Introduction
Ovarian cancer is the third most common
gynecological reproductive malignant tumor, after cervical and
uterine cancer. In 2019, ~22,530 new cases of ovarian cancer and
13,980 cancer-related deaths were reported in the United States,
accounting for ~5% of all tumor mortality rates (1). Ovarian cancer can be divided into
three categories according to the cells from which it originated:
Epithelial ovarian cancer (EOC), germ cell ovarian cancer and
interstitial ovarian cancer (2,3).
Most of the malignant types of ovarian cancer are classified as the
EOC subtype, accounting for >90% of cases (4). At present, the main treatment
strategy of ovarian cancer is surgery, in combination with
chemotherapy, radiotherapy and/or hormone therapy. Molecular
targeted therapy and biological targeted therapy have gradually
become novel and effective treatments. Understanding the molecular
triggers for EOC is essential to develop effective targeted
therapies for this deadly disease.
Inositol monophosphatase 2 (IMPA2) is located on
human chromosome 18 (18p11.21). IMPA2 encodes a phosphatase, with a
molecular weight of 32 kDa, which dephosphorylates inositol
monophosphate into inositol. IMPA2 is a rate-limiting catalyst for
phosphatidylinositol synthesis. In the central nervous system,
mutations in IMPA2 cause changes in the calcium homeostasis of
neurons and induce bipolar disorder (5-7).
Notably, the involvement of IMPA2 in cancer has been reported. For
example, Ulger et al (8)
analyzed differentially expressed genes in the human promyelocytic
leukemia HL-60 cell line and normal leukocytes using microarray
technology. Their study revealed that IMPA2 was highly expressed in
the HL-60 cell line. In addition, Zhang et al (9) revealed a novel function of IMPA2,
which was significantly upregulated in cervical cancer and acted as
a novel oncogene in cervical cancer through the MAPK signaling
pathway. Another study also determined that suppression of IMPA2
negatively enhanced mTORC1 activity by inhibiting the
phosphorylation of AKT/mTORC1 and inhibited autophagy in clear cell
renal cell cancer (10). However,
the role and the underlying mechanisms of IMPA2 in EOC are largely
unknown.
In the present study, the function role of IMPA2 in
EOC was investigated. IMPA2 knockdown not only reduced in
vitro cell proliferation, migration and invasion, but also
suppressed in vivo tumorigenesis of the EOC cell lines.
IMPA2 downregulation also resulted in dysregulation of various
signaling pathway and genes, including inhibition of AKT/mTOR
signaling and epithelial-mesenchymal transition (EMT). Therefore,
IMPA2 could function as an oncogene in EOC.
Materials and methods
IMPA2 transcript abundance analysis
based on The Cancer Genome Atlas (TCGA) database
IMPA2 mRNA expression levels in ovarian cancer were
analyzed using the Gemini website (http://gemini.cancer-pku.cn/). A total of 309 tumor
tissues and 97 normal tissues were included. The expression levels
of IMPA2 were also analyzed in multiple cancer tissues from the
UALCAN website (http://ualcan.path.uab.edu/analysis.html).
Cell culture
The human epithelial ovarian cancer (EOC) cell lines
SKOV3 (HTB-77), ES-2 (CRL-1978), OVCAR3 (HTB-161) and HEY
(CRL-3252) were purchased from American Type Culture Collection.
The SKOV3 and ES-2 cell lines were cultured in McCoy's 5A medium
(Thermo Fisher Scientific, Inc.). The OVCAR3 and HEY cell lines
were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.). All the medium was supplemented with 10% FBS and 1%
antibiotics (both Gibco; Thermo Fisher Scientific, Inc.). The cells
were cultured at 37˚C in a humidified incubator with 5%
CO2.
Lentivirus-mediated IMPA2 knockdown in
the EOC cell lines
The 2nd generation pGCSIL-GFP lentivirus system
(Shanghai Genechem Co., Ltd.) was used to knockdown the expression
levels of IMPA2 in the ES-2 and SKOV3 cell lines. Two targeted
sequences of IMPA2 were ligated into the vector. Lentivirus was
packaged by transfecting 10 µg of pGCSIL-GFP vectors, as well as 5
µg of pHelper1.0 and 5 µg of Helper2.0 vectors, into the 293T cell
line (Procell Life Science & Technology Co., Ltd.) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 72 h at 37˚C. The lentivirus was purified by
ultracentrifugation at 80,000 g for 2 h at 4˚C. The lentivirus was
used to infect the ES-2 and SKOV3 cell lines at a MOI of 1:20.
Immunofluorescence was used to determine infection efficacy and
selection was performed by flow sorting (Thermo Fisher Scientific,
Inc.). Reverse transcription-quantitative (RT-qPCR) and western
blot analyses were used to detect knockdown efficiency. The
sequences of short hairpin control (shCtrl), shIMPA2-1 and
shIMPA2-2 were 5'-TTCTCCGAACGTGTCACGT-3',
5'-GCCACAGTCATCATCAGAGAA-3' and 5'-GCTCATAGCTCAGGCCTTACA-3',
respectively.
Total RNA isolation and RT-qPCR
Total RNA was extracted from the EOC cell lines
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. RNA was reverse
transcribed at 42˚C for 60 min using the M-MLV kit (Promega
Corporation). cDNA was detected on a 7500 real-time system (Thermo
Fisher Scientific, Inc.), using SYBR Green mix (Takara
Biotechnology Co., Ltd.). The qPCR steps were as follows: step 1:
95˚C for 30 sec; and step 2: 95˚C for 5 sec, 60˚C for 34 sec,
number of cycles 40. The following primer sequences were used:
IMPA2 forward, 5'-GGCATCGTGATAGACACTTC-3' and reverse,
5'-CATCCCGCCCATAGTTAATC-3'; GAPDH forward,
5'-GTATGACAACAGCCTCAAGAT-3' and reverse,
5'-GTCCTTCCACGATACCAAAG-3'. The relative gene expression was
analyzed by using the 2-ΔΔCq method (11).
