Introduction
Cholangiocarcinoma (CCA) is an epithelial cell
malignancy arising from multiple locations along the biliary tree
with features of cholangiocyte differentiation; it is the second
most common hepatic malignancy after hepatocellular carcinoma (HCC)
(1). Over past decades, the
mortality of CCA has gradually increased worldwide (2). The well-accepted classification of
CCA is based on the anatomical location, referring to intrahepatic
CCA (iCCA), perihilar CCA (pCCA) or distal CCA (dCCA) (3). iCCA refers to CCA located proximally
to the second-degree bile ducts, while pCCA refers to CCA between
the second-degree bile ducts and the insertion of the cystic duct,
whereas dCCA pertains to CCA between the origin of the cystic duct
and ampulla of Vater (4).
Clinically, each subtype has been demonstrated to have a distinct
epidemiology, prognosis and clinical management. According to a
previous study, pCCA, dCCA and iCCA account for ~50, ~40 and ~10%
of CCA cases, respectively, with certain rare cases of mixed CCA
(5).
The worldwide incidence of CCA is not homogeneous
due to the variable prevalence of risk factors. Similar to HCC,
several studies have highlighted hepatitis B virus (HBV) and
hepatitis C virus (HCV) as key factors for the development of CCA
(6,7). Apart from HBV and HCV, there are
still certain non-viral risk factors that may also induce CCA, such
as diabetes, alcoholism and dyslipidemia (8,9). A
combination of CT and MRI with magnetic resonance
cholangiopancreatography is recommended for the diagnosis of CCA.
Occasionally, the cancer antigen 19-9 (CA 19-9) is applied as a
potential serum biomarker for CCA diagnosis, with CA 19-9 levels
>1,000 U/ml indicating the presence of metastatic disease
(10,11). However, the CA 19-9 is not specific
for CCA and the diagnostic rate is far from satisfactory. At
present, resection is still the mainstay of effective therapy for
CCA (12). At the same time, liver
transplantation has been suggested as an alternative option for a
subset of cirrhotic patients. However, the source of liver would be
a major issue. Accumulating research has investigated the molecular
pathogenesis of CCA. A study by Farshidfar et al (13) implied that isocitrate
dehydrogenase, fibroblast growth factor receptor, encoding Family A
receptors as well as biofilm-associated protein families were
closely associated with the formation of CCA based on a
comprehensive whole-exome and transcriptome sequencing analysis of
a large cohort of patients with CCA. To date, several chemical
therapeutic medicines targeting relevant signaling cascades (such
as those regulating tumor cell proliferation, cell migration,
survival maintenance as well as governance of cell fate) have been
identified and confirmed in clinical trials (1). However, misclassification of CCA
remains a critical concern for clinical administration. At the same
time, based on the issues of therapeutic resistance and genetic
heterogeneity of the disorder, there is still no curative therapy
for CCA. If diagnosed at the early stage, patients with CCA have a
6-30% longer 5-year survival rate compared with those diagnosed at
an advanced stage (14).
Unfortunately, effective biomarkers for early diagnosis of CCA are
out of scope at this moment due to the ‘silent’ clinical
characteristics of this disorder (most patients of CCA are
asymptomatic in the early stage), which makes it difficult to
identify. All of these limitations restrict the sensitivity of
cytological as well as other pathological diagnostic approaches in
the clinic.
Encoded by the FOSB gene, the FBJ murine
osteosarcoma viral oncogene homolog B (FOSB) protein shares
structural similarities with the other members within the Fos
family and is a key component protein of the activator protein-1
(AP-1) transcription factor. As a tumor-related gene enhancer
element, AP-1 has been suggested to be closely associated with
several types of cancer (15). The
components of AP-1 complex have critical roles in cancer cell
viability, proliferation and migration (16). Yet, the functions of FOSB in the
development and progression of CCA remain to be fully
elucidated.
Due to the high misdiagnosis rate as well as
atypical symptoms of CCA in the early stage, current modalities for
establishing a diagnosis of CCA are insufficient, so that the
identification of effective biomarkers may be another beneficial
future direction. In the present study, a systematically
differentially expressed gene (DEG) profile for patients with CCA
was generated. The underlying molecular mechanisms and external
functions of potential target genes were also evaluated by
functional experiments, which all together provided a valuable
reference for future clinical manipulation.
Materials and methods
Data sources
In the present study, 3 independent data sources
were utilized for DEG analysis of CCA: i) The open public data from
the Gene Expression Omnibus (GEO) database; dataset no. GSE132305
(ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132305).
This dataset included formalin-fixed, paraffin-embedded samples
(182 primary CCA and 38 non-tumor bile duct tissues) from patients
treated by surgical resection. The whole-genome expression profiles
of the above samples were evaluated by the Affymetrix Human Genome
U133 Plus 2.0 Array chip platform for differential gene analysis.
ii) The second data source was the CCA cell line HUCCT1, as
previously reported (17), which
was cultured for use in a cell proliferation assay. iii) As the
third data source, clinical patients with CCA were recruited.
Differential gene expression
analysis
The limma package in R was used for differentially
expressed mRNA analysis, with an absolute value of the
log-transformed differential expression multiple (Log2FC)>1 and
P<0.05 as the selection criteria.
Functional enrichment analysis
The ‘clusterProfiler’ package in R was used for Gene
Ontology (GO) enrichment analysis in the categories Biological
Process, Molecular Function and Cellular Component and for Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
P<0.05 was considered to indicate significant enrichment of the
corresponding entries. The final results were visualized using the
on-line tool from pathview (pathview.uncc.edu).
