Introduction
Cervical cancer is the most common malignant tumor
of the female genital tract worldwide (1). Persistent high-risk human
papillomavirus (HPV) oncoproteins E6 and E7 are the main pathogenic
factors of cervical cancer (2). At
the molecular level, the HPV E6 and E7 proteins directly activate
Akt, and this pathway is further stimulated in cervical cancer
cells by amplifications and mutations of the PI3K genes. As it has
a key role in the control of HPV gene expression and development of
cervical cancer, the PI3K/AKT/mammalian target of rapamycin (mTOR)
pathway may have potential as a therapeutic target for cervical
cancer (3-5).
The tumor microenvironment (TME) contains stromal
cells and immune cells that shape cancer development and impact
responses to tumor therapy (6).
Cancer cell proliferation, angiogenesis and metastasis also
contribute to the establishment of an immunosuppressive
environment. These factors are associated with tumor progression
and poor clinical outcomes (7,8).
However, factors that contribute to immunosuppression in the TME
are poorly defined.
Epithelial-mesenchymal transition (EMT) plays an
important role in tumor development from initiation to metastasis.
EMT contributes to the majority of the hallmarks of cancer and
continues to be an attractive target for cancer therapy (9). Classical EMT is characterized by the
phenotype change of epithelial cells to cells with mesenchymal
properties, but EMT is also associated with multiple other
molecular processes, including tumor immune evasion (10). Immunosuppression occurs as a direct
consequence of the EMT program or develops through some additional,
still-uncharacterized signaling channels (11).
The PI3K/AKT/mTOR pathway can promote migration and
induce EMT in numerous types of tumors, including cervical cancer
(8,12). EMT-related changes in the
expression of various receptor tyrosine kinases (RTKs) (13) have been reported, although the
proliferation and survival dependence of specific RTKs under
different conditions remains to be fully elucidated. The expression
of an EGFR family member-ERBB3(14) is associated with the epithelial
phenotype of the cell line and the sensitivity to EGFR inhibition.
ERBB3 heterodimerizes (15) with
additional EGFR family members after stimulation with various
ligands, including neuregulin (16). ERBB3 contains multiple binding
sites for p85, which is the regulatory subunit of PI3K (17). This allows direct recruitment and
activation of PI3K signals via ERBB3(18). Although changes in ERBB3 expression
have been observed, the functional consequences of these changes
and the relationship with downstream signals after EMT (19) have not been fully described.
These targets involved in cervical cancer are not
functionally exclusive; rather, they are intertwined and
reciprocal, and together they form intricate TME networks to meet
context-specific needs for cellular function. To improve
understanding of the correlation between TME and prognosis of
cervical cancer, the present study assessed cervical cancer cell
lines and tissues to reveal the roles of ERBB3 in the EMT induction
of TME harboring immunosuppression, migration and invasion of
cervical cancer, and to explore whether PI3K/AKT/mTOR signaling is
involved in this process.
The current study intended to explore how ERBB3
mediates the PI3K/AKT/mTOR pathway and changes the tumor immune
microenvironment to affect the EMT status of cervical cancer, which
may provide further understanding of MMPs involved in
immunotherapy.
Materials and methods
Data collection and preprocessing
RNAseq data in the transcripts per million (TPM)
format from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) and The
Genotype-Tissue Expression (GTEx) (https://gtexportal.org/) were uniformly processed
using the Toil process (20).
Through extraction of the cervical squamous cell carcinoma and
endocervical adenocarcinoma (CESC) data in TCGA and corresponding
normal tissue data in GTEx, the 306 cervical tumor samples were
classified as the malignant group and the three samples adjacent to