Western blot analysis
Total protein was extracted from the ES-2 and SKOV3
cell lines using RIPA lysis buffer (Beyotime Institute of
Biotechnology). The concentration of the protein was measured using
a BCA kit. Briefly, 30 µg of protein were separated on 10 or 12%
SDS-gels by SDS-PAGE, followed by transferring onto a PVDF
membrane. After blocking with 5% skimmed milk at 25˚C for 1 h, the
membrane was incubated with the primary antibodies (1:1,000) at 4˚C
overnight, followed by the secondary antibodies (1:5,000) at room
temperature for 2 h. The protein expression levels were
subsequently detected using an ECL Western Blotting Substrate kit
(Thermo Fisher Scientific, Inc.). The antibody against IMPA2 was
purchased from Sigma-Aldrich; Merck KGaA (cat. no. HPA029561). The
antibodies against Snail (cat. no. 3879), mTOR (cat. no. 2983),
phosphorylated (p)-AKT (cat. no. 4060) were purchased from Cell
Signaling Technology, Inc. The antibodies against Twist (cat. no.
ab50887), AKT (cat. no. ab8805) and p-mTOR (cat. no. ab109268) were
purchased from Abcam. The antibody against GAPDH (cat. no.
sc-32233) was purchased from Santa Cruz Biotechnology, Inc. The
secondary antibodies were purchased from Cell Signaling Technology,
Inc. (anti-mouse IgG HRP-linked, product no. 7076; anti-rabbit IgG
HRP-linked, product no. 7074). Western blotting results were
semi-quantified using ImageJ software (1.8.0.172; National
Institutes of Health).
MTT assay
An equal number (3,000) of ES-2 and SKOV3 cells were
seeded into 96-well plates. Cell viability was detected between
days 1 and 5. Briefly, the cells were washed with PBS three times.
The cells were then incubated with MTT solution (5 mg/ml) at 37˚C
for 3 h. The culture medium and MTT were removed and 100 µl
dimethyl sulfoxide was added to each well. The plates were shaken
for 5 min and the optical density was measured at 490 nm to
determine the viable cells.
Multiparametric high-content
screening
Multiparametric high-content screening was used to
determine cell proliferation. After transfection with the
lentivirus, the ES-2 and SKOV3 cells (2,000 cells/well) were seeded
into 96-well plates at equal concentration for 5 days. The cell
number was measured by automatically detecting the intensity and
distribution of fluorescence.
Colony formation assay
Equal numbers of EOC cells (1.0x103
cells/well) were seeded into 6-well plates. After colonies had
formed, the cells were washed with PBS three times, followed by
fixation with methanol at room temperature for 15 min and staining
with 2% Giemsa solution (Thermo Fisher Scientific, Inc.) at room
temperature for 10 min. Images of the colonies (≥50 cells) were
captured and calculated using an optical camera.
Apoptosis detection
Apoptosis was detected using Annexin V-FITC staining
(Thermo Fisher Scientific, Inc.). Briefly, a total of
106 ES-2 and SKOV3 cells were washed with PBS and
stained with 5 µl of Annexin V and 2 µl of PI for 15 min at 4˚C.
Apoptosis cells were detected using a FACScan flow cytometer (BD
Biosciences) and the data were analyzed by Cellquest Software
version 3.3 (BD Biosciences).
Transwell assay
Migration and invasion were determined using
Transwell and Matrigel assays with or without Matrigel,
respectively. For cell invasion, the upper chambers were precoated
with Matrigel (Corning, Inc.) at 4˚C overnight. The cells were
seeded (4.0x105 cells/well) onto the upper surface of
8.0-µm pore chambers. A total of 600 µl McCoy's 5A medium,
containing 30% FBS, was added to the lower chambers. The cells were
washed with PBS and fixed with 4% paraformaldehyde at room
temperature for 15 min, 24 h later. Following staining with 0.1%
crystal violet at room temperature for 20 min, the cells on the
lower surface of the chambers were washed with clean water and
images were captured under an optical microscope, and the
magnification was x100.
Xenograft tumorigenesis assay
The animal experiments were conducted in May 2021 at
the Affiliated Tumor Hospital Xinjiang University and were approved
by the Ethics Committee of the Affiliated Tumor Hospital Xinjiang
University (approval no. G-202117). A total of 10, female BALB/C
nude mice (4-weeks-old; weighing 18-22 g) were purchased from
Beijing Vital River Laboratory Animal Technology Co., Ltd. and
maintained at 18-23˚C with 40-60% humidity, under a 12-h light/dark
cycle, with free access to food and water. The mice were
subcutaneously injected with shCtrl and shIMPA2 ES-2 cells
(5x106 per mouse). The mice were euthanized using
CO2 (a flow rate of 50% chamber volume/min) on day 26,
as indicated by the AMVA Guidelines for the Euthanasia of Animals
(2020 Edition) (12). The tumor
volume and weight were measured. The volume of the tumors was
calculated using the following calculation: Volume=0.5 (length x
width2).
