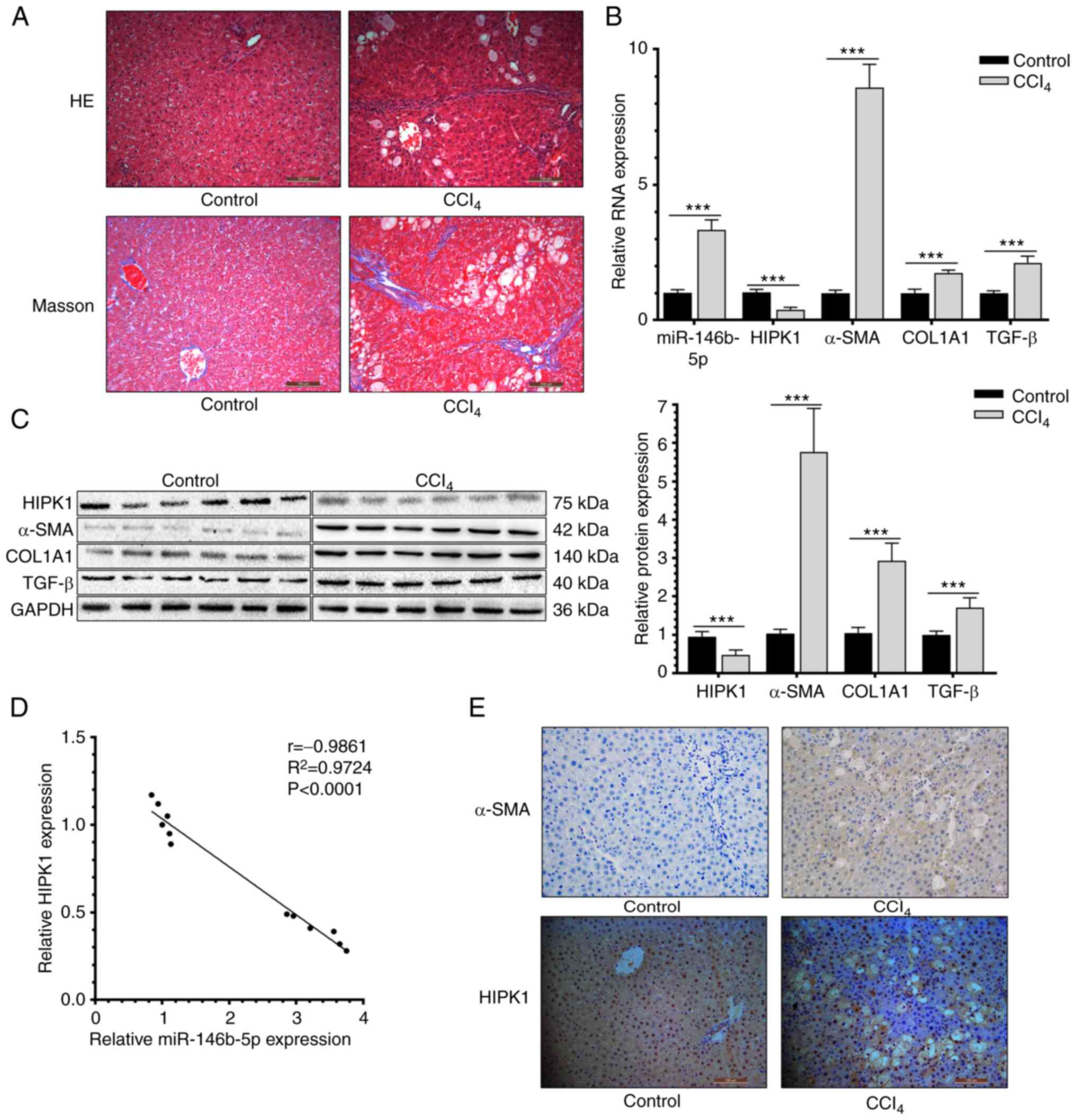 | Figure 1miR-146b-5p is upregulated and HIPK1
is downregulated in rats with HF. (A) HE staining and Masson
staining of normal and HF rats hepatic tissues (x200
magnification). (B) Relative miR-146b-5p, HIPK1, α-SMA, COL1A1 and
TGF-β RNA expression of normal and HF rats hepatic tissues. (C)
Relative HIPK1, α-SMA, COL1A1 and TGF-β protein expression of
normal and HF rats hepatic tissues. (D) Correlation between HIPK1
and miR-146b-3p expression. (E) α-SMA and HIPK1 expression in
normal and HF rats hepatic tissues (x200 magnification).