Protein-protein interaction (PPI)
network construction
The Search Tool for the Retrieval of INteracting
Genes and proteins (STRING) database (https://STRING-db.org/; version 11.0) was utilized to
establish the key protein interactions (18). Cytoscape (cytoscape.org, version 3.7.2) was used to visualize
the PPI network. At the same time, the cytoHubba plug-in in
Cytoscape was utilized for target gene selection based on the
algorithm method of maximum neighborhood component (19).
Cell transfection and Cell Counting
Kit-8 (CCK-8) proliferation assay
The FOSB-low expression group (FOSB-LE) was obtained
by transfection of FOSB-small interfering (si)RNA following the
protocol of a previous study (20). FOSB-siRNA
(5'-GCCAACCACAATTCAATGAAT-3') was synthesized by Merck China.
HUCCT1 cells (Sunncell Bio) were cultured in DEME (Thermo Fisher
Scientific, Inc. cat. no. 11965118) at 37˚C, 5% CO2 to
obtain a final 60-80% confluence. Negative control siRNA (NC-siRNA,
the NC siRNA scrambled sequence for siRNAs, Merck; cat. no. SIC002)
and FOSB-siRNA were transfected with Lipofectamine® (Thermo Fisher
Scientific, Inc. cat. no. L3000015) according to the manufacturer's
protocol. The next day, the transfection complex medium was
replaced with complete DMEM (Thermo Fisher Scientific, Inc., cat.
no. 10565018). At the same time, FOSB overexpression (FOSB-OE) was
achieved by transfection of 100 ng/ml FOSB-RNA plasmid (Hanbio,
Inc. cat. no. 202103886). The empty vector was used as a negative
control.
For the cell proliferation/viability assay, the
HUCCT1 cells were seeded and cultured with DMEM with 10% FBS+1%
penicillin-streptomycin at 1x103 cells/well. The
cultured cells with 100 ng/ml NC-siRNA or FOSB-siRNA or FOSB-RNA to
reach a concentration of 2x103 cells/well for all groups
and allowed to grow for another 24 h. The CCK-8 solution (10 µl;
cat. no. HY-K0301; MedChemExpress) was added to the medium and
cells were incubated for 2 h. The absorbance at 450 nm was measured
using a spectrophotometer and the final results were calculated.
The experiments were repeated three times independently.
Subjects and reverse
transcription-quantitative PCR (RT-qPCR)
All patients with CCA in the present study were
randomly selected from Tianjin Nankai Hospital (Tianjin, China)
from January 2018 to January 2021 based on the following inclusion
criteria: i) Age of 40-70 years; ii) confirmed by either CT or MRI
measurement; iii) confirmed by pathology examination. At the same
time, all individuals with chronic hepatobiliary disorder and/or
the presence of a malignancy other than CCA were excluded. To
eliminate inter-tumor error between different TNM stages of CCA,
only those participants with TNM stage II CCA were selected for
subsequent analysis. Based on these criteria, 12 patients with CCA
were recruited, including 8 males and 4 females, and the average
age was 62.32±5.86 years. The demographic and clinicopathological
characteristics of patients are shown in Table SI.
The present study was officially approved by the
ethics committee of Tianjin Nankai Hospital (Tianjin, China). The
cancerous as well as paracancerous tissue samples of the clinical
patients were processed for the gene/protein expression analyses
(RT-qPCR and western blot analysis).
For RT-qPCR, the CCA cancerous and paracancerous
tissues were dissected, snap-frozen and stored in liquid nitrogen.
The total RNA was extracted using Rapid RNA extraction kit (Thermo
Fisher Scientific, Inc.; cat. no. AM9775) according to the
manufacturer's protocol. Subsequently, real-time PCR was performed
with BrightGreen 2X qPCR MasterMix-Low ROX (Thermo Fisher
Scientific, Inc.; cat. no. B21703). The RT-qPCR reaction system was
set up as follows: 5 µl 2 X Master Mix (Thermo Fisher Scientific,
Inc.; cat. no. 14001014), 0.5 µl 10 µmol/l PCR upstream and
downstream specific primers, 2 µl cDNA template, with
double-distilled water to 10 µl. PCR was carried out at 95˚C for 10
min; 95˚C for 10 sec, 60˚C for 1 min, 40 cycles; 95˚C for 15 sec,
60˚C for 60 sec, 95˚C for 15 sec. Relative expression was
calculated using the 2-ΔΔCq method with GAPDH as the
endogenous reference gene (21).
The reaction was performed in an Applied Biosystems 7300 PULAS
system (Thermo Fisher Scientific, Inc.). The primer sequences were
FOSB was as follows: Forward, 5'-GCTGCAAGATCCCCTACGAAG-3'
and reverse, 5'-ACGAAGAAGTGTACGAAGGGTT-3' and GAPDH:
Forward, 5'-GGAGCGAGATCCCTCCAAAAT-3' and reverse,
5'-GGCTGTTGTCATACTTCTCATGG-3'.
Western blot analysis
The proteins of CCA cancerous and paracancerous
tissues were extracted using RIPA Lysis Buffer (strong; cat. no.
HY-K1001; MedChemExpress; 150-250 µl cold RIPA Lysis Buffer per 20
mg of tissue sample) and the total protein concentration was
determined using the Pierce BCA protein assay kit (Thermo Fisher
Scientific, Inc.).