Statistical analysis
Statistical significance was analyzed using GraphPad
Prism 8.0 (GraphPad Software, Inc.) software. The experiments were
conducted for three independent repeats. The statistical results
are presented as the mean ± standard error of mean (SEM). The
differences between two groups were analyzed using an unpaired
Student's t-test. Statistical differences between ≥3 groups were
determined using one-way ANOVA followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
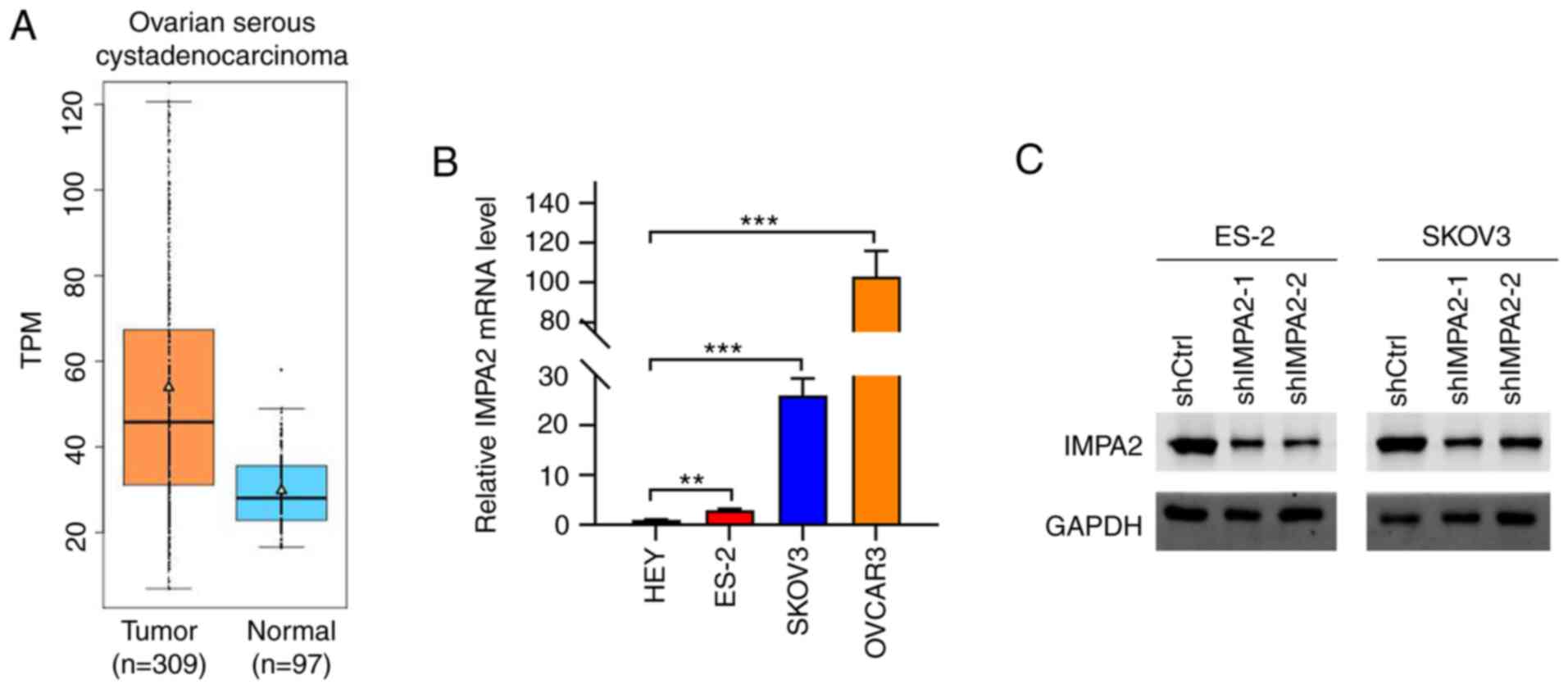
IMPA2 is upregulated in ovarian cancer
tissues
The expression level of IMPA2 was analyzed in
ovarian cancer tissues using the Gemini website. The results showed
that IMPA2 was significantly upregulated in ovarian cancer tissues
(Fig. 1A). The expression level of
IMPA2 was also analyzed in multiple cancer tissues from the UALCAN
website (http://ualcan.path.uab.edu/analysis.html). It was
determined that IMPA2 was upregulated in other cancers, such as
cervical squamous cell carcinoma and endocervical adenocarcinoma
(CESC) (Fig. S1). The mRNA
expression level of IMPA2 was then detected in different ovarian
cancer cell lines and the results showed that the ES-2, SKOV3 and
OVCAR3 cell lines had relatively higher mRNA expression levels of
IMPA2 compared with that in the HEY cell line (Fig. 1B).
Lentivirus-mediated IMPA2 knockdown in
EOC cells
To investigate the role of upregulated IMPA2 in EOC,
a lentivirus was constructed to knock down the expression level of
IMPA2 in the EOC cells. As revealed in Fig. 1B, the expression of IMPA2 was
markedly higher in OVCAR3 cells as compared with SKOV3 and ES-2
cells. It was difficult to knock down IMPA2 in OVCAR3 cells. Thus,
IMPA2 expression was knocked down in the SKOV3 and ES-2 cell lines.
Western blot analysis results indicated that IMPA2 expression
levels were efficiently downregulated in the SKOV3 and ES-2 cell
lines transfected with shIMPA2-1 and shIMPA2-2, respectively
(Fig. 1C).
