Introduction
Lung cancer is a major cause of mortality in
middle-aged and elderly patients (1). In addition, it has one of the highest
incidence rates among all tumor types (1). A total of 2.1 million newly diagnosed
and 1.8 million deaths were reported in 2018 from 322
population-based registries in 71 countries, making it the number
one cause of cancer-associated mortalities (2). Among all lung cancer subtypes,
adenocarcinoma is the most invasive and heterogeneous, with an
abnormally high tumor mutation burden (3). Despite advances in the development of
lung cancer treatment methods over the past decade, the prevention,
early diagnosis and management of patients with lung cancer remain
challenging (4).
The complement pathway is an integral part of the
innate immune system that serves to clear microbes and impaired
cells by driving inflammation (5).
This process in turn recruits innate and adaptive immune cells to
attack the cell membrane of pathogens (6). Activation of the complement pathway
serves an important role in the development of tumors (7). It can be activated by classical,
lectin or alternative pathways, all of which converge onto lead the
activation of C3b. For example, lung cancer could be activated by
the classical complement pathway (6). This then forms the membrane attack
complex to mediate cell lysis (7).
Complement C3 and C4 are key components in this pathway that are
important for complement activation (8). C3 is the mediator molecule in the
process of complement activation, whilst C4 is the terminal
by-product, the level of which provides an indication of complement
activation in the body (8).
Previous studies have revealed the presence of
complement-associated proteins such as complement factor H in the
tumor microenvironment, in which tumor cells (such as lung cancer
cells) can exhibit multiple effects (such as activating the
complement) on complement proteins (9,10).
The complement-activated lectin pathway plays an important role in
human solid tumors, including those of the female reproductive
system, the lungs and the digestive tract (11). Therefore, the present study
selected these two molecules as markers.
Accumulating evidence suggest that the complement
system can serve a role in tumor progression by promoting cancer
cell angiogenesis, proliferation and antitumor immunity (11,12).
The presence of data supporting complement activation and C5b-9 in
deposition-related data in multiple types of malignancies, such as
lung and pancreatic cancer, support this notion (13). To the best of our knowledge,
Niculescu et al (14) first
identified abnormal complement activation and elevated sC5b-9
levels in patients with breast cancer. However, the association
between serum complement C3b and C4d levels and the prognosis of
patients with NSCLC combined with lymph node metastasis remains
unclear.
The present study examined serum complement C3b and
C4d expression levels in patients with NSCLC combined with lymph
node metastasis before exploring their potential as prognostic
factors in such patients. A predictive model of mortality risk was
constructed based on complement C3b and C4d expression levels. This
was presented through Nomograms, which can be readily calculated
and would provide a beneficial tool to support the decision-making
of clinicians.
Materials and methods
Patients
The present study included data from 132 patients
with NSCLC collected from the First Affiliated Hospital of Soochow
University (Suzhou, China) and Sichuan Cancer Hospital (Chengdu,
China) between February 1, 2010 and April 1, 2019. The median age
of the patients was 65 years (range, 57-69 years), and the cohort
included 45 (34.1%) men. In addition, data from 50 patients [mean
age, 64.5±10.92; male, 16 (32.0%); female, 34 (68.0%)] with NSCLC
from the Sichuan Cancer Hospital between June 2012 and May 2019
were collected as an external validation cohort for subsequent
modeling with the same inclusion and exclusion criteria as for the
132 patients above. NSCLC was diagnosed by pathological analysis.
Patients who lacked information on complement composition data,
those with SLE and renal dysfunction, and those who withdrew from
treatment or had missing follow-up information were excluded
(Fig. 1). Patient data, including
age, sex, serum carcinoembryonic antigen (CEA) levels, body mass
index, albumin levels, lymphocyte count, C-reactive protein (CRP)
level, neutrophils, hemoglobin, prognostic nutritional index (PNI),
platelet count, neutrophil-lymphocyte ratio, surgery, staging of
documented lung cancer, radiotherapy, tyrosine kinase inhibitor
application, diabetes mellitus, Karnofsky performance status (KPS)
score (15), smoking, heart
failure, hyperlipidemia (plasma total cholesterol concentration
>5.17 mmol/l OR plasma triacylglycerol concentration >2.3
mmol/l), were all selected for analysis. The data distributions of
C3 and C4 were tested for normality and were revealed to be skewed
by normality test. According to these statistical principles, data
from skewed distributions are suitable for analysis using the
median (16). Therefore, the
median was selected as the cut-off value of continuous variables.
Informed written consent was obtained from all patients or their
immediate family members. All research programs are in line with
the guidelines of the Ethics Committee of Soochow University and
followed the Declaration of Helsinki. Only the medical records of
the 182 patients in total were collected from the hospital
database.
The inclusion criteria were as follows: i) Patients
can understand the study and agree to sign a written informed
consent document; ii) patients are aged 18-75 years and must have a
life expectancy of >3 months; iii) patients must have a
confirmed histological or cytological diagnosis of NSCLC; iv)
Eastern Cooperative Oncology Group score standard of 0-2; v)
patients must have normal organ and marrow function within 2 weeks
prior to the study. Normal organ and marrow function was defined as
absolute neutrophil count >1,500/ml; platelets >100,000/ml;
total bilirubin within normal institutional limits (1.71-17.1
µmol/l); aspartate transaminase/alanine aminotransferase <2.5X
institutional upper limit of normal; creatinine ≤1.5X institutional
upper limit of normal; and urine dipstick for proteinuria of
<1+. If urine dipstick is >1+, a 24-h urine for protein must
demonstrate <500 mg protein in 24 h to allow participation in
the present study. Exclusion criteria: i) Women who were pregnant
due to concerns their complement values may be affected by the
fetus; ii) if during the treatment, a serious active infection from
which an intravenous injection of antibiotics was required; iii)
the patient has symptoms of brain metastases or suffers from severe
mental or cognitive impairment; iv) patients who had congestive
heart failure, arrhythmia, myocardial infarction, unstable angina,
stroke or transient ischemic attack in 6 months; and v) patients
with other malignancies within 5 years, except for those with
cervical carcinoma in situ, skin squamous cell carcinoma of
the skin or the basic control of skin basal cell carcinoma.