Subsequently, 50 µg protein of each sample was
subjected to 0.1% SDS-PAGE and then transferred onto a PVDF
membrane (cat. no. IPFL00010; EMD Millipore). The membranes were
blocked with 5% nonfat dry milk powder at 37˚C for 1.5 h. The
corresponding primary antibodies (1:1,000 dilution) were added and
then incubated at 4˚C overnight. The membrane was washed
extensively with PBS containing 0.1% Tween-20 and then incubated
with horseradish peroxidase-labeled secondary antibodies (1:10,000)
at room temperature for 1 h. After washing with PBS, the membrane
was processed with ECL developer (cat. no. 6883P3; Univ-bio) and
images were acquired with an ultrasensitive multifunctional imager.
FOSB, GAPDH and secondary antibodies were purchased from Abcam
(cat. no. ab252237, ab9485 and ab205718 respectively).
Uniform Manifold Approximation and
Projection (UMAP) algorithm analysis
The analysis was performed as previously described
(22) to convert mRNA information
of high-dimensional data into two dimensional data. The analysis
was conducted using online UMAP photography tool (cloudtutu.com/#/index).
Statistical analysis
All experiments were repeated 3 times independently.
SAS version 19.0 (SAS Institute, Inc.) was used for data analysis.
The continuous variables were tested for normality of distribution
and Student's t-test was applied to identify significant
differences between groups. For the non-continuous variables, the
Wilcoxon rank-sum test was conducted to analyze differences between
the groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
Establishment of a differential gene
expression profile for patients with CCA
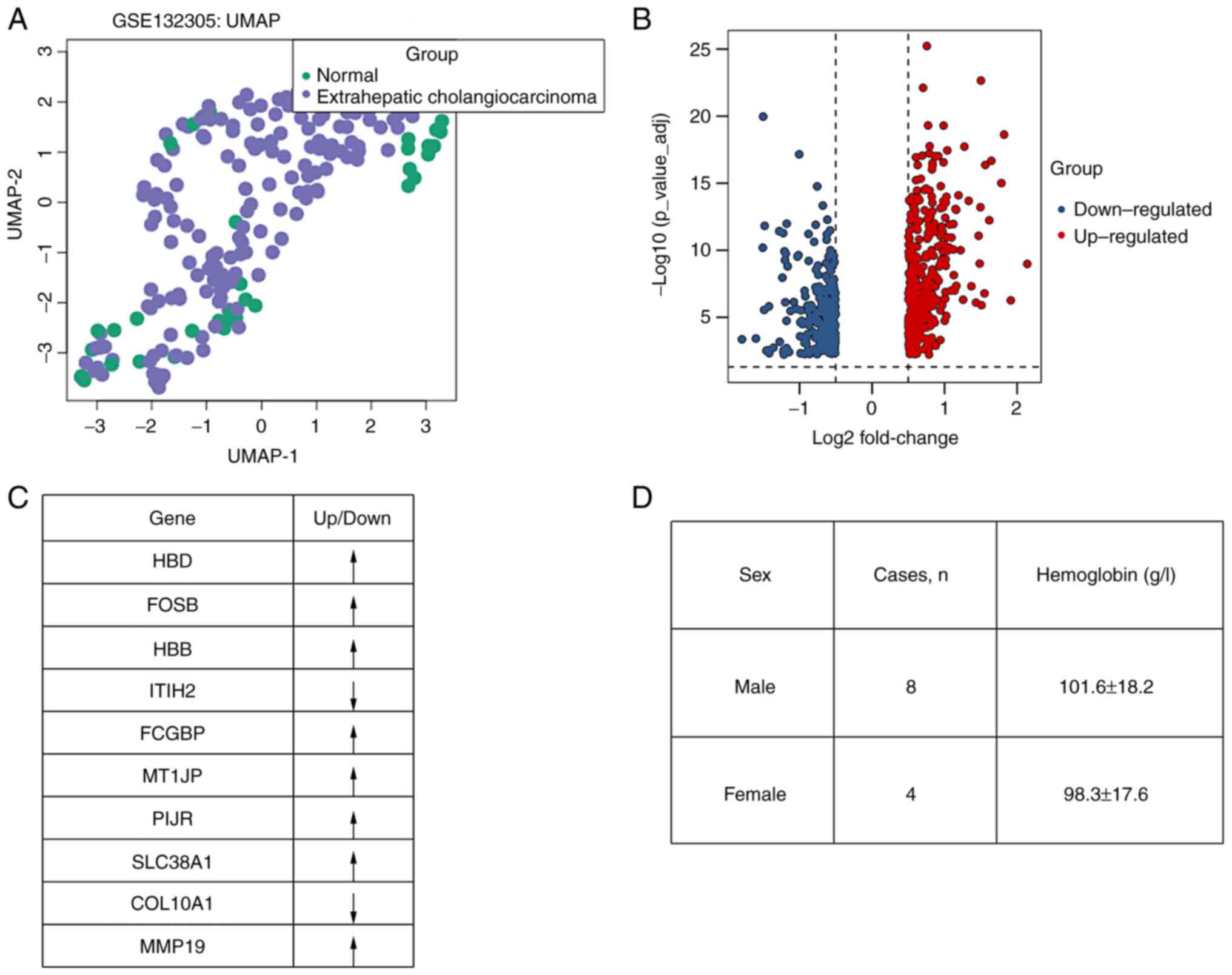
In the present study, the data from GSE132305 were
first analyzed, based on the UMAP algorithm analysis. When dealing
with large-sample genomic data, the UMAP may help reduce the
dimension of the data to achieve data visualization and identify
internal genetic relationships. At present, UMAP has been widely
used in the study of population genetics. In the present study, it
was demonstrated that the overall expression pattern between
primary CCA and non-tumor bile duct was significantly different via
the UMAP method (Fig. 1A).