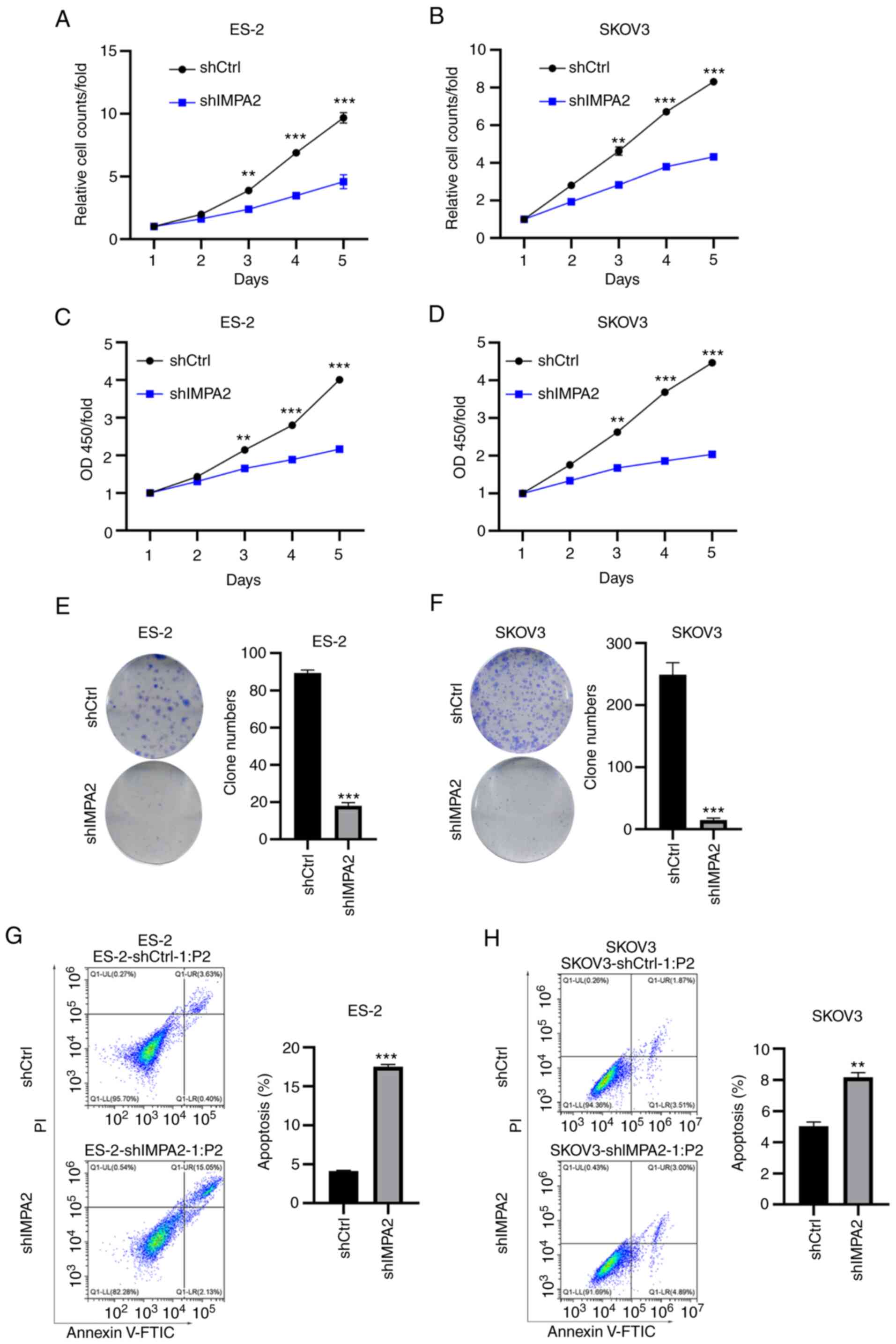
IMPA2 knockdown suppresses the
proliferation and colony formation in the EOC cells
Next, cell proliferation and colony formation assays
were performed in the SKOV3 and ES-2 cell lines, transfected with
shCtrl and shIMPA2. Multiparametric high-content screening was
performed to detect cell proliferation of the SKOV3 and ES-2 cell
lines for 5 days. It was determined that cell proliferation was
significantly inhibited following knockdown of IMPA2 expression in
the ES-2 cell lines (Fig. 2A).
Consistent results were found in the SKOV3 cell line following
knockdown of IMPA2 expression (Fig.
2B). To validate the results, an MTT assay was performed and
the results showed that knockdown of IMPA2 expression led to a
decrease in cell viability in the ES-2 and SKOV3 cell lines,
respectively (Fig. 2C and D). Likewise, colony formation abilities
of the ES-2 (Fig. 2E) and SKOV3
(Fig. 2F) cell lines were markedly
inhibited following transfection with shIMPA2. Next, Annexin V/PI
staining was performed to analyze apoptosis in the EOC cell lines
transfected with shCtrl and shIMPA2 using flow cytometry. The
results showed that knockdown of IMPA2 expression enhanced
apoptosis in both the ES-2 (Fig.
2G) and SKOV3 (Fig. 2H) cell
lines. Collectively, IMPA2 knockdown showed notable anticancer
effects in the EOC cell lines.