Complement C3b and C4d detection
Blood samples were collected from patients with
NSCLC combined with lymph node metastasis at the time of diagnosis.
Peripheral blood samples were collected and anti-coagulated with
EDTA. Samples were then centrifuged at 800 x g for 10 min at room
temperature to collect the supernatants. All blood plasma specimens
were stored at -80˚C in a specimen refrigerator for further study.
Complement detection was performed within 3 days after plasma
collection. According to the manufacturer's protocols, the
complement C3b and C4d levels in plasma were detected using ELISA
kits (cat. nos. WLS11421 and ZY-E67-44H; Shanghai Yuanye
Biotechnology Co., Ltd.).
Statistical analysis
NCSS-PASS software version 10.0 (NCSS, LLC) was used
for sample size assessment. Power was set to 0.99 and α to 0.5.
P<0.05 was considered to indicate a statistically significant
difference. Missing values (≤5.0%) were estimated by the random
forest method using the ‘mice’ package (17) in RStudio (R version 3.5.0; RStudio,
Inc.) (18). Categorical variables
were represented as proportions and matched using the χ2
test. Commonly and skewed distributed variables were presented as
the median with interquartile range. Group comparisons were
performed using either one-way ANOVA or Kruskal-Wallis test
followed by Tukey's test for each of the pairwise comparisons.
Survival data was displayed by Kaplan-Meier (KM)
curves on a cumulative basis and compared using a log-rank test.
The univariate and multivariate survival responses of OS were
adjusted using Cox regression models to estimate OS. Forest plots
were used for the visualization of the importance of prognosis by
the covariate. Using Harrell's regression modeling R package of
‘rms’ (R software, version 5.1-2; https://www.rdocumentation.org/
packages/rms/versions/5.1-2).
To establish the prognostic risk, risk factors were
identified using Cox multifactor regression models (variants with
P-values <0.05 were included in the model). The weight of each
variant was quantified, before nomograms were generated and
internal validation was performed using 1,001 bootstrapping (R
version 3.5.0, ‘rms’ package) (19). A calibration test of 5-year OS to
an ideal curve estimated the concordance of the derived model.
Log-rank tests and KM curves were applied to analyze the
associations of C3b and C4d with survival outcomes. Spearman's
correlation test was performed to analyze the association between
PNI and C3b as well as C4d. C-statistics was calculated by ‘rms’
package in R software. A dot plot was created based on the accuracy
of the predictions, with different colors used to indicate correct
and incorrect predictions, and to calculate the percentage correct.
Statistical analyses were performed using RStudio (R version 3.5.0)
with the following R packages: ‘rms’, ‘ggplot2’, ‘risk regression’,
‘PredictABLE’ and ‘survminer’ (20-22).
Results
Baseline characteristics
The characteristics of the patients included in the
present study are listed in Table
I. Specifically, the present study included 132 patients who
suffered from NSCLC combined with lymph node metastasis diagnosed
between February 2010 and April 2019. By the follow-up endpoint
(December 2021), the overall mortality rate was 70.0%. The median
serum CEA and CRP levels were 8.28 ng/ml and 3.80 µmol/l,
respectively. A total of 23 (17.4%) patients in the included
population were diagnosed with stage I, 19 (14.3%) with stage II,
30 (22.7%) with stage III and 60 (45.4%) with stage IV according to
TNM staging (23). In terms of
treatment, 59 (45.0%) patients underwent surgery and 34 (26.0%)
patients received radiation therapy. The KPS score was also
evaluated, with 116 (88.0%) patients obtaining a score of ≥80. The
comorbidities of these patients were also examined. There were 12
(9.0%) with type II diabetes mellitus and nine (7.0%) cases of
hyperlipidemia. A total of 52 (39.0%) patients had hypertension. In
addition, 64 (48.0%) of all patients were smokers.
 | Table IStudy participant characteristics at
enrollment. |
Table I
Study participant characteristics at
enrollment.