The expression in the gene panel of 182 patients
with primary CCA was compared with that of 38 non-tumor bile duct
tissue with genes that had an absolute value of (Log2FC)>1 and
P<0.05 considered DEGs. All of the DEGs are listed in Table SII. A negative value of LogFC
indicated downregulation, while a positive value of LogFC suggested
upregulation. Based on these criteria, 676 DEGs were identified in
this CCA cohort, including 277 downregulated and 399 upregulated
ones (Fig. 1B). All of the DEGs
were further sorted based on the absolute value of Log2FC, and it
was revealed that hemoglobin subunit delta gene (HBD),
FOSB, hemoglobin subunit beta gene (HBB),
ITIH2, FCGBP, MT1JP, PIJR,
SLC38A1, COL10A1 and MMP19 were the most
significant DEGs (with the largest absolute value of Log2FC,
presented as Fig. 1C). Since two
potential genes (HBB and HBD) represent the members
of hemoglobin, the hemoglobin levels of 12 recruited patients with
CCA were also evaluated. Both male and female patients displayed
decreased levels of hemoglobin (Fig.
1D) (compared with normal ranges of hemoglobin levels: 120-160
g/l for males and 110-150 g/l for females).
GO and KEGG enrichment analysis
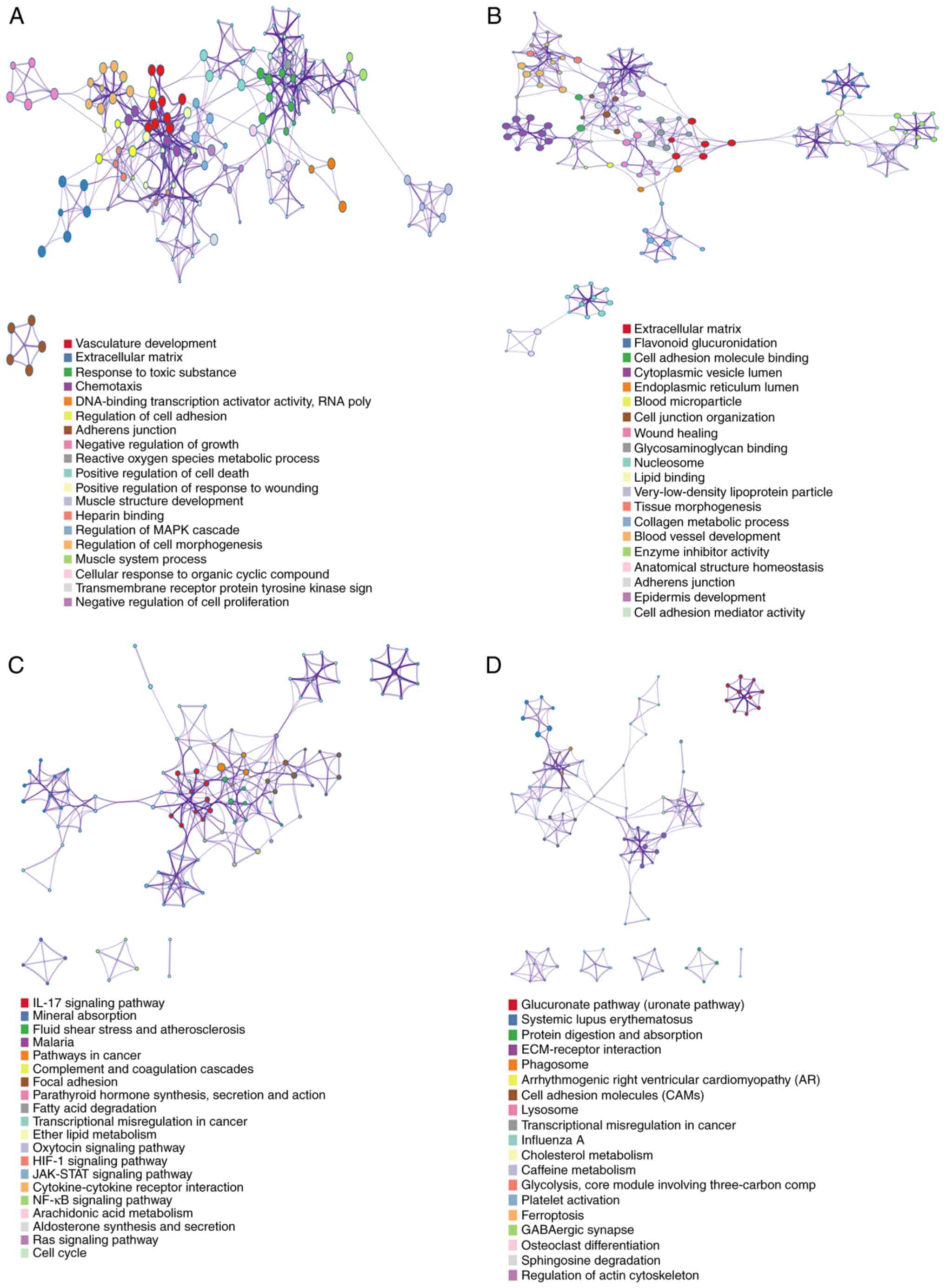
The GO enrichment analysis is able to determine the
functions and biological processes the gene products may be
involved in. GO enrichment analysis may provide an understanding of
the biological functions and metabolic pathways affected by
differential gene enrichment, which is particularly important in
mechanistic research. Metabolism processes were suggested to be
closely associated with significantly up-regulated DEGs, suggesting
these processes could be enhanced in CCA formation (including
vasculature development; Fig. 2A).