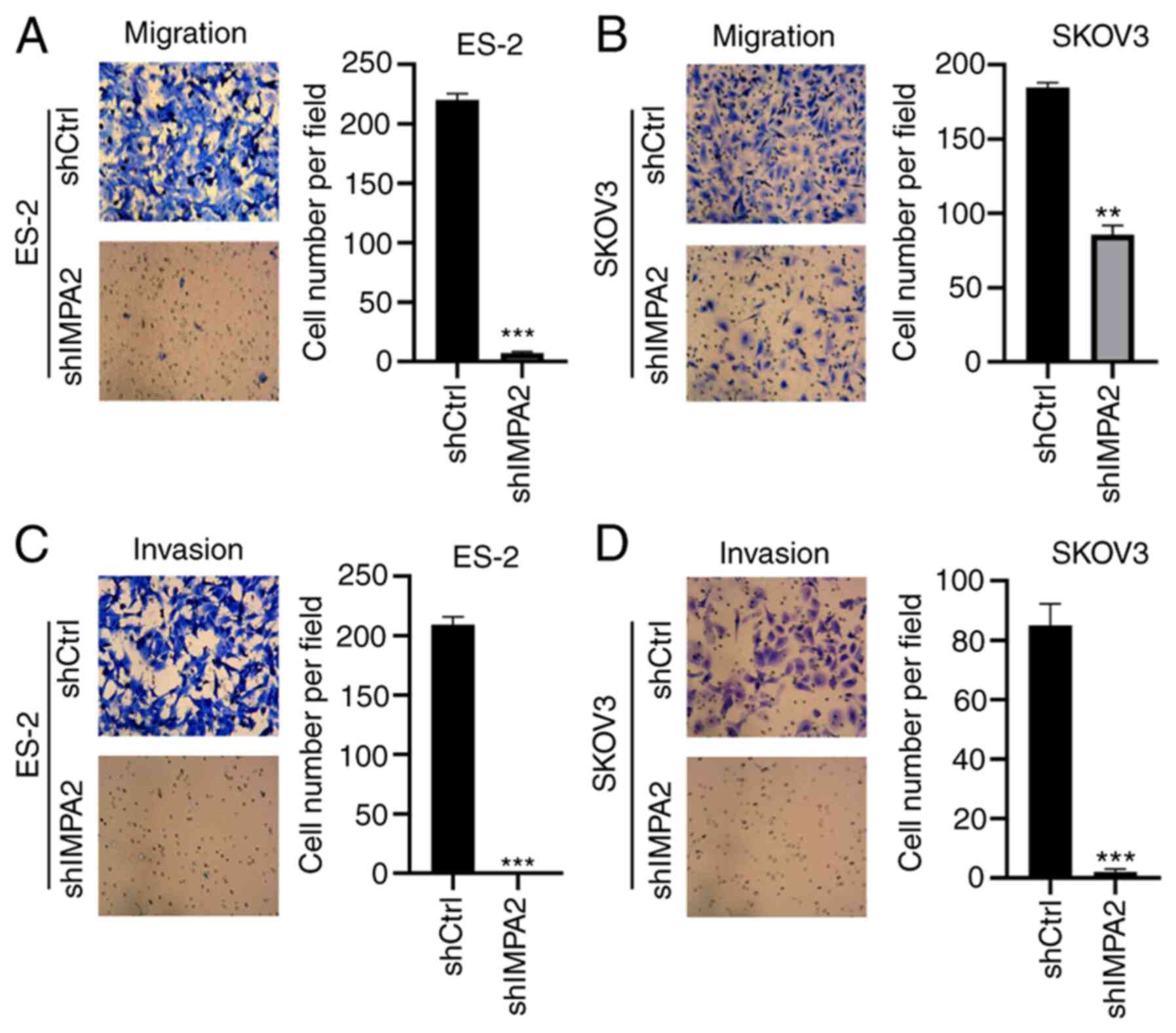
Knockdown of IMPA2 expression inhibits
migration and invasion in the EOC cell lines
To investigate the role of IMPA2 on migration and
invasion in the EOC cell lines, the EOC cells transfected with
shCtrl and shIMPA2 were subjected to Transwell invasion and
migration assays. Firstly, Transwell migration assays were
performed to analyze migration. Compared with that in the ES-2 cell
line transfected with shCtrl, the ES-2 cell line transfected with
shIMPA2 exhibited decreased migratory ability (Fig. 3A). Similar results were found in
the SKOV3 cell line following knockdown of IMPA2 expression
(Fig. 3B). Furthermore, Matrigel
assays were used to analyze invasion. Consistently, knockdown of
IMPA2 expression significantly suppressed the invasion of the ES-2
and SKOV3 cells (Fig. 3C and
D). Taken together, knockdown of
IMPA2 expression inhibited the migration and invasion of the EOC
cell lines.
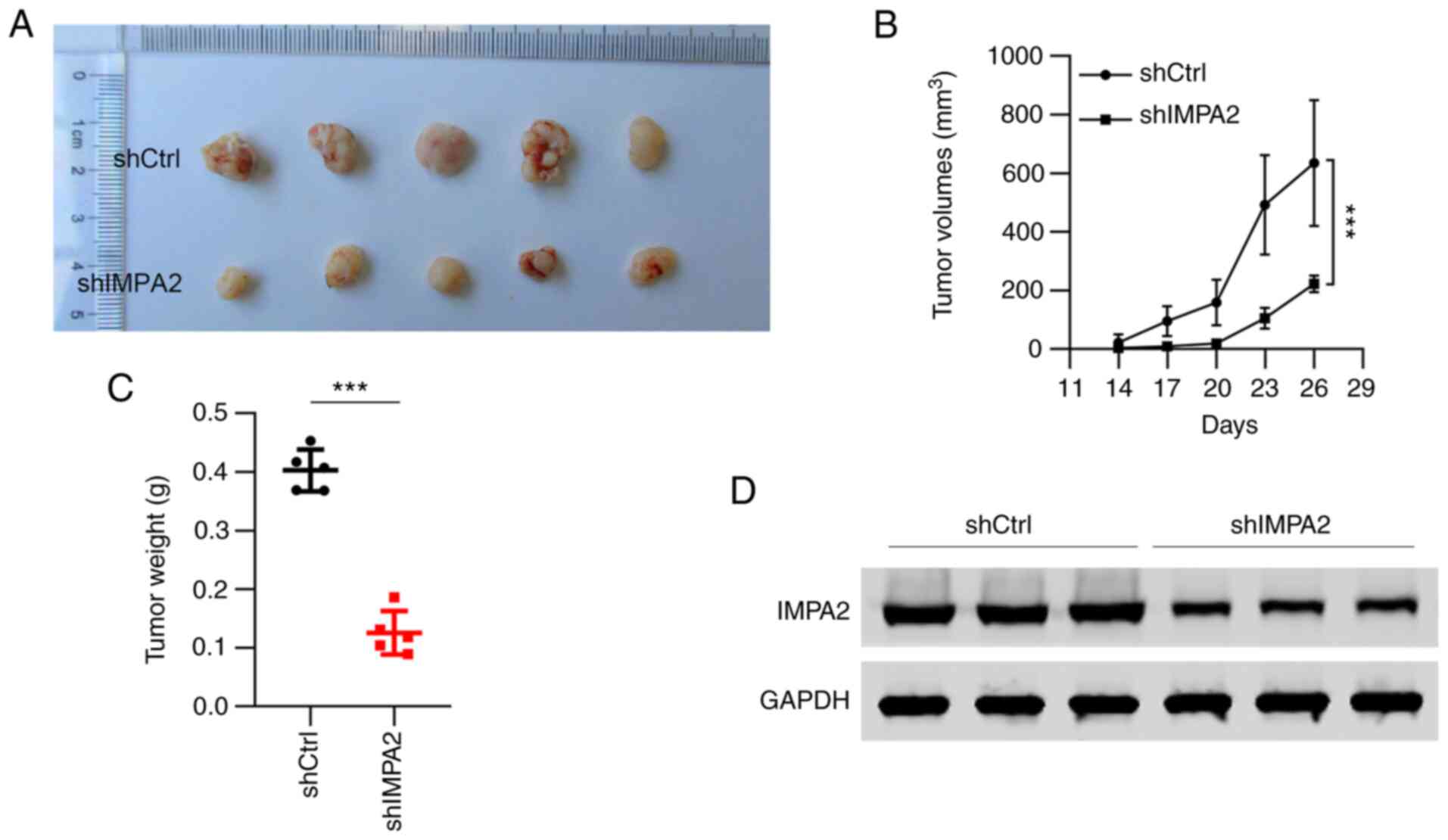
Knockdown of IMPA2 expression
suppresses xenograft tumorigenesis in the EOC cell lines
The aforementioned results have demonstrated the
in vitro role of IMPA2 knockdown in the EOC cell lines;
therefore, the in vivo effect was subsequently investigated.