| Variables | Total (n=132) | Stage I (n=23) | Stage II
(n=19) | Stage III
(n=30) | Stage IV
(n=60) | P-value |
|---|
| Median age (IQR),
years | 65.00
(57.00-69.00) | 66.00
(61.00-71.00) | 63.00
(59.50-66.50) | 65.00
(58.25-69.00) | 63.00
(53.75-69.00) | 0.5 |
| Sex, n (%) | | | | | | 0.101 |
|
Male | 45(34) | 11(48) | 9(47) | 6(20) | 19(32) | |
|
Female | 87(66) | 12(52) | 10(53) | 24(80) | 41(68) | |
| Surgery, n (%) | | | | | | <0.001 |
|
No | 73(55) | 5(22) | 4(21) | 14(47) | 50(83) | |
|
Yes | 59(45) | 18(78) | 15(79) | 16(53) | 10(17) | |
| Radiation, n
(%) | | | | | | 0.426 |
|
No | 98(74) | 19(83) | 15(79) | 19(63) | 45(75) | |
|
Yes | 34(26) | 4(17) | 4(21) | 11(37) | 15(25) | |
| Chemotherapy, n
(%) | | | | | | 0.262 |
|
AP | 106(80) | 22(96) | 16(84) | 22(73) | 46(77) | |
|
DP | 17(13) | 1(4) | 1(5) | 6(20) | 9(15) | |
|
EP | 5(4) | 0 (0) | 1(5) | 0 (0) | 4(7) | |
|
GP | 1(1) | 0 (0) | 0 (0) | 0 (0) | 1(2) | |
|
NP | 1(1) | 0 (0) | 0 (0) | 1(3) | 0 (0) | |
|
TP | 2(2) | 0 (0) | 1(5) | 1(3) | 0 (0) | |
| Target therapy
(tyrosine kinase inhibitors), n (%) | | | | | | 0.082 |
|
No | 88(67) | 20(87) | 14(74) | 19(63) | 35(58) | |
|
Yes | 44(33) | 3(13) | 5(26) | 11(37) | 25(42) | |
| Karnofsky
Performance Status, n (%) | | | | | | 0.13 |
|
50 | 2(2) | 0 (0) | 1(5) | 0 (0) | 1(2) | |
|
60 | 3(2) | 0 (0) | 0 (0) | 1(3) | 2(3) | |
|
70 | 11(8) | 2(9) | 1(5) | 1(3) | 7(12) | |
|
80 | 21(16) | 1(4) | 4(21) | 3(10) | 13(22) | |
|
90 | 51(39) | 8(35) | 4(21) | 14(47) | 25(42) | |
|
100 | 44(33) | 12(52) | 9(47) | 11(37) | 12(20) | |
| Smoking, n (%) | | | | | | 0.014 |
|
No | 68(52) | 15(65) | 15(79) | 12(40) | 26(43) | |
|
Yes | 64(48) | 8(35) | 4(21) | 18(60) | 34(57) | |
| Hypertension, n
(%) | | | | | | 0.103 |
|
No | 80(61) | 13(57) | 10(53) | 14(47) | 43(72) | |
|
Yes | 52(39) | 10(43) | 9(47) | 16(53) | 17(28) | |
| Diabetes, n
(%) | | | | | | 0.016 |
|
No | 120(91) | 17(74) | 19(100) | 27(90) | 57(95) | |
|
Yes | 12(9) | 6(26) | 0 (0) | 3(10) | 3(5) | |
| Hyperlipemia, n
(%) | | | | | | 0.315 |
|
No | 123(93) | 20(87) | 18(95) | 27(90) | 58(97) | |
|
Yes | 9(7) | 3(13) | 1(5) | 3(10) | 2(3) | |
| OS Status, n
(%) | | | | | | <0.001 |
|
Alive | 40(30) | 17(74) | 13(68) | 3(10) | 7(12) | |
|
Deceased | 92(70) | 6(26) | 6(32) | 27(90) | 53(88) | |
| Median OS time
(IQR), months | 25.10
(10.65-61.30) | 61.30
(23.70-73.80) | 61.50
(50.30-69.45) | 26.00
(11.10-51.45)a,b | 14.20
(8.23-34.95)c,d | <0.001 |
| Mean ± SD body mass
index | 23.03±3.25 | 23.18±3.02 | 23.10±3.16 | 22.56±3.46 | 23.19±3.30 | 0.842e |
| Median (IQR) serum
carcinoembryonic antigen, ng/ml | 8.28 (2.57,
39.05) | 6.96 (2.19,
51.80) | 6.02 (2.85,
27.73) | 3.89 (2.18,
16.64) | 11.30 (2.84,
40.16) | 0.334f |
| Mean ± SD
C-reactive protein, µmol/l | 5.98±5.46 | 5.19±5.87 | 2.14±3.05 | 5.24±5.66 | 7.68±5.09 |
<0.001e |
| Mean ± SD serum
albumin, g/l | 40.95±4.86 | 41.66±4.93 | 43.15±5.33 | 41.25±4.82 | 39.84±4.50 | 0.052e |
| Median (IQR)
neutrophils, 109/l | 4.34
(3.32-5.53) | 3.45
(3.05-4.34) | 4.44
(2.67-5.05) | 3.98
(3.57-4.64) | 5.01
(3.66-6.38)c | 0.006f |
| Median (IQR)
lymphocytes, 109/l | 1.77
(1.26-2.33) | 2.31
(1.75-2.52) | 2.34
(2.00-2.58) | 1.56
(1.21-1.99)a,b | 1.54
(1.10-1.95)c,d |
<0.001e |
| Mean ± SD
hemoglobin, g/l | 132.98±16.72 | 132.39±18.23 | 136.11±15.17 | 131.70±17.76 | 132.85±16.35 | 0.836e |
| Median (IQR)
platelets, 109/l | 216.00
(174.00-259.25) | 207.00
(154.50-240.50) | 204.00
(190.50-243.00) | 222.00
(187.75-252.50) | 219.50
(171.00-272.00) | 0.696f |
| Median (IQR)
prognostic nutritional index | 48.85
(44.95-53.38) | 47.45
(45.27-54.67) | 51.40
(48.17-55.08) | 49.55
(45.01-53.24) | 48.05
(44.83-51.46) | 0.162f |
| Median (IQR)
neutrophil lymphocyte ratio | 2.52
(1.71-3.95) | 1.71
(1.16-2.21) | 2.05
(1.22-2.58) | 2.45
(1.82-3.65) | 3.27
(2.24-4.90)c |
<0.005e |
| Median (IQR) C3,
µmol/l | 366.10
(201.32-448.69) | 461.85
(374.92-500.84) | 444.03
(395.73-498.42) | 247.26
(171.94-405.44)a,b | 316.56
(186.92-404.25)c,d |
<0.001e |
| Median (IQR) C4,