On the other hand, multiple metabolic processes were indicated to
be connected with significantly down-regulated DEGs, indicating
these processes should be inhibited in CCA formation (including
extracellular matrix (ECM); Fig.
2B).
KEGG enrichment analysis comprises a collection of
manually drawn pathway maps, which represent the current knowledge
of the potential related molecular interaction, reaction and
relation networks for DEGs. In the present study, the pathways
associated with the 676 DEGs were investigated using the KEGG
enrichment method. The upregulated genes supported by
differentially expression analysis were categorized into distinct
signaling cassettes (including IL-signaling pathway and mineral
absorption; Fig. 2C).
Downregulated genes were grouped into signaling cassettes
(including glucuronate signaling pathway, protein digestion and
absorption; Fig. 2D).
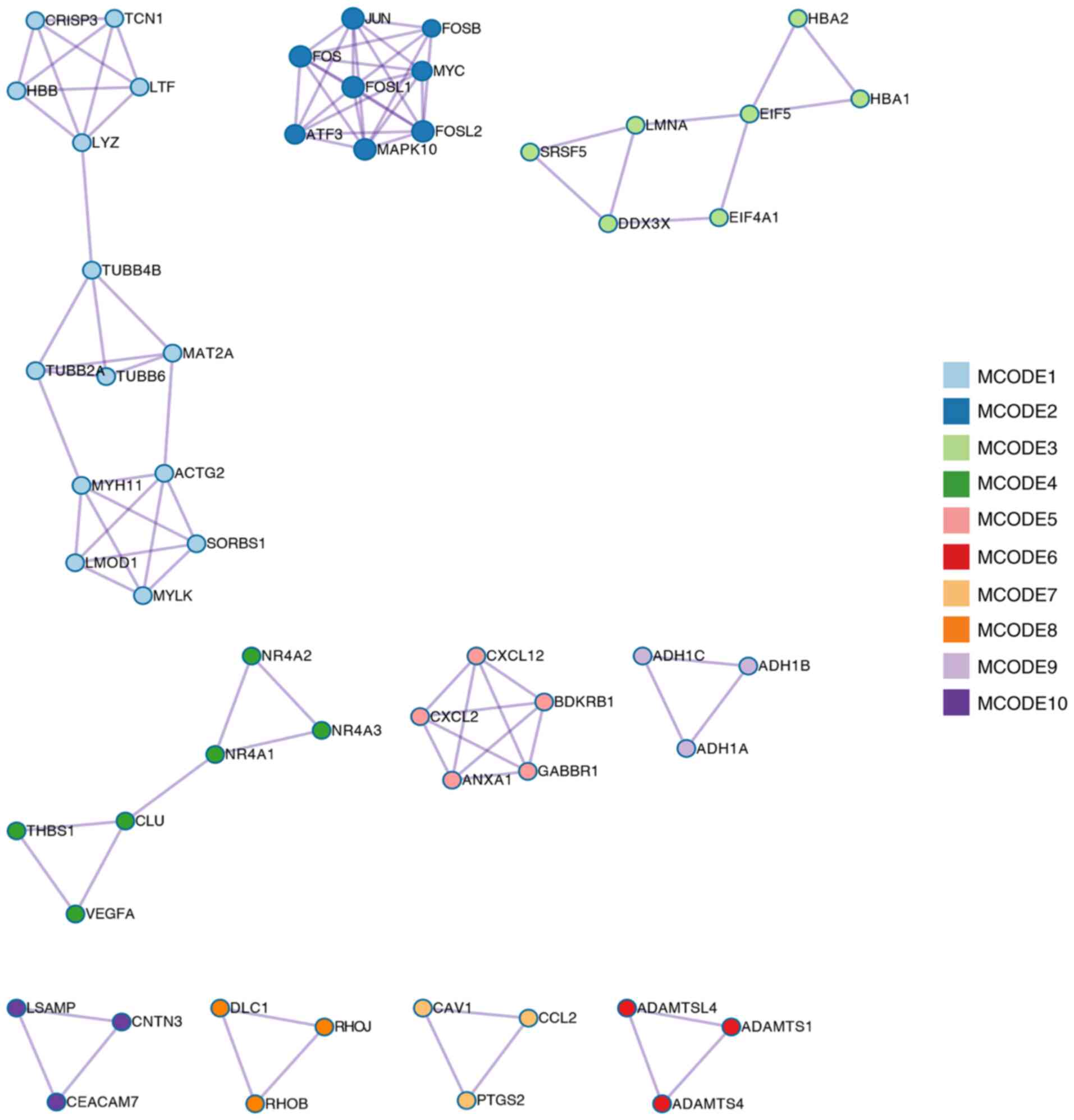
PPI network construction
The STRING database was used to construct a PPI
network for the 676 DEGs and the gene interactions with a
confidence score ≥0.4 were selected for visualization with
Cytoscape software. Based on this criterion, 10 MCODEs (MCODE1-10)
were identified as significant clustering modules (Fig. 3), among which MCODE1 and 2 were
more obvious compared with the others. MCODE1 included HBB protein,
which interacted with 3 differentially expressed candidate proteins
(so that the degree of HBB=3). At the same time, MCODE2 included
FOSB, which was associated with the most differentially expressed
candidate proteins (score=7).
According to the DEG analysis, HBD,
FOSB and HBB were suggested as potential biomarkers
for CCA (Fig. 1C). Probably due to
the abnormal expression of HBD and HBB, patients with CCA exhibited
hemoglobin level changes (Fig.
