Introduction
Colorectal cancer (CRC) is the third most common
cancer worldwide and metastatic CRC (mCRC) continues to be
associated with a poor prognosis (1). Therapeutic strategies for mCRC have
improved over the past decades (2). However, there is an urgent need for
novel therapeutic strategies for the patients who exhibit cancer
progression even after treatment with cytotoxic chemotherapy and
targeted agents (3). KRAS mutation
represents one of the most prevalent genetic alterations in cancers
(4). In CRC tumors, 85-90% of KRAS
mutations occur in exon 2, with codon 12 and 13 being the most
predominant mutations (5).
Anti-EGFR antibodies, such as cetuximab or panitumumab, have been
demonstrated to be effective against RAS oncogene wild-type mCRC.
However, patients will eventually develop drug resistance and
further disease progression will occur regardless of the initial
efficacy (6).
Therefore, a concentrated research effort has been
made to elucidate the mechanism underlying the acquisition of
resistance to anti-EGFR therapy by CRC. The occurrence of tumor
somatic mutations in the RAS/RAF/MAPK, PI3K/PTEN/AKT and Janus
kinase/STAT signaling pathways, have been reported to be potential
therapeutic targets in CRC (7).
However, therapeutic approaches proposed for overcoming resistance
to anti-EGFR therapy have rarely been demonstrated to confer
significant clinical benefits (8,9).
Increasing evidence indicates that the tumor microenvironment
(TME), including tumor-stromal cell interactions, also contributes
to changes in tumor characteristics during tumor initiation and
progression (10,11). These characteristics of the TME,
including helping the formation of cancer stem cells (CSCs), are
responsible for the development and maintenance of tumors and
resistance to cytotoxic drugs (12).
Furthermore, based on the Hierarchy (CSC) Theory,
which suggests CSCs are more likely to generate a tumor, it has
been reported that CSCs have a longer life span and a greater
ability to self-renew compared with non-stem cells (13). Therefore, it can be hypothesized
that alterations in the TME prompt cetuximab resistance in CRC
cells and that CSCs may be the intrinsic driving force for
activating or inducing cetuximab resistance.
It has previously been demonstrated that certain
specific exosomes, which are considered to be the main group of
extracellular vesicles, are biologically active lipid-bilayer
vesicles that are naturally released from different types of normal
or tumor cells (14). Exosomes are
lipid bilayer membrane vesicles (50-100 nm) derived from the
luminal membrane of multivesicular bodies and are secreted via the
fusion of multivesicular bodies with the plasma membrane or via
budding from the membrane (15).
These vesicles contain nucleic acids, proteins and lipids, which
thereby allows for the transfer of genetic material and enable the
exchange of information between cells within the microenvironment
(10).
Our previous study has demonstrated that exosomes
serve a key role in transmitting multidrug resistance (MDR) between
tumor cells (16). However, few
studies have been reported that the features that contribute to the
transmissibility of drug resistance via exosomes in
cetuximab-acquired resistant CRC (17). Furthermore, drug resistance often
occurs following chemotherapy combined with cetuximab therapy in
the clinic (18). Therefore, there
is need for further investigations into the mechanism of anti-EGFR
therapy resistance to support the development of novel therapeutic
strategies.
In the present study, the potential association
between exosomes derived from MDR cells and the response to
anti-EGFR treatment in CRC cell lines was investigated.
Furthermore, the possible effects of exosomes on sensitivity to
anti-EGFR treatment were explored in vivo and in
vitro. This suggested that inhibition of the secretion of
exosomes from MDR cells could potentially represent a rational
therapeutic strategy to prevent and overcome cetuximab resistance
in patients with mCRC.
Materials and methods
The Cancer Genome Atlas (TCGA) gene
expression data
The TCGA database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
created by the National Cancer Institute and the National Human
Genome Research Institute, has characterized over 20,000 primary
cancer and matched normal samples spanning 33 cancer types
worldwide. The TCGA-colon adenocarcinoma study dataset (accession
no. phs000178) was chosen to produce a survival curve for CRC. The
gene expression profile data (mRNA) of patients with colon cancer
were downloaded from TCGA database, and the gene expression profile
data was transformed with lg2(x + 1). Samples with missing data and
large differences were removed, 372 colon cancer samples were
identified, and the expression status of the KRAS gene in patients
with colon cancer was analyzed. According to the median KRAS mRNA
expression, the samples were divided into the KRAS mutant group and
KRAS wild-type group.
Patients and sample collection
A total of 60 patients with CRC patients were
enrolled in the study and fresh tissue samples were collected and
fixed with 10% formalin solution at 23˚C for 48 h after obtaining
written informed consent between January 2020 and January 2022 at
Shuguang Hospital, Shanghai University of Traditional Chinese
Medicine (Shanghai, China). All patients with CRC were diagnosed
according to Chinese Society of Clinical Oncology diagnosis and
treatment guidelines for colorectal cancer 2018(19). The inclusion criteria were as
follows: i) Pathologically confirmed colon cancer; ii) stage I to
III in TNM pathological staging according to the American Joint
Committee on Cancer (20); iii)
men or women aged 18-75 years; iv) Karnofsky's performance scoring
≥70(21); v) no strict heart,
liver, kidney or hematopoietic system disease or other factor
affecting drug evaluation; and vi) volunteers who had given
informed consent. Patients with any of the following conditions
were excluded: i) Did not meet the inclusion criteria; ii) mental
disorder, pregnancy or lactation; and iii) incomplete information.
The patients in the CRC group were aged 45-75 years, with a mean
age of 52±11 years. The patients in the advanced CRC group were
aged 45-75 years, with a mean age of 54±14 years (Table SI). Among the 18 patients with
early-stage CRC (age range, 45-75 years), with a mean age of 52±11
years enrolled in the present study, 8 were male and 10 were
female. Of the 32 patients with advanced CRC (the majority of
patients were diagnosed in advanced stages, which refers to stage
II and III colon cancer) (22,23)
(age range, 45-75 years; mean age, 54±14 years), 20 were male and
12 were female. Tumor samples and paired adjacent tissues were
collected from all patients. Written informed consent was obtained
from every patient.
The present study was approved by the Medical Ethics
and Human Clinical Trial Committee of the affiliated hospital,
Shuguang Hospital, Shanghai University of Traditional Chinese
Medicine (Shanghai, China).
Establishment of the CRC/MDR cell
line
The human HCT116, LoVo, Caco-2, HT-29 and SW480 CRC
cell lines were purchased from The Cell Bank of Type Culture
Collection of The Chinese Academy of Sciences. The HCT116 and LoVo
cells were grown at 37˚C in a 5% CO2 humidified
atmosphere in RPMI 1640 (HyClone; Cytiva) supplemented with 10%
(v/v) heat-inactivated fetal calf serum (Gibco; Thermo Fisher
Scientific, Inc.), 2 mM glutamine, 100 units/ml penicillin and 100
µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.).
The HT-29, Caco-2 and SW480 cells were grown at 37˚C in a 5%
CO2 humidified atmosphere in F12K (HyClone; Cytiva),
DMEM (HyClone; Cytiva) and L-15 medium (HyClone; Cytiva),
respectively, supplemented with 10% (v/v) heat-inactivated fetal
calf serum (Gibco; Thermo Fisher Scientific, Inc.), 2 mM glutamine,
100 units/ml penicillin and 100 µg/ml streptomycin (Invitrogen;
Thermo Fisher Scientific, Inc.). The HCT116/oxaliplatin (L-OHP)
cells were established by gradually increasing the concentration of
L-OHP in parental cells (HCT116) first. Subsequently, the cells
were intermittently treated with a high-dose concentration
(9.6-19.2 µg/ml) of L-OHP (24).
Then they were routinely maintained in RPMI 1640 medium containing
5,000 ng/ml L-OHP (Sigma-Aldrich; Merck KGaA) as previously
reported (25). The HCT116/L-OHP
cells were designated as CRC/MDR cells (Table I) and maintained at 37˚C in a 5%
CO2 humidified atmosphere in the L-OHP-containing medium
containing RPMI 1640 supplemented with 10% (v/v) heat-inactivated
fetal calf serum (Gibco; Thermo Fisher Scientific, Inc.); 2 mM
glutamine, 100 units/ml penicillin and 100 µg/ml streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.), and 5 µg/ml L-OHP
(Sigma-Aldrich; Merck KGaA). A cell proliferation analysis assay
was performed as described subsequently to detect the
IC50 values of chemotherapeutic drugs in MDR HCT116/MDR
cells and parental HCT116 cells, which were treated with
chemotherapeutic drugs with increasing concentrations (0, 125, 250,
500 and 1,000 µg/ml), including VCR, cDDP, 5-Fu and MMC, for 48 h
at 37˚C. However, CRC/MDR cells were cultured in L-OHP-free media
for 1 week at 37˚C prior to subsequent experimentation to make sure
the cells were all in the same environment (26,27).
The human CRC Caco-2 and HT-29 cell lines are usually considered as
KRAS wild-type (28,29) and were purchased from The Cell Bank
of Type Culture Collection of The Chinese Academy of Sciences with
a statement of authentication.
 | Table ISensitivity to chemotherapy in HCT116
and HCT116/MDR cells. |
Table I
Sensitivity to chemotherapy in HCT116
and HCT116/MDR cells.
| | IC50
(µg/ml) | |
|---|
| Chemotherapeutic
drugs | HCT116 | HCT116/MDR | Resistance
factor |
|---|
| L-OHP | 19.84±1.42 |
157.48±16.73a | 7.94 |
| cDDP | 3.32±0.37 |
26.38±1.92a | 7.95 |
| 5-Fu | 7.14±1.21 |
33.29±3.64a | 4.66 |
| MMC | 2.55±0.35 |
12.50±2.49a | 4.90 |
Exosome isolation and
characterization
The CRC/MDR cells were cultured in exosome-depleted
complete medium (Serum-free Media for exosome culture; Umibio)
while CRC cells were cultured in normal medium for 48 h at 4˚C.
Exosomes were extracted from tumor cells, including CRC and CRC/MDR
cells via ultracentrifugation. Briefly, the supernatant was
obtained via centrifugation at 2,000 x g for 30 min at 4˚C and
10,000 x g for 30 min at 4˚C as previously reported (30). The medium was then filtered using a
0.22-µm filter, followed by ultracentrifugation at 120,000 x g for
70 min at 4˚C.
The exosome-enriched pellets were suspended in 50 µl
PBS. For exosome TEM observation, 5 µl exosomes were loaded onto
formvar carbon-coated grids, and were fixed with 1% glutaraldehyde
solution at 4˚C overnight. Subsequently, the exosomes were
negatively stained with 50 µl 2% aqueous phosphotungstic acid for
60 sec at 23˚C. The exosomes were imaged with a transmission
electron microscope at 80 kV. The number and size distribution of
the exosomes were detected using a NanoSight LM10 Nanoparticle
Characterization System (Malvern Instruments, Inc.). The exosomal
protein concentration was quantified via the BCA method and
exosome-associated protein marker HSP70 and CD81 expression was
analyzed via western blotting.