µmol/l | 408.56
(315.79-652.06) | 665.84
(605.91-704.94) | 642.61
(480.41-672.95) | 411.74
(365.68-574.30) | 331.41
(278.67-482.72)c,d |
<0.001e |
Regression analysis
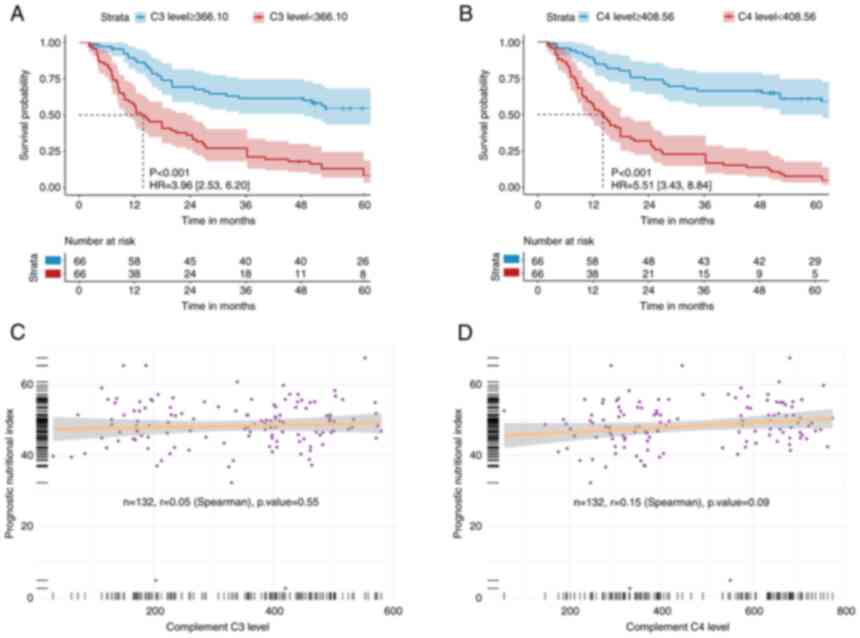
According to single-factor analysis, C3b
levels (≤366.10 µmol/l, median) were a predictor of
cancer-associated mortality [hazard ratio (HR) 3.96; 95% confidence
interval (CI) 2.53-6.20; P<0.001; Table II]. KM curves revealed that those
in the low C3b group had an increased cumulative incidence of
mortality compared with patients in their high-level group
(log-rank P<0.001; Fig. 2A). In
addition, patients with low C4d levels (≤408.56 µmol/l, median)
demonstrated a higher incidence of mortality on the survival curve
compared with patients in the high-level group (P<0.001;
Fig. 2B). The correlation between
complement C3b/C4d levels and neutrophil-lymphocyte ratio (NLR)
levels was next investigated as both were continuous variables, but
no statistically significant correlation could be found (Fig. 2C and D).
 | Table IICox regression analysis of hazard
ratios in terms of patients with NSCLC with digestive disease
(univariate analysis). |
Table II
Cox regression analysis of hazard
ratios in terms of patients with NSCLC with digestive disease
(univariate analysis).
| | Non-adjustment | Model 1+ |
|---|
| Variation | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| Age, ≥65 vs. <65
years | 1.15
(0.76-1.73) | 0.507 | - | - |
| Sex, male vs.
female | 1.77
(1.13-2.78) | 0.013 | - | - |
| Surgery, yes vs.
no | 0.26
(0.17-0.41) | <0.001 | 0.27
(0.17-0.43) | <0.001 |
| Radiation therapy,
yes vs. no | 1.21
(0.77-1.88) | 0.405 | 1.26
(0.80-1.96) | 0.315 |
| Target therapy, yes
vs. no | 1.13
(0.74-1.73) | 0.561 | 1.06
(0.69-1.65) | 0.784 |
| Smoking, yes vs.
no | 2.28
(1.50-3.47) | <0.001 | 2.17
(1.26-3.74) | 0.005 |
| Hypertension, yes
vs. no | 1.23
(0.81-1.86) | 0.328 | 1.23
(0.80-1.89) | 0.356 |
| Diabetes, yes vs.
no | 1.15
(0.55-2.37) | 0.714 | 1.29
(0.62-2.69) | 0.492 |
| Hyperlipemia, yes
vs. no | 0.73
(0.32-1.66) | 0.45 | 0.72
(0.31-1.68) | 0.451 |
| Body mass index,
<24.0 vs. ≥24.0 | 0.88
(0.58-1.35) | 0.572 | 0.83
(0.54-1.27) | 0.39 |
| Stage of non-small
cell lung cancer, IV+III vs. II+I | 5.98
(3.23-11.07) | <0.001 | 5.90
(3.16-11.03) | <0.001 |
| Serum
carcinoembryonic antigen level, >8.28 ng/ml vs. ≤8.28 ng/ml | 1.09
(0.73-1.65) | 0.665 | 1.13
(0.75-1.70) | 0.565 |
| Serum C-reactive
protein level, >3.80 µmol/l vs. ≤3.80 µmol/l | 3.10
(2.01-4.78) | <0.001 | 2.90
(1.87-4.50) | <0.001 |
| Chemotherapy, AP
vs. others | 0.64
(0.39-1.04) | 0.07 | 0.67
(0.41-1.11) | 0.121 |
| Albumin level,
>40.95 g/l vs. ≤40.95 g/l | 0.45
(0.30-0.69) | <0.001 | 0.44
(0.29-0.68) | <0.001 |
| Neutrophils count,
>4.34x109/l vs. ≤4.34x109/l | 2.03
(1.34-3.08) | 0.001 | 2.07
(1.37-3.15) | 0.001 |
| Lymphocytes count,
>1.77x109/l vs. ≤1.77x109/l | 0.27
(0.17-0.43) | <0.001 | 0.28
(0.18-0.45) | <0.001 |
| Hemoglobin level,
>133 g/l vs. ≤133 g/l | 0.72
(0.47-1.08) | 0.114 | 0.54
(0.35-0.85) | 0.008 |
| Platelet count,
>216x109/l vs. ≤216x109/l | 1.66
(1.10-2.51) | 0.017 | 1.73
(1.14-2.62) | 0.011 |
| Prognostic
nutritional index score, >48.9 vs. ≤48.9 | 0.58
(0.38-0.88) | 0.01 | 0.54
(0.35-0.82) | 0.004 |
| Neutrophil
lymphocyte ratio, >2.52 vs. ≤2.52 | 3.62
(2.33-5.62) | <0.001 | 3.49
(2.25-5.44) | <0.001 |
| Complement C4
level, ≤408.56 vs. >408.56 µmol/l | 5.51
(3.43-8.84) | <0.001 | 5.52
(3.41-8.93) | <0.001 |
| Complement C3
level, ≤366.10 vs. >366.10 µmol/l | 3.96
(2.53-6.20) | <0.001 | 3.76
(2.38-5.92) | <0.001 |
| Karnofsky
Performance Status, ≥80 vs. <80 | 0.45
(0.26-0.79) | 0.005 | 0.45
(0.25-0.79) | 0.005 |
Subsequently, Besides C3b and C4d, albumin level,
sex, PNI score, neutrophil and platelet counts, NLR, NSCLC stage,
surgery, KPS score and smoking status were associated with
mortality (Table II). After
correction for age and sex, patients with low C3b and C4d also
exhibited a higher mortality incidence compared with those with
high C3b and C4d levels.