***P<0.001. miR, microRNA; HIPK1, homeodomain
interacting protein kinase 1; HF, hepatic fibrosis; HE, hematoxylin
and eosin; α-SMA, α-smooth muscle actin; COL1A1, collagen type I α
1 chain; TGF-β, transforming growth factor β. |
miR-146b-5p upregulates the fibrosis
markers in HSCs
HSC-T6 cells were induced with 0.1 µg/ml LPS
treatment and then the upregulated mRNA level of α-SMA validated
the activation of HSCs (Fig. 2A).
The present study first examined the transfection efficacy of
miR-146b-5p mimics and inhibitor in HSC-T6 cells by RT-qPCR
(Fig. 2B). As revealed by the MTT
assay, overexpression of miR-146b-5p enhanced the viability of
LPS-induced HSC-T6 cells and knockdown of miR-146b-5p reduced the
cell viability (Fig. 2C). In
addition, transfection of miR-146b-5p mimics upregulated the mRNA
and protein levels of HF markers α-SMA, COL1A1 and TGF-β and
downregulated the level of HIPK1. Knockdown of miR-146b-5p led to
an opposite trend (Fig. 2D and
E). It can be concluded that
miR-146b-5p stimulated the activation of HSCs and upregulated the
fibrosis markers in HSCs.