1D). According to a literature search, the roles of FOSB in CCA
progression have remained largely elusive. Of note, the PPI
interaction network demonstrated that FOSB protein was a central
regulatory factor and is expected to have a pivotal role in the
disorder. Accordingly, the present study further focused on the
functions of FOSB to provide verification in independent
experimental systems.
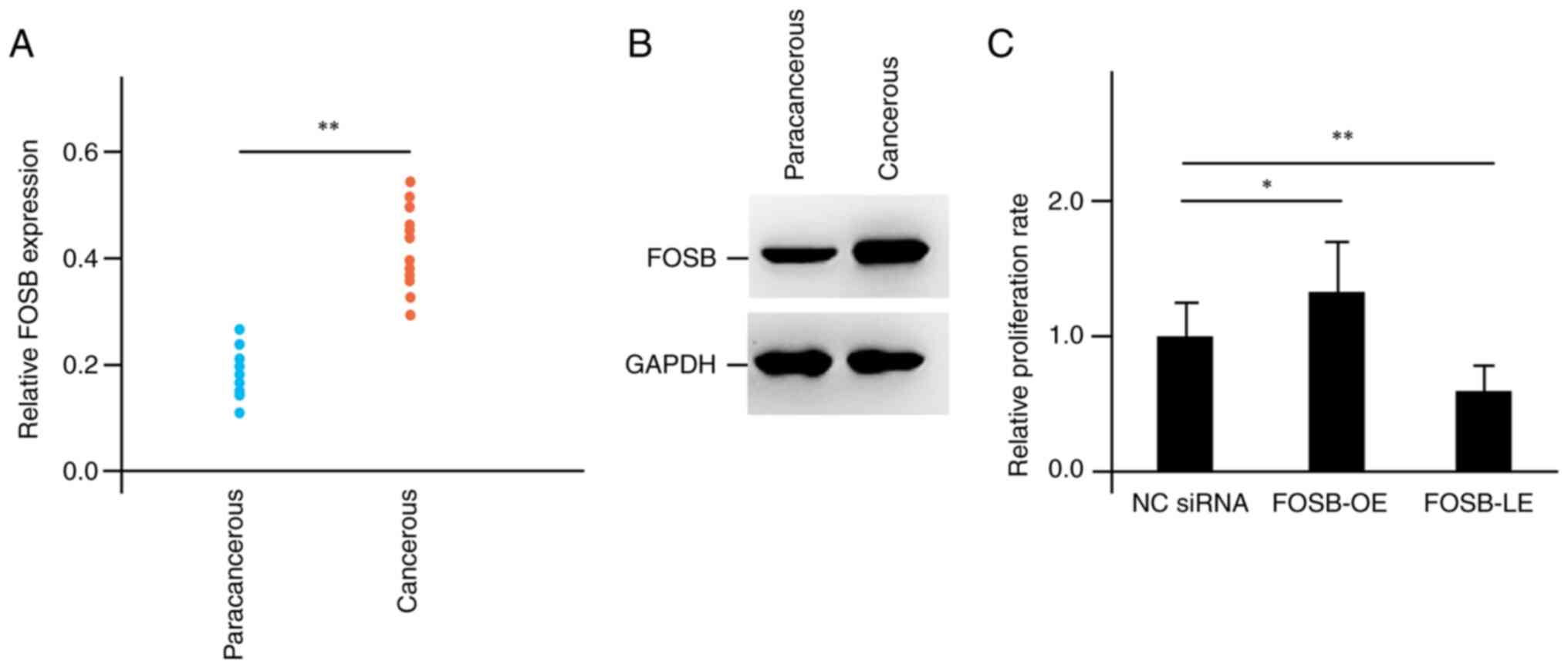
Investigation of the functions of FOSB
in the formation of CCA
Based on the previous analysis, FOSB was
suggested as a potential biomarker for CCA formation. Next, the
present study sought to further evaluate the functions of
FOSB in-depth using recruited clinical patients as well as
the CCA cell line HUCCT1. Compared with paracancerous tissues, the
CCA cancerous tissues exhibited enhanced RNA expression levels of
FOSB according to the RT-qPCR results (Fig. 4A). At the same time, the protein
activity of FOSB in the CCA cancerous tissues was also elevated as
determined by western blot analysis (Fig. 4B). These outcomes were in line with
the previous analysis. Since carcinogenesis is generally associated
with changes in cell proliferation, a CCK-8 assay was performed on
the HUCCT1 cells. FOSB-LE was achieved by transfection of
FOSB-siRNA, while FOSB-OE was obtained by transfection of
FOSB-RNA overexpression plasmid. Compared with the control
group (negative control siRNA-transfected, NC-siRNA), the cell
viability was significantly reduced in the FOSB-LE group, while it
was markedly increased in the FOSB-OE group, supporting the
function of FOSB as an oncogene in CCA (Fig. 4C).