Complement C3b levels (HR, 2.23; 95% CI, 1.20-4.14;
P=0.012) and C4d levels (HR, 2.14; 95% CI, 1.14-4.00; P=0.017) were
also positively associated with the risk of mortality after
correction by Cox multifactorial regression analysis (Table III). In addition, surgery,
albumin level and PNI score were independent risk factors for OS in
patients with NSCLC.
 | Table IIIMultivariate analysis of the
different risk factors for overall survival. |
Table III
Multivariate analysis of the
different risk factors for overall survival.
| Variation | Hazard ratio (95%
CI) | P-value |
|---|
| Sex, male vs.
female | 1.23
(0.61-2.46) | 0.568 |
| Surgery, yes vs.
no | 0.33
(0.18-0.60) | <0.001 |
| Smoking, yes vs.
no | 1.22
(0.66-2.24) | 0.524 |
| Stage of non-small
cell lung cancer, IV+III vs. II+I | 1.34
(0.64-2.80) | 0.436 |
| Serum C-reactive
protein level, >3.80 µmol/l vs. ≤3.80 µmol/l | 1.67
(0.91-3.05) | 0.097 |
| Albumin level,
>40.95 µmol/l vs. ≤40.95 µmol/l | 0.48
(0.25-0.91) | 0.026 |
| Neutrophils count,
>4.34x109/l vs. ≤4.34x109/l | 0.92
(0.52-1.65) | 0.786 |
| Lymphocytes count,
>1.77x109/l vs. ≤1.77x109/l | 0.55
(0.30-1.01) | 0.052 |
| Platelet count,
>216x109/l vs. ≤216x109/l | 0.97
(0.61-1.54) | 0.905 |
| Prognostic
nutritional index score, >48.9 vs. ≤48.9 | 1.94
(1.03-3.67) | 0.042 |
| Neutrophil
lymphocyte ratio, >2.52 vs. ≤2.52 | 1.08
(0.53-2.21) | 0.823 |
| Complement C4
level, ≤408.56 vs. >408.56 µmol/l | 2.14
(1.14-4.00) | 0.017 |
| Complement C3
level, ≤366.10 vs. >366.10 µmol/l | 2.23
(1.20-4.14) | 0.012 |
| Karnofsky
Performance Status, ≥80 vs. <80 | 0.69
(0.36-1.32) | 0.266 |
Predictive model construction and
validation
Subsequently, the independent risk factors (factors
that were statistically significant after correction for
multi-factor COX regression analysis) calculated by the
multi-factor analysis were used to construct a prognostic model for
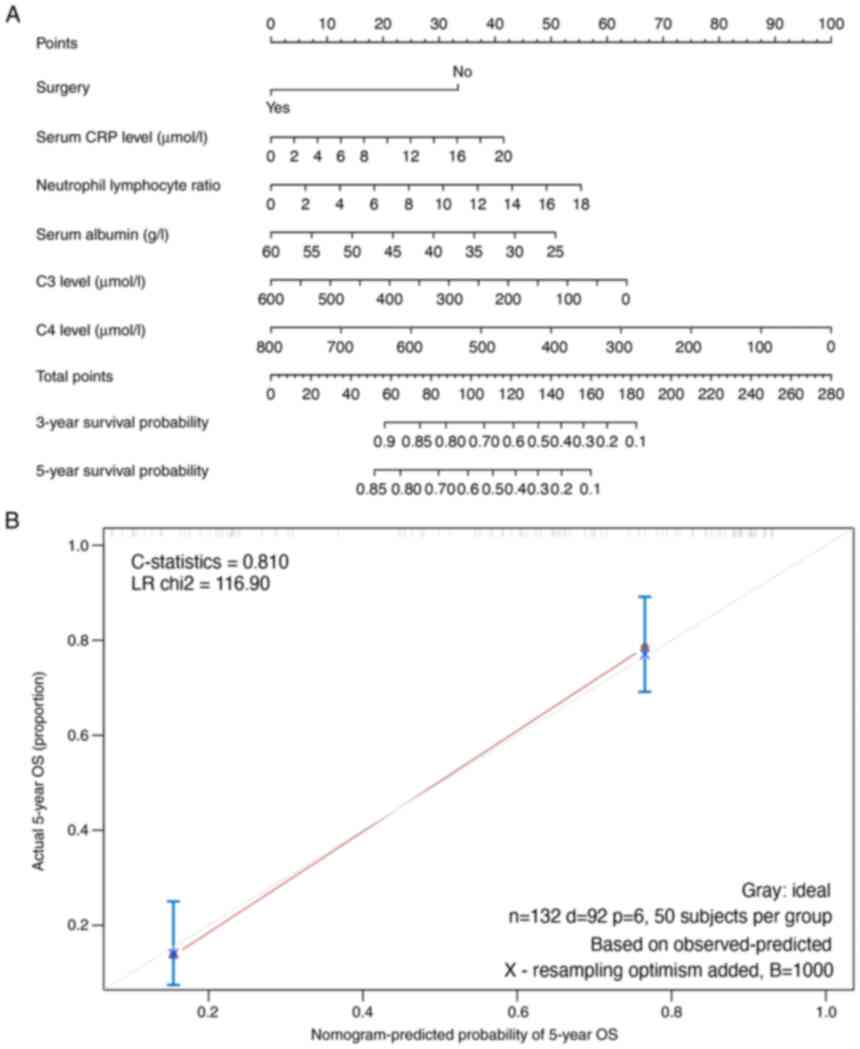
mortality risk from NSCLC, using a Nomogram (Fig. 3A). This predictive model was
validated internally using the bootstrap validation method. For
validation, nomogram had a C-statistics (effect sizes that reflect
prediction accuracy) of 0.810 for predicting mortality risk. In the
validation cohort of 50 patients, the nomogram had an estimated
C-statistics of 0.810 for OS, which also demonstrated a suitable
calibration curve in estimation in Fig. 3B. Overall, 50 patients were
collected from both hospitals as the external validation cohort for
the model. The validation revealed that the prediction accuracy of
the present constructed model was 77.0% (the number of correct
predictions divided by the total number of points) (Fig. S1). Lower complement levels of C4d
were revealed in patients with gastrointestinal lymph node
metastases compared with those in patients without metastases in
these 50 validated cases (362.1±117.3 vs. 584.5±136.7;
P<0.05).
Discussion
The present study examined the levels of complement
proteins, namely complement C3b and C4d, in patients with NSCLC
combined with gastrointestinal lymph node metastasis between
February 2010 and April 2019. Patients in the low-level C3b or C4d
expression group displayed a lower OS compared with patients in the
low-level group. Multivariate estimation revealed that C3b and C4d
levels were independent risk factors for overall mortality.
Subsequently, the independent risk factors (C3b, C4d, surgery,
albumin level and PNI score) calculated using this multivariate
analysis were incorporated into a predictive mortality risk model,
specifically as a nomogram. After internal validation, it was found
to be accurate in predicting the mortality possibility.
Tumor development is a complex biological process
that involves numerous genes (24). During this process, the immune
system serves an important role (25). Complement is a part of the immune
system that connects the adaptive and innate immune responses
(26). Previous studies have