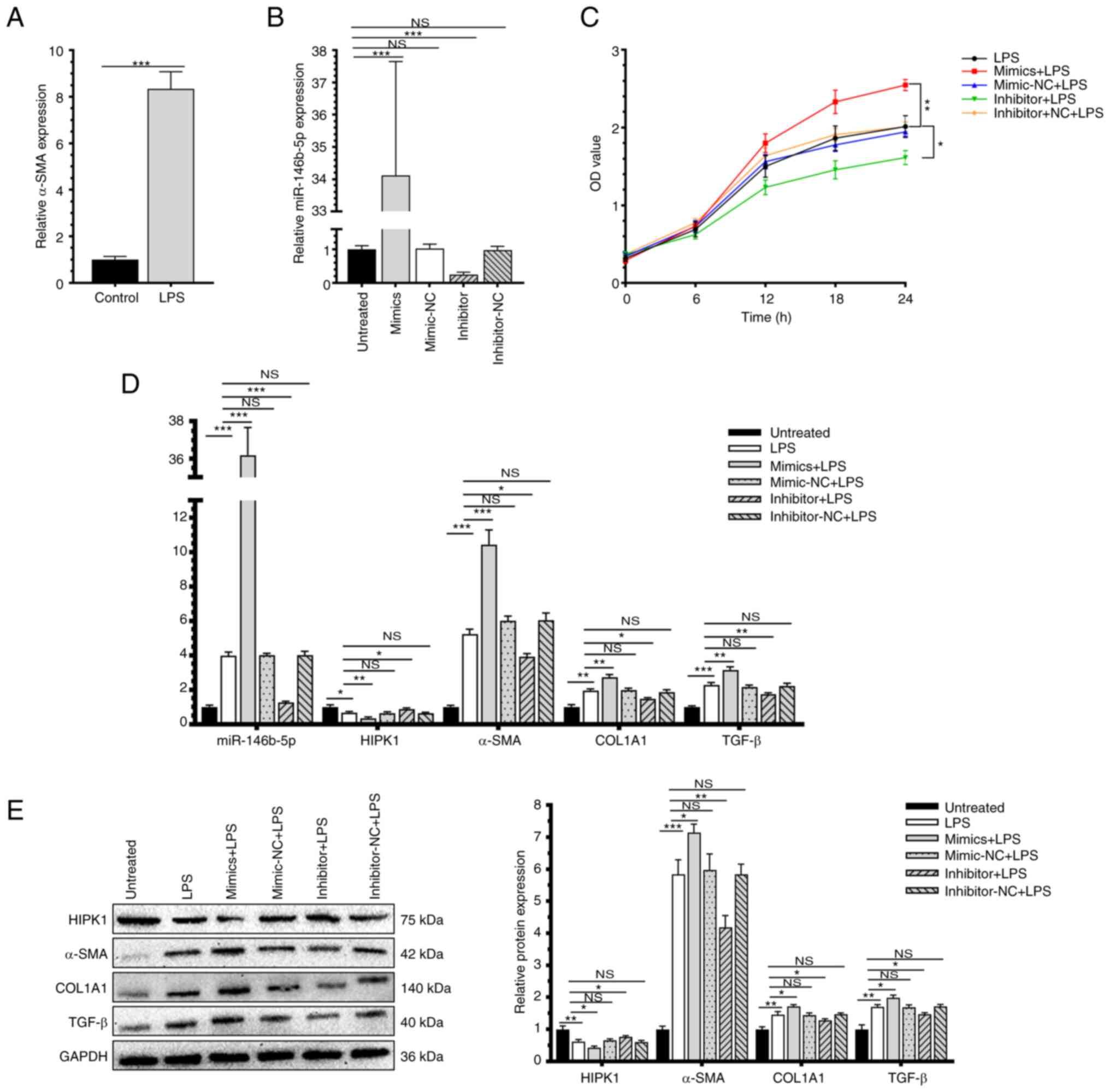 | Figure 2miR-146b-5p upregulates the fibrosis
markers in HSCs. (A) α-SMA is upregulated in vitro cell
model with LPS treatment. (B) miR-146b-5p transfection efficiency.
(C) Cell viability of HSC-T6 transfected with miR-146b-5p mimics,
inhibitor or NCs and treated with LPS (compared with untreated
group). (D) Relative miR-146b-5p, HIPK1, α-SMA, COL1A1 and TGF-β
RNA expression. (E) Relative HIPK1, α-SMA, COL1A1 and TGF-β protein
expression. *P<0.05; **P<0.01;
***P<0.001. miR, microRNA; HSCs, hepatic stellate
cells; α-SMA, α-smooth muscle actin; LPS, lipopolysaccharide; NCs,
negative controls; HIPK1, homeodomain interacting protein kinase 1;
COL1A1, collagen type I α 1 chain; TGF-β, transforming growth
factor β; NS, no significance. |
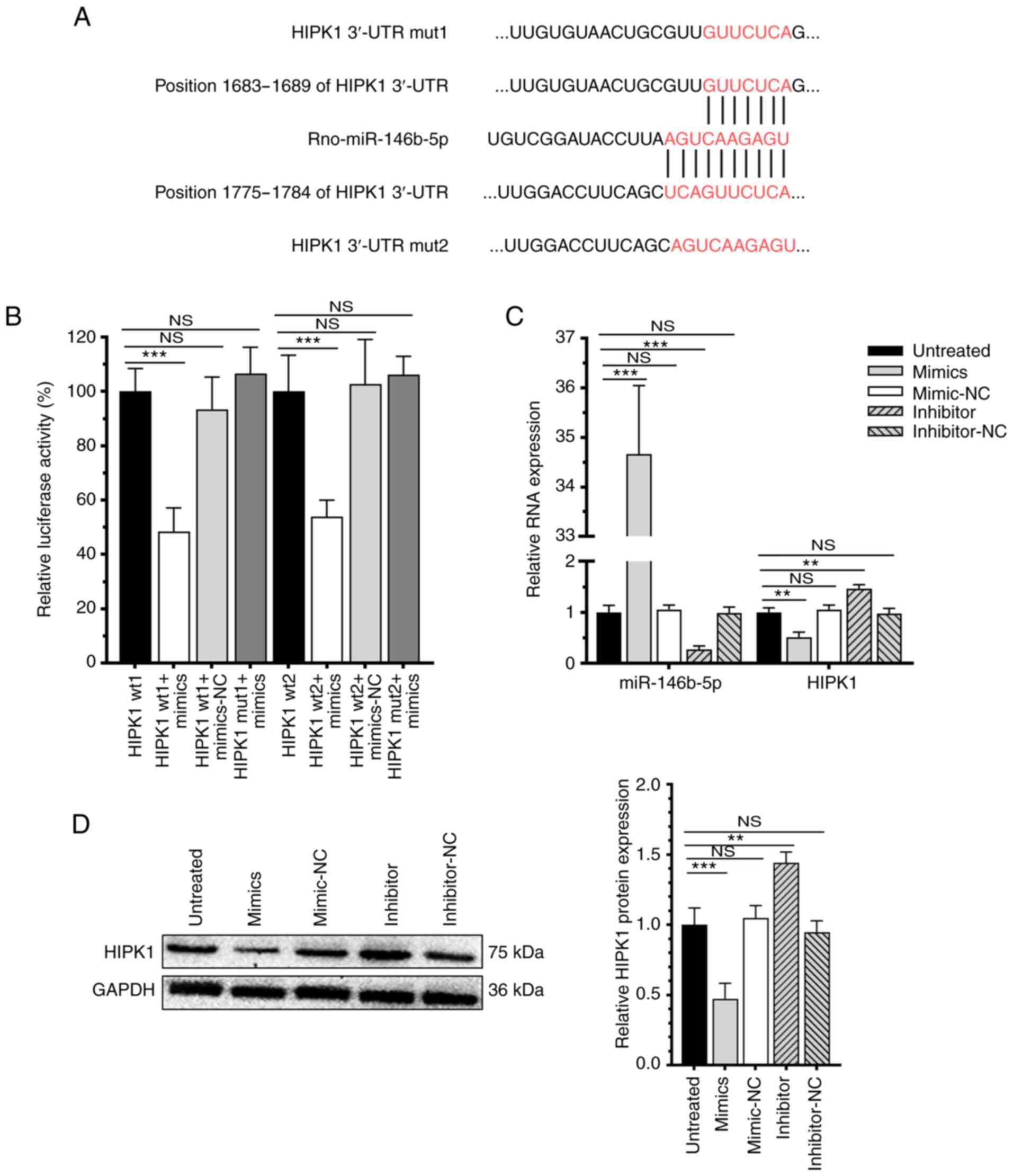
miR-146b-5p directly targets HIPK1 and
downregulates its expression level
Using TargetScan 7.2 and StarBase 3.0, miR-146b-5p
was predicted to bind to the HIPK1 and HIPK2 3'UTR (Figs. 3A and S1). Subsequently, the dual-luciferase
reporter assay showed that overexpression of miR-146b-5p