Western blotting
Tumor cell (HT-29 and Caco-2 cells) proteins were
extracted using RIPA lysis buffer (Beyotime Institute of
Biotechnology) as previously reported (31). Protein concentration was quantified
using the BCA method. A total of 40 µg protein for each group was
separated via SDS-PAGE on a 12% gel and transferred to 0.45 µm PVDF
membranes (Merck KGaA). Membranes were blocked with 5% BSA
(Beyotime Institute of Biotechnology) for 2 h at 23˚C and then
incubated with primary antibodies against the following proteins at
4˚C overnight: HSP70 (1:1,000; cat. no. 4876; Cell Signaling
Technology, Inc.), CD81 (1:1,000; cat. no. ab79559; Abcam), EGFR
(1:1,000; cat. no. 66455; ProteinTech Group, Inc.), phosphorylated
(p)-EGFR (1:1,000; cat. no. 18986; ProteinTech Group, Inc.), AKT
(1:1,000; cat. no. 4691; Cell Signaling Technology, Inc.), p-AKT
(1:1,000; cat. no. 13038; Cell Signaling Technology, Inc.), Sox2
(1:1,000; cat. no. 3579; Cell Signaling Technology, Inc.) and
programmed death-ligand 1 (PD-L1; 1:1,000; cat. no. 66248;
ProteinTech Group, Inc.). GAPDH rabbit monoclonal antibody
(1:1,000; cat. no. 5174; Cell Signaling Technology, Inc.) and
tubulin rabbit monoclonal antibody (1:1,000; cat. no. 2148; Cell
Signaling Technology, Inc.) were used as the internal control. The
membranes were washed three times with TBS with 0.1% Tween-20
(TBST) and incubated at 37˚C for 1 h with HRP-conjugated
anti-rabbit secondary antibody (1:2,000; cat. no. ab97051; Abcam)
and HRP-conjugated anti-mouse secondary antibody (1:2,000; cat. no.
ab6728; Abcam). Then, the membranes were washed three times with
TBST. Bands were visualized via chemiluminescence using
Immobilon® ECL Ultra Western HRP Substrate (Merck KGaA)
according to the manufacturer's protocol and a ChemiScope 6200
Touch (Clinx Science Instruments Co., Ltd.). The data were analyzed
using ImageJ software (version 1.8.0; National Institutes of
Health).
Sphere formation assay
The cells were transferred to ultra-low attachment
plates (Corning, Inc.) in serum-free DMEM (HyClone; Cytiva)
containing 10 ng/ml Fibroblast Growth Factor (basic) (FUJIFILM Wako
Pure Chemical Corporation), 10 mg/ml human insulin (CSTI), 100
mg/ml human transferrin (Roche Diagnostics) and 100 mg/ml BSA
(Nacalai Tesque, Inc.) and incubated at 37˚C in a 5% CO2
incubator for 10 days. The number of cell spheres, defined as
spherical, non-adherent cell clusters <100 µm in diameter, was
quantified and the spheres were imaged using an inverted light
microscope and analysed using ImageJ software (version 1.8.0;
National Institutes of Health).
Reverse transcription-quantitative PCR
(RT-qPCR)
RNA was extracted from HT-29 and Caco-2 cells
(1x106) using TRIzol® (Takara Bio, Inc.) as
previously reported (25). Total
RNA was reverse transcribed into complementary DNA using the
SuperScript™ IV First-Strand Synthesis System (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Then, the mixture system, including SYBR Green qPCR Master mix
(Thermo Fisher Scientific, Inc.), cDNA templates and primers were
applied for qPCR to detect the mRNA expression according to the
manufacturer's instructions. The thermocycling conditions were:
95˚C for 3 min, followed by 40 cycles of 95˚C for 15 sec and 60˚C
for 15 sec. Primer sequences for target genes, CD133, Nanog, Oct-4,
CD29, CD44, Sox2 and GAPDH are presented in Table SII. GAPDH was used as the
reference gene. The mRNA expression levels were quantified using
the CFX Connect Real-Time PCR Detection System (Bio-Rad
Laboratories, Inc.) and were analyzed using the CFX management
software v2.0 (Bio-Rad Laboratories, Inc.). Relative quantification
of mRNA was performed using the 2-ΔΔCq method (32).
Cell proliferation analysis
HT-29 and Caco-2 cells were seeded into 96-well
plates (5,000 cells/well) and exposed to increasing concentrations
of cetuximab (0, 125, 250, 500 and 1,000 µg/ml; Merck & Co.,
Inc.) for 24, 48 and 72 h at 37˚C. The proliferative ability of
cells was determined using the Cell Counting Kit-8 (CCK-8; Med Chem
Express) assay for 2 h according to the manufacturer's protocol and
is presented as cell proliferation (%). Absorbance was quantified
at 450 nm using a Cytation 5 Cell Imaging Multi-Mode Reader (BioTek
Instruments, Inc.). The half-maximal inhibitory concentration
(IC50) of drugs was determined using GraphPad Prism
version 8.0 (GraphPad Software, Inc.).
Colony formation assays
The HT-29 and Caco-2 cells were seeded in six-well
plates (~100 cells/well). Cells were treated with different
exosomes for 72 h as previously reported (33). Subsequently, cells were washed
twice with PBS, cultured in RPM1640 medium and DMEM (HyClone;
Cytiva) and allowed to form colonies at 37˚C for 14 days. Media
were replaced every 4-5 days. Cell colonies were defined as cell
populations >0.5 mm in diameter and the number of cell colonies
was counted manually. Colonies were washed three times with PBS,
and were then fixed with 95% ethanol at room temperature for 20
min. Colonies were stained with crystal violet (2%) at 37˚C for 30
min. Following extensive washing with PBS, the cells were observed
under a light microscope (Olympus Soft Imaging Solutions GmbH) and
five fields were selected randomly for colony counting. The colony
formation rate was then calculated using the following equation:
Colony formation rate=(number of clones)/(number of seeded cells)
x100%.
Sphere formation assay for tumor stem
cells
For formation of spheres, cells were cultured in
NeuroCult NS-A basal serum-free medium (human; Stemcell
Technologies, Inc.) supplemented with 20 µg/ml heparin (Stemcell
Technologies, Inc.), 20 ng/ml hEGF (R&D Systems, Inc.), 10
ng/ml hFGF-b (PeproTech, Inc.) and NeuroCult NS-A Proliferation
Supplements (Stemcell Technologies, Inc.). This combination of
medium is usually used for stem cells. Cells were seeded at low
densities in 12-well low-adhesion plates (1 ml per well). The cells
were cultured and then seeded at clonal density in low-adhesion
plates. The spheroids were grown until Day 7 and images were
captured at a magnification of x100 using an inverted light
microscope (first-generation). Thereafter, the first-generation
spheroids were dissociated to single cells, and equal numbers of
live cells were cultured. Subsequently, the cells were plated at
clonal density, resulting in second-generation spheroids. The
number of second-generation spheroids was quantified using ImageJ
software (version 1.8.0; National Institutes of Health). As
previously reported, 7 days were considered to be one cycle
(34). Viable spheres were those
active spheres which had the ability to grow just like cells. These
viable spheres defined as SP cells possessing stem cell
characteristics, such as proliferation, self-renewal and
differentiation (35).
Tumor mouse model
HT-29 cells were harvested in serum-free PBS and 100
µl cell suspensions (1x107 cells/ml) were injected
subcutaneously into 20 female BALB/c nude mice (age, 6-8 weeks;
weight, ~20.0±0.6 g; Shanghai SLAC Laboratory Animal Co. Ltd.;
license no. SCXK2017-0005). The animal experiment was approved by
the Institutional Animal Care and Use Committee of Shuguang
Hospital, Shanghai University of Traditional Chinese Medicine
(approval no. PZSHUTCM200724027). The animals were kept under
specific pathogen-free conditions. The room temperature was 20˚C,
the relative humidity was 60%. The light/dark cycle was 12/12 h.
The animals had free access to drinking water and food normally,
and were fasting for 12 h before the experiment. When the tumors
reached an average volume of 100 mm3, the mice were
randomly divided into four groups (n=5) as follows: i) Saline (0.2
ml per mouse) intraperitoneal injection daily; ii) 0.05 g/kg
cetuximab solution intraperitoneal injection every other day; iii)
100 µl CRC exosomes peritumoral injection then 0.05 g/kg cetuximab
solution intraperitoneal injection every other day; iv) 100 µl
CRC/MDR exosomes peritumoral injection then 0.05 g/kg cetuximab
solution intraperitoneal injection every other day. The length and
width of tumors were recorded every 3 days. After 27 days from the
first day as marked as day 0, according to the Guideline of
assessment for humane endpoints in animal experiment (RB/T
173-2018), the diameter of tumors was <1.2 cm. The humane
endpoints in the present study were that the animals showed no
activity, including grooming, food intake of <1 g per day, or
>20% body weight loss over 3 days (the maximum body weight loss
over the entire experimental period was <15% and the maximum
body weight loss over 3 days was <12%), or the diameter of
tumors was >1.2 cm. The animals were sacrificed via cervical
dislocation in deep anesthesia (5% isoflurane; 5 l/min for 1 min)
and were surgically removed primary tumors and weighed. Tumor
volumes were quantified using the following formula: length x
width2 x 0.5.
TUNEL assay
The TUNEL assay was performed using a DeadEnd™
Colorimetric TUNEL System kit (Promega Corporation) to detect the
apoptosis of the subcutaneous tumors according to the
manufacturer's protocol (16). The
peeled tumor was washed with xylene twice for 5 min each. The
tumors were fixed with 10% neutral formalin at 23˚C for 3 h,
dehydrated in gradient alcohol, embedded in paraffin, and sliced
into 4-µm thick slices, and heated at 65˚C for 1 h. The tissues
were dewaxed, washed with running water and repaired with pH 8.0
EDTA antigen repair solution in a water bath at 98˚C for 25 min.
The tissue was treated with Proteinase K working liquid for 15-30
min, and the mixture of the TUNEL reaction was prepared according
to the instructions. TUNEL working solution was added to the
tissues and these were incubated in the dark at 37˚C for 60 min.
The samples were rinsed with PBS three times. Subsequently, 50 µl
converter-POD was added to the specimen after the slide was dried.
The slide was covered and reacted at 37˚C in a dark wet box, and
rinsed three times within 30 min. Subsequently, 50 µl 5 mg/ml DAPI
substrate was added and tissues were incubated at 25˚C for 3 min.