cancer from TCGA together with the 10 normal cervix tissues from
GTEx were classified as the non-malignant group. The RNAseq data in
TPM format were log2 transformed for expression comparison between
samples.
Related hub genes selection
A total of 14 EMT-related genes were selected based
on the article ‘Guidelines and definitions for research on
epithelial-mesenchymal transition’ written by the EMT International
Association (TEMTIA) in 2020(21).
When searching for pathway related genes, GeneCards (https://www.genecards.org/) was used; the key words
‘PI3K/AKT’ and ‘PI3K/AKT/mTOR’ were searched for and genes with a
relevance score >4.0 were selected.
Gene Expression Profiling Interactive
Analysis (GEPIA)
GEPIA (http://gepia.cancer-pku.cn/index.html) is a
user-friendly web portal for gene expression analysis based on TCGA
and GTEx data. In the current study, expression analysis of ERBB3
was evaluated using the project ID of TCGA-CESC. In the module
‘Expression DIY’ of GEPIA, the expression of ERBB3 between
pan-cancer and normal tissue samples was studied with the option of
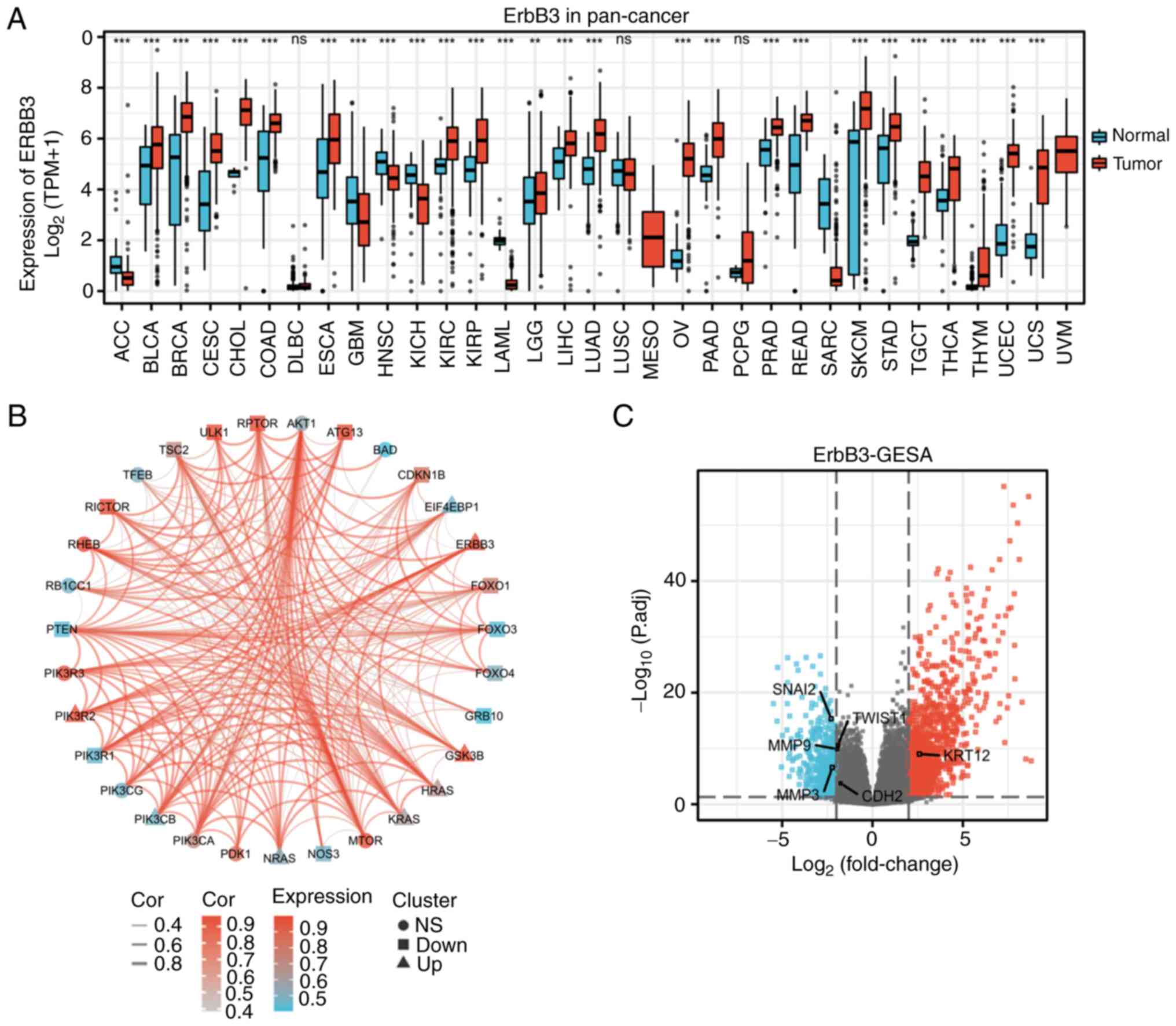
matching normal TCGA data and GTEx data (Fig. 1A).
Differentially-expressed genes
(DEGs)
Batch correction, normalization and difference
analysis of RNA-seq data from GSE63514, GSE9750, and GSE44001 were
performed to screen for DEGs in CESC samples. GSE63514(22), GSE9750(23) and GSE44001 (24,25)
were obtained from the NCBI Gene Expression Omnibus database
(ncbi.nlm.nih.gov/geo). The GSE63514
dataset used the GPL570 [HG-U133 Plus 2] Affymetrix Human Genome
U133 Plus 2.0 Array platform, which contained 28 cervical cancer
tissue samples and 24 normal samples. The GSE9750 dataset used the
GPL96 Affymetrix Human Genome U133A Array platform and included 33
cervical cancer tissue samples that were primarily marked by HPV16
or HPV18, and 21 normal cervical samples. The GSE44001 dataset used
the GPL14951 Illumina HumanHT-12 WG-DASL V4.0 R2 expression
beadchip, which contained 300 cervical cancer tissue samples.
GSE63514 and GSE9750 were set as the reference group and GSE44001
as the test group, and the R software limma package was used to
identify DEGs between the groups (26). A total of 13,473 DEGs, including
6,514 downregulated and 6,595 upregulated genes, were identified in
cervical cancer. The results were visualized using R software
(version 3.6.3) (statistical analysis and visualization) with the R
package ggplot2 [version 3.3.3] (27) to generate a volcano plot (Fig. 1C), which identified important
genes.
Genomic alteration types and
alteration frequency analysis
Genomic alteration types (missense mutation with
putative driver or unknown significance, amplification and no
alterations) and alteration frequency of 14 EMT-associated genes
and 30 PI3K/AKT/mTOR pathway-associated genes were obtained from
the cBioPortal for Cancer Genomics (http://www.cbioportal.org), using the ‘OncoPrint’
module and ‘Cancer Types Summary’ module for visualization.
Immune cell infiltration
estimation
For the immune infiltration analysis, transcriptome
or other omics data was used to calculate the score of immune cells
in the tissue through algorithms, and inferred the infiltration of
immune cells in the tissue. Single-sample Gene Set Enrichment
Analysis (GSEA) in immune infiltration, which uses the markers of
each type of immune cells (28),
was used as the gene set to calculate the enrichment of each type
of immune cells in each sample.
Gene Ontology (GO) Term and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis and GSEA
GO and KEGG (29,30)
analyses were applied to explore the biological functions of target
genes in CESC. GO analysis is a powerful bioinformatics tool to
determine the biological processes (BPs), cellular components (CCs)
and molecular functions (MFs) related to ERBB3. GSEA was utilized
to investigate the potential mechanisms of ERBB3. GO, KEGG and GSEA
were performed using the R (version 3.6.4) package ClusterProfiler
(31). P<0.1 and q<0.2 were
selected as the cut-off level.