An equal amount of ES-2 cells transfected with shCtrl and shIMPA2
were subcutaneously injected into female BALB/c nude mice. All of
the 5 mice injected with shCtrl ES-2 cells developed tumors,
whereas the mice injected with shIMPA2 cells formed small tumors
(Fig. 4A). The results of the
tumor volumes and tumor weight showed that IMPA2 knockdown
significantly reduced the tumorigenicity of the ES-2 cells in the
nude mice (Fig. 4B and C). In addition, the expression of IMPA2
in the tumors was detected, and a decreased expression of IMPA2 was
found in the tumors treated with shIMPA2 cells (Fig. 4D), which indicated that knockdown
of IMPA2 inhibited tumor growth in vivo.
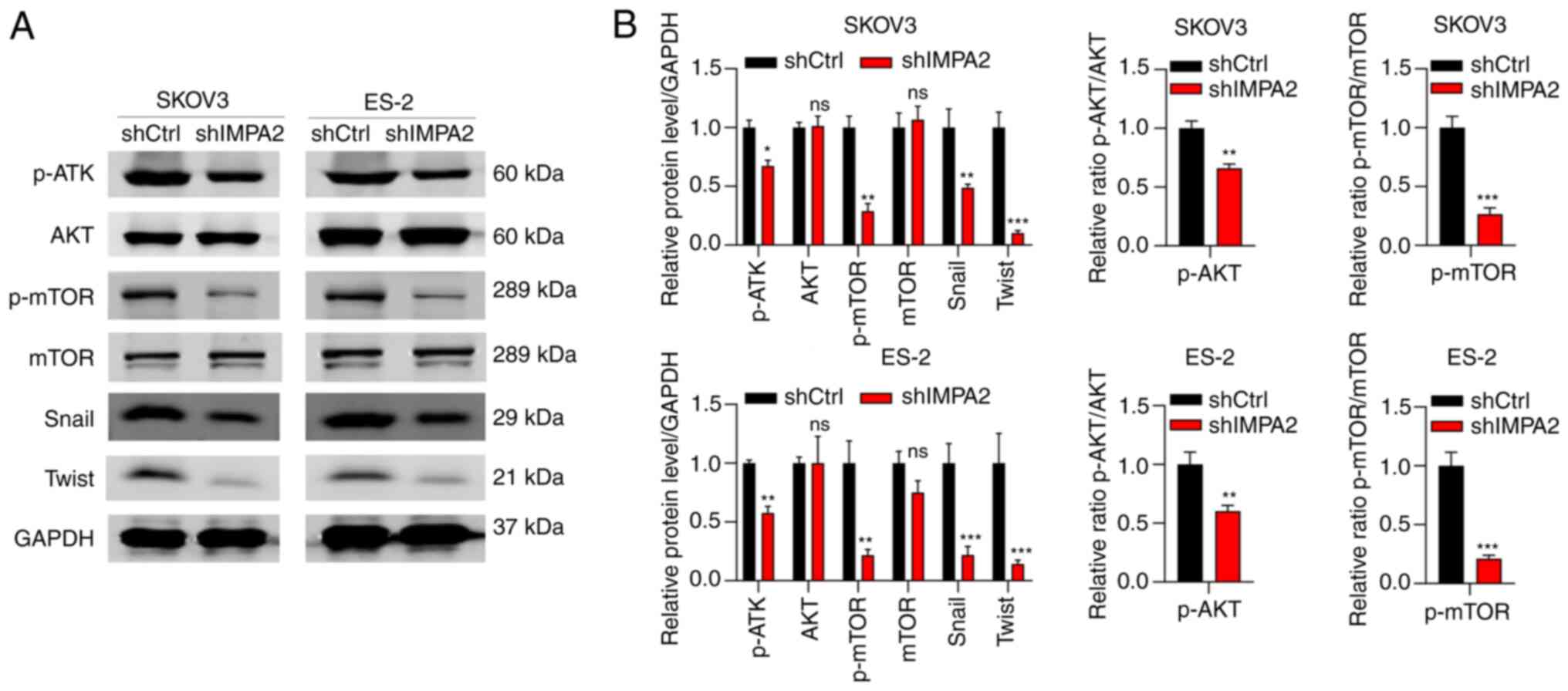
IMPA2 regulates AKT/mROR signaling
pathway and is involved in EMT
To determine the downstream effectors of IMPA2, the
SKOV3 cells transfected with shCtrl and shIMPA2 were analyzed using
western blot analysis. It was found that knockdown of IMPA2
expression reduced the phosphorylation of AKT and mTOR. No changes
in the protein expression level of total AKT and mTOR were found in
the cells with IMPA2 knockdown expression (Fig. 5). Subsequently, whether IMPA2
regulated the protein expression levels of invasion- and
EMT-related markers was analyzed. The results showed that IMPA2
knockdown resulted in decreased protein expression levels of Twist
and Snail. These results suggested that IMPA2 regulates the
AKT/mTOR oncogenic signaling pathway and is involved in EMT
process.