Discussion
CCA represents a heterogeneous group of malignancies
that probably occur at any location along the biliary tree of the
human body. For the last decades, researchers have been trying to
obtain breakthroughs in the discovery of early diagnostic
biomarkers for CCA. However, the research progress is far from
satisfactory. The well-accepted hypothesis of the
adenoma-dysplasia-carcinoma sequence observed in other cancers has
not yet been fully approved in CCA, based on the fact that cells of
varying origin may cause the disease (1). The present study first systematically
analyzed the DEG profile for patients with CCA using an open public
data source. Two primary target genes identified (HBD and
HBB) belong to the hemoglobin gene family. Of note,
iron-deficiency anemia is a relatively common presenting feature
for multiple gastrointestinal malignancies. Ahmad et al
(23) reported on a case of a
Caucasian, 84-year-old female presenting with recurrent, severe
iron-deficiency anemia, who was eventually diagnosed with
intra-hepatic CCA. Since hemoglobin genes were also significantly
differentially expressed in CCA in the present analysis, the
hemoglobin levels were measured in the recruited patients. Compared
with the normal range of hemoglobin levels (120-160 g/l for males
and 110-150 g/l for females), patients with CCA displayed decreased
level of hemoglobin. Hemoglobin is characterized as a key protein
consisting of 2 α-like type and 2 β-like type chains. A variation
on the globin genes results in an error in the production of the
coded chains, which initiate hemoglobinopathies, such as
thalassemia and sickle cell diseases (22). In the present case, elevated
expression levels of HBB and HBD were observed, which is expected
to lead to enhanced activity of hemoglobin. However, this was not
the case. It was speculated that the decreased hemoglobin was
either caused by other internal negative inhibitory signaling
events or a consequence of unknown protective molecular mechanisms
associated with CCA. Further investigation in this context is
required. The function of hemoglobin is heterogeneous among
individuals and anemia has been indicated to be closely associated
with distinct diseases. To this end, the present study focused on
another potential biomarker gene for CCA diagnosis, namely
FOSB.
The AP-1 protein family consists of diverse factors,
including FOSB, FOS-related antigen 1, FRA2, c-Jun and Jun-B, which
have key roles in the transcriptional regulation of numerous genes
related to cell proliferation, differentiation, migration and
metastasis, as well as survival (20). To date, FOSB has been suggested to
be involved in the progression of several types of cancer. For
instance, FOSB was observed to be unmethylated in non-small cell
lung cancer and identified as a novel predictive biomarker for
NSCLC prognosis (24). At the same
time, FOSB was indicated to be required for the migration and
invasion of prostate cancer cells, which was dependent on the
regulation of TGF-β1(20). Another
study reported that SERPINE1-FOSB fusion was identified as a
consistent genetic alteration in pseudomyogenic
hemangioendothelioma, suggesting FOSB as a useful diagnostic
biomarker for the disease (25).
In the present study, GO enrichment analysis indicated that ECM
processes were markedly enriched in CCA. Accumulating evidence has
demonstrated the connection between FOSB and ECM formation. An
in vitro study supported that the DNA-binding activity of
FOSB was enhanced after stretch stimulation of smooth muscle cells
(26). FOSB silencing attenuated
the expression of the profibrotic factors tenascin C and connective
tissue growth factor, which all together indicated the role of FOSB
as a mechanosensitive regulator of ECM production in smooth muscle
(27). More interestingly, FOSB
has also been implied to be a key regulator of matrix
metalloproteinases (MMPs). For instance, one of the MMP members,
MMP-9, is a central factor in several processes of tumor formation,
such as angiogenesis, cell migration, as well as invasiveness based
on the fact that it is able to almost fully degrade the basement
membrane and ECM components. A study by Li et al (28) suggested that FOSB increased the
expression of MMP-9 directly in MCF-7 breast cancer cells and
overexpression of FOSB enhanced the cellular viability and
decreased cell apoptosis. The present study supported these
conclusions. Compared with paracancerous tissues, both RNA and
protein expression levels of FOSB were increased in CCA cancerous
tissues. Furthermore, FOSB also inhibited the proliferation of the
CCA cell line HUCCT1 according to the CCK-8 assay results. Based on
the PPI network established, FOSB potentially interacted with 7
differentially expressed targets, which are JUN, MYC, FOSL2, FOS,
FOSL1, ATF3 and MAPK10. JUN, FOSL2, FOS, FOSL1, as well as ATF3 all
belong to the AP-1 gene family, whereas FOSB is associated with
them to a certain extent. AP-1 family elements are regulatory
sequences within genomes, which direct the selective expression of
particular genes in order to obtain their highly specialized forms
and functions. Previously, AP-1 members were reported to be
activated by the Ras/MAPK pathway in nearly all cell types
(16). However, how the regulatory
sequence elements associated with CCA remain elusive, providing a
worthwhile direction for future study.
In summary, in this integrated study, the DEGs in
patients with CCA were elucidated in-depth and several candidate
genes (particularly FOSB) were proposed for early diagnosis.
The present study provided a direction for the future clinical
management of CCA.
Supplementary Material
The demographic and
clinicopathological characteristics of the patients
Differentially expressed gene
profiling for cholangiocarcinoma.
Acknowledgements
Not applicable.
Funding
Funding: This study was funded by the National Natural Science
Foundation of China (grant no. 82070687 to BZ).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SW collected data and contributed to the conception.
LY performed the experiments, and analyzed and interpreted the
data. XS analyzed and interpreted the data as well as supervised
the work. BZ designed the experiments and wrote the manuscript. XS
and LY confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
This study was officially approved by the ethics
committee of Tianjin Nankai Hospital (Tianjin, China). Informed
consent was obtained from all individual participants included in
the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Saha SK, Zhu AX, Fuchs CS and Brooks GA:
Forty-Year trends in cholangiocarcinoma incidence in the U.S.:
Intrahepatic disease on the rise. Oncologist. 21:594–599.