specifically reduced the luciferase activity of pGL3-HIPK1-WT, but
the luciferase activity of pGL3-HIPK1-MuT was not affected
(Fig. 3B), confirming the binding
between miR-146b-5p and HIPK1. Moreover, transfection of
miR-146b-5p mimics significantly downregulated the mRNA and protein
levels of HIPK1 and transfection of the miR-146b-5p inhibitor
upregulated the mRNA and protein levels of HIPK1 (Fig. 3C and D). Collectively, miR-146b-5p could target
the HIPK1 3'UTR and downregulate HIPK1 through inhibiting the
transcription.
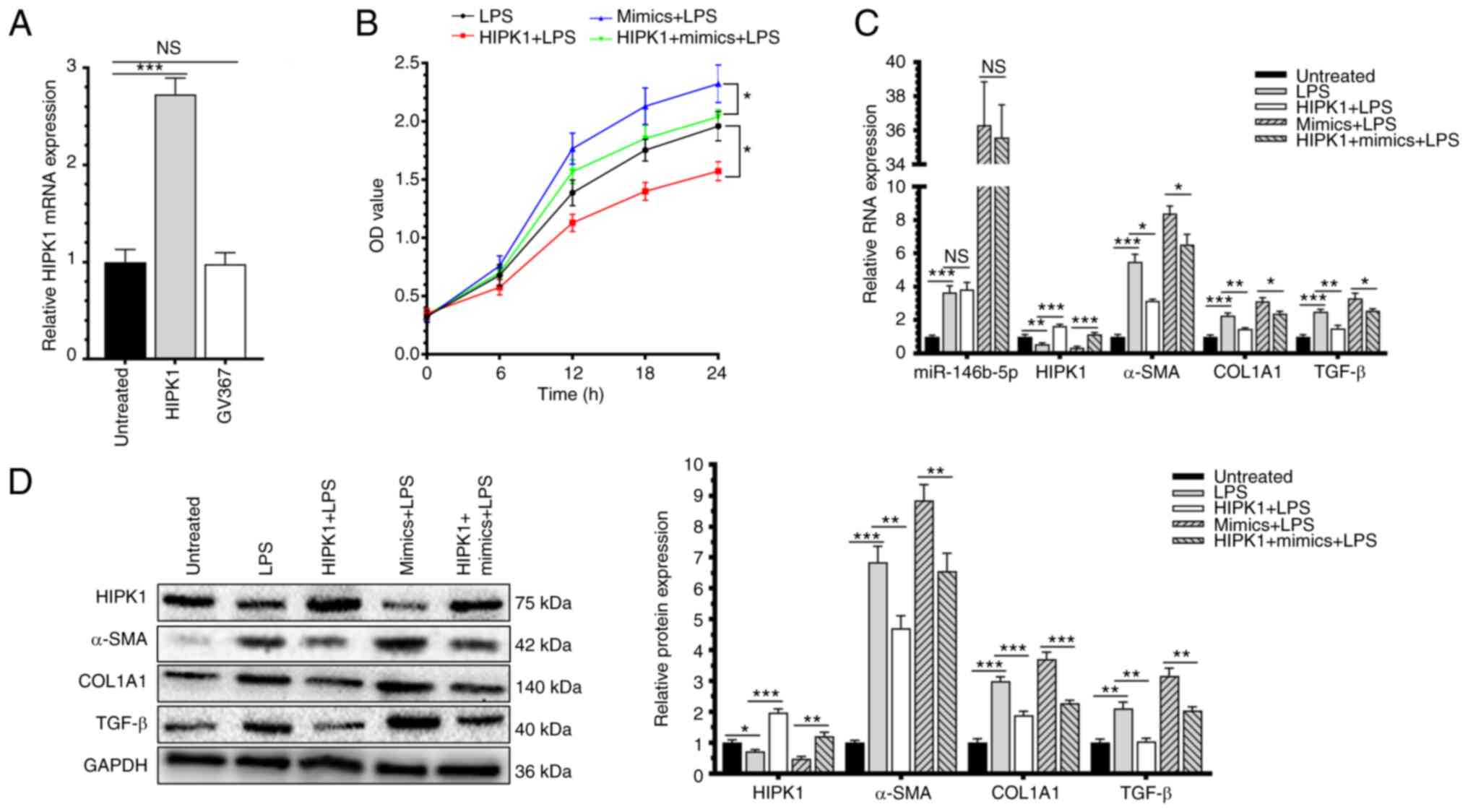
Overexpression of HIPK1 effectively
decreased the effect of miR-146b-5p in HSC-T6 activation
To further explore the role of HIPK1 in the
progression of HF, its level was intervened by lentivirus
transfection (Fig. 4A). After
co-intervention of HIPK1 and miR-146b-5p in HSC-T6 cells, they were
activated by LPS induction. Overexpression of HIPK1 significantly
reduced the viability of LPS-induced HSC-T6 cells and
interestingly, it could effectively decrease the positive effect of
overexpressed miR-146b-5p in promoting the cell viability (Fig. 4B). In addition, the upregulated
mRNA and protein levels of α-SMA, COL1A1 and TGF-β in LPS-induced
HSC-T6 cells overexpressing miR-146b-5p were markedly decreased by
the overexpression of HIPK1 (Fig.
4C and D). Therefore, HIPK1
was responsible for the regulatory effect of miR-146b-5p in the
activation of HSC-T6, thus may contribute to fibrosis
progression.
Discussion
HF is the most common type of chronic liver disease
and it is a scar formation process following liver injury (14). It is characterized by excessive
deposition of ECM resulting from the activation of HSCs (15). A damaged liver structure, formation
of nodules, scar repair and activation of multiple signaling
pathways eventually lead to irreversible liver cirrhosis and even
liver cancer (4). So far, liver
transplantation and drug treatment are available in severe liver
diseases, although their clinical applications are markedly limited
by high medical costs and low survival rate (16). Therefore, it is important to search
for potential targets that can regulate HSC activation, which is
conductive to the clinical management of HF.