Discussion
As the major subtype of malignant ovarian cancer,
EOC threatens the health of women globally. Since effective
treatment options are very limited, the 5-year survival rate in
patients with EOC is poor. Novel biomarkers and targeted therapy
for EOC are constantly being developed. In the present study, it
was demonstrated that IMPA2 was required for the malignant
proliferation of the EOC cells in vitro and in vivo.
Firstly, the mRNA expression levels of IMPA2 were higher in ovarian
cancer tissues compared with in normal tissues based on TCGA data.
Secondly, knockdown of IMPA2 significantly suppressed the
proliferation, growth, migration and invasion of the EOC cell
lines. Then, IMPA2 knockdown also blocked the tumorigenesis of the
ES-2 cell lines in vivo. These primary results indicated
that IMPA2 was associated with tumor growth in the EOC cell
lines.
Inositol monophosphatase (IMPase) is a critical
enzyme that catalyzes the production of free myo-inositol by
dephosphorylating myo-inositol monophosphate. Expression of IMPase
is pivotal for cellular function as its product, myo-inositol, is
the primary substrate for phospholipids of the cell membrane
(13). The IMPase family is
comprised of two members in mammals, IMPA1 and IMPA2. Previous
studies have shown that genetic variants in IMPA2 are associated
with the risk of brain disorders. Ohnishi et al (14) found that single nucleotide
polymorphisms in the promoter of IMPA2 were associated with the
incidence of bipolar disorder in Japanese cohorts. However, the
authors further demonstrated that the transgenic mice with
upregulation of IMPA2 expression behaved normally and exhibited no
signs of manic changes (15).
Recently, the significance of IMPA2 has been investigated in
carcinogenesis. For example, IMPA2 expression was decreased in
clear cell renal cell cancer (ccRCC) tissues in a grade-dependent
manner. IMPA2 was also negatively regulated by microRNA-25 and its
reduction promoted the metastasis of ccRCC cells (16). By contrast, IMPA2 functions as an
oncogene in cervical cancer. Overexpression of IMPA2 in cervical
cancer tissues enhanced the malignant growth of the cancer cells
(17). The role of IMPA2 appears
controversial and may be dependent on the type of cancer. In the
present study, knockdown experiments on the IMPA2 gene were
performed in the EOC cell lines. It was found that knockdown of
IMPA2 expression reduced the proliferation and colony formation in
the ES-2 and SKOV3 cell lines. Apoptosis was enhanced following
knockdown of IMPA2 expression. Furthermore, the migratory and
invasive abilities of the ES-2 and SKOV3 cell lines were
significantly reduced following IMPA2 knockdown. Notably, IMPA2
interference also exhibited notable anticancer effects on tumor
growth in the EOC cells in nude mice. The results from the present
study suggested that IMPA2 was essential for the proliferation,
growth, migration and invasion of the EOC cell lines.
It is well-known that activation of the
PI3K/AKT/mTOR signaling pathway plays an important role in the
development of various types of cancer, including EOC (18). Inhibitors of this signaling pathway
are candidate drugs for management of this malignancy (19). Inhibition of PI3K/AKT also enhances
the sensitivity of ovarian cancer to cisplatin treatment (20). In addition, a previous study
reported that IMPA2 is involved in the phosphatidylinositol
signaling pathway (21), thus,
whether IMPA2 was also involved in the phosphatidylinositol
signaling pathway in EOC cancer was investigated. In the present
study, it was found that IMPA2 knockdown reduced the
phosphorylation and activity of AKT and mTOR. This suggests that
IMPA2 knockdown inhibited the growth of EOC cells, partly by
suppressing the AKT/mTOR pathway.
EMT is important for physiological and pathological
function in mammals. Dysregulation of EMT contributes to the
development of human diseases, such as carcinogenesis (22,23).
Since the functional results of the present study revealed that
IMPA2 knockdown inhibited the capacities of invasion and migration
in EOC cells, it was theorized that IMPA2 may be related with EMT.
Based on the results of the present study, EMT was significantly
regulated by IMPA2 knockdown, including downregulation of Twist and
Snail protein expression levels. Amongst these, Snail is an
essential transcription factor and associated with prognosis in
patients with EOC (24). The
results indicated that IMPA2 was involved in the EMT process.
In summary, it was demonstrated for the first time,
to the best of our knowledge, that IMPA2 functions as an oncogene
in EOC. IMPA2 knockdown in the EOC cell lines notably blocked the
proliferation, migration, invasion and tumorigenesis of the EOC
cells. The present study indicated that IMPA2 promoted EOC
development partly by regulating the AKT/mTOR pathway and the EMT
process.
Supplementary Material
The mRNA expression levels of IMPA2
were analyzed in multiple types of cancer. IMPA2, inositol
monophosphatase 2; TCGA, The Cancer Genome Atlas.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TA, GAbdurexit and GAbliz initiated and designed the
study. TA and GT conducted the experiments. YZ and GAbduxkur
conducted the data analysis. GAbdurexit and GAbliz wrote the
original manuscript. All the authors revised and reviewed the
manuscript. TA and GAbliz confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The animal experiments were approved (approval no.
G-202117) by the Ethics Committee of the Affiliated Tumor Hospital
Xinjiang University (Urumqi, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Henderson JT, Webber EM and Sawaya GF:
Screening for ovarian cancer: Updated evidence report and
systematic review for the US preventive services task force. JAMA.