2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rizvi S, Khan SA, Hallemeier CL, Kelley RK
and Gores GJ: Cholangiocarcinoma-evolving concepts and therapeutic
strategies. Nat Rev Clin Oncol. 15:95–111. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Razumilava N and Gores GJ: Combination of
gemcitabine and cisplatin for biliary tract cancer: A platform to
build on. J Hepatol. 54:577–578. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
DeOliveira ML, Cunningham SC, Cameron JL,
Kamangar F, Winter JM, Lillemoe KD, Choti MA, Yeo CJ and Schulick
RD: Cholangiocarcinoma: Thirty-one-year experience with 564
patients at a single institution. Ann Surg. 245:755–762.
2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Welzel TM, Mellemkjaer L, Gloria G, Sakoda
LC, Hsing AW, El Ghormli L, Olsen JH and McGlynn KA: Risk factors
for intrahepatic cholangiocarcinoma in a low-risk population: A
nationwide case-control study. Int J Cancer. 120:638–641.
2007.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Donato F, Gelatti U, Tagger A, Favret M,
Ribero ML, Callea F, Martelli C, Savio A, Trevisi P and Nardi G:
Intrahepatic cholangiocarcinoma and hepatitis C and B virus
infection, alcohol intake, and hepatolithiasis: A case-control
study in Italy. Cancer Causes Control. 12:959–964. 2001.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tyson GL and El-Serag HB: Risk factors for
cholangiocarcinoma. Hepatology. 54:173–184. 2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Palmer WC and Patel T: Are common factors
involved in the pathogenesis of primary liver cancers? A
meta-analysis of risk factors for intrahepatic cholangiocarcinoma.
J Hepatol. 57:69–76. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Charatcharoenwitthaya P, Enders FB,
Halling KC and Lindor KD: Utility of serum tumor markers, imaging,
and biliary cytology for detecting cholangiocarcinoma in primary
sclerosing cholangitis. Hepatology. 48:1106–1117. 2008.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Patel AH, Harnois DM, Klee GG, LaRusso NF
and Gores GJ: The utility of CA 19-9 in the diagnoses of
cholangiocarcinoma in patients without primary sclerosing
cholangitis. Am J Gastroenterol. 95:204–207. 2000.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Endo I, Gonen M, Yopp AC, Dalal KM, Zhou
Q, Klimstra D, D'Angelica M, DeMatteo RP, Fong Y, Schwartz L, et
al: Intrahepatic cholangiocarcinoma: Rising frequency, improved
survival, and determinants of outcome after resection. Ann Surg.
248:84–96. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Farshidfar F, Zheng S, Gingras MC, Newton
Y, Shih J, Robertson AG, Hinoue T, Hoadley KA, Gibb EA, Roszik J,
et al: Integrative genomic analysis of cholangiocarcinoma
identifies distinct IDH-Mutant molecular profiles. Cell Rep.
18:2780–2794. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Patel T: Worldwide trends in mortality
from biliary tract malignancies. BMC Cancer. 2(10)2002.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tang C, Jiang Y, Shao W, Shi W, Gao X, Qin
W, Jiang T, Wang F and Feng S: Abnormal expression of FOSB
correlates with tumor progression and poor survival in patients
with gastric cancer. Int J Oncol. 49:1489–1496. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Vierbuchen T, Ling E, Cowley CJ, Couch CH,
Wang X, Harmin DA, Roberts CWM and Greenberg ME: AP-1 transcription
factors and the BAF complex mediate signal-dependent enhancer
selection. Mol Cell. 68:1067:–1082.e12. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Iwaki J, Kikuchi K, Mizuguchi Y,
Kawahigashi Y, Yoshida H, Uchida E and Takizawa T: MiR-376c
down-regulation accelerates EGF-dependent migration by targeting
GRB2 in the HuCCT1 human intrahepatic cholangiocarcinoma cell line.
PLoS One. 8(e69496)2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47(D1):D607–D613.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Barrett CS, Millena AC and Khan SA: TGF-β
effects on prostate cancer cell migration and invasion require
FosB. Prostate. 77:72–81. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Becht E, McInnes L, Healy J, Dutertre CA,
Kwok IWH, Ng LG, Ginhoux F and Newell EW: Dimensionality reduction
for visualizing single-cell data using UMAP. Nat Biotechnol: Dec 3,
2018 (Epub ahead of print).
|
|
23
|
Ahmad SS, Basheer FT, Idris SF, Hariraj R,
Mathialagan R and Douds A: Cholangiocarcinoma presenting as
hemobilia and recurrent iron-deficiency anemia: A case report. J
Med Case Rep. 4(133)2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kim DS, Lee WK and Park JY: Association of
FOSB exon 4 unmethylation with poor prognosis in patients with
late-stage non-small cell lung cancer. Oncol Rep. 43:655–661.
2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Hung YP, Fletcher CD and Hornick JL: FOSB
is a useful diagnostic marker for pseudomyogenic
hemangioendothelioma. Am J Surg Pathol. 41:596–606. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Alhendi AMN, Patrikakis M, Daub CO, Kawaji
H, Itoh M, de Hoon M, Carninci P, Hayashizaki Y, Arner E and
Khachigian LM: Promoter usage and dynamics in vascular smooth
muscle cells exposed to fibroblast growth factor-2 or
interleukin-1β. Sci Rep. 8(13164)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Van Doren SR: Matrix metalloproteinase
interactions with collagen and elastin. Matrix Biol. 44-46:224–231.
2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li H, Li L, Zheng H, Yao X and Zang W:
Regulatory effects of ΔFosB on proliferation and apoptosis of MCF-7
breast cancer cells. Tumour Biol. 37:6053–6063. 2016.PubMed/NCBI View Article : Google Scholar
|