Biological functions of miRNAs in tumors (17), atherosclerosis (18), metabolic syndrome (19) and other diseases have gradually
emerged. With increased clinical research on miRNAs, their vital
involvement in liver diseases has been recognized. Accumulating
evidence has shown that miRNAs are able to regulate the activation
and proliferation of HSCs and thus they mediate the progression of
HF. Riaz et al (20)
reported that miR-188-5p alleviates the severity of HF through
inhibiting the activation and proliferation of HSCs via the
PTEN/AKT pathway. Li et al (21) proved that miR-34c promotes HSC
activation and the progression of HF by targeting the key enzyme
ACSL1 that affects fatty acid synthesis. Overexpression of
miR-494-3p suppresses HSC activation through inhibiting
proliferation and inducing apoptosis by targeting TRAF3(22). miR-146b has been extensively
analyzed in pancreatic cancer (23), colorectal cancer (24), thyroid cancer (25) and gallbladder cancer (26), while its biological function in the
progression of HF has been rarely reported. Ge et al
(27,28) revealed that miR-146b promotes the
activation and proliferation of HSCs and the
HMGB1/p65/miR-146b/HNF1A axis exerts a key effect on regulating HSC
functions and HF. In the present study, miR-146b-5p was
significantly upregulated in liver tissues of rats with HF than in
liver tissues of normal rats. In addition, both the mRNA and
protein levels of HF markers α-SMA, COL1A1 and TGF-β were
upregulated. Overexpression of miR-146b-5p markedly enhanced
LPS-induced activated HSC-T6 cells and correspondingly, the mRNA
and protein levels of HF markers α-SMA, COL1A1 and TGF-β were
upregulated. As expected, transfection of the miR-146b-5p inhibitor
resulted in an opposite trend. Consistent with previous findings,
the results of the present study showed that miR-146b-5p was able
to induce the activation of HSCs and upregulate HF markers; thus
suggesting that miR-146b-5p may serve an important role during the
progression of HF.
HIPK1 is a regulator of transcription factors
containing the homology domains (29). The tumor suppressor role of HIPK1
has been revealed in numerous types of tumors due to its regulatory
effects on cell apoptosis and DNA damage repair (30). Palkina et al (31) validated that miR-3065-5p inhibits
proliferation and alters cell cycle distribution of melanoma cells
by targeting HIPK1. Rey et al (10) proposed that HIPK1 is significantly
upregulated in colorectal cancer (CRC) in a stage-dependent manner,
which restricts the uncontrolled growth of CRC cells by activating
the p53 signaling pathway. A number of efforts have been made to
explore the role of HIPK1 in tumors, kidney diseases and diabetes
(32). Its function in HF has not
yet been reported. The present study showed that compared with rats
in the control group, the mRNA and protein levels of HIPK1 in rat
liver tissues from the HF group were significantly downregulated.
In addition, the miR-146b-5p level was negatively correlated with
the level of HIPK1 in rats liver and as predicted by using
TargetScan 7.2 and StarBase 3.0, miR-146b-5p was able to target the
HIPK1 3'UTR and regulate its expression, indicating a potential
binding relationship between them. Subsequently, it was confirmed
by the dual-luciferase reporter assay that miR-146b-5p could
directly target with HIPK1. Transfected with miR-146b-5p
significantly downregulated the expression of HIPK1. Notably,
overexpression of HIPK1 by lentivirus transfection markedly
decreased the effects of miR-146b-5p in enhancing the viability and
upregulating the HF markers α-SMA, COL1A1 and TGF-β in LPS-induced
activated HSC-T6 cells. In addition, it is worth noting that the
present study was based on the rats HF model and combined this with
cell experiments to research the molecular mechanism. Although it
has a certain correlation with human HF, the present study did not
involve any samples of clinical patients; and according to the
TargetScan 7.2 prediction, miR-146b-5p could also directly target
with HIPK2; however, this potential association was not
investigated in the present study; moreover, since the miR-146b-5p
is a miRNA, which does not encode any protein, the histology of
liver sections in hepatic fibrosis cannot show the interaction of
miR-146b-5p and HIPK1. The present study also did not verify the
reversal effect of miR-146b-5p on HF in rat model, which is one of
its the limitations. Therefore, further in-depth research is needed
to explore its clinical application. Taken together, miR-146b-5p
promoted the activation of HSCs and contributed to the progression
of HF by targeting and downregulating HIPK1.
miR-146b-5p was significantly upregulated during the
progression of HF. Overexpressed miR-146b-5p could significantly
activate HSCs, upregulate α-SMA, COL1A1 and TGF-β and thus
contribute to the progression of HF via directly targeting and
downregulating HIPK1. miR-146b-5p is a potential biomarker and
therapeutic target for HF.