revealed that the complement system is involved in the development
of lung and pancreatic cancer, as well as metastasis (27,28).
Osther et al (29)
previously indicated that the complement system may be activated
through the lectin pathway in patients with pancreatic cancer. It
has also been revealed that complement-converting enzymes C5 and
C3b are involved in lung cancer development, since these two
mediators may affect all three known routes of complement
activation pathways (30,31). CRP is an important biological
marker of inflammation, which in turn correlates with complement
activation (21). Therefore,
increased CRP levels are frequently accompanied with increased
complement activation and C4 levels (32).
Previous studies have demonstrated that a number of
complement components can be used as biomarkers for lung cancer
diagnosis and determination of prognosis (33,34).
Complement components have recently been regarded as biomarkers in
predicting mortality risk in NSCLC (35). Oner et al (36) demonstrated that C3b and C4d levels
are aberrantly expressed in patients with lung cancer, which were
then proposed to be biomarkers for long-term survival prediction in
these patients. As a component of the complement component, sC5b-9
has been used as a therapeutic target in various complement
activation-related diseases, such as thrombosis and viral
infections (37-39).
A number of studies have revealed that aberrant complement
activation accompanied by elevated sC5b-9 levels can be seen in
infection and inflammation (40,41).
However, to the best of our knowledge, few studies have examined
complement components C3b and C4d as potential indicators of
disease prognosis in cancer, especially NSCLC combined with lymph
node metastasis. Therefore, the present study attempted to test the
viability of complement C3b and C4d as potential biomarkers to
predict the long-term risk of mortality in patients with NSCLC
combined with lymph node metastasis.
The present study first examined the expression
levels of complement C3b and C4d in patients. Univariate analysis
first demonstrated that low levels of complement C3b and C4d were
strong predictors of cancer-associated mortality. In addition, sex,
serum CRP, albumin, neutrophil, platelet count, PNI, NLR, lung
cancer stage, surgery, smoking and KPS scores were associated with
mortality. Subsequent multivariate analysis indicated that C3b,
C4d, surgery, albumin and PNI were independent risk factors of
NSCLC.
Nomograms are intuitive methods for visualizing risk
prediction models (42,43). They have been widely used to
predict survival and tumorigenesis risk (44,45).
Recently, several studies have successfully developed risk
prediction models combining miRNA expression levels with different
clinical indicators of colon or esophageal cancer (46-48).
However, few studies have used complement levels combined with
other clinical risk factors of patients with lung cancer to build
prognostic models. The present study developed a risk model capable
of individualizing the long-term predictive risk of patients with
lung cancer based on C3b and C4d and a number of
clinicopathological characteristics. The model displayed accuracy
in assessing the mortality possibility in patients. Therefore, to
the best of our knowledge, this is the first prediction model that
considered clinicopathological variables parallel to complement
levels. Depending on this model, high-risk, low-survival patients
at high risk can be selected for specific therapies.
There are limitations with the present study. The
role of complement C3b and C4d needs to be validated in in
vitro experiments. Therefore, studies on the molecular
mechanisms of complement activation in patients with NSCLC combined
with lymph node metastasis should also be continued. The predictive
map also needs to be validated using a larger sample size. In
addition, a prospective study should be launched before predictive
models can be carried out.