319:595–606. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lheureux S, Braunstein M and Oza AM:
Epithelial ovarian cancer: Evolution of management in the era of
precision medicine. CA Cancer J Clin. 69:280–304. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Berney DM, Stoneham S, Arora R, Shamash J
and Lockley M: Ovarian germ cell tumour classification: Views from
the testis. Histopathology. 76:25–36. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian cancer statistics, 2018. CA Cancer J Clin. 68:284–296.
2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tomioka Y, Jimenez E, Salagre E, Arias B,
Mitjans M, Ruiz V, Sáiz P, García-Portilla MP, de la Fuente L,
Gomes-da-Costa SP, et al: Association between genetic variation in
the myo-inositol monophosphatase 2 (IMPA2) gene and age at onset of
bipolar disorder. J Affect Disord. 232:229–236. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jimenez E, Arias B, Mitjans M, Goikolea
JM, Roda E, Sáiz PA, García-Portilla MP, Burón P, Bobes J, Oquendo
MA, et al: Genetic variability at IMPA2, INPP1 and GSK3beta
increases the risk of suicidal behavior in bipolar patients. Eur
Neuropsychopharmacol. 23:1452–1462. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bloch PJ, Weller AE, Doyle GA, Ferraro TN,
Berrettini WH, Hodge R and Lohoff FW: Association analysis between
polymorphisms in the myo-inositol monophosphatase 2 (IMPA2) gene
and bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry.
34:1515–1519. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ulger C, Toruner GA, Alkan M, Mohammed M,
Damani S, Kang J, Galante A, Aviv H, Soteropoulos P, Tolias PP, et
al: Comprehensive genome-wide comparison of DNA and RNA level scan
using microarray technology for identification of candidate
cancer-related genes in the HL-60 cell line. Cancer Genet
Cytogenet. 147:28–35. 2003.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang K, Liu L, Wang M, Yang M, Li X, Xia
X, Tian J, Tan S and Luo L: A novel function of IMPA2, plays a
tumor-promoting role in cervical cancer. Cell Death Dis.
11(371)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kuei CH, Lin HY, Lee HH, Lin CH, Zheng JQ,
Chen KC and Lin YF: IMPA2 downregulation enhances mTORC1 activity
and restrains autophagy initiation in metastatic clear cell renal
cell carcinoma. J Clin Med. 9(956)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Underwood W and Anthony R: AVMA guidelines
for the euthanasia of animals: 2020 edition, 2020.
|
|
13
|
Agranoff BW and Fisher SK: Inositol,
lithium, and the brain. Psychopharmacol Bull. 35:5–18.
2001.PubMed/NCBI
|
|
14
|
Ohnishi T, Yamada K, Ohba H, Iwayama Y,
Toyota T, Hattori E, Inada T, Kunugi H, Tatsumi M, Ozaki N, et al:
A promoter haplotype of the inositol monophosphatase 2 gene (IMPA2)
at 18p11.2 confers a possible risk for bipolar disorder by
enhancing transcription. Neuropsychopharmacology. 32:1727–1737.
2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ohnishi T, Watanabe A, Ohba H, Iwayama Y,
Maekawa M and Yoshikawa T: Behavioral analyses of transgenic mice
harboring bipolar disorder candidate genes, IMPA1 and IMPA2.
Neurosci Res. 67:86–94. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lin YF, Chou JL, Chang JS, Chiu IJ, Chiu
HW and Lin YF: Dysregulation of the miR-25-IMPA2 axis promotes
metastatic progression in clear cell renal cell carcinoma.
EBioMedicine. 45:220–230. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bang S, Li J, Zhang M, Cui R, Wu X, Xin Z,
Ma D, Zhang J and Zhang H: The clinical relevance and function of
krüppel-Like factor 16 in breast cancer. Cancer Manag Res.
12:6373–6383. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bast RC Jr and Mills GB: Dissecting
‘PI3Kness’: The complexity of personalized therapy for ovarian
cancer. Cancer Discov. 2:16–18. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wu YH, Huang YF, Chen CC, Huang CY and
Chou CY: Comparing PI3K/Akt Inhibitors Used in Ovarian Cancer
Treatment. Front Pharmacol. 11(206)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Xing F, Sun C, Luo N, He Y, Chen M, Ding
S, Liu C, Feng L and Cheng Z: Wogonin increases cisplatin
sensitivity in ovarian cancer cells through inhibition of the
phosphatidylinositol 3-Kinase (PI3K)/Akt Pathway. Med Sci Monit.
25:6007–6014. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sjøholt G, Ebstein RP, Lie RT, Berle JØ,
Mallet J, Deleuze JF, Levinson DF, Laurent C, Mujahed M, Bannoura
I, et al: Examination of IMPA1 and IMPA2 genes in manic-depressive
patients: Association between IMPA2 promoter polymorphisms and
bipolar disorder. Mol. Psychiatry. 9:621–629. 2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Piera-Velazquez S and Jimenez SA:
Endothelial to mesenchymal transition: Role in physiology and in
the pathogenesis of human diseases. Physiol Rev. 99:1281–1324.
2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Dongre A and Weinberg RA: New insights
into the mechanisms of epithelial-mesenchymal transition and
implications for cancer. Nat Rev Mol Cell Biol. 20:69–84.
2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yoshida J, Horiuchi A, Kikuchi N, Hayashi
A, Osada R, Ohira S, Shiozawa T and Konishi I: Changes in the
expression of E-cadherin repressors, Snail, Slug, SIP1, and Twist,
in the development and progression of ovarian carcinoma: The
important role of Snail in ovarian tumorigenesis and progression.
Med Mol Morphol. 42:82–91. 2009.PubMed/NCBI View Article : Google Scholar
|