Supplementary Material
Prediction of the targeting
association between HIPK2 and rno-miR-146b-5p. HIPK1, homeodomain
interacting protein kinase 2.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by Jiangxi Outstanding
Talents Fund (grant no. 20192BcB23022), National Natural Science
Foundation Youth Project (grant no. 81900561) and National Natural
Science Foundation Regional Project (grant nos. 81760115 and
81960121).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JX, NC and TX made substantial contributions to
conception, design and acquisition of data. ZH and XS made
substantial contributions to the analysis and interpretation of the
data. All authors contributed to the writing of the article and
have read and approved the manuscript. JX and TX confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
The animal research of the present study was
approved by the Medical Research Ethics Committee of the First
Affiliated Hospital of Nanchang University (ethics approval no.
2020117).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hernandez-Gea V and Friedman SL:
Pathogenesis of liver fibrosis. Annu Rev Pathol. 6:425–456.
2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hou W and Syn WK: Role of metabolism in
hepatic stellate cell activation and fibrogenesis. Front Cell Dev
Biol. 6(150)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lee UE and Friedman SL: Mechanisms of
hepatic fibrogenesis. Best Pract Res Clin Gastroenterol.
25:195–206. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jung YK and Yim HJ: Reversal of liver
cirrhosis: Current evidence and expectations. Korean J Intern Med.
32:213–228. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mekala S, Tulimilli SV, Geesala R,
Manupati K, Dhoke NR and Das A: Cellular crosstalk mediated by
platelet-derived growth factor BB and transforming growth factor
beta during hepatic injury activates hepatic stellate cells. Can J
Physiol Pharmacol. 96:728–741. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Nakamura M, Kanda T, Sasaki R, Haga Y,
Jiang X, Wu S, Nakamoto S and Yokosuka O: MicroRNA-122 inhibits the
production of inflammatory cytokines by targeting the PKR activator
PACT in human hepatic stellate cells. PLoS One.
10(e0144295)2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yu F, Chen B, Fan X, Li G, Dong P and
Zheng J: Epigenetically-regulated MicroRNA-9-5p suppresses the
activation of hepatic stellate cells via TGFBR1 and TGFBR2. Cell
Physiol Biochem. 43:2242–2252. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li X, Luo Y, Yu L, Lin Y, Luo D, Zhang H,
He Y, Kim YO, Kim Y, Tang S and Min W: SENP1 mediates TNF-induced
desumoylation and cytoplasmic translocation of HIPK1 to enhance
ASK1-dependent apoptosis. Cell Death Differ. 15:739–750.
2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Rey C, Soubeyran I, Mahouche I, Pedeboscq
S, Bessede A, Ichas F, De Giorgi F and Lartigue L: HIPK1 drives p53
activation to limit colorectal cancer cell growth. Cell Cycle.
12:1879–1891. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
van der Laden J, Soppa U and Becker W:
Effect of tyrosine autophosphorylation on catalytic activity and
subcellular localisation of homeodomain-interacting protein kinases
(HIPK). Cell Commun Signal. 13(3)2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kondo S, Lu Y, Debbas M, Lin AW, Sarosi I,
Itie A, Wakeham A, Tuan J, Saris C, Elliott G, et al:
Characterization of cells and gene-targeted mice deficient for the
p53-binding kinase homeodomain-interacting protein kinase 1
(HIPK1). Proc Natl Acad Sci USA. 100:5431–5436. 2003.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kisseleva T and Brenner DA: Mechanisms of
fibrogenesis. Exp Biol Med (Maywood). 233:109–122. 2008.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Iwaisako K, Jiang C, Zhang M, Cong M,
Moore-Morris TJ, Park TJ, Liu X, Xu J, Wang P, Paik YH, et al:
Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl
Acad Sci USA. 111:E3297–E3305. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jesudian A, Desale S, Julia J, Landry E,
Maxwell C, Kallakury B, Laurin J and Shetty K: Donor factors
including donor risk index predict fibrosis progression, allograft
loss, and patient survival following liver transplantation for
hepatitis C virus. J Clin Exp Hepatol. 6:109–114. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Winkle M, El-Daly SM, Fabbri M and Calin
GA: Noncoding RNA therapeutics-challenges and potential solutions.