The tissues were rinsed with PBS for 10 min at 25˚C. Subsequently,
the tissue was sealed with Anti-Fade Mounting Medium (ab104135;
Abcam). A drop of glycerin was added to observe the apoptotic cells
(200-500 cells in total) and capture images with a 20x objective
and 10x eyepiece. Three random fields of view per sample were
observed. The observation and capture of digital images was
performed using a Nikon E80i fluorescence microscope (Nikon
Corporation).
Immunohistochemistry (IHC)
Tumor tissue samples were fixed with 4%
paraformaldehyde solution for 3 h at 23˚C, and then embedded with
paraffin after gradient dehydration. Paraffin-embedded tumor tissue
samples (5-µm-thick sections) were selected for IHC. Slices were
placed in an oven for 30 min for dewaxing and were then hydrated
with gradient alcohol. Subsequently, the slides were washed again
with PBS and incubated with the prefabricated avidin peroxidase
macromolecular complex for 30 min. The peroxidase reaction was
completed by incubation in PBS containing 0.01% hydrogen peroxide
at room temperature for ~5 min. The paraffin sections were
incubated in the sealing solution [10% donkey serum (Gibco; Thermo
Fisher Scientific, Inc.) + 5% skim milk + 4% BSA + 0.1%
TritonX-100] for 10 min, then incubated with antibodies at 4˚C
overnight. The primary antibodies for IHC were p-EGFR (1:750; cat.
no. 18986; ProteinTech Group, Inc.), p-AKT (1:300; cat. no. 4060;
Cell Signaling Technology, Inc.), Sox2 (1:300; cat. no. 14962; Cell
Signaling Technology, Inc.) and PD-L1 (1:300; cat. no. 66248;
ProteinTech Group, Inc.). The slides were washed again with PBS.
The slices were incubated with Polymer helper for 20 min, and then
washed with PBS for 5 min three times. Subsequently, the samples
were incubated with an HRP-conjugated goat anti-rabbit IgG
secondary antibody (1:2,000; cat. no. ab6702; Abcam) at 37˚C for 1
h. DAB (cat. no. ab64238; Abcam) was added immediately after the
liquid around the tissue dried, slides were incubated at room
temperature for 10 min and the reaction was terminated with water
washing. The slices were dehydrated and made transparent after
counter-staining with hematoxylin for 5 min at room temperature.
Results of staining were assessed using a Nikon E80i light
microscope (Nikon Corporation).
Statistical analysis
SPSS version 21.0 (IBM Corp.) was used for the
statistical analysis. DFS was compared using a two-sided log-rank
test. Hazard ratios with 95% confidence intervals were calculated
using the Cox proportional hazards model. The Kaplan-Meier method
was used to draw survival curves, and a log-rank test was carried
out to compare the survival rates. The Wilcoxon test was used for
continuous variables and Pearson's χ2 test for
categorical variables. All data obtained in the present study are
presented as the mean ± SD from at least three independent
experiments. Statistical analyses were performed using GraphPad
Prism 8.0 (GraphPad Software, Inc.). A two-sided unpaired Student's
t-test with Benjamini-Hochberg correction was used to compare two
groups. Differences among three or more groups were analyzed using
one-way ANOVA followed by Tukey's post-hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Activation of the EGFR signaling
pathway is associated with a poor prognosis in cetuximab-treated
patients with CRC
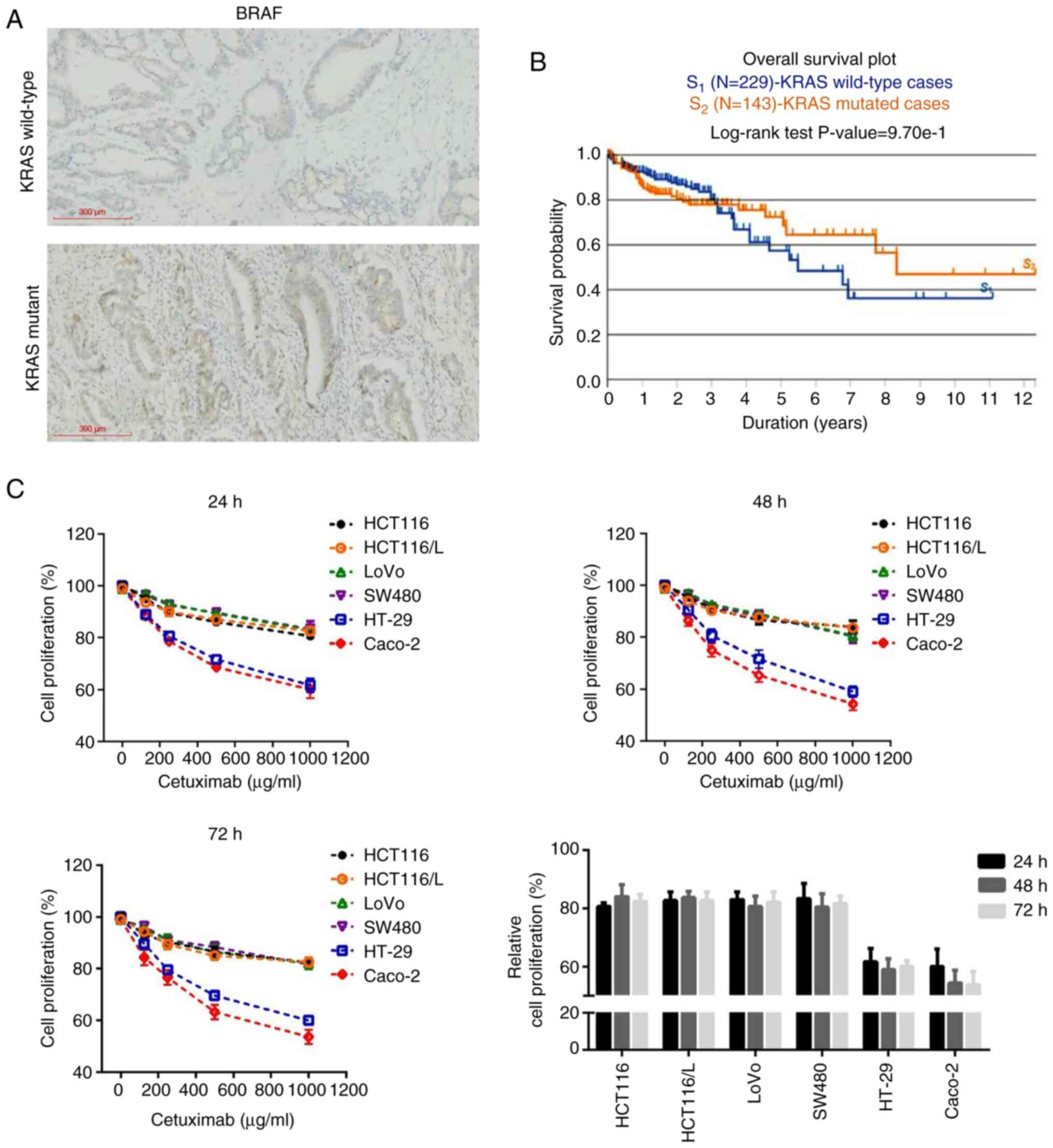
According to IHC scoring (36) results, 78.3% (47/60) and 20.0%
(12/60) of tumors were defined as presenting high and low BRAF
protein expression levels, respectively (Fig. 1A). Analysis of the TCGA gene
expression data demonstrated that the overall survival rate at 3
years postdiagnosis in patients with KRAS wild-type (n=229) was
markedly higher than the overall survival rate in patients with the
KRAS mutation (n=143) (P<0.05). However, 3-12 years
postdiagnosis, the overall survival rates in patients with KRAS
wild-type were lower than overall survival rates in patients with
the KRAS mutation (Fig. 1B). As
previously reported, KRAS-mutant CRC is associated with a poorer
prognosis compared with KRAS wild-type CRC (37).
The dose and time-dependent effects of cetuximab on
cell proliferation were assessed using the CCK-8 assay to explore
the sensitivity of different CRC cell lines to cetuximab. Caco-2
and HT-29 cell lines with cetuximab treatment exhibited a marked
cetuximab-sensitive phenotype compared with the other CRC cell
lines (HCT116, HCT116/L, LoVo and SW480 cells) (Fig. 1C). It should be noted that although
the cell proliferation was slightly increased at 72 h compared with
48 h in HT-29 cells, the difference was too small to be
statistically significant. Anti-EGFR antibody treatment was
effective in the KRAS wild-type Caco-2 and HT-29 cell lines, which
were used as cetuximab-sensitive cells in the subsequent
experiments.
Exosomes derived from CRC/MDR cells
could increase resistance to cetuximab in vitro
Increasing evidence suggests that exosomes secreted
by cancer cells are key mediators of cell-to-cell communication
(16,38) and have the capacity to considerably
modify the TME, which impacts disease progression. Compared with
HCT116 cells, HCT116/MDR cells showed chemotherapeutic drug
resistance (Table I), and the
difference was statistically significant (P<0.01). The exosomes
secreted from CRC/MDR cells and their parental sensitive CRC cells
were analyzed for their phenotype (purity and shape) using TEM and
for size and particle number using the LM10 Nanoparticle
Characterization System (Fig. 2A).
A previous study reported that the particles were determined to be
cup-shaped membrane-bound vesicles with a diameter of ~100 nm
(39). The mean diameter of these
nanovesicles ranged between 90 and 120 nm for exosomes from both
CRC/MDR cells and their parental sensitive CRC cells (data not
shown), which was consistent with the characteristic features of
exosomes.
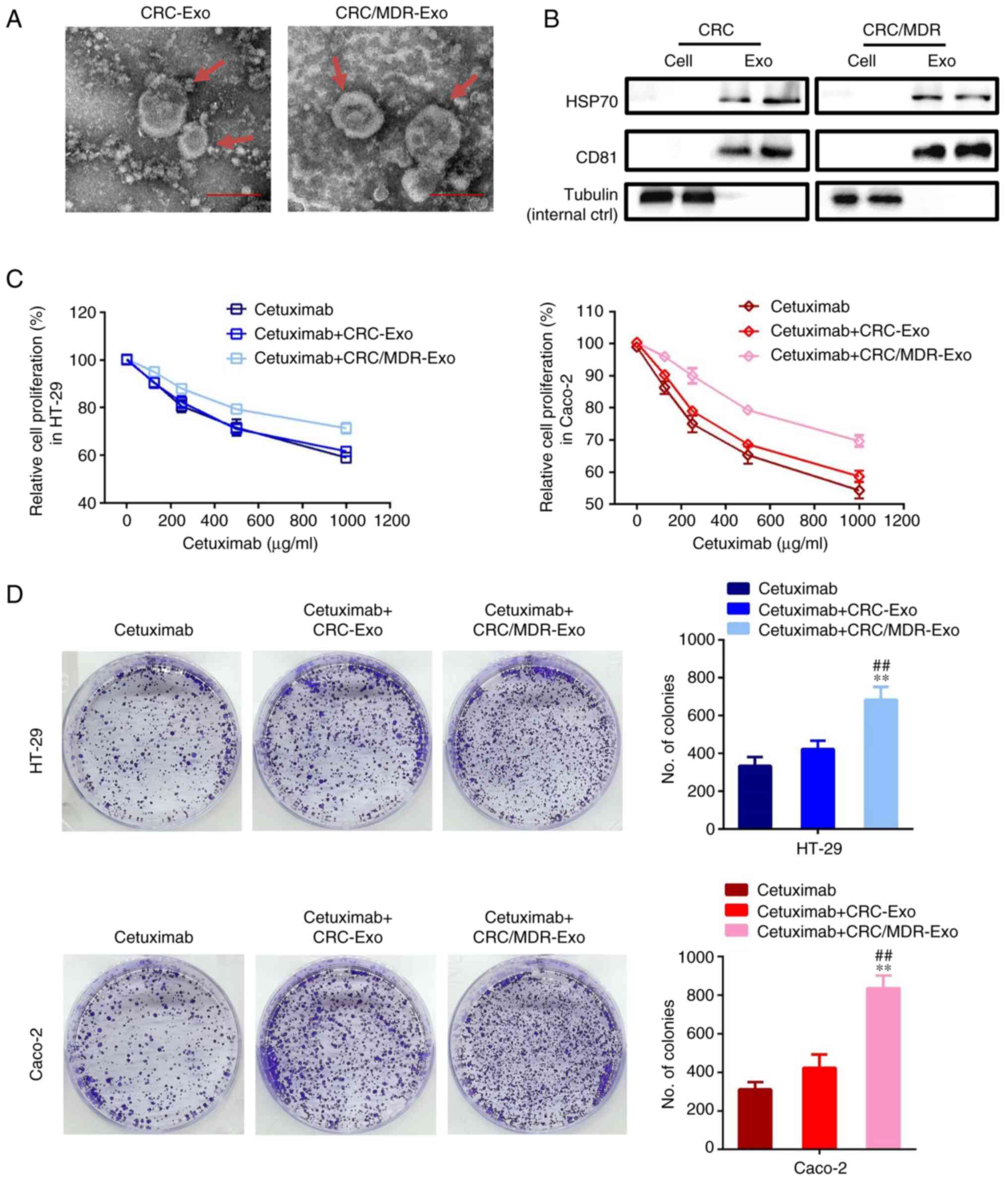 | Figure 2Exosomes derived from CRC/MDR cells
increase cetuximab resistance in cetuximab-sensitive cells. (A)
Exosomes isolated from the cell culture medium of CRC and CRC/MDR
cells were analyzed for phenotype (purity and shape) using TEM. Red
arrows indicate representative exosome examples. Scale bar, 100 nm.
(B) Western blotting was performed to detect the exosome markers,
HSP70 and CD81. Tubulin was used as an internal control. The
surface markers of exosomes could not be observed in normal cells.
By contrast, tubulin can be observed in cells, while HSP70 and CD81
cannot be observed in cells. (C) A Cell Counting Kit-8 assay was
used to assess the effect of cetuximab on the proliferation of
HT-29 and Caco-2 cells pre-treated with exosomes derived from
CRC/MDR cells or CRC/MDR cells for 72 h in relation to controls,
which were not treated with exosomes. The concentrations of
cetuximab used for the drug dose-response curve analysis of the
indicated cells were 0, 125, 250, 500 and 1,000 µg/ml. (D) HT-29
and Caco-2 cells were treated with exosomes derived from CRC cells
and CRC/MDR cells for colony formation analysis. Cells were imaged
using a light microscope fitted with a digital camera. The
differences among the several groups were analyzed using one-way
ANOVA followed by Duncan's test. Data are presented as the mean ±
SD from at least three experiments. **P<0.01 vs.
cetuximab; ##P<0.01 vs. cetuximab + CRC-Exo. CRC,
colorectal cancer; exo, exosome; MDR, multidrug resistance. |
The results of the present study also revealed that
the mean sizes of exosomes from CRC/MDR cells and their parental
sensitive cells were all between 100 and 200 nm, whereas the
particle numbers for the exosomes were all >2.0x107
particles/ml (data not shown), although without significant
differences. Furthermore, the exosome markers HSP70 and CD81 were
detected via western blotting. The exosomes from both CRC/MDR cells
and their parental sensitive CRC cells (referred to as CRC/MDR-Exo
and CRC Exo), showed the typical exosomal marker proteins (40) of HSP70 and CD81 (Fig. 2B).
The effect of the exosomes of CRC/MDR cells on the
spread of drug resistance of cetuximab to KRAS-wild-type cell lines
was investigated. HT-29 and Caco-2 cells were incubated with
CRC-exosomes or CRC/MDR-exosomes and treated with cetuximab at
different concentrations for cell proliferation analysis.
CRC/MDR-exosome treatment markedly reduced the sensitivity of HT-29
and Caco-2 cells to cetuximab (Fig.
2C). Similar efficiency was also noted in the clone formation
assay of HT-29 and Caco-2 cells. The clone formation assay showed
that compared with the cetuximab group, the CRC-exosome treatment
group exhibited no significant differences in colony numbers in
HT-29 cells or Caco-2 cells, while the colony formation numbers of
both cell lines in the CRC/MDR-exosome group were increased
compared with those in the cetuximab group (Figs. 2D and S1).
Exosomes derived from CRC/MDR cells
regulate the protein expression levels of EGFR-associated proteins
in HT-29 and Caco-2 cells
As demonstrated in a previous study (41), the RAS-RAF signaling pathway is one
of the downstream signaling pathways of EGFR, and even if the KRAS
gene is mutated in the RAS-RAF signaling pathway, the phenomenon of
targeted resistance is not entirely due to the expression of EGFR
and p-EGFR. The western blotting results showed that, compared with
the control group, the intervention of CRC-exosomes had no
significant effect on the expression of EGFR, p-EGFR and Akt in
Caco-2 and HT-29 cells (Fig. 3A,
C and D). However, the CRC/MDR-exosome
intervention had significant effect on the expression of Sox2 and
PD-L1 in Caco-2 cells (Fig.
3D).
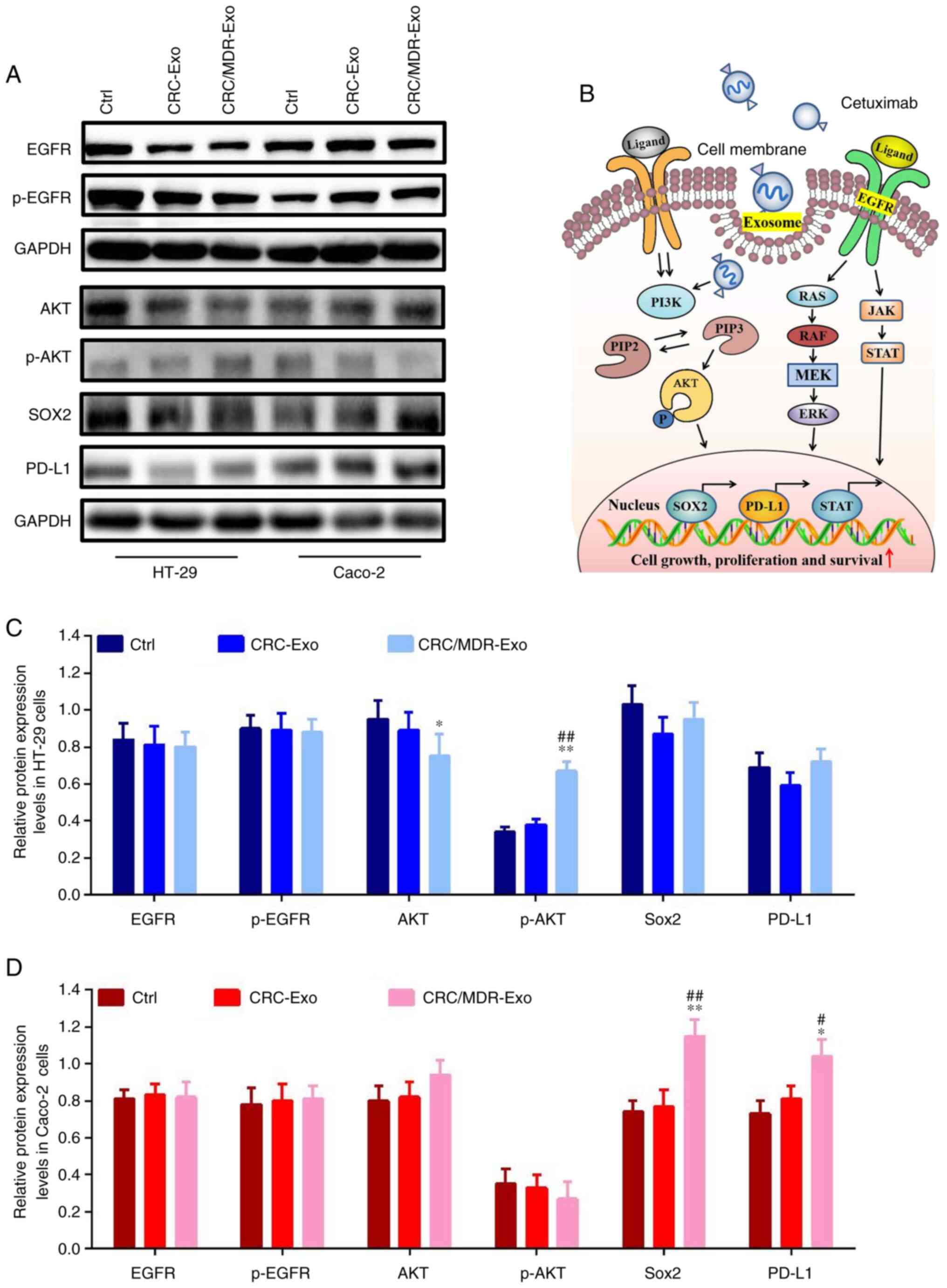 | Figure 3Exosomes derived from CRC/MDR cells
regulate EGFR-related protein expression levels in HT-29 and Caco-2
cells. (A) Western blotting of EGFR, p-EGFR, AKT, p-AKT, Sox2 and
PD-L1 proteins in HT-29 and Caco-2 cells with the indicated
treatments. (B) Proposed working model. In KRAS wild-type CRC
cells, the activation of the PI3K/AKT signaling pathway increased
cell proliferation and survival, increased the expression and
transcription of Sox2 and PD-L1 and resulted in cetuximab
resistance. Western blotting quantitative assays of EGFR, p-EGFR,
AKT, p-AKT, Sox2 and PD-L1 proteins in (C) HT-29 and (D) Caco-2
cells with the indicated treatments. The differences among the
several groups were analyzed using one-way ANOVA followed by
Duncan's test. Data are presented as the mean ± SD from at least
three experiments. *P<0.05 and **P<0.01
vs. cetuximab; #P<0.05 and ##P<0.01 vs.
cetuximab + CRC-Exo. CRC, colorectal cancer; exo, exosome; MDR,
multidrug resistance; Ctrl, Control; p, phosphorylated; PIP2,
phosphatidylinositol-4, 5-bisphosphate; PIP3,
phosphatidylinositol-3,4,5-triphosphate; JAK, Janus kinase; PD-L1,
programmed death-ligand 1. |
The phosphorylation levels of AKT were significantly
elevated in the CRC/MDR-exosomes group compared with the
CRC-exosomes group in HT-29 cells. However, CRC/MDR-exosome
treatment demonstrated no significant effect on the phosphorylation
levels of AKT in Caco-2 cells (Fig.
S2). Therefore, HT-29 cells were selected for subcutaneous
xenograft in the animal model.
Based on the aforementioned results and other
studies (16,42), we hypothesized that
CRC/MDR-exosomes could activate the phosphorylation of components
of the PI3K/AKT signaling pathway in HT-29 cells and regulate the
Sox2-mediated activity of stem cells in Caco-2 cells. These effects
together potentially directed the inhibitory effect of cetuximab
resistance in KRAS wild-type cells (Fig. 3B). Furthermore, the present study
demonstrated that CRC/MDR-exosomes significantly regulated the
protein expression levels of the key EGFR signaling pathway protein
AKT in KRAS-wild type cells compared with that in the CRC exosome
treatment group.
Exosomes derived from CRC/MDR cells
increase tumorigenesis-associated CSC gene expression
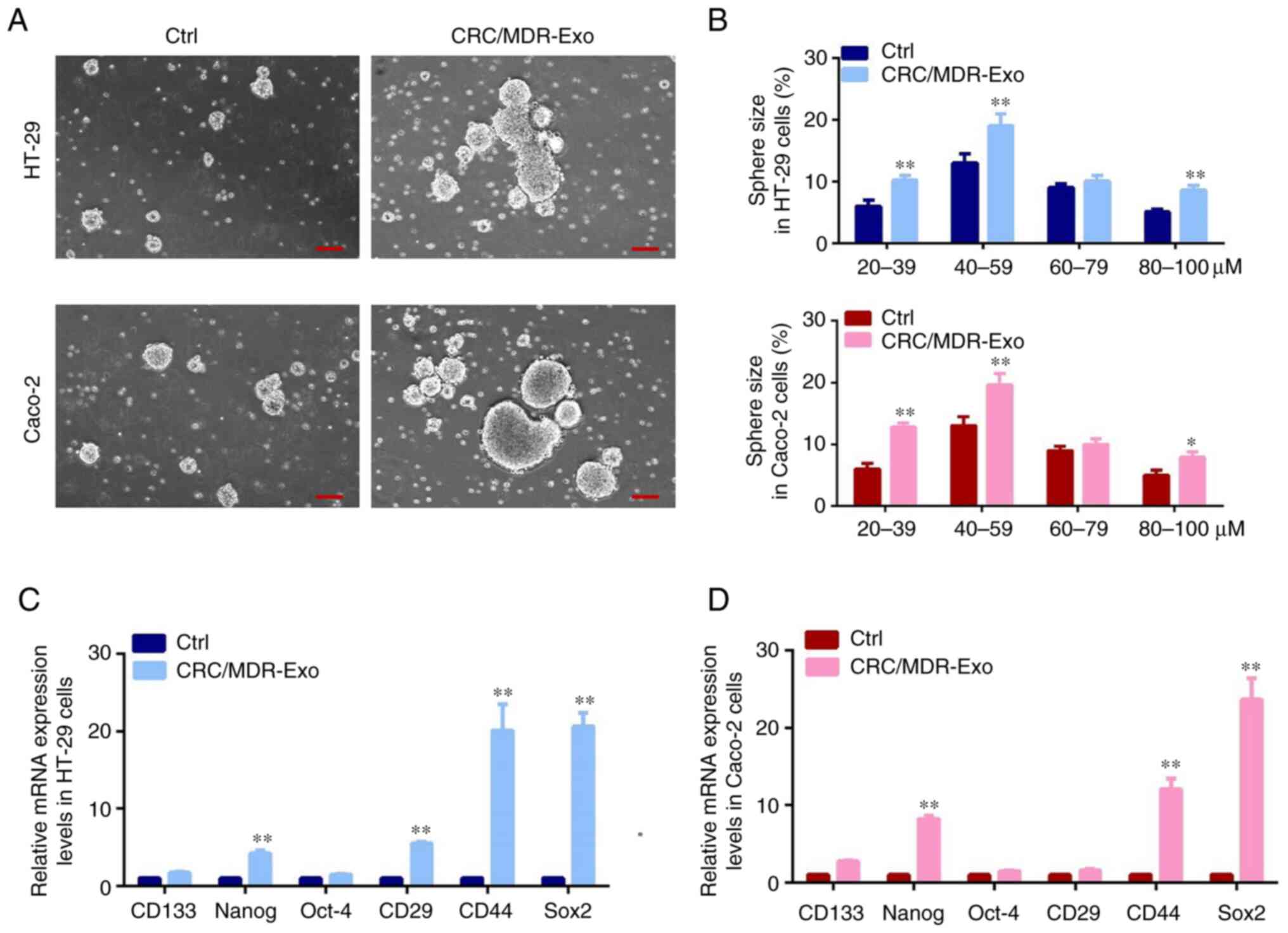
Cell proliferation is a fundamental step in
tumorigenesis progression, which can be analyzed via
sphere-formation. In the present study, the number and size of
spheres were quantified and analyzed using stem cell medium to test
sphere-forming capacity (Fig. 4A
and B). Previous studies reported
that a clear distinction could be observed in the capacity of cells
to form spheres under different treatment conditions (43,44).
Consistent with previous studies, spheres were counted as 10-100 µM
size range, and in this size range the spheres had more capability
to grow. The present study demonstrated that CRC/MDR-exosomes
caused an increase of heterogeneous cell populations containing a
number of sphere-forming subpopulations in HT-29 cells and Caco-2
cells compared with the cells without exosomes treatment,
especially in the range of 20 to 39, 40 to 59, and 80 to 100 µM
size compared with the control group under the same culture
conditions. Furthermore, the present study compared the mRNA
expression levels of stem-cell-associated markers, such as CD133,
Nanog, Oct-4, CD29, CD44 and Sox2, via RT-qPCR to assess whether
the spheres formed had the features of tumor-initiating cells.
Results of the present study also showed that higher expression
levels of Nanog, CD44 and Sox2 were observed under CRC/MDR-exosomes
treatment as compared with the normal culture condition without
exosome treatment. The expression levels of CD29 were also
significantly higher in CRC/MDR-exosome treatment cells compared
with cells without exosome treatment in HT-29 cells (Fig. 4C and D). The aforementioned results suggested
that sphere forming cells may exhibit increased stemness following
treatment with CRC/MDR-exosomes.
Effects of exosomes derived from
CRC/MDR cells in vivo
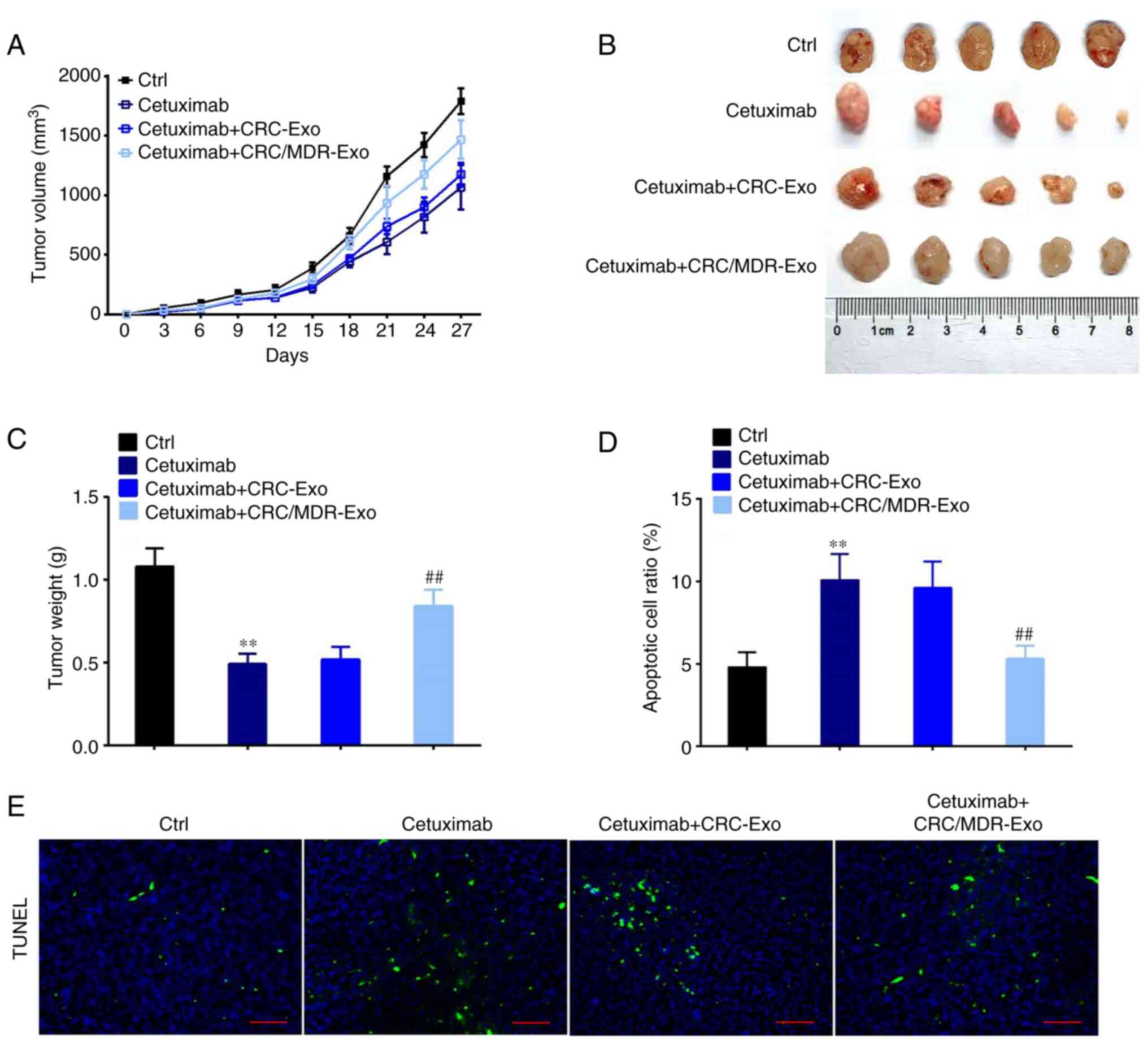
Subcutaneous xenograft of HT-29 cells in nude mouse
models was performed to assess whether CRC/MDR-exosomes inhibited
the anti-tumor effects of cetuximab in vivo. Treatment with
exosomes derived from CRC/MDR cells also reduced the inhibitory
effect of cetuximab on the tumor growth as compared with cetuximab
alone, whereas the exosomes derived from CRC cells greatly
recovered the inhibitory effect of cetuximab on the tumor growth
(Fig. 5A). Final tumor volume and
weight assessment demonstrated that tumors derived from the
cetuximab + CRC/MDR-exosomes group were significantly heavier than
those in the group of exosomes derived from CRC cells with
cetuximab treatment (Fig. 5B and
C).
Subsequently, the TUNEL assay was performed to
observe the apoptotic tumor cells in the subcutaneous xenograft of
HT-29 cells with different treatments (Fig. 5D and E). The treatment of cells with exosomes
derived from CRC/MDR cells followed by incubation with cetuximab
demonstrated significantly fewer apoptotic tumor cells in the
xenograft tumor compared with the cetuximab + CRC-exosome group.
However, the TUNEL assay showed the most apoptotic tumor cells in
the cetuximab alone group.
Effects of CRC/MDR-exosomes on the
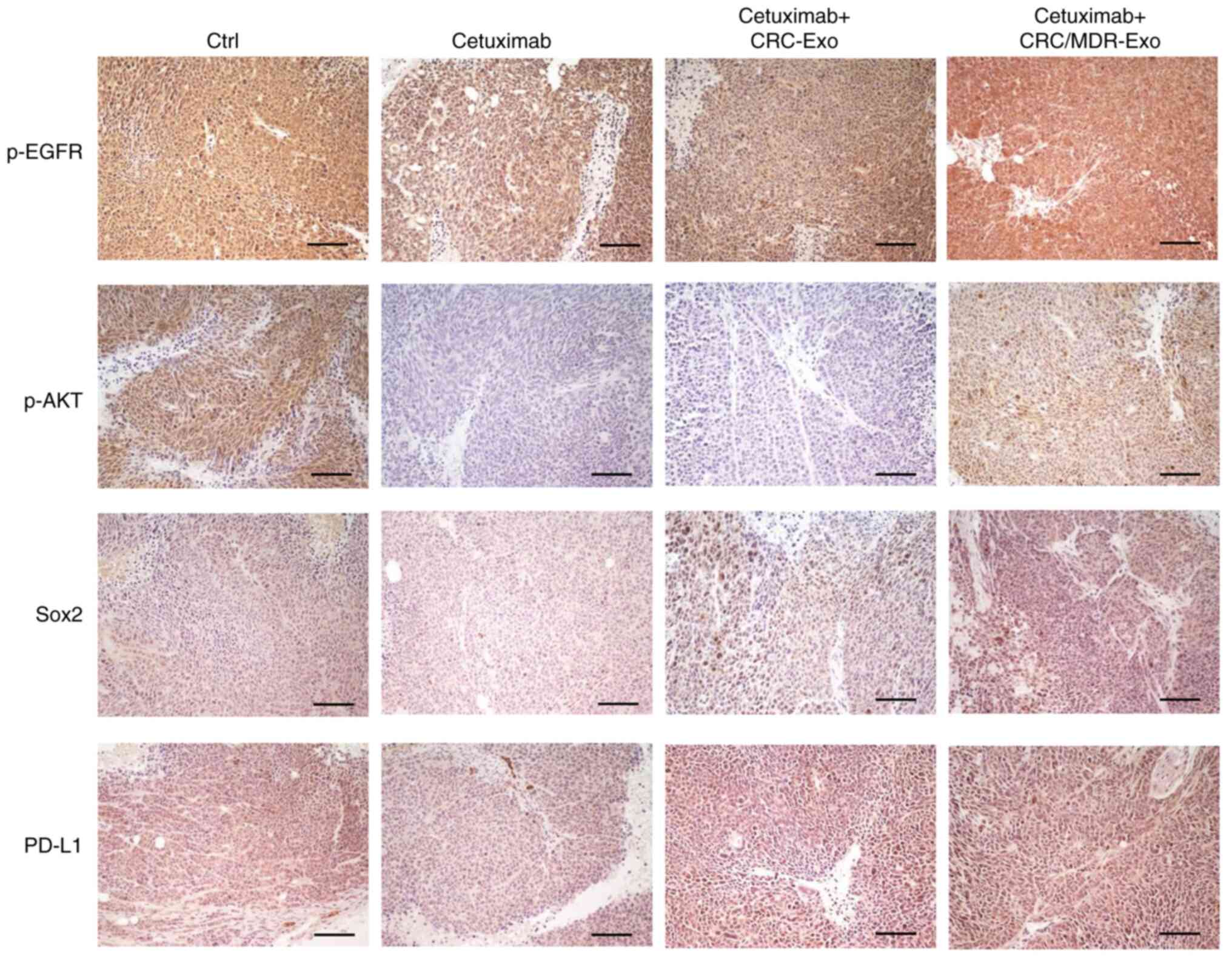
protein expression levels of EGFR-associated proteins in vivo
The protein expression levels of p-EGFR, p-AKT, Sox2
and PD-L1 in the xenograft tumor tissues were detected via IHC. The
images obtained via IHC demonstrated that treatment with exosomes
derived from CRC-MDR cells followed by incubation with cetuximab
demonstrated higher expression levels (based on the trend of brown
staining in each group) of Sox2 and PD-L1 in the xenograft tumor
compared with cetuximab alone. This suggested that exosomes from
CRC/MDR cells activated cetuximab-sensitive cells and enhanced cell
stemness via stimulation of the PI3k/AKT signaling pathway, which
conferred cetuximab resistance in CRC cells (Fig. 6 and Table SIV).
Discussion
Cetuximab is one of the most widely used EGFR
inhibitors in the treatment of patients with mCRC and wild-type
KRAS status and use in combination with chemotherapy is a standard
first-line treatment regimen (8).
However, primary or acquired resistance to cetuximab often occurs
during targeted therapy (45). To
optimize individualized cetuximab therapy in patients with CRC, it
is essential to identify the possible mechanism of the response to
cetuximab therapy. Previous studies of drug resistance to EGFR
inhibitors have focused on understanding resistance mechanisms to
EGFR kinase inhibitors and the findings from those studies have
been applied to develop the next generation of clinical trials for
CRC treatment (41,46). However, there has been limited
exploration of the mechanisms of acquired cetuximab resistance and
the mechanism of acquired cetuximab resistance has been neglected
in the TME.
The TME originates from the idea of ‘seed and soil’,
which was proposed by Stephen Paget (47). The ‘seed and soil’ hypothesis holds
that metastasis depends on the interaction between the ‘seed’
(cancer cell) and the ‘soil’ (host microenvironment). Accumulating
evidence has demonstrated that exosomes support the development of
drug resistance in cancer cells via the secretion of different
proteins and nucleic acids in the TME, which can establish drug
resistance in nearby or distant cancer cells (10,29,48).
It was therefore hypothesized that one mechanism by which MDR cells
containing exosomes might facilitate tumor niche development is via
the alteration of the tumor stroma. Therefore, the present study
aimed to investigate the potential regulatory effect of MDR cells
on CRC cells that were sensitive to cetuximab.
TCGA analysis demonstrated that patients with KRAS
wild-type had longer progression-free survival than those with
KRAS-mutant. Previous data have shown that mCRC lesions harboring
KRAS and BRAF mutations are highly associated with a poor prognosis
and poor objective response to cetuximab therapy (49). Furthermore, an increasing trend
(brown staining indicating positive cells) of
serine/threonine-protein kinase BRAF protein expression levels was
also identified in patients with mCRC with a lack of response to
cetuximab therapy (KRAS mutant group). The results of the present
study were in accordance with the findings of a previous study,
which reported that cetuximab and panitumumab anti-EGFR therapy
significantly improved the survival of patients with KRAS wild-type
memorial sloan-kettering cancer center, but was ineffective in the
KRAS mutant group (50).
TEM and the assessment of the protein expression
levels of exosome specific proteins were performed to characterize
exosomes obtained from the culture medium of CRC and CRC/MDR cells.
It was demonstrated that exosomes derived from CRC/MDR cells
significantly increased resistance to cetuximab treatment in
previously cetuximab-sensitive CRC cells. These results were in
agreement with the previously reported observation of
chemotherapy-induced drug resistance in CRC (51). Furthermore, it has been reported
that even with targeted therapeutic agents, such as cetuximab or
panitumumab, resistance has also occurred in patients with mCRC
(52) and has been implicated as a
selection process for CSC-like cells (53).
It has been reported that CSCs maintain the vitality
of tumor cell populations via self-renewal and infinite
proliferation, which suggests that CSCs can participate in
tumorigenesis, invasion and metastasis and have critical
significance in chemoradiotherapy resistance (54). Generally, tumor stem cells in
organs can be distinguished by surface markers, such as CD24 ligand
for P-selectin, CD44 hyaluronan receptor, CD133 five-transmembrane
glycoprotein expressed on the cell surface and epithelial cell
adhesion molecule, as a side population of cells with ABC
transporters and aldehyde dehydrogenase activity (55). In the present study, both HT-29 and
Caco-2 cells consistently demonstrated several CSC-like features,
such as an enhanced ability to generate tumor spheres in
vitro and enhanced tumorigenic ability in vivo.
Aberrant Sox2 expression is associated with numerous
malignancies and has well-characterized roles in tumor growth,
metastasis and drug resistance (56). Emerging evidence has reported that
inhibition of PI3K signaling decreases the protein expression
levels of Sox2 in CSCs, which suggests that PI3K/mTOR inhibition
may successfully circumvent cetuximab resistance via the
downregulation of Sox2 and the modulation of downstream
transcriptional programs (57,58).
In the present study, Sox2 and PD-L1 protein expression levels in
Caco-2 cells were related to the response to CRC/MDR-exosomes.
Meanwhile, based on the morphology of sphere samples, cell stemness
was demonstrated to be markedly increased in cells treated with
exosomes from CRC/MDR cells compared with untreated controls, which
suggested that anti-EGFR therapy resistance could be related to the
change of the TME after chemotherapy resistance. The TME in CRC may
potentially disrupt the downstream signal transduction of anti-EGFR
therapy, which renders the wild-type KRAS CRC resistant to
cetuximab. The present study revealed that CRC/MDR-exosomes could
activate the phosphorylation of components of the PI3K/AKT
signaling pathway in HT-29 cells and regulate the Sox2-mediated
activity of stem cells in Caco-2 cells. Numerous clinical
investigations have reported that even with targeted therapeutic
agents such as cetuximab or panitumumab, resistance has developed
in patients with mCRC and therefore has been implicated as a
selection process for CSC-like cells (49,59).
In our previous study, it was reported that elevated
protein levels of PI3K and AKT were observed in MDR colorectal
cancer cells compared with in sensitive cancer cells (57). This suggests a major resistance
mechanism in PI3K/AKT-activated protein kinase (MAPK)-targeted
therapies in CRC. Moreover, the previous study suggests that the
activation of a non-canonical and independent PI3K/AKT signaling
pathway, which involved the overexpression of Sox2 and PD-L1
mediated the activation of tumor stem cells in CRC (25). However, in the present study that
Sox2 and PD-L1 protein expression levels demonstrated different
responses to CRC/MDR-exosomes in different KRAS wild-type cells. A
new mechanism that promotes cetuximab resistance progression via
CRC/MDR-exosome regulation of the activation of the PI3K/AKT
signaling pathway and Sox2 and PD-L1 protein expression levels was
proposed in the present study.
It was previously demonstrated that exosome delivery
from cisplatin-resistant gastric cancer cells may induce
chemoresistance phenotypes in sensitive cancer cells (16). The present study demonstrated that
MDR/CRC-exosomes can change the cell proliferation and apoptotic
phenotype of cetuximab-sensitive CRC cells. Furthermore, the
CRC-exosomes demonstrated a smaller effect on the cell apoptotic
phenotype of cetuximab-sensitive CRC cells compared with the
MDR/CRC-exosomes. To the best of our knowledge, the present study
is the first to document the transmissibility of cetuximab
resistance via CRC/MRD-exosomes, not only by the enhanced
generation of tumor spheres in vitro, but also via enhanced
tumorigenic ability in vivo. Furthermore, the translational
regulation of Sox2 and PD-L1 via the PI3K/AKT signaling pathway was
also demonstrated as a response potentially mediated by
CRC/MDR-exosomes.
However, the major limitations of the present study
include that downstream regulation of PI3K/AKT in
cetuximab-sensitive cells was not investigated following treatment
with CRC/MDR-exosomes. Furthermore, in clinical applications
cetuximab is administered to patients with mCRC, in combination
with irinotecan when irinotecan-based therapy has failed (60). It has been reported that patients
with KRAS mutant could not benefit from cetuximab; however, a
recent clinical study has reported that patients with RAS wild-type
tumors derived a significant benefit from the addition of cetuximab
to FOLFIRI (fluorouracil, leucovorin, and irinotecan, the
first-line treatment of mCRC) (61). Therefore, it is also important to
study chemotherapy resistance in KRAS mutants. Although certain
factors in drug resistance have previously been widely studied, the
findings of the present study present important new possibilities
for future clinical application.
Supplementary Material
Exosomes derived from CRC/MDR cells
increase cetuximab resistance in cetuximab-sensitive cells. HT-29
and Caco-2 cells were treated with exosomes derived from CRC cells
and CRC/MDR cells for colony formation analysis. Cells were imaged
using a microscope fitted with a digital camera. Data are presented
as the mean ± SD from at least three experiments.
**P<0.01 vs. cetuximab; ##P<0.01 vs.
cetuximab + CRC-Exo. CRC, colorectal cancer; exo, exosome; MDR,
multidrug resistance; Ctrl, control.
Exosomes derived from CRC/MDR cells
regulate the phosphorylation levels of EGFR and AKT proteins in
HT-29 and Caco-2 cells. Western blotting assays of the ratio of
p-EGFR to EGFR and p-AKT to AKT proteins in the in HT-29 and Caco-2
cells treated with the indicated treatments. **P<0.01
vs. cetuximab; ##P<0.01 vs. cetuximab + CRC-Exo. CRC,
colorectal cancer; p, phosphorylated; exo, exosome; MDR, multidrug
resistance; Ctrl, control.
Information of patients with
colorectal cancer.
Primers for reverse
transcription-quantitative PCR.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant nos. 81874399, 81973808 and
82174459), the Science Foundation for Shanghai Committee of Science
Project (grant nos. 21S21901400 and 19411972000), Shanghai
Municipal Health Commission (grant no. 202140198).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZWe and ZWa performed the experiments. ZL, MZ, YZ,
QC, LZ and QT analyzed the data and prepared figures. HS and HZ
conceived and designed the experiments, analyzed the data and
critically revised the manuscript. ZWe and HS confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Institutional Animal
Care and Use Committee of Shuguang Hospital, Shanghai University of
Traditional Chinese Medicine (approval no. PZSHUTCM200724027;
Shanghai, China). The present study was approved by the Medical
Ethics and Human Clinical Trial Committee of the affiliated
hospitals, Shanghai University of Traditional Chinese Medicine
(approval no. 2020-824-31; Shanghai, China). All of the patients or
their parents/guardians provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mizukami T, Izawa N, Nakajima TE and
Sunakawa Y: Targeting EGFR and RAS/RAF signaling in the treatment
of metastatic colorectal cancer: From current treatment strategies
to future perspectives. Drugs. 79:633–645. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Weng J, Li S, Zhu Z, Liu Q, Zhang R, Yang
Y and Li X: Exploring immunotherapy in colorectal cancer. J Hematol
Oncol. 15(95)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kim SY and Kim TW: Current challenges in
the implementation of precision oncology for the management of
metastatic colorectal cancer. ESMO Open. 5(e000634)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Beckler MD, Higginbotham JN, Franklin JL,
Ham AJ, Halvey PJ, Imasuen IE, Whitwell C, Li M, Liebler DC and
Coffey RJ: Proteomic analysis of exosomes from mutant KRAS colon
cancer cells identifies intercellular transfer of mutant KRAS. Mol
Cell Proteomics. 12:343–355. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bellier J, Nokin MJ, Caprasse M, Tiamiou
A, Blomme A, Scheijen JL, Koopmansch B, MacKay GM, Chiavarina B,
Costanza B, et al: Methylglyoxal scavengers resensitize
KRAS-mutated colorectal tumors to cetuximab. Cell Rep.
30:1400–1416. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tak E, Kim M, Cho Y, Choi S, Kim J, Han B,
Kim HD, Jang CS, Kim JE, Hong YS, et al: Expression of
neurofibromin 1 in colorectal cancer and cetuximab resistance.
Oncol Rep. 47(15)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Woolston A, Khan K, Spain G, Barber LJ,
Griffiths B, Gonzalez-Exposito R, Hornsteiner L, Punta M, Patil Y,
Newey A, et al: Transcriptomic determinants of therapy resistance
and immune landscape evolution during Anti-EGFR treatment in
colorectal cancer. Cancer Cell. 36:35–50. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Martinelli E, Ciardiello D, Martini G,
Troiani T, Cardone C, Vitiello PP, Normanno N, Rachiglio AM,
Maiello E, Latiano T, et al: Implementing anti-epidermal growth
factor receptor (EGFR) therapy in metastatic colorectal cancer:
Challenges and future perspectives. Ann Oncol. 31:30–40.
2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Mangiapane LR, Nicotra A, Turdo A,
Gaggianesi M, Bianca P, Di Franco S, Sardina DS, Veschi V, Signore
M, Beyes S, et al: PI3K-driven HER2 expression is a potential
therapeutic target in colorectal cancer stem cells. Gut.
71:119–128. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yang YN, Zhang R, Du JW, Yuan HH, Li YJ,
Wei XL, Du XX, Jiang SL and Han Y: Predictive role of
UCA1-containing exosomes in cetuximab-resistant colorectal cancer.
Cancer Cell Int. 18(164)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang Y, Huo L, Wei Z, Tang Q and Sui H:
Hotspots and frontiers in inflammatory tumor microenvironment
research: A scientometric and visualization analysis. Front
Pharmacol. 13(862585)2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Touil Y, Igoudjil W, Corvaisier M, Dessein
AF, Vandomme J, Monté D, Stechly L, Skrypek N, Langlois C, Grard G,
et al: Cancer cells escape 5FU chemotherapy-induced cell death by
entering stemness and quiescence associated with the c-Yes/YAP
axis. Clin Cancer Res. 20:837–846. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Mallini P, Lennard T, Kirby J and Meeson
A: Epithelial-to-mesenchymal transition: What is the impact on
breast cancer stem cells and drug resistance. Cancer Treat Rev.
40:341–348. 2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yin Z, Yu M, Ma T, Zhang C, Huang S,
Karimzadeh MR, Momtazi-Borojeni AA and Chen S: Mechanisms
underlying low-clinical responses to PD-1/PD-L1 blocking antibodies
in immunotherapy of cancer: A key role of exosomal PD-L1. J
Immunother Cancer. 9(e001698)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ji Q, Zhou L, Sui H, Yang L, Wu X, Song Q,
Jia R, Li R, Sun J, Wang Z, et al: Primary tumors release
ITGBL1-rich extracellular vesicles to promote distal metastatic
tumor growth through fibroblast-niche formation. Nat Commun.
11(1211)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sun MY, Xu B, Wu QX, Chen WL, Cai S, Zhang
H and Tang QF: Cisplatin-resistant gastric cancer cells promote the
chemoresistance of cisplatin-sensitive cells via the exosomal
RPS3-mediated PI3K-Akt-Cofilin-1 signaling axis. Front Cell Dev
Biol. 9(618899)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tan Z, Gao L, Wang Y, Yin H, Xi Y, Wu X,
Shao Y, Qiu W, Du P, Shen W, et al: PRSS contributes to cetuximab
resistance in colorectal cancer. Sci Adv.
6(eaax5576)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cremolini C, Rossini D, Dell'Aquila E,
Lonardi S, Conca E, Del Re M, Busico A, Pietrantonio F, Danesi R,
Aprile G, et al: Rechallenge for patients with RAS and BRAF
wild-type metastatic colorectal cancer with acquired resistance to
first-line cetuximab and irinotecan: A phase 2 single-arm clinical
trial. JAMA Oncol. 5:343–350. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chinese Society Of Clinical Oncology Csco
Diagnosis And Treatment Guidelines For Colorectal Cancer Working
Group. Diagnosis and treatment guidelines for colorectal cancer
working group CSOCOC: Chinese society of clinical oncology (CSCO)
diagnosis and treatment guidelines for colorectal cancer 2018
(English version). Chin J Cancer Res. 31:117–134. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A (eds): AJCC cancer staging manual. 7th
edition. Springer, New York, NY, 2010.
|
|
21
|
Karnofsky DA, Abelmann WH, Craver LF and
Burchenal JH: The use of the nitrogen mustards in the palliative
treatment of carcinoma-with particular reference to bronchogenic
carcinoma. Cancer. 1:634–656. 1948.
|
|
22
|
Junjie P, Ji Z and Fangqi L: Chinese
expert consensus on the diagnosis and treatment of locally advanced
rectal cancer. Chin J Cancer. 1:41–80. 2017.(In Chinese).
|
|
23
|
National Health Commission of the People's
Republic of China. Chinese Protocol of Diagnosis and Treatment of
Colorectal Cancer (2020 edition). Zhonghua Wai Ke Za Zhi.
58:561–585. 2020.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
24
|
Lu H, Sun J, Xu JH and Fan ZZ:
Establishment of HCT116/L-OHP in oxaliplatin-resistant cells in
human colon cancer. Tumors. 31:675–681. 2011.
|
|
25
|
Sui H, Pan SF, Feng Y, Jin BH, Liu X, Zhou
LH, Hou FG, Wang WH, Fu XL, Han ZF, et al: Zuo Jin Wan reverses
P-gp-mediated drug-resistance by inhibiting activation of the
PI3K/Akt/NF-κB pathway. BMC Complement Altern Med.
14(279)2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sui H, Zhou L, Zhang Y, Huang JP, Liu X,
Ji Q, Fu XL, Wen HT, Chen ZS, Deng WL, et al: Evodiamine suppresses
ABCG2 mediated drug resistance by inhibiting p50/p65/NF-κB pathway
in colorectal cancer. J Cell Biochem. 117:1471–1481.
2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang Z, Sun X, Feng Y, Liu X, Zhou L, Sui
H, Ji Q, E Q, Chen J, Wu L and Li Q: Dihydromyricetin reverses
MRP2-mediated MDR and enhances anticancer activity induced by
oxaliplatin in colorectal cancer cells. Anticancer Drugs.
28:281–288. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kim SA, Park H, Kim KJ, Kim JW, Sung JH,
Nam M, Lee JH, Jung EH, Suh KJ, Lee JY, et al: Cetuximab resistance
induced by hepatocyte growth factor is overcome by MET inhibition
in KRAS, NRAS, and BRAF wild-type colorectal cancers. J Cancer Res
Clin Oncol. 148:2995–3005. 2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chu YC, Tsai TY, Yadav VK, Deng L, Huang
CC, Tzeng YM, Yeh CT and Chen MY: 4-Acetyl-Antroquinonol B improves
the sensitization of cetuximab on both kras mutant and wild type
colorectal cancer by modulating the expression of
Ras/Raf/miR-193a-3p signaling axis. Int J Mol Sci.
22(7508)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li F, Zhan L, Dong Q, Wang Q, Wang Y, Li
X, Zhang Y and Zhang J: Tumor-derived exosome-educated hepatic
stellate cells regulate lactate metabolism of hypoxic colorectal
tumor cells via the IL-6/STAT3 pathway to confer drug resistance.
Onco Targets Ther. 13:7851–7864. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Sun MY, Zhang H, Tao J, Ni ZH, Wu QX and
Tang QF: Expression and biological function of rhotekin in gastric
cancer through regulating p53 pathway. Cancer Manag Res.
11:1069–1080. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wang T, Dong J, Yuan X, Wen H, Wu L, Liu
J, Sui H and Deng W: A new chalcone derivative C49 reverses
doxorubicin resistance in MCF-7/DOX cells by inhibiting
P-glycoprotein expression. Front Pharmacol.
12(653306)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liu L, Salnikov AV, Bauer N,
Aleksandrowicz E, Labsch S, Nwaeburu C, Mattern J, Gladkich J,
Schemmer P, Werner J and Herr I: Triptolide reverses
hypoxia-induced epithelial-mesenchymal transition and stem-like
features in pancreatic cancer by NF-κB downregulation. Int J
Cancer. 134:2489–2503. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wang J, Guo LP, Chen LZ, Zeng YX and Lu
SH: Identification of cancer stem cell-like side population cells
in human nasopharyngeal carcinoma cell line. Cancer Res.
67:3716–3724. 2007.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Elston CW and Ellis IO: Pathological
prognostic factors in breast cancer. I. The value of histological
grade in breast cancer: Experience from a large study with
long-term follow-up. Histopathology. 19:403–410. 1991.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Bauman JE and Grandis JR: Targeting
secondary immune responses to cetuximab: CD137 and the outside
story. J Clin Invest. 124:2371–2375. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Whiteside TL: Exosomes and tumor-mediated
immune suppression. J Clin Invest. 126:1216–1223. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Binenbaum Y, Fridman E, Yaari Z, Milman N,
Schroeder A, David GB, Shlomi T and Gil Z: Transfer of miRNA in
macrophage-derived exosomes induces drug resistance in pancreatic
adenocarcinoma. Cancer Res. 78:5287–5299. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Witwer KW, Buzás EI, Bemis LT, Bora A,
Lässer C, Lötvall J, Nolte-'t Hoen EN, Piper MG, Sivaraman S, Skog
J, et al: Standardization of sample collection, isolation and
analysis methods in extracellular vesicle research. J Extracell
Vesicles. 27(2)2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Yonesaka K, Zejnullahu K, Okamoto I, Satoh
T, Cappuzzo F, Souglakos J, Ercan D, Rogers A, Roncalli M, Takeda
M, et al: Activation of ERBB2 signaling causes resistance to the
EGFR directed therapeutic antibody cetuximab. Sci Transl Med.
3(99ra86)2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Duan H, Liu Y, Gao Z and Huang W: Recent
advances in drug delivery systems for targeting cancer stem cells.
Acta Pharm Sin B. 11:55–70. 2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhao JJ, Lin J, Zhu D, Wang X, Brooks D,
Chen M, Chu ZB, Takada K, Ciccarelli B, Admin S, et al: miR-30-5p
functions as a tumor suppressor and novel therapeutic tool by
targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res.
74:1801–1813. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Yun JH, Kim KA, Yoo G, Kim SY, Shin JM,
Kim JH, Jung SH, Kim J and Nho CW: Phenethyl isothiocyanate
suppresses cancer stem cell properties in vitro and in a xenograft
model. Phytomedicine. 30:42–49. 2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Han Y, Peng Y, Fu Y, Cai C, Guo C, Liu S,
Li Y, Chen Y, Shen E, Long K, et al: MLH1 deficiency induces
cetuximab resistance in colon cancer via Her-2/PI3K/AKT signaling.
Adv Sci (Weinh). 7(2000112)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Li H, Zhou L, Zhou J, Li Q and Ji Q:
Underlying mechanisms and drug intervention strategies for the
tumour microenvironment. J Exp Clin Cancer Res.
40(97)2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ribatti D, Mangialardi G and Vacca A:
Stephen Paget and the ‘seed and soil’ theory of metastatic
dissemination. Clin Exp Med. 6:145–149. 2006.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lièvre A, Bachet JB, Boige V, Cayre A,
Corre DL, Buc E, Ychou M, Bouché O, Landi B, Louvet C, et al: Kras
mutations as an independent prognostic factor in patients with
advanced colorectal cancer treated with cetuximab. J Clin Oncol.
26:374–379. 2008.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Formica V, Sera F, Cremolini C, Riondino
S, Morelli C, Arkenau HT and Roselli M: KRAS and BRAF mutations in
Stage II and III colon cancer: A systematic review and
meta-analysis. J Natl Cancer Inst. 114:517–527. 2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Cai MH, Xu XG, Yan SL, Sun Z, Ying Y, Wang
BK and Tu YX: Regorafenib suppresses colon tumorigenesis and the
generation of drug resistant cancer stem-like cells via modulation
of miR-34a associated signaling. J Exp Clin Cancer Res.
37(151)2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Hsu HC, Thiam TK, Lu YJ, Yeh CY, Tsai WS,
You JF, Hung HY, Tsai CN, Hsu A, Chen HC, et al: Mutations of
KRAS/NRAS/BRAF predict cetuximab resistance in metastatic
colorectal cancer patients. Oncotarget. 7:22257–22270.
2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Lu H, Chen I, Shimoda LA, Park Y, Zhang C,
Tran L, Zhang H and Semenza GL: Chemotherapy-induced Ca2+ release
stimulates breast cancer stem cell enrichment. Cell Rep.
18:1946–1957. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Cioffi M, D'Alterio C, Camerlingo R,
Tirino V, Consales C, Riccio A, Ieranò C, Cecere SC, Losito NS,
Greggi S, et al: Identification of a distinct population of
CD133+CXCR4+ cancer stem cells in ovarian cancer. Sci Rep.
5(10357)2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Steinbichler TB, Dudás J, Skvortsov S,
Ganswindt U, Riechelmann H and Skvortsova II: Therapy resistance
mediated by cancer stem cells. Semin Cancer Biol. 53:156–167.
2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Wuebben EL and Rizzino A: The dark side of
SOX2: Cancer- a comprehensive overview. Oncotarget. 8:44917–44943.
2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Keysar SB, Le PN, Miller B, Jackson BC,
Eagles JR, Nieto C, Kim J, Tang B, Glogowska MJ, Morton JJ, et al:
Regulation of head and neck squamous cancer stem cells by PI3K and
SOX2. J Natl Cancer Inst. 109(djw189)2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Bouhaddou M, Lee RH, Li H, Bhola NE,
O'Keefe RA, Naser M, Zhu TR, Nwachuku K, Duvvuri U, Olshen AB, et
al: Caveolin-1 and Sox-2 are predictive biomarkers of cetuximab
response in head and neck cancer. JCI Insight.
6(e151982)2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Rothenberg SM, Concannon K, Cullen S,
Boulay G, Turke AB, Faber AC, Lockerman EL, Rivera MN, Engelman JA,
Maheswaran S and Haber DA: Inhibition of mutant EGFR in lung cancer
cells triggers SOX2-FOXO6-dependent survival pathways. Elife.
4(e06132)2015.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Liu K, Lin B, Zhao M, Yang X, Chen M, Gao
A, Liu F, Que J and Lan X: The multiple roles for Sox2 in stem cell
maintenance and tumorigenesis. Cell Signal. 25:1264–1271.
2013.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Cunningham D, Humblet Y, Siena S, Khayat
D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype
C, et al: Cetuximab monotherapy and cetuximab plus irinotecan in
irinotecan-refractory metastatic colorectal cancer. N Engl J Med.
351:337–345. 2004.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Van Cutsem E, Lenz HJ, Köhne CH, Heinemann
V, Tejpar S, Melezínek I, Beier F, Stroh C, Rougier P, van Krieken
JH and Ciardiello F: Fluorouracil, leucovorin, and irinotecan plus
cetuximab treatment and RAS mutations in colorectal cancer. J Clin
Oncol. 33:692–700. 2015.PubMed/NCBI View Article : Google Scholar
|