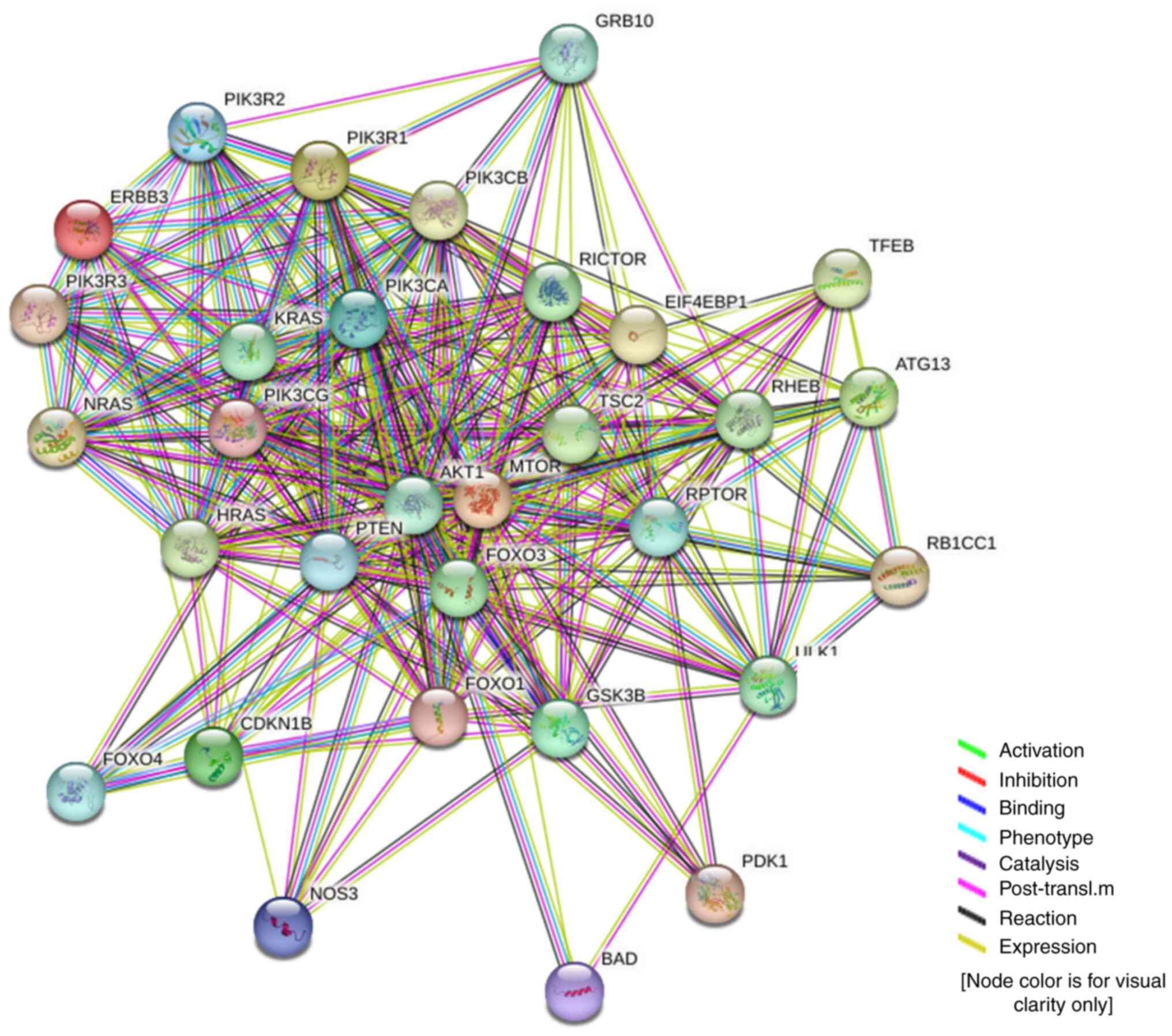
Protein interactions and biological
processes
The direct and indirect relationship between ERBB3
and 30 hub genes in the PI3K/AKT/mTOR signaling pathway were
analyzed using the online tool STRING (https://string-db.org).
Reverse transcription-quantitative PCR
(RT-qPCR)
The MecDNA-HUtrC007Ce01 commercial chip (cat. no.
8*R100-M-20190104; Shanghai Outdo Biotech Co., Ltd.) contains cDNA
reverse transcribed from RNA extracted from six cervical cancer
cell lines: CaSki, MS751, ME180, C33A, SiHa and HeLa. According to
the manufacturer's instructions, qPCR was performed using
SYBR® Premix Ex Taq™ II (Tli RNaseH Plus; RR820Q,;
Takara Bio, Inc.). Briefly, following the addition of 20 µl qPCR
MasterMix into each well, the Axigen PlateMax Ultraclear Sealing
Film (UC-500) was used to seal the chip, and it was placed on ice
for 15 min to fully dissolve the freeze-dried cDNA. The chip was
then centrifuged at 1,750 x g for 3 min at a temperature ramping
rate of 2˚C/sec. qPCR was performed using a Roche
LightCycler® 480II (Roche Diagnostics) with the
following program: Initial denaturation (95˚C, 30 sec); 40 cycles
of denaturation (95˚C, 5 sec), annealing (60˚C, 30 sec) and
elongation (95˚C, 5 sec); final elongation (60˚C, 1 min) and a
final hold (60˚C). The fold-change of gene expression was
calculated using 2-(ΔCq experimental group-ΔCq control group)
(32). β-actin was used as an
internal control and primers are as follows: ERBB3 Forward,
5'-GACCCAGGTCTACGATGGGAA-3'; ERBB3 reverse,
5'-GTGAGCTGAGTCAAGCGAG-3'; human β-actin forward,
5'-GAAGAGCTACGAGCTGCCTGA-3'; human β-actin reverse,
5'-CAGACAGCACTGTGTTGGCG-3' (product length, 191 bp).
Risk survival analysis
Kaplan-Meier curves (33) can describe the survival status of
each group of patients or the survival status of each group of
experimental animals. The present study analyzed RNA-seq data from
TCGA-CESC cohort (n=304) and selected the median as the cutoff
value. Hazard ratio (HR) is defined as the ratio of the two risk
rates. When HR is >1, the research object is a risk factor; when
HR is <1, the research object is a protective factor; when HR=1,
the research object has no effect on survival time. As an outcome
index, overall survival (OS) refers to the time to death. The
prognostic data come from a Cell article (34). Data filtering: control/normal (not
all items have control/normal) were removed and clinical
information was retained. For the. nomogram chart, on the basis of
multifactor regression analysis, the ruler score was set to
characterize the situation of each variable in the multifactor
regression model, and finally the total score was calculated to
predict the probability of event occurrence (35).
Transcriptomics analysis
The key regulators in the PI3K/AKT/mTOR pathway were
searched using TRRUST (https://www.grnpedia.org/trrust/), a reliable,
intuitive tool for human and mouse transcriptional regulatory
networks (36).
Statistical analysis
Software R (version 3.6.3) (37) was used for statistical analysis and
visualization. For differential analysis of single gene expression,
the R package of ggplot2 (version 3.3.3) (38) was used for visualization. For
multigene association analysis, we used the R package of igraph
(version 1.2.6) (39) and ggraph
package (version 2.0.5) (40). For
GO-KEGG analysis and GSEA, the R package of ggplot2 and cluster
Profiler package was used (33).
Visualization of Kaplan-Meier OS analysis was based on the use of
the R package of survminer (0.4.9 version) (41) and for statistical analysis of
survival data the survival package (3.2-10 version) was used.