In conclusion, the present study demonstrated that
complement C3b and C4d are independent risk factors for the
prediction of mortality in patients with NSCLC combined with
gastrointestinal lymph node metastasis. In addition, a nomogram
based on C3b and C4d levels was demonstrated to be accurate for
assessing overall mortality.
Supplementary Material
Prediction accuracy of the external
data for the new model. CPP, correct predicted percentage; PCA,
principal component analysis.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YW conducted the interpretation and analysis of
data. YL conceived the study. MX and HZ analyzed the data. YL and
YW confirm the authenticity of all the raw data. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
Consent was obtained from all patients or their
immediate family members. All research programs are in line with
the guidelines of the Ethics Committee of Soochow University and
follow the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000-14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ding L, Getz G, Wheeler DA, Mardis ER,
McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan
MB, et al: Somatic mutations affect key pathways in lung
adenocarcinoma. Nature. 455:1069–1075. 2008.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mulshine JL and D'Amico TA: Issues with
implementing a high-quality lung cancer screening program. CA
Cancer J Clin. 64:352–363. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bowry SK, Kircelli F, Himmele R and
Nigwekar SU: Blood-incompatibility in haemodialysis: Alleviating
inflammation and effects of coagulation. Clin Kidney J. 14 (Suppl
4):i59–i71. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ain D, Shaikh T, Manimala S and
Ghebrehiwet B: The role of complement in the tumor
microenvironment. Fac Rev. 10(80)2021.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Afshar-Kharghan V: The role of the
complement system in cancer. J Clin Invest. 127:780–789.
2017.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Sacks SH and Zhou W: The role of
complement in the early immune response to transplantation. Nat Rev
Immunol. 12:431–442. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Kochanek DM, Ghouse SM, Karbowniczek MM
and Markiewski MM: Complementing cancer metastasis. Front Immunol.
9(1629)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zirakzadeh AA, Sherif A, Rosenblatt R,
Bergman EA, Winerdal M, Yang D, Cederwall J, Jakobsson V,
Hyllienmark M, Winqvist O and Marits P: Tumour-associated B cells
in urothelial urinary bladder cancer. Scand J Immunol.
91(e12830)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cedzynski M and Swierzko AS: Components of
the lectin pathway of complement in solid tumour cancers. Cancers
(Basel). 14(1543)2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kemper C and Köhl J: Back to the
future-non-canonical functions of complement. Semin Immunol.
37:1–3. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kolev M, Le Friec G and Kemper C:
Complement-tapping into new sites and effector systems. Nat Rev
Immunol. 14:811–820. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Niculescu F, Rus HG, Retegan M and Vlaicu
R: Persistent complement activation on tumor cells in breast
cancer. Am J Pathol. 140:1039–1043. 1992.PubMed/NCBI
|
|
15
|
Razvi Y, Chan S, Zhang L, Tsao M, Barnes
E, Danjoux C, Sousa P, Zaki P, McKenzie E, DeAngelis C and Chow E:
A review of the rapid response radiotherapy program in patients
with advanced cancer referred for palliative radiotherapy over two
decades. Support Care Cancer. 27:2131–2134. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Barakat A, Mittal A, Ricketts D and Rogers
BA: Understanding survival analysis: Actuarial life tables and the
Kaplan-Meier plot. Br J Hosp Med (Lond). 80:642–646.
2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Schlosser P, Knaus J, Schmutz M, Dohner K,
Plass C, Bullinger L, Claus R, Binder H, Lubbert M and Schumacher
M: Netboost: Boosting-supported network analysis improves
high-dimensional omics prediction in acute myeloid leukemia and
huntington's disease. IEEE/ACM Trans Comput Biol Bioinform.
18:2635–2648. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kleinendorst RWD, Barzaghi G, Smith ML,
Zaugg JB and Krebs AR: Genome-wide quantification of transcription
factor binding at single-DNA-molecule resolution using
methyl-transferase footprinting. Nat Protoc. 16:5673–5706.
2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
LaFreniere LS, Newman MG and Graham JW:
Parental support and monitoring influences on adolescent alcohol
use: A peer selection mediation model. Ment Health Addict Res.
6(10)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Min SH and Zhou J: Smplot: An R package
for easy and elegant data visualization. Front Genet.
12(802894)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang H, Wu J, Xie K, Fang T, Chen C, Xie
H, Zhou L and Zheng S: Precise engineering of prodrug cocktails
into single polymeric nanoparticles for combination cancer therapy:
Extended and sequentially controllable drug release. ACS Appl Mater
Interfaces. 9:10567–10576. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Niu C, Wu D, Li AJ, Qin KH, Hu DA, Wang
EJ, Tucker AB, He F, Huang L, Wang H, et al: Identification of a
prognostic signature based on copy number variations (CNVs) and
CNV-modulated gene expression in acute myeloid leukemia. Am J
Transl Res. 13:13683–13696. 2021.PubMed/NCBI
|
|
23
|
Kandathil A, Kay FU, Butt YM, Wachsmann JW
and Subramaniam RM: Role of FDG PET/CT in the eighth edition of TNM
staging of non-small cell lung cancer. Radiographics. 38:2134–2149.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Pisani G and Baron B: Nuclear paraspeckles
function in mediating gene regulatory and apoptotic pathways.