Nat Rev Drug Discov. 20:629–651. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ali Sheikh MS, Alduraywish A, Almaeen A,
Alruwali M, Alruwaili R, Alomair BM, Salma U, Hedeab GM, Bugti N
and Abdulhabeeb IAM: Therapeutic value of miRNAs in coronary artery
disease. Oxid Med Cell Longev. 2021(8853748)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Fodor A, Lazar AL, Buchman C, Tiperciuc B,
Orasan OH and Cozma A: MicroRNAs: The link between the metabolic
syndrome and oncogenesis. Int J Mol Sci. 22(6337)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Riaz F, Chen Q, Lu K, Osoro EK, Wu L, Feng
L, Zhao R, Yang L, Zhou Y, He Y, et al: Inhibition of miR-188-5p
alleviates hepatic fibrosis by significantly reducing the
activation and proliferation of HSCs through PTEN/PI3K/AKT pathway.
J Cell Mol Med. 25:4073–4087. 2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li B, Liu J, Xin X, Zhang L, Zhou J, Xia
C, Zhu W and Yu H: MiR-34c promotes hepatic stellate cell
activation and liver fibrogenesis by suppressing ACSL1 expression.
Int J Med Sci. 18:615–625. 2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li H, Zhang L, Cai N, Zhang B and Sun S:
MicroRNA-494-3p prevents liver fibrosis and attenuates hepatic
stellate cell activation by inhibiting proliferation and inducing
apoptosis through targeting TRAF3. Ann Hepatol.
23(100305)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhou M, Gao Y, Wang M, Guo X, Li X, Zhu F,
Xu S and Qin R: MiR-146b-3p regulates proliferation of pancreatic
cancer cells with stem cell-like properties by targeting MAP3K10. J
Cancer. 12:3726–3740. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang D, Feng M, Ma X, Tao K and Wang G:
Transcription factor SP1-induced microRNA-146b-3p facilitates the
progression and metastasis of colorectal cancer via regulating
FAM107A. Life Sci. 277(119398)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liu X, Fu Q, Bian X, Fu Y, Xin J, Liang N,
Li S, Zhao Y, Fang L, Li C, et al: Long non-coding RNA MAPK8IP1P2
inhibits lymphatic metastasis of thyroid cancer by activating hippo
signaling via sponging miR-146b-3p. Front Oncol.
10(600927)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ouyang B, Pan N, Zhang H, Xing C and Ji W:
miR146b5p inhibits tumorigenesis and metastasis of gallbladder
cancer by targeting Tolllike receptor 4 via the nuclear factor-κB
pathway. Oncol Rep. 45(15)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ge S, Wu X, Xiong Y, Xie J, Liu F, Zhang
W, Yang L, Zhang S, Lai L, Huang J, et al: HMGB1 inhibits HNF1A to
modulate liver fibrogenesis via p65/miR-146b signaling. DNA Cell
Biol. 39:1711–1722. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ge S, Zhang L, Xie J, Liu F, He J, He J,
Wang X and Xiang T: MicroRNA-146b regulates hepatic stellate cell
activation via targeting of KLF4. Ann Hepatol. 15:918–928.
2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kim YH, Choi CY, Lee SJ, Conti MA and Kim
Y: Homeodomain-interacting protein kinases, a novel family of
co-repressors for homeodomain transcription factors. J Biol Chem.
273:25875–25879. 1998.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Meng L, Zhao X and Zhang H: HIPK1
interference attenuates inflammation and oxidative stress of acute
lung injury via autophagy. Med Sci Monit. 25:827–835.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Palkina N, Komina A, Aksenenko M, Moshev
A, Savchenko A and Ruksha T: miR-204-5p and miR-3065-5p exert
antitumor effects on melanoma cells. Oncol Lett. 15:8269–8280.
2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Conte A and Pierantoni GM: Update on the
regulation of HIPK1, HIPK2 and HIPK3 protein kinases by microRNAs.
MicroRNA. 7:178–186. 2018.PubMed/NCBI View Article : Google Scholar
|