Wilcoxon rank sum test was used to assess differences in gene
expression. Spearman's rank correlation coefficient was used to
assess the significance of correlations. The qPCR data are
presented as the mean ± standard deviation of three experiments,
and qPCR and RNA-seq data were analyzed using one-way ANOVA
followed by Tukey's multiple comparison test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Genomics
ERBB3 single-gene DEG in CESC. In the CESC
group, the average level of the normal group was 3.598±1.642, while
the average level of the tumor group was 5.539±0.902. The
difference was statistically significant (P<0.001) (Fig. 1A). Closely associated genes to
ERBB3 in the PI3K/AKT/mTOR pathway were PIK3CA, PIK3R2, PIK3R3,
ATG13, MTOR, RICTOR, RHEB and GSK3B (Fig. 1B).
The total number of gene identifications (IDs) after
removing the null value was 35,905. Among them, 1,706 IDs met the
|log2(FC)|>2 and P<0.05 threshold. Under this threshold,
there were 1,221 IDs with high expression (positive logFC) and 485
with low expression (negative logFC). The genes that met this
threshold and were significantly associated with EMT included MMP3
and SNAI2 (downregulated genes), and KRT12 (upregulated gene)
(Fig. 1C). The expression of seven
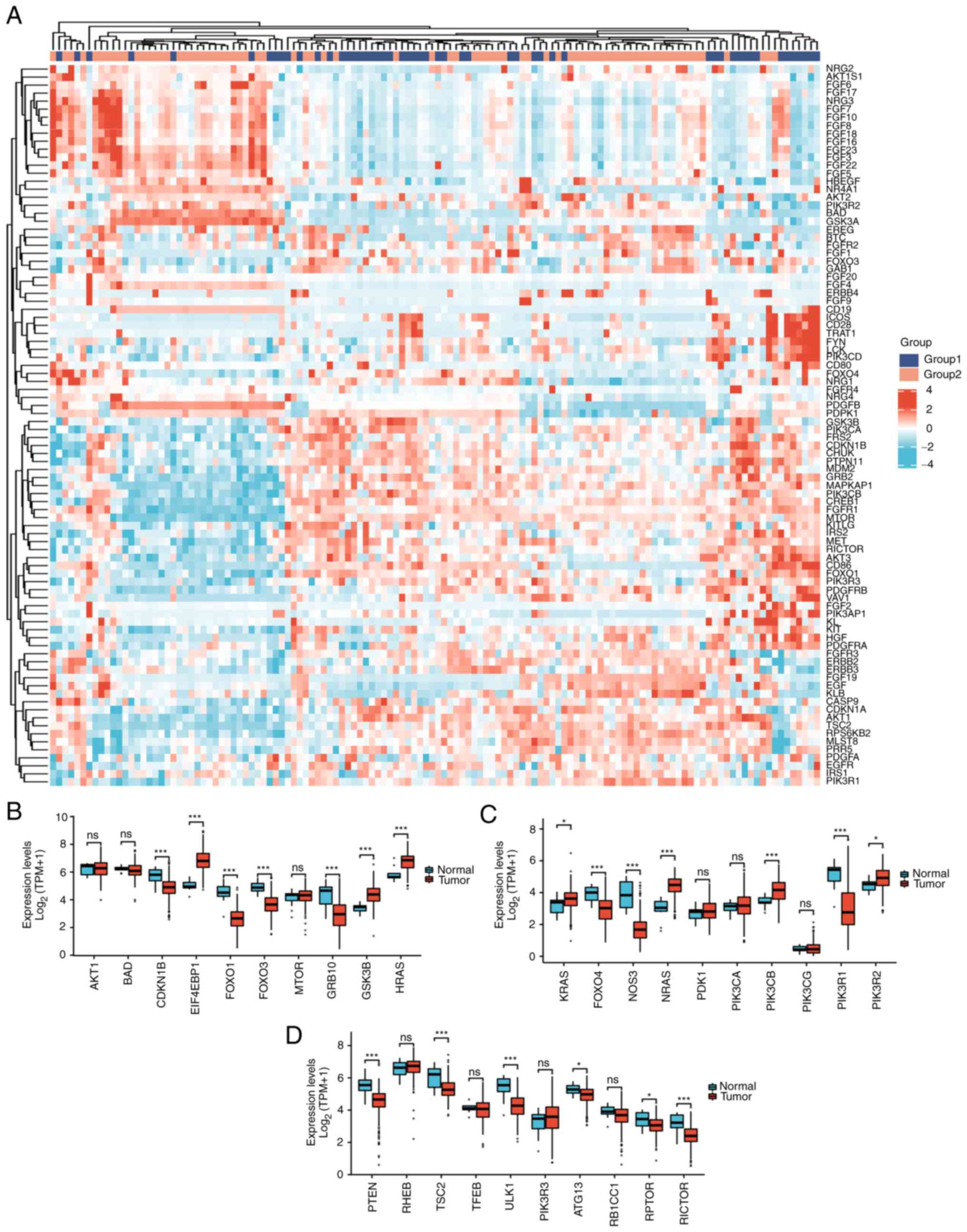
hub genes in the PI3K/AKT/mTOR pathway were increased in cervical
cancer: EIF4EBP1, GSK3B, HRAS, KRAS, NRAS, PIK3CB and PIK3R2
(Fig. 2).