Noncoding RNA Res. 4:128–134. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
de Matos AL, Franco LS and McFadden G:
Oncolytic viruses and the immune system: The dynamic duo. Mol Ther
Methods Clin Dev. 17:349–358. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Arthur CM, Chonat S, Fasano R, Yee MEM,
Josephson CD, Roback JD and Stowell SR: Examining the role of
complement in predicting, preventing, and treating hemolytic
transfusion reactions. Transfus Med Rev. 33:217–224.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Noh EM, Kim JM, Lee HY, Song HK, Joung SO,
Yang HJ, Kim MJ, Kim KS and Lee YR: Immuno-enhancement effects of
platycodon grandiflorum extracts in splenocytes and a
cyclophosphamide-induced immunosuppressed rat model. BMC Complement
Altern Med. 19(322)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Cserhalmi M, Papp A, Brandus B, Uzonyi B
and Józsi M: Regulation of regulators: Role of the complement
factor H-related proteins. Semin Immunol. 45(101341)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Osther K, Fornvik K, Liljedahl E, Salford
LG and Redebrandt HN: Upregulation of C1-inhibitor in pancreatic
cancer. Oncotarget. 10:5703–5712. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ramos AA, Castro-Carvalho B, Prata-Sena M,
Malhão F, Buttachon S, Dethoup T, Kijjoa A and Rocha E: Can
marine-derived fungus Neosartorya siamensis KUFA 0017
extract and its secondary metabolites enhance antitumor activity of
doxorubicin? An in vitro survey unveils interactions against lung
cancer cells. Environ Toxicol. 35:507–517. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Boonruang S, Prakobsri K, Pouyfung P,
Prasopthum A, Rongnoparut P and Sarapusit S: Structure-activity
relationship and in vitro inhibition of human cytochrome CYP2A6 and
CYP2A13 by flavonoids. Xenobiotica. 50:630–639. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Cedeno DL, Tilley DM, Vetri F, Platt DC
and Vallejo R: Proteomic and phosphoproteomic changes of
MAPK-related inflammatory response in an animal model of
neuropathic pain by differential target multiplexed SCS and
low-rate SCS. J Pain Res. 15:895–907. 2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yokoyama S, Hamada T, Higashi M, Matsuo K,
Maemura K, Kurahara H, Horinouchi M, Hiraki T, Sugimoto T, Akahane
T, et al: Predicted prognosis of pancreatic cancer patients by
machine learning. Clin Cancer Res. 26:2411–2421. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Muntel J, Gandhi T, Verbeke L, Bernhardt
OM, Treiber T, Bruderer R and Reiter L: Surpassing 10 000
identified and quantified proteins in a single run by optimizing
current LC-MS instrumentation and data analysis strategy. Mol
Omics. 15:348–360. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li J, Cao Z, Mi L, Xu Z and Wu X:
Complement sC5b-9 and CH50 increase the risk of cancer-related
mortality in patients with non-small cell lung cancer. J Cancer.
11:7157–7165. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Oner F, Savaş I and Numanoğlu N:
Immunoglobulins and complement components in patients with lung
cancer. Tuberk Toraks. 52:19–23. 2004.PubMed/NCBI
|
|
37
|
Ruggenenti P, Di Marco F, Cortinovis M,
Lorini L, Sala S, Novelli L, Raimondi F, Gastoldi S, Galbusera M,
Donadelli R, et al: Eculizumab in patients with severe coronavirus
disease 2019 (COVID-19) requiring continuous positive airway
pressure ventilator support: Retrospective cohort study. PLoS One.
16(e0261113)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Keshari RS, Silasi R, Popescu NI, Regmi G,
Chaaban H, Lambris JD, Lupu C, Mollnes TE and Lupu F: CD14
inhibition improves survival and attenuates thrombo-inflammation
and cardiopulmonary dysfunction in a baboon model of Escherichia
coli sepsis. J Thromb Haemost. 19:429–443. 2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Mezo B, Horvath O, Sinkovits G, Veszeli N,
Kriván G and Prohászka Z: Validation of early increase in
complement activation marker sC5b-9 as a predictive biomarker for
the development of thrombotic microangiopathy after stem cell
transplantation. Front Med (Lausanne). 7(569291)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Palikhe A, Sinisalo J, Seppanen M, Haario
H, Meri S, Valtonen V, Nieminen MS and Lokki ML: Serum complement
C3/C4 ratio, a novel marker for recurrent cardiovascular events. Am
J Cardiol. 99:890–895. 2007.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Iltumur K, Karabulut A, Toprak G and
Toprak N: Complement activation in acute coronary syndromes. APMIS.
113:167–174. 2005.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Iasonos A, Schrag D, Raj GV and Panageas
KS: How to build and interpret a nomogram for cancer prognosis. J
Clin Oncol. 26:1364–1370. 2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Balachandran VP, Gonen M, Smith JJ and
DeMatteo RP: Nomograms in oncology: More than meets the eye. Lancet
Oncol. 16:e173–e180. 2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Yang Y, Qu A, Zhao R, Hua M, Zhang X, Dong
Z, Zheng G, Pan H, Wang H, Yang X and Zhang Y: Genome-wide
identification of a novel miRNA-based signature to predict
recurrence in patients with gastric cancer. Mol Oncol.
12:2072–2084. 2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kawai K, Ishihara S, Yamaguchi H, Sunami
E, Kitayama J, Miyata H and Watanabe T: Nomogram prediction of
metachronous colorectal neoplasms in patients with colorectal
cancer. Ann Surg. 261:926–932. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Lv Y, Duanmu J, Fu X, Li T and Jiang Q:
Identifying a new microRNA signature as a prognostic biomarker in
colon cancer. PLoS One. 15(e0228575)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhang L, Chen J, Wang L, Chen L, Du Z, Zhu
L, Cui M, Zhang M and Song L: Linc-PINT acted as a tumor suppressor
by sponging miR-543 and miR-576-5p in esophageal cancer. J Cell
Biochem. 120:19345–19357. 2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Yang Y, Qu A, Wu Q, Zhang X, Wang L, Li C,
Dong Z, Du L and Wang C: Prognostic value of a hypoxia-related
microRNA signature in patients with colorectal cancer. Aging
(Albany NY). 12:35–52. 2020.PubMed/NCBI View Article : Google Scholar
|