Introduction
Cancer stem cells (CSCs) are a minor population of
cells within tumors that show high self-renewal ability and have
the capacity for differentiation, resulting in cancer cell
heterogeneity and thus playing a key role in tumor development
(1). In a minimum definition, CSCs
exhibit stem cell-like self-renewal properties and high
differentiation ability (2,3).
Previous studies have shown that CSCs or cancer initiating cells
are significantly correlated with poor prognosis (4,5) and
resistance to tumor treatments including radiation and chemotherapy
(6,7). CSCs represent a small population that
are in a quiescent state; thus, they show resistance to some
chemotherapies that target the cell cycle and remain in tumors
after treatment. CSCs are thus considered to be the cells that
initiate recurrence (8). Research
has focused on CSC markers as novel targets for cancer
treatment.
Various molecules associated with clinical prognosis
or recurrence have been reported as CSC markers. CD44 has been well
established as a CSC marker in numerous tumor types (9). In our previous study, we showed that
induction of CD44, a CSC marker in head and neck squamous cell
carcinoma, caused resistance to apoptosis in response to DNA damage
(10). We also showed that
combination chemotherapy inhibited tumor recurrence in a head and
neck squamous cell carcinoma xenograft mouse model, possibly
through suppressing the expression of CD44(11).
Several studies have explored the mechanism of drug
resistance acquisition in tumors. Some reports indicated that drug
resistance is acquired after expression of drug excretion pumps,
including multidrug resistance mutation 1 (MDR1) and ATP-binding
cassette subfamily G member 2 (ABCG2) (12,13).
ABCG2 is significantly expressed and drug resistant inside
population cells, which exhibit characteristics of CSCs of head and
neck squamous cell carcinoma (14,15).
Clinically, ABCG2 positivity is significantly correlated with poor
prognosis in right-sided colon cancer (16). CSCs have been demonstrated to
express drug efflux transporters. Notably, MDR1 and ABCG2 are also
CSC markers in several types of tumors including head and neck
cancer (16,17).
Various CSC markers related to clinical prognosis
have been reported. However, the association between these CSC
markers and the characteristics of CSCs at the cellular level is
unknown. Whether the developed cancer cells that acquire
multidrug-resistance express CSC markers is unknown, and whether
the markers are associated with CSC characteristics is unclear.
Cisplatin is used as the first-line chemotherapy
treatment of head and neck squamous cell carcinoma in clinical
practice (18). In this study, we
investigated how CSC markers are related to the characteristics of
CSCs such as self-renewal ability, pluripotency, drug resistance,
and cell invasion ability in cisplatin-resistant HNSCC cells.
Materials and methods
Cell culture and generation of
cisplatin-resistant cells
The HNSCC cell line HSC-3 was obtained from Riken
Cell Bank (Ibaraki, Japan). Cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Life Technologies Japan Ltd.,
Yokohama, Japan) supplemented with 10% fetal bovine serum (FBS;
Life Technologies Japan Ltd.) and antibiotics (100 U/ml penicillin
and 100 µg/ml streptomycin 25 µg/ml) at 37˚C in a humidified
atmosphere containing 5% CO2.
To generate cisplatin-resistant cells, HSC-3 cells
were initially cultured in medium containing 1 µM cisplatin; the
cisplatin concentration was then slowly increased up to 100 µM. The
generated cell line was viable in medium containing cisplatin at
200 µM for over 2 days.
Cell viability assay
The viability of cells treated with various drugs
was measured using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide with a
Cell Proliferation Kit I (Roche Diagnostics, Mannheim, Germany)
following the manufacturer's instructions. The absorbance was
measured at 595 nm with a TECAN SpectraFluor plus XFluor4 software
(Tecan Japan Co., Ltd.). All data are presented as the means ±
standard deviations of at least three independent experiments. To
calculate the half maximal inhibitory concentration
(IC50), a cell proliferation curve was created from the
cell viability assay; the value that inhibited cell proliferation
by 50% was calculated. The IC50 value was determined
from the results from at least three experiments.
Immunofluorescence
Cells were cultured for 48 h in six-well covered
glass chamber slides. After two washes with phosphate-buffered
saline (PBS) containing 1% bovine serum albumin (Sigma-Aldrich),
cell surface Fc receptors were blocked with IgG (Santa Cruz
Biotechnology Inc.) on ice for 15 min. The cells were then stained
with a 1:100 dilution of a fluorescein isothiocyanate
(FITC)-conjugated anti-CD44 monoclonal antibody (BD Biosciences) or
an isotype-matched FITC-conjugated IgG control antibody (BD
Biosciences) for 30 min at 37˚C. After washing, the cells were
analyzed using an ECLIPSE Ti-U microscope equipped with an
Intensilight C-HGFI illumination system (Nikon Co., Ltd.). Digital
images were processed with NIS Elements D version 4.0 imaging
software (Nikon Co., Ltd.) and Adobe Photoshop elements 10 version
10.0 (Adobe Systems).
Efflux pump assay
Cells (1x105 cells) were plated in
96-well tissue culture plates, wall black/clear bottom
(Sigma-Aldrich) and cultured overnight. After cells achieved 70%
confluency, the medium was aspirated and cells were washed twice
with PBS(+) with glucose buffer: PBS with
CaCl2.·2H2O (0.9 mM),
MgCl2·12H20 (0.33 mM) and glucose (10 mM).
Next, 100 µl PBS(+) with glucose buffer was added per well and
cells were incubated for 1 h at 37˚C. The buffer was aspirated and
replaced with 100 µl Calcein-AM (4 mM solution; PromoKine,
Heidelberg, Germany) in DMEM and the plate was incubated for 1 h at
37˚C. After aspiration of the medium, cells were washed with
ice-cold PBS(+) and lysed in 100 µl 1% SDS/PBS for 10 min in the
dark at room temperature. Calcein incorporated into cells,
indicated by intense yellow-green fluorescence, was assessed using
TECAN SpectraFluor plus XFluor4 software (Tecan Japan Co., Ltd.,
Kawasaki, Japan). All data are presented as the means ± standard
deviations of at least three independent experiments.
Immunoblot analysis
Whole-cell extracts were obtained with a lysis
buffer (10x Cell Lysis buffer; Cell Signaling Technology, Beverly,
MA, USA) supplemented with 1 mM PMSF and one tablet of protease
inhibitor cocktail (Complete, EDTA-free; Roche Diagnostics GmbH).
Protein concentrations were assayed, and equal amounts of protein
were subjected to 8% SDS-polyacrylamide gel electrophoresis,
followed by immunoblotting with anti-CD44 mouse monoclonal
antibody, anti-cleaved PARP rabbit monoclonal antibody, anti-SOX9
rabbit monoclonal antibody, anti-E-cadherin mouse monoclonal
antibody, anti-Vimentin rabbit monoclonal antibody, anti-FGF9
rabbit monoclonal antibody (all from Cell Signaling Technology),
anti-breast cancer resistance protein (ABCG2) rabbit polyclonal
antibody and anti-β-actin antibody (Sigma-Aldrich). Membranes were
then incubated with corresponding peroxidase-conjugated secondary
antibodies (anti-rabbit IgG antibody or anti-mouse IgG antibody;
Cell Signaling Technology), and the positive bands were visualized
by chemoluminescence (Clarity™ Western ECL substrate; Bio-Rad). The
images were developed with an ImageQuant™ LAS500 Imaging System (GE
Healthcare Bio-Sciences AB). For semi-quantitative analysis of
protein expression, the protein bands were quantified by
densitometry using Image J (National Institutes of Health). The
expression of the target protein relative to the expression of
β-actin was calculated.
Flow cytometry analysis
Cells were harvested by trypsinization, washed with
PBS (-), and centrifuged; cell pellets were resuspended in FACS
buffer (PBS containing 0.5% bovine serum albumin). The cells were
stained for 30 min at 4˚C with a FITC-conjugated anti-human CD44
antibody (BD Biosciences, San Jose, CA, USA) or an isotype-matched
FITC-conjugated IgG control antibody (BD Biosciences). Data
acquisition and analysis were performed using an EC800 Flow
Cytometry Analyzer with EC800 analysis software (Sony Biotechnology
Inc.). All data are presented as the means ± standard deviations of
at least three independent experiments.
Real-time polymerase chain
reaction
Total RNA was purified using the RNeasy Mini kit
(Qiagen), and 600 ng of total RNA was used for reverse
transcription with an iScript™ Advanced cDNA Synthesis Kit
(Bio-Rad). The real-time polymerase chain reaction contained 1 µl
of cDNA sample diluted 20x, 1 µl each of forward and reverse
primers (final, 500 nM), 7 µl of nuclease-free water, and 10 µl of
SsoAdvanced SYBR Green Supermix (Bio-Rad). The Bio-Rad PrimePCR
assay system was used with the following primers: PrimePCR™SYBR
Green Assay: ABCG2, human; or PrimePCR™SYBR Green Assay: CD44,
human. As a control, we used PrimePCR™SYBR Green Assay: GAPDH,
human. The reaction cycles were an initial 5 min at 95˚C, followed
by 45 cycles of 95˚C for 10 sec and 72˚C for 10 sec. The reactions
and absolute quantification analyses were performed with a Thermal
Cycler Dice Real Time System TP800 (Takara Bio).
Small interfering RNA (siRNA)
transfection
siRNAs specifically targeting human ABCG2 (siRNAs #1
and #2, IDs s18056 and s18057), CD44 (siRNA #1 and #2, IDs s2681
and s2682), and SOX9 (siRNAs #1 and #2, IDs s13306 and s532658)
were obtained from Life Technologies (Carlsbad, CA, USA). Cells
were plated in six-well plates (1x105 per well) and
transfected with siRNAs for 3 days in antibiotic-free media using
siRNA Lipofectamine RNAiMAX and OPTI-MEM I reduced serum medium
(Invitrogen, Carlsbad, CA, USA), in accordance with the
Lipofectamine protocol. After 72 h of transfection, the cells were
used for experiments. siRNA transfection was confirmed by immuno
blotting (Fig. S1). The efficacy
of knockdown by siRNAs was determined by calculating the inhibition
of protein expression with the following results: siSOX9#1 90.1%,
siSOX9#2 93%, siABCG2#1 12%, siABCG2#2 61%, siCD44#1 81%, and
siCD44#2 83% (Fig. S1).
Microarray analysis
Total RNA was isolated from HSC-3 and R HSC-3 cell
lines using the RNeasy Mini kit (Qiagen) following the
manufacturer's instruction. The cRNA was amplified, labeled using
an Agilent low-Input QuickAmp labeling Kit, One-color (Agilent
Technologies) and then hybridized to a 60K Agilent 60-mer
oligomicroarray (SurePrint G3 Human GE microarray 8X60K Ver3.0;
Agilent Technologies) following the manufacturer's instructions.
The hybridized microarray slides were scanned using an Agilent
scanner. Relative hybridization intensities and background
hybridization values were calculated using Agilent Feature
Extraction Software (9.5.1.1). The raw signal intensities and flags
for each probe were calculated from hybridization intensities
(gProcessedSignal) and spot information (gIsSaturated) following
the procedures recommended by Agilent (Flag criteria on GeneSpring
Software; absent (A): ‘Feature is not positive and significant’ and
‘Feature is not above background’, marginal (M): ‘Feature is not
Uniform’, ‘Feature is Saturated’, and ‘Feature is a population
outlier’, and Present (P): others). The raw signal intensities of
all samples were normalized by quantile algorithm with
‘preprocessCore’ library package (19) on Bioconductor software (20). To identify up- and down-regulated
genes, we calculated Z-scores (21) and ratios (non-log scaled
fold-change) from the normalized signal intensities of each probe
for comparison between control and experiment samples. We
established the following criteria to identify up- and
down-regulated genes: up-regulated genes, Z-score ≥2.0 and ratio
≥1.5-fold; and down-regulated genes, Z-score ≤-2.0 and ratio
≤0.66.
Cell invasion assay
Cell invasion analysis was performed using BD Falcon
culture inserts (Becton Dickinson and Company, Franklin Lakes, NJ,
USA), which contain 8-µm pores in polyethylene terephthalate
membranes. Briefly, 1x105 cells were resuspended in
serum-free medium and added to the upper Transwell chamber in a
12-well plate. The lower chamber was filled with serum-free DMEM or
DMEM containing 10% fetal bovine serum. After 24 h, cells remaining
on the upper surface of the membrane were removed with a cell
scraper. Invaded cells to the membrane were fixed with cold 6.0%
(v/v) glutaraldehyde for 30 min and stained with 0.5% (w/v) crystal
violet. Cells were counted in 10 high-power microscope fields. All
data are presented as the means ± standard deviations of at least
three independent experiments.
Statistical analysis
All quantitative data are presented as the mean ±
SD. Data from multiple groups were evaluated using one-way analysis
of variance followed by Tukey-Kramer test in Fig. 1D, 3A, 3B
and 4B. A paired student's T test
was performed to compare two groups in Fig. 1B, 1C, 2A,
2B and 2D. Values of P<0.05 were considered
statistically significant. All statistical analyses were performed
using Statistics Analysis software for Mac Ver. 3.0 (Esumi Co.
Ltd., Tokyo, Japan).
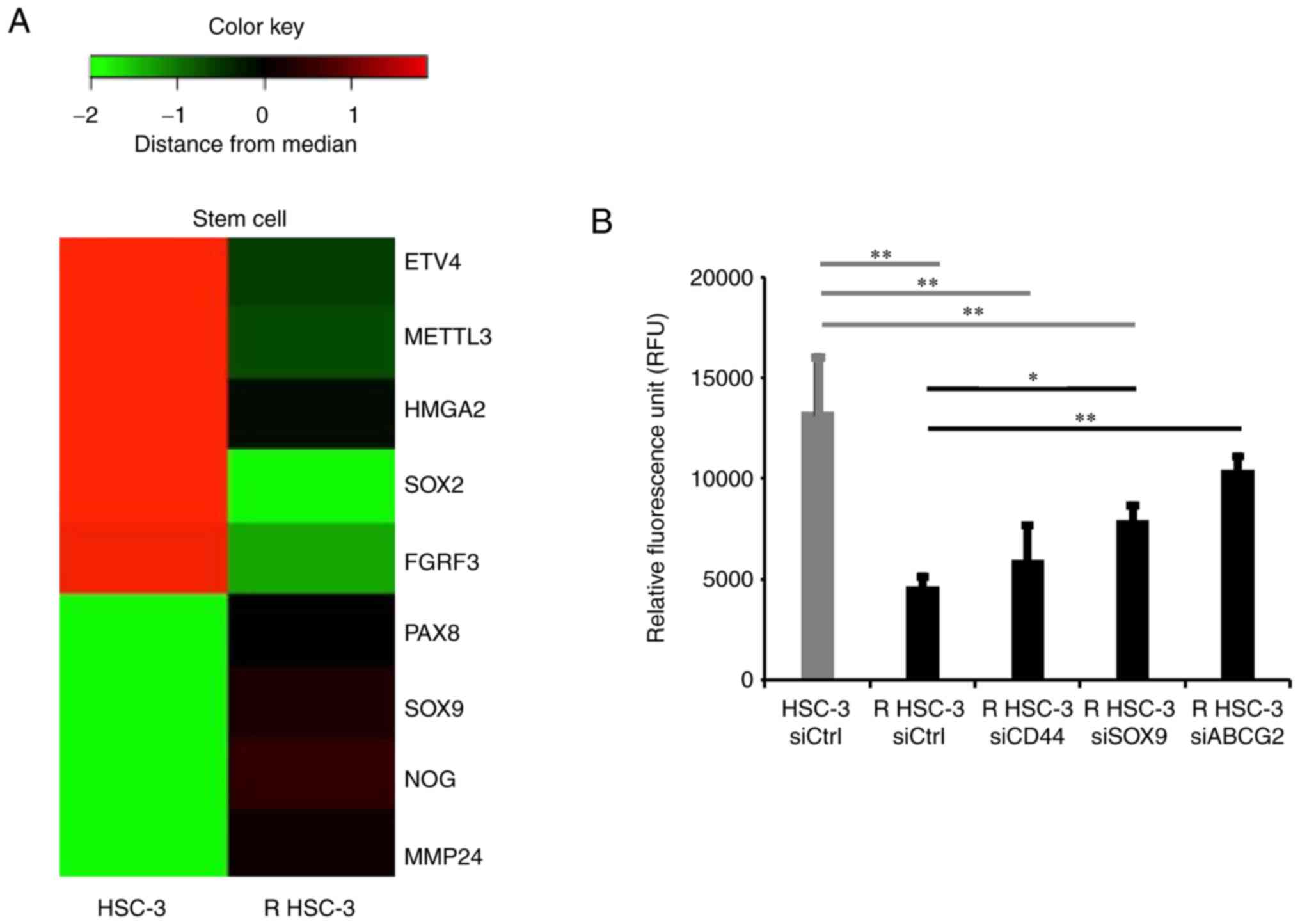 | Figure 4Evaluation of stem cell-related genes
by microarray analysis and improvement of drug excretion ability in
acquired multidrug-resistant head and neck squamous cell carcinoma
cells. (A) Stem cell-related gene expression in HSC-3 and R HSC-3
cells shown on a heat map. (B) Efflux pump assay in cells with
CD44, SOX9 and ABCG2 knockdown. *P<0.05 and
**P<0.01. R HSC-3, drug-resistant HSC-3; ABCG2,
ATP-binding cassette subfamily G member 2; SOX9, SRY-box
transcription factor 9; si, short interfering; Ctrl, control; ETV4,
ets variant transcription factor 4; METTL3, methyltransferase like
3; HMGA2, high mobility group AT-hook 2; SOX2, SRY-box
transcription factor 2; FGFR3, fibroblast growth factor receptor 3;
PAX8, paired box 8; NOG, noggin; MMP24, matrix metallopeptidase
24. |
Results
Cisplatin-resistant head and neck
squamous cell carcinoma cells acquire multidrug resistance
In this study, we used the human head and neck
squamous cell carcinoma cell line HSC-3. To establish
cisplatin-resistant cells, we cultured cells in the presence of a
low concentration (1 µM) of cisplatin and continued the cell
culture with gradually increasing concentrations of cisplatin.
Finally, we acquired a cell group that maintained survival even
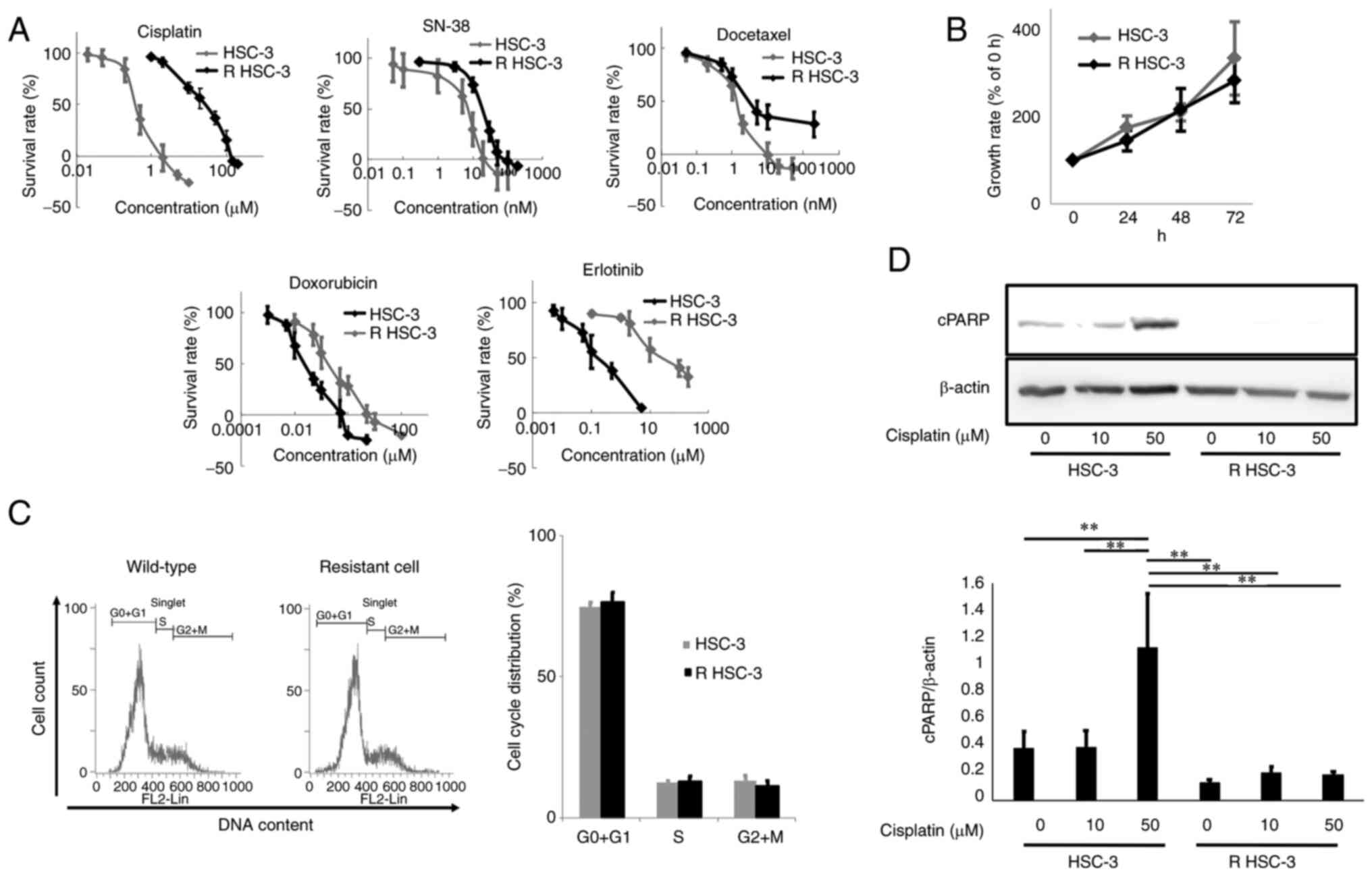
with cisplatin 100 µM. The half maximal inhibitory concentration
(IC50) of cisplatin in the parental HSC-3 cells was 0.7±0.2 µM,
whereas the IC50 of cisplatin-resistant HSC-3 cells was 20.3±6.3 µM
(Fig. 1A and Table I). Furthermore, the
cisplatin-resistant HSC-3 cells were also resistant to other drugs
with different mechanisms of action (Table I). Therefore, we named this
multidrug-resistant cell line as resistant HSC-3 cells (R
HSC-3).
 | Table IHalf maximal inhibitory concentration
(IC50) of chemotherapy reagents in HSC-3 and R HSC-3 cells. |
Table I
Half maximal inhibitory concentration
(IC50) of chemotherapy reagents in HSC-3 and R HSC-3 cells.
| Treatment | Unit | IC50 | HSC-3 R HSC-3 |
|---|
| Cisplatin | mM | 0.7±0.2 | 20.3±6.3 |
| SN-38 | nM | 7.1±1.7 | 22.2±3.7 |
| Docetaxel | nM | 0.9±0.2 | 6.6±1.2 |
| Erlotinib | mM | 0.2±0.04 | 31.9±2.0 |
| Doxorubicin | nM | 26±6 | 223±16 |
We evaluated the cell proliferation rate of R HSC-3
cells and found no significant difference in cell proliferation
compared with HSC-3 cells (Fig.
1B). Most anti-cancer agents that target the cell cycle exert
their effects in cancer cells with abnormal proliferation; these
agents do not work for CSCs. CSCs remain in G0 phase (quiescent
phase) (22,23) and therefore targeting the quiescent
CSCs remains challenging (24).
We also performed cell cycle analysis and found no
significant difference in the cell cycle distribution of the
drug-resistant and parental cells (Fig. 1C). Notably, R HSC-3 cells treated
with cisplatin showed reduced expression of cleaved PARP, which is
a marker of apoptosis, compared with the parental HSC-3 cells
(Fig. 1D). These data indicated
that the R HSC-3 cells showed no significant difference in cell
proliferation and cell cycle distribution compared with the
parental strain but failed to undergo apoptosis in response to
cisplatin.
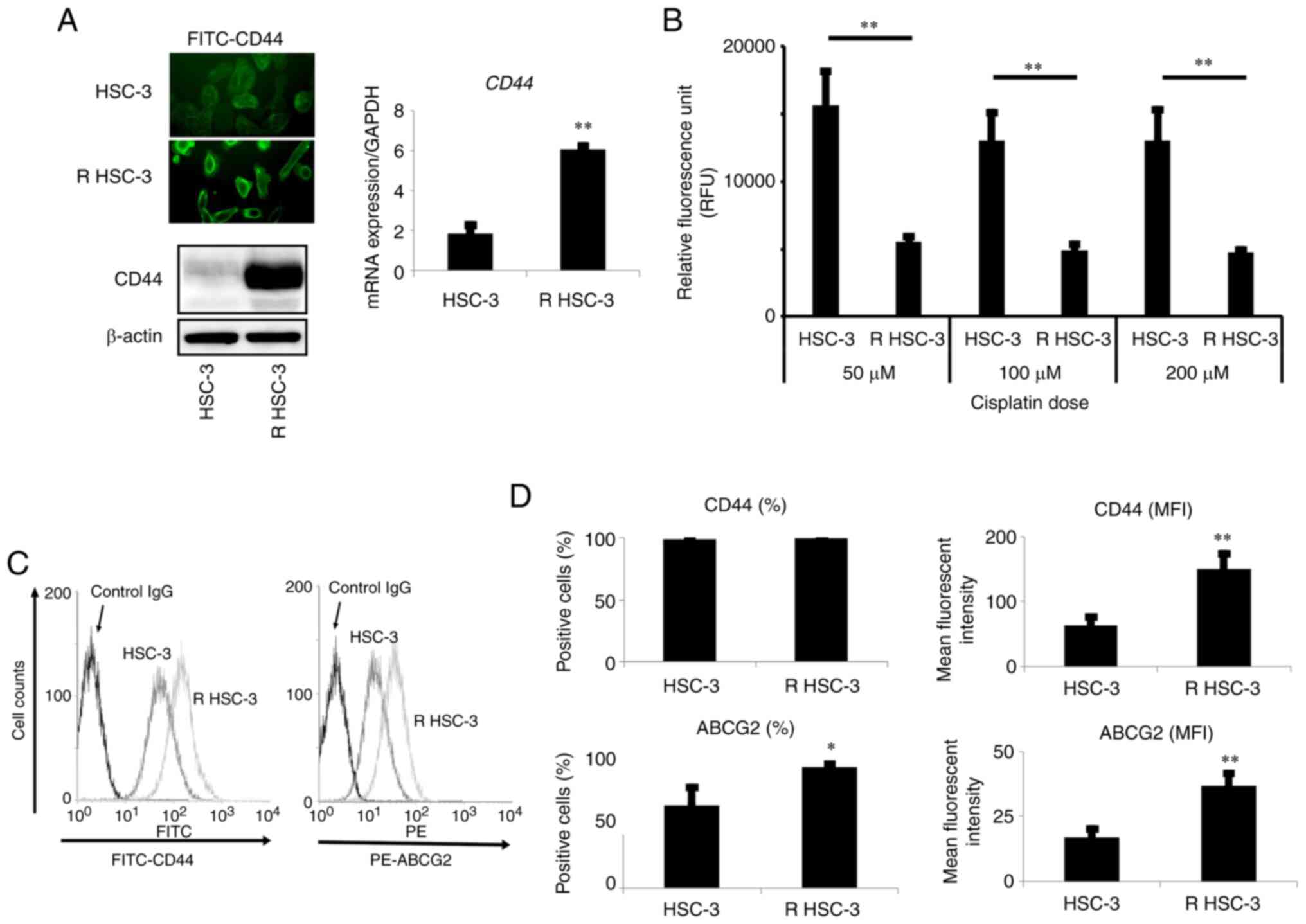
Increased drug excretion in the
multidrug-resistant head and neck squamous cell carcinoma cell
line
The expression of drug transporters is closely
associated with drug resistance of cancer cells (25), and previous studies indicated that
one of the mechanisms for acquiring drug resistance in cancer cells
is the aberrant activity of drug efflux pumps in cancer cells
(26). Therefore, we next examined
the drug efflux function of R HSC-3 cells using calcein-AM efflux
assays. In this assay, cells are cultured with cisplatin in the
presence of calcein-AM; the uptake of calcein-AM, monitored by
fluorescence imaging, reflects the amount of cisplatin in cells. In
R HSC-3 cells cultured with cisplatin concentrations ranging from
50 to 200 µM, the intracellular fluorescence did not increase,
indicating that the drug excretion ability of R HSC-3 cells was
significantly higher (P<0.01) than that of parental cells
(Fig. 2B). We also found that the
ABCG2 transporter was expressed at higher levels in the resistant
cells compared with the parental cells (Fig. 2C-D).
We evaluated the expression of CD44, a marker of
cancer stem cells in cancer including head and neck cancer, and
found that the mRNA and protein levels of CD44 were increased in R
HSC-3 cells compared with levels in parental HSC-3 cells (Fig. 2A). Flow cytometric analysis showed
that the number of CD44-positive cells in HSC-3 cells was 98%, but
that in R HSC-3 was not significantly different from 100%. However,
while the mean fluorescence intensity (MFI) was 63.1 in HSC-3
cells, the MRI in R HSC-3 was significantly increased to 150.8
(P<0.01) (Fig. 2C and D).
Together, these results showed that head and neck
squamous cell carcinoma cells with acquired multidrug resistance
exhibited increased drug efflux function and elevated expression of
the ABCG2 transporter compared with the parental cells. The
drug-resistant cells also exhibited increased expression of
CD44.
Multidrug-resistant head and neck
squamous cell carcinoma cells exhibit characteristics of CSCs
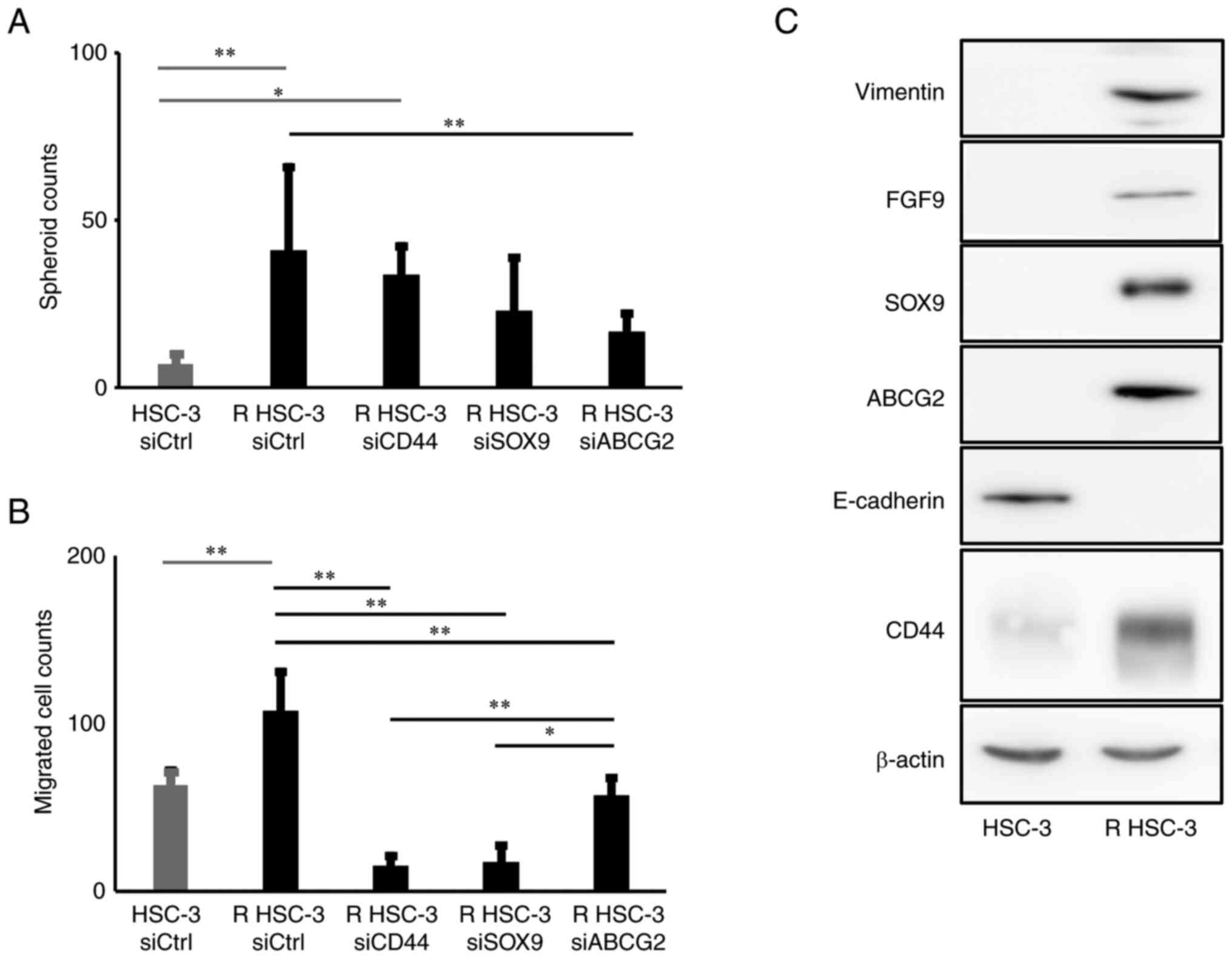
Next, we analyzed the characteristics of R HSC-3
cells. To evaluate the self-renewal ability of cells, we performed
spheroid formation assays and found that the formation of spheroids
was significantly higher in R HSC-3 cells than in HSC-3 cells
(P<0.01) (Fig. 3A). R HSC-3
also showed significantly increased cell invasion (P<0.01)
(Fig. 3B). Additionally, R HSC-3
cells showed low E-cadherin and high vimentin expression that
characterized an epithelial-mesenchymal transition-phenotype
compared with the parental cells (Fig.
3C). These results indicated that the multidrug-resistant head
and neck squamous cell carcinoma cells showed enhanced spheroid
formation and invasion ability compared with the parental cell
line.
Increased expression of SOX9 in
multidrug-resistant head and neck squamous cell carcinoma
cells
To evaluate the expression of stem cell-related
genes in R HSC-3 cells, we performed microarray analysis. We found
that the stem cell-related genes PAX8, SOX9,
NOG, and MMP24 were significantly increased and
ETV4, METTL3, HMGA2, SOX2, and
FGFR3 were significantly decreased (Table II, Fig. 4A). Although we tried to confirm
protein level, only SOX9 expression was detected. Immunoblot showed
that the expression of SOX9 was significantly increased in R HSC-3
cells compared with parental cells (Fig. 3C). We therefore speculated that
SOX9 may also be involved in the characteristics of CSCs in the
multidrug-resistant head and neck squamous cell carcinoma
cells.
 | Table IIUp- and downregulated stem cell
related genes in HSC3 vs. R HSC-3 cells by microarray analysis. |
Table II
Up- and downregulated stem cell
related genes in HSC3 vs. R HSC-3 cells by microarray analysis.
| GeneSymbol | Description |
Compare1_Zscore | Compare1_ratio |
|---|
| MMP24 | Homo sapiens matrix
metallopeptidase 24 (membrane-inserted), mRNA [NM_006690] | 4.804045 | 170.8623 |
| NOG | Homo sapiens
noggin, mRNA [NM_005450] | 4.185133 | 34.99634 |
| PAX8 | Homo sapiens paired
box 8, transcript variant PAX8A, mRNA [NM_003466] | 2.236811 | 10.71776 |
| SOX9 | Homo sapiens SRY
(sex determining region Y)-box 9, mRNA [NM_000346] | 2.315078 | 7.151924 |
| ETV4 | Homo sapiens ets
variant 4, transcript variant 2, mRNA [NM_001079675] | -2.73807 | 0.097956 |
| FGFR3 | Homo sapiens
fibroblast growth factor receptor 3, transcript variant 1, mRNA
[NM_000142] | -3.53021 | 0.110719 |
| HMGA2 | Homo sapiens high
mobility group AT-hook 2, transcript variant 1, mRNA
[NM_003483] | -2.68699 | 0.207217 |
| METTL3 | Homo sapiens
methyltransferase like 3, mRNA [NM_019852] | -2.72488 | 0.147527 |
| SOX2 | Homo sapiens SRY
(sex-determining region Y)-box 2, mRNA [NM_003106] | -3.90926 | 0.014163 |
SOX9 has a cell-autonomous effect in Sertoli cells.
Furthermore, previous studies revealed that SOX9 upregulates FGF9
to generate a SOX9/fibroblast growth factor 9 (FGF9) feed-forward
loop (27). FGF family (including
FGF9) signals regulate a variety of cellular processes during
embryogenesis and carcinogenesis (28). We found that FGF9 expression was
increased in R HSC-3 cells (Fig.
3C).
SOX9, CD44, and ABCG2 are involved in
the characteristics of CSCs
We next examined the roles of CD44, SOX9 and ABCG2
in the drug excretion ability, spheroid formation ability, and cell
invasion ability of R HSC-3 cells using siRNA-mediated silencing.
We found that drug excretion ability was significantly decreased by
knockdown of SOX9 (P<0.05) and by knockdown of ABCG2 (P<0.01)
in R HSC-3 cells; in contrast, knockdown of CD44 had no impact on
drug excretion activity (Fig. 4B).
Furthermore, no significant change was observed in spheroid
formation in R HSC-3 cells with knockdown of SOX9 or CD44, but the
knockdown of ABCG2 reduced spheroid formation ability (Fig. 3A). We observed a significant
decrease (P<0.01) in migration of cells with knockdown of CD44,
SOX9, and ABCG2 compared with controls (Fig. 3B).
Discussion
In this study, we generated a drug-resistant head
and neck squamous cell carcinoma cell line. We examined whether the
multidrug-resistant cells exhibited characteristics of CSCs and
which CSC-related molecules are associated with the characteristics
of CSCs.
HSC-3 cells were continuously cultured in the
presence of cisplatin and acquired resistance to cisplatin. We
initially aimed to create cisplatin-resistant head and neck
squamous cell lines in several cell lines, including HSC-2, HSC-3,
and SAS. However, the cell lines other than HSC-3 gradually died as
the concentration of cisplatin increased, and we only obtained
cisplatin-resistant HSC-3 cells. The cisplatin-resistant strain
also acquired cross-resistance, including resistance to SN-38,
docetaxel, doxorubicin, and the molecular-targeted drug erlotinib.
The multidrug-resistant cell line did not show any significant
changes in cell proliferation compared with the parental cell line.
However, the drug excretion ability, cell migration ability, and
spheroid-forming ability were enhanced. In addition, the
expressions of ABCG2 and CD44 were increased in the drug-resistant
cells, and microarray analysis revealed that the expression of SOX9
was also increased. These findings indicated that the
multidrug-resistant head and neck squamous cell carcinoma cells
exhibited characteristics of cancer stem cells. We further examined
the roles of CD44, SOX9, and ABCG2 in the drug excretion ability,
cell invasion ability, and spheroid formation ability of
multidrug-resistant head and neck squamous carcinoma cells. Our
results showed that ABCG2 is involved in spheroid formation, SOX9
and ABCG2 are involved in the drug excretion ability, and CD44,
SOX9, and ABCG2 are involved in cell invasion ability of R HSC-3
cells.
CSCs exhibit self-renewal ability and drug tolerance
and contribute to tumor metastasis and recurrence (29). However, the relationship between
the acquisition of drug resistance and the expression of CSC
markers and the characteristics of CSC such as cell invasion and
spheroid formation has not been clarified. In this study, we
created a multidrug-resistant head and neck squamous cell carcinoma
cell line and analyzed stem cell-related genes by microarray
analysis. The results revealed significantly increased expressions
of PAX8, SOX9, NOG, and MMP24 genes in
drug-resistant cells. The molecules were examined by western
blotting (data not shown), but only SOX9 was detected. Although
this study newly verified the involvement of SOX9 in the
characteristics of CSCs, PAX8, NOG, and MMP24
will also need further detailed verification in the future. The
stem cell-related genes that were significantly reduced by
microarray analysis were ETV4, METTL3, HMGA2,
SOX2 and FGFR3 genes. Future studies should examine
why these CSC-related genes tended to decrease with the acquisition
of multidrug resistance. In this study, we investigated
relationship between expression of CD44, ABCG2, and SOX9 and CSCs
properties such as drug excretion ability, cell invasion ability,
and spheroid-forming ability. Our results showed that SOX9 and
ABCG2 are involved in drug excretion ability, ABCG2 is involved in
spheroid formation, and CD44, SOX9, and ABCG2 are involved in cell
invasion ability of drug-resistant cells. SOX9 forms a feed-forward
loop with FGF9, and we confirmed that FGF9 was also expressed in R
HSC-3 cells. These results indicated that SOX9 and FGF9 may operate
through a feed-forward loop in head and neck squamous cell
carcinoma to promote metastasis and progression.
Early studies showed that the ATP-binding proteins
MDR-1 and ABCG2 were involved in drug excretion of cancer cells
(30,31). ABCG2-positive cells exhibit drug
resistance and epithelial-mesenchymal transition phenotypes
(32). Subsequent reports
identified ABCG2 as one of the CSC markers for head and neck
squamous cell carcinoma (33). In
this study, we found that ABCG2, which is involved in drug
excretion ability, plays a role in spheroid formation and invasion
ability. CD44 is a CSC marker for head and neck squamous cell
carcinoma and is significantly associated with prognosis (34,35).
Our previous study reported that CD44 is involved in the inhibition
of apoptosis induction and induction of CD44 degradation by
chemotherapy may be effective in inhibiting tumor recurrence
(10,11). We found that CD44 is involved in
cell invasion activity of drug-resistant cells. SOX9 is a member of
the SRY-related high-mobility group of box transcription factors.
Studies showed that SOX9 is mutated in skeletal malformations, XY
transsexuals, and campomelic dysplasia characterized by neonatal
lethality (36), and SOX9 is
important for the differentiation of embryonic stem cells into
salivary glands (37).
Furthermore, SOX9 is associated with prognosis in lung cancer and
breast cancer and a risk factor for chemotherapy failure in head
and neck cancer (38,39). Our study revealed that SOX9 is
involved in drug excretion and cell invasion in R HSC-3 cells.
These findings indicated that SOX9 may be a CSC marker of
HNSCC.
In this study, we created a multidrug-resistant head
and neck squamous cell carcinoma cell line and showed that
multidrug-resistant cells exhibited characteristics of CSCs.
Furthermore, analysis of the multidrug-resistant squamous cell
carcinoma cells showed that not only CD44 and ABCG2 but also SOX9
may be new predictors of prognosis for head and neck cancer. We
also showed that ABCG2, CD44, and SOX9 play roles in various CSC
activities. However, this study was performed using in vitro
experiments. Therefore, in vivo experiments will be required
to evaluate whether these molecules affect tumor growth and drug
resistance in mice.
Supplementary Material
CD44, SOX9 and ABCG2 expression levles
in cells with siRNA transfection. Changes in the expression of
CD44, SOX9 and ABCG2 by siRNA transfection were confirmed using
western blotting. R HSC.3, drug-resistant HSC-3; ABCG2, ATP-binding
cassette subfamily G member 2; si, short interfering; SOX9, SRY-box
transcription factor 9.
Acknowledgements
Not applicable.
Funding
Funding: This investigation was supported in part by JSPS
KAKENHI (grant nos. 17K11691, 17K11890 and 20K09947).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KM contributed to experimental design, performed the
majority of experiments and drafted the manuscript. NU conducted
experimental design of all experiments and data analysis and
drafted the manuscript. MA performed some experiments and data
analysis. MM participated in some experiments and data analysis. EO
contributed to experimental conceptualization and drafted the
manuscript. NU and EO confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
Dr Naoki Umemura, https://orcid.org/0000-0002-3249-782X; Dr Makoto
Adachi, https://orcid.org/0000-0002-9382-7052; Dr Emika
Ohkoshi, https://orcid.org/0000-0001-9700-9973.
References
|
1
|
Scadden DT: Cancer stem cells refined. Nat
Immunol. 5:701–703. 2004.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Visvader JE and Lindeman GJ: Cancer stem
cells: Current status and evolving complexities. Cell Stem Cell.
10:717–728. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Magee JA, Piskounova E and Morrison SJ:
Cancer stem cells: Impact, heterogeneity, and uncertainty. Cancer
Cell. 21:283–296. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chen D, Wu M, Li Y, Chang L, Yuan Q,
Ekimyan-Salvo M, Deng P, Yu B, Yu Y, Dong J, et al: Targeting BMI1+
cancer stem cells overcomes chemoresistance and inhibits metastases
in squamous cell carcinoma. Cell Stem Cell. 20:621–634.
2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
White RA, Neiman JM, Reddi A, Han G,
Birlea S, Mitra D, Dionne L, Fernandez P, Murao K, Bian L, et al:
Epithelial stem cell mutations that promote squamous cell carcinoma
metastasis. J Clin Invest. 123:4390–4404. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q,
Hjelmeland AB, Dewhirst MW, Bigner DD and Rich JN: Glioma stem
cells promote radioresistance by preferential activation of the DNA
damage response. Nature. 444:756–760. 2006.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Meacham CE and Morrison SJ: Tumour
heterogeneity and cancer cell plasticity. Nature. 501:328–337.
2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Li J, Condello S, Thomes-Pepin J, Ma X,
Xia Y, Hurley TD, Matei D and Cheng JX: Lipid desaturation is a
metabolic marker and therapeutic target of ovarian cancer stem
cells. Cell Stem Cell. 20:303–314, e305. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Codd AS, Kanaseki T, Torigo T and Tabi Z:
Cancer stem cells as targets for immunotherapy. Immunology.
153:304–314. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ohkoshi E and Umemura N: Induced
overexpression of CD44 associated with resistance to apoptosis on
DNA damage response in human head and neck squamous cell carcinoma
cells. Int J Oncol. 50:387–395. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Nanbu T, Umemura N, Ohkoshi E, Nanbu K,
Sakagami H and Shimada J: Combined SN-38 and gefitinib treatment
promotes CD44 degradation in head and neck squamous cell carcinoma
cells. Oncol Rep. 39:367–375. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Pena-Solorzano D, Stark SA, Konig B,
Sierra CA and Ochoa-Puentes C: ABCG2/BCRP: Specific and nonspecific
modulators. Med Res Rev. 37:987–1050. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen Z, Shi T, Zhang L, Zhu P, Deng M,
Huang C, Hu T, Jiang L and Li J: Mammalian drug efflux transporters
of the ATP binding cassette (ABC) family in multidrug resistance: A
review of the past decade. Cancer Lett. 370:153–164.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lu BC, Li J, Yu WF, Zhang GZ, Wang HM and
Ma HM: Elevated expression of Nrf2 mediates multidrug resistance in
CD133+ head and neck squamous cell carcinoma stem cells. Oncol
Lett. 12:4333–4338. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Guan GF, Zhang DJ, Zheng Y, Wen LJ, Yu DJ,
Lu YQ and Zhao Y: Significance of ATP-binding cassette transporter
proteins in multidrug resistance of head and neck squamous cell
carcinoma. Oncol Lett. 10:631–636. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hu J, Li J, Yue X, Wang J, Liu J, Sun L
and Kong D: Expression of the cancer stem cell markers ABCG2 and
OCT-4 in right-sided colon cancer predicts recurrence and poor
outcomes. Oncotarget. 8:28463–28470. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xiong B, Ma L, Hu X, Zhang C and Cheng Y:
Characterization of side population cells isolated from the colon
cancer cell line SW480. Int J Oncol. 45:1175–1183. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Posner MR, Hershock DM, Blajman CR,
Mickiewicz E, Winquist E, Gorbounova V, Tjulandin S, Shin DM,
Cullen K, Ervin TJ, et al: Cisplatin and fluorouracil alone or with
docetaxel in head and neck cancer. N Engl J Med. 357:1705–1715.
2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5(R80)2004.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Quackenbush J: Microarray data
normalization and transformation. Nat Genet. 32 Suppl:S496–S501.
2002.PubMed/NCBI View
Article : Google Scholar
|
|
22
|
Takeishi S and Nakayama KI: Role of Fbxw7
in the maintenance of normal stem cells and cancer-initiating
cells. Br J Cancer. 111:1054–1059. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Takeishi S, Matsumoto A, Onoyama I, Naka
K, Hirao A and Nakayama KI: Ablation of Fbxw7 eliminates
leukemia-initiating cells by preventing quiescence. Cancer Cell.
23:347–361. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Saito Y, Uchida N, Tanaka S, Suzuki N,
Tomizawa-Murasawa M, Sone A, Najima Y, Takagi S, Aoki Y, Wake A, et
al: Induction of cell cycle entry eliminates human leukemia stem
cells in a mouse model of AML. Nat Biotechnol. 28:275–280.
2010.PubMed/NCBI View
Article : Google Scholar
|
|
25
|
Kukal S, Guin D, Rawat C, Bora S, Mishra
MK, Sharma P, Paul PR, Kanojia N, Grewal GK, Kukreti S, et al:
Multidrug efflux transporter ABCG2: Expression and regulation. Cell
Mol Life Sci. 78:6887–6939. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Fletcher JI, Haber M, Henderson MJ and
Norris MD: ABC transporters in cancer: More than just drug efflux
pumps. Nat Rev Cancer. 10:147–156. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Moniot B, Declosmenil F, Barrionuevo F,
Scherer G, Aritake K, Malki S, Marzi L, Cohen-Solal A, Georg I,
Klattig J, et al: The PGD2 pathway, independently of FGF9,
amplifies SOX9 activity in Sertoli cells during male sexual
differentiation. Development. 136:1813–1821. 2009.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chaffer CL, Dopheide B, Savagner P,
Thompson EW and Williams ED: Aberrant fibroblast growth factor
receptor signaling in bladder and other cancers. Differentiation.
75:831–842. 2007.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Giancotti FG: Mechanisms governing
metastatic dormancy and reactivation. Cell. 155:750–764.
2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Modok S, Mellor HR and Callaghan R:
Modulation of multidrug resistance efflux pump activity to overcome
chemoresistance in cancer. Curr Opin Pharmacol. 6:350–354.
2006.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kage K, Tsukahara S, Sugiyama T, Asada S,
Ishikawa E, Tsuruo T and Sugimoto Y: Dominant-negative inhibition
of breast cancer resistance protein as drug efflux pump through the
inhibition of S-S dependent homodimerization. Int J Cancer.
97:626–630. 2002.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Suresh R, Ali S, Ahmad A, Philip PA and
Sarkar FH: The role of cancer stem cells in recurrent and
drug-resistant lung cancer. Adv Exp Med Biol. 890:57–74.
2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Nieh S, Jao SW, Yang CY, Lin YS, Tseng YH,
Liu CL, Lee TY, Liu TY, Chu YH and Chen SF: Regulation of tumor
progression via the Snail-RKIP signaling pathway by nicotine
exposure in head and neck squamous cell carcinoma. Head Neck.
37:1712–1721. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang J, Wu Y, Gao W, Li F, Bo Y, Zhu M, Fu
R, Liu Q, Wen S and Wang B: Identification and characterization of
CD133+CD44+ cancer stem cells from human laryngeal squamous cell
carcinoma cell lines. J Cancer. 8:497–506. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lee Y, Shin JH, Longmire M, Wang H, Kohrt
HE, Chang HY and Sunwoo JB: CD44+ cells in head and neck squamous
cell carcinoma suppress T-cell-mediated immunity by selective
constitutive and inducible expression of PD-L1. Clin Cancer Res.
22:3571–3581. 2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wagner T, Wirth J, Meyer J, Zabel B, Held
M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, et al:
Autosomal sex reversal and campomelic dysplasia are caused by
mutations in and around the SRY-related gene SOX9. Cell.
79:1111–1120. 1994.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tanaka J, Mabuchi Y, Hata K, Yasuhara R,
Takamatsu K, Kujiraoka S, Yukimori A, Takakura I, Sumimoto H,
Fukada T, et al: Sox9 regulates the luminal stem/progenitor cell
properties of salivary glands. Exp Cell Res.
382(111449)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Khorani K, Schwaerzler J, Burkart S, Kurth
I, Holzinger D, Flechtenmacher C, Plinkert PK, Zaoui K and Hess J:
Establishment of a plasticity-associated risk model based on a
SOX2- and SOX9-related gene set in head and neck squamous cell
carcinoma. Mol Cancer Res. 19:1676–1687. 2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Al-Zahrani KN, Abou-Hamad J, Pascoal J,
Labreche C, Garland B and Sabourin LA: AKT-mediated phosphorylation
of Sox9 induces Sox10 transcription in a murine model of
HER2-positive breast cancer. Breast Cancer Res.
23(55)2021.PubMed/NCBI View Article : Google Scholar
|