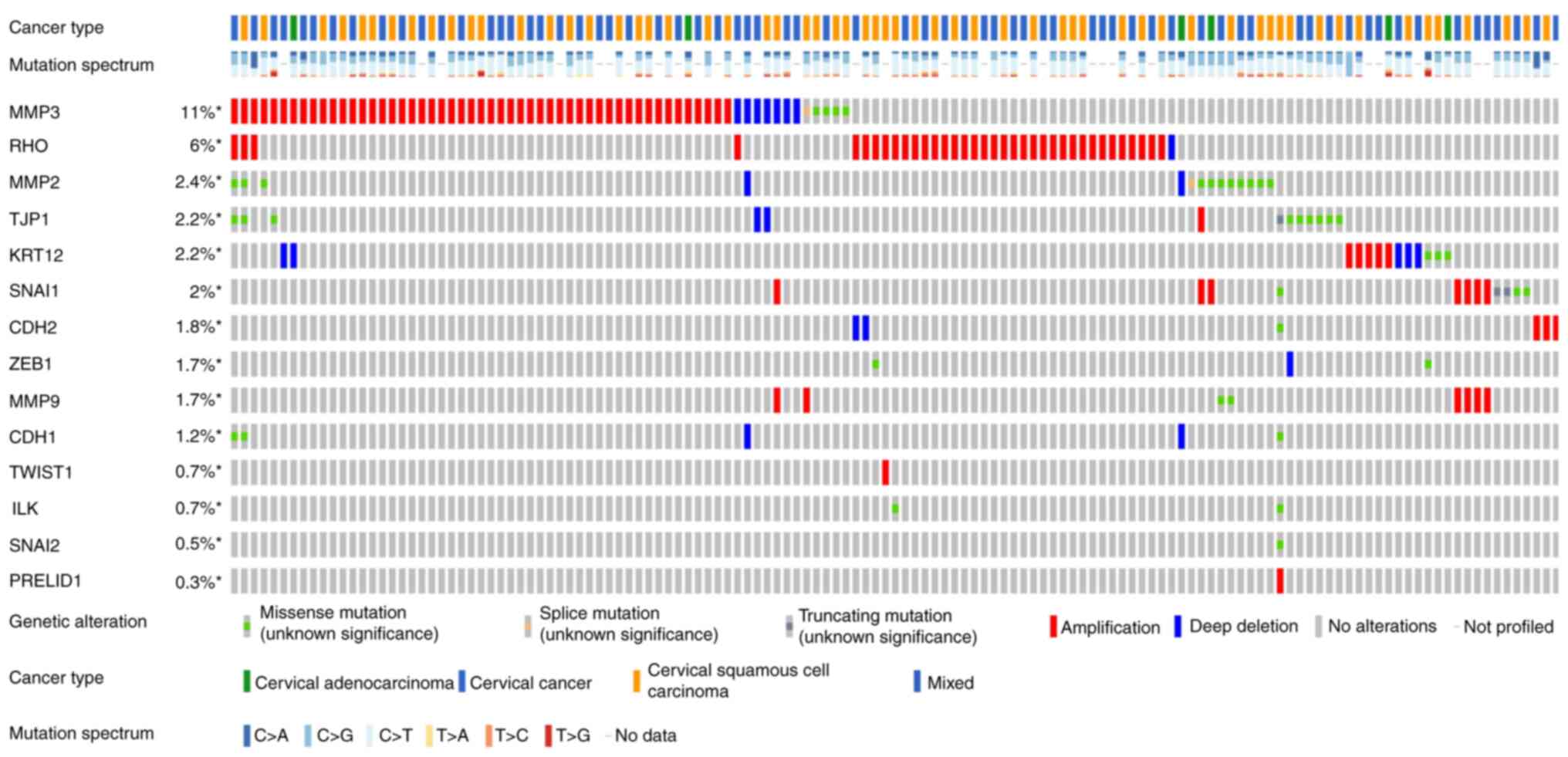
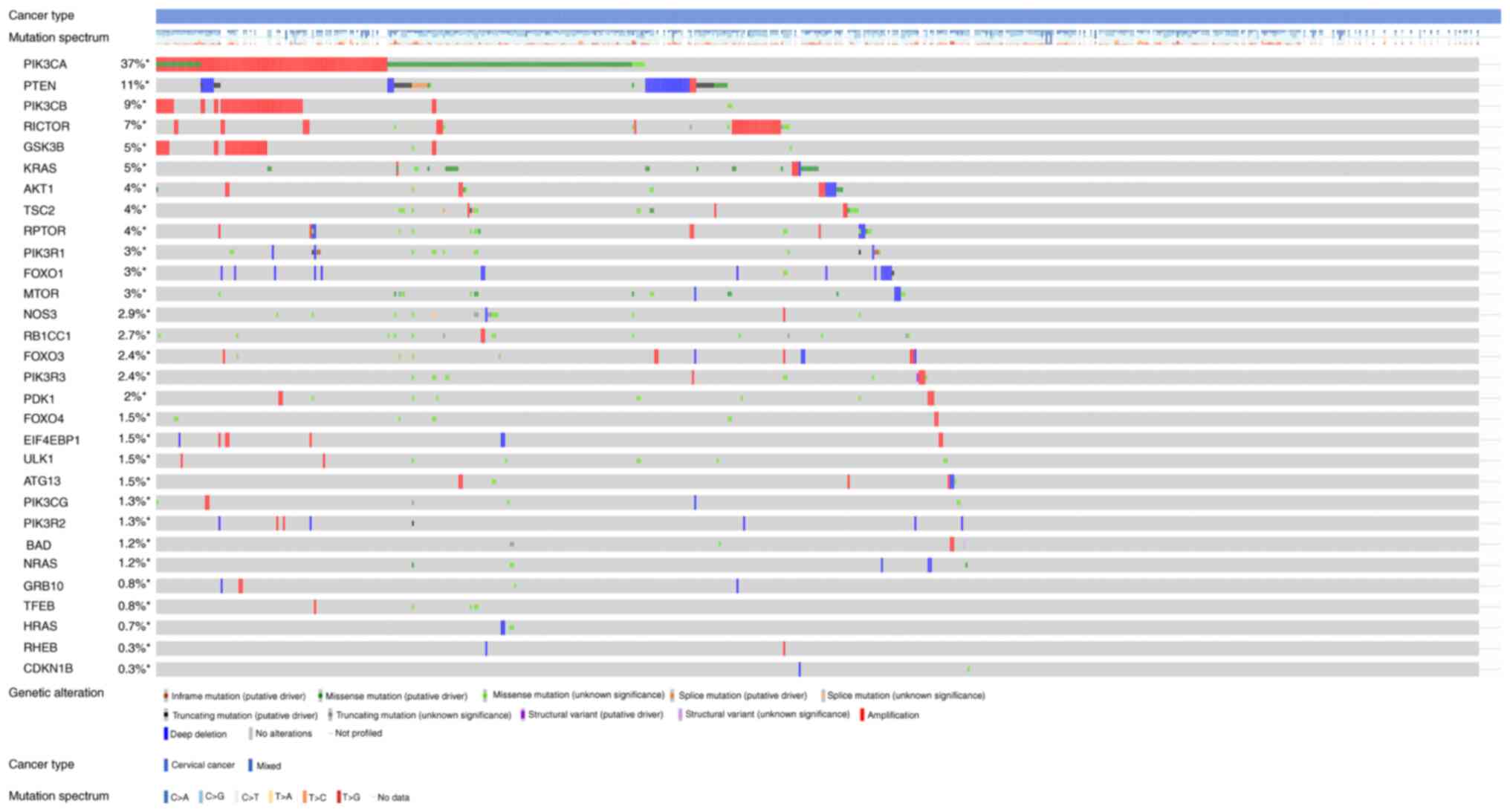
Figs. 3 and
4 show the somatic variation
pattern in cervical cancer. These schematics represented the
distribution of the number of protein-altering somatic mutations
and copy number variations in 607 samples of cervical cancer. The
highest frequencies of mutations among the EMT-related genes were
revealed in MMP3 (11%, including amplification, deep deletion and
missense mutation); PIK3CA (37%, including amplification and
missense mutation); and PTEN (11%, including deep deletion and
truncating mutation). PIK3CA mutation status assesses the hotspot
mutations of the PIK3CA gene (42).
Selection of EMT-related genes with GO
and KEGG analyses
Among 30 PI3K/AKT/mTOR pathway-related genes,
upregulated genes in cervical cancer were as follows: EIF4EBP1
(P<0.001), GSK3B (P<0.001), HRAS (P<0.001), KRAS
(P<0.05), NRAS (P<0.001), PIK3CB (P<0.001), and PIK3R2
(P<0.001); downregulated genes included CDKN1B (P<0.001),
FOXO1 (P<0.001), FOXO3 (P<0.001), GRB10 (P<0.001), FOXO4
(P<0.001), NOS3 (P<0.001), PIK3R1 (P<0.001), PTEN
(P<0.001), TSC2 (P<0.001), ULK1 (P<0.001), ATG13
(P<0.05), RPTOR (P<0.05) and RICTOR (P<0.001). Therefore,
it is of certain significance to study this pathway in relation to
carcinogenesis and prognosis of cervical cancer (Fig. 2B-D).
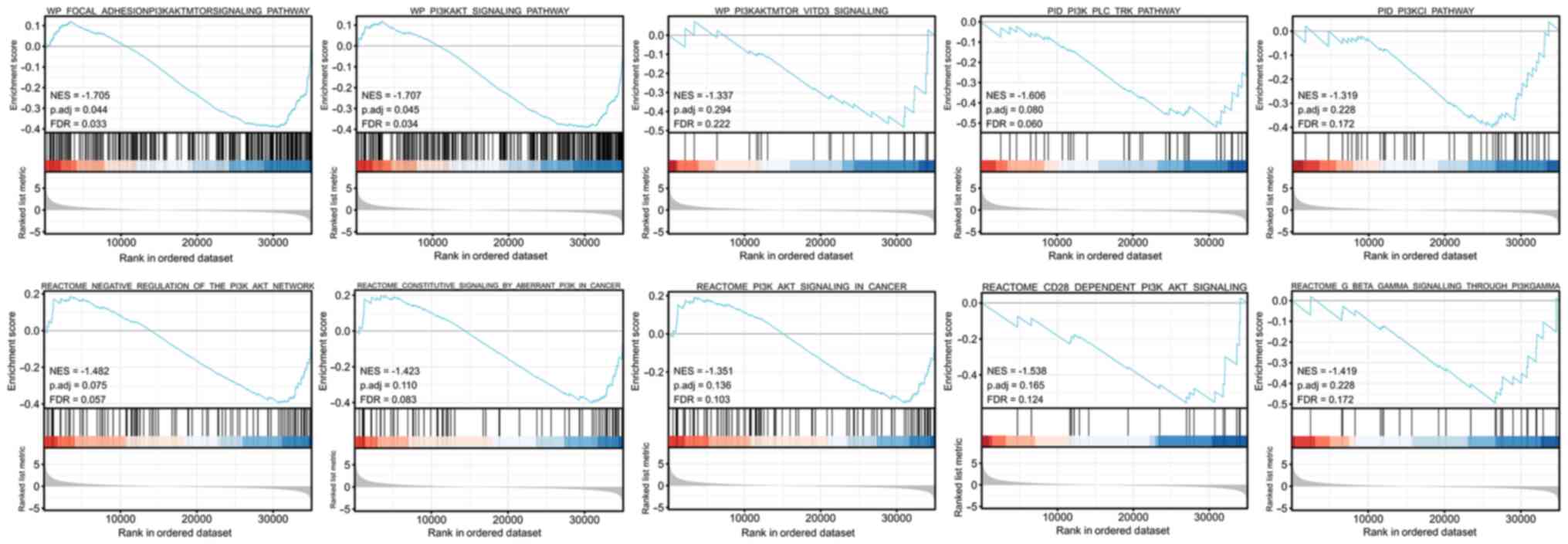
It was revealed that in cervical cancer the ERBB3
gene was enriched in the PI3K/AKT signaling pathway (NES=-1.707,
P=0.045, FDR=0.034; Fig. 5).
TWIST1, tight junction protein 1 (TJP1), MMP9, MMP3
and vimentin (VIM) (Fig. 6) with
all annotated functional molecules were compared using
hypergeometric distribution tests to determine which functional
roles were involved in that stack. The function of genes was
divided into three categories: BPs, CCs and MFs.
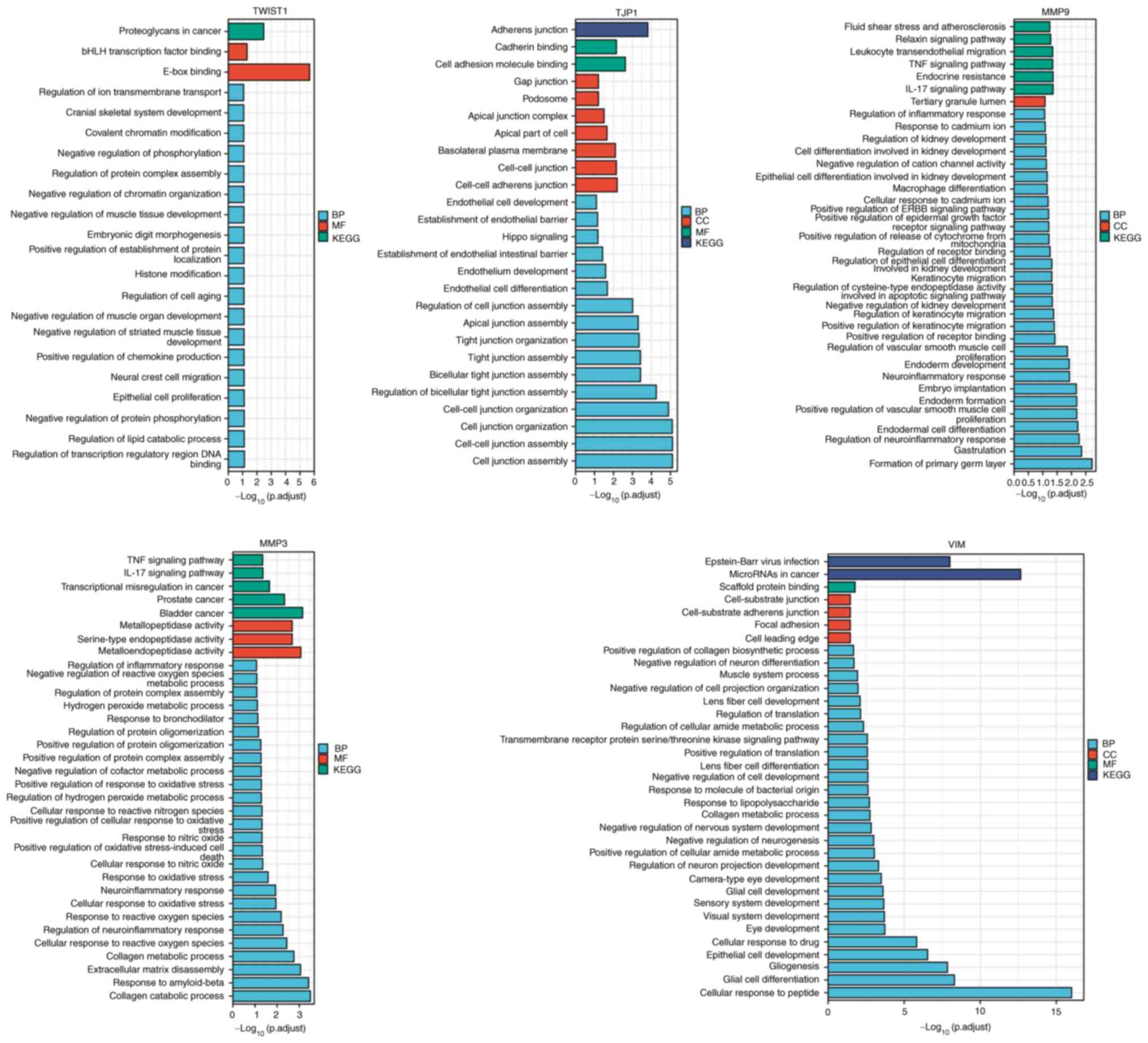 | Figure 6GO-KEGG cluster analysis. The x-axis
represents ‘-log(adj. P)’; the greater the value, the stronger the
significance. The y-axis represents the GO term. Each color
represents an enrichment, including BP, CC, MF and KEGG. KEGG,
Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; BP,
biological processes; CC, cellular components; MF, molecular
functions; TJP1, tight junction protein 1; VIM, vimentin. |
Significant differences in EMT-related gene
expression in cervical cancer were revealed. The following gene
expression levels were upregulated: E-cadherin (CDH1), VIM, TWIST1,
MMP3 and MMP9. The following gene expression levels were
downregulated: N-cadherin (CDH2), SNAI2, MMP2, zinc finger
E-box-binding homeobox 1 (ZEB1), integrin-linked protein kinase
(ILK), RHO, TJP1 and SNAIL1 (Fig.
7). Further study on the correlation degree of ERBB3- and
EMT-related factors revealed the following results: R=0.307,
P<0.001 for KRT12; R=0.323, P<0.001 for TJP1; R=-0.407,
P<0.001 for TWSIT1; and R=-0.306, P<0.001 for MMP9 (Fig. 8).
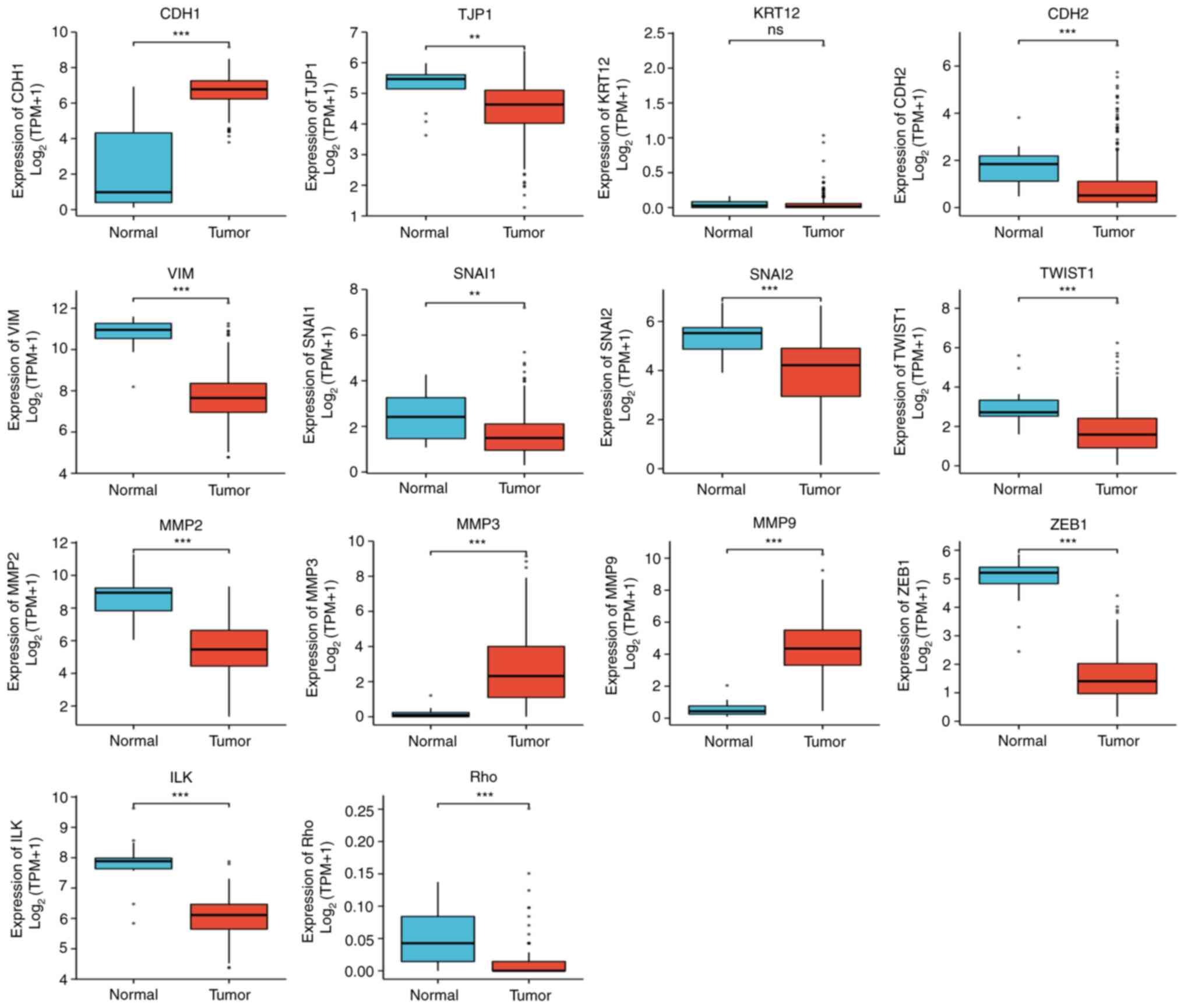 | Figure 7The following significant differences
in the expression of epithelial-mesenchymal transition
related-factors in CESC were found: CDH1↑, CDH2↓, VIM↑, SNAI2↓,
TWIST1↑, MMP2↓, MMP3↑, MMP9↑, ZEB1↓, ILK↓, RHO↓, TJP1↓ and Snail1↓
(statistical method, Wilcoxon rank-sum test).
**P<0.01 and ***P<0.001. Ns, not
significant; CDH1, E-cadherin; KRT12, cytokeratin 12; TJP1, tight
junction protein 1; CDH2, N-cadherin; VIM, vimentin; ZEB1, zinc
finger E-box-binding homeobox 1; ILK, integrin-linked protein
kinase. |
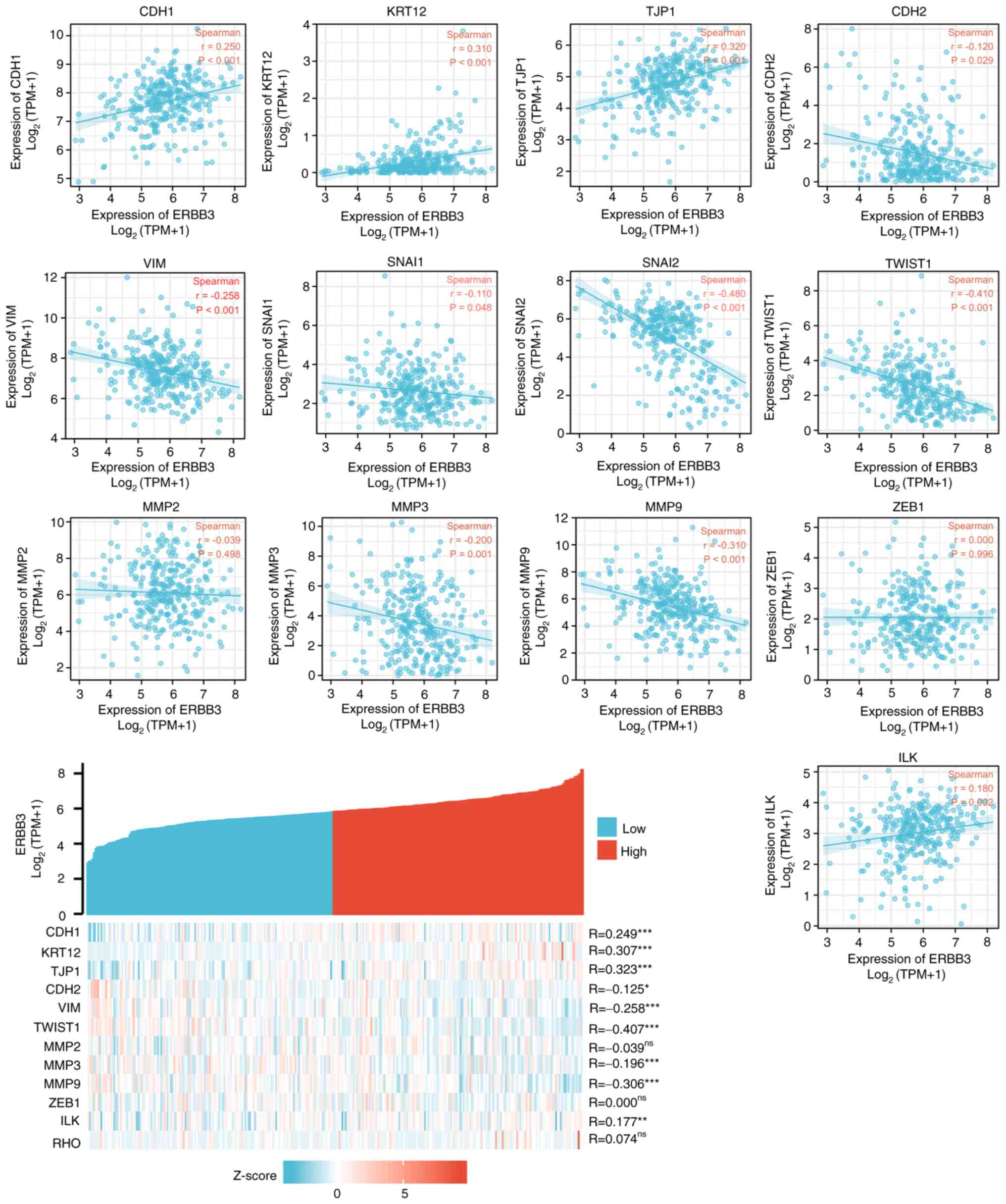 | Figure 8Correlation between ERBB3 and factors
associated with epithelial-mesenchymal transition status.
Statistically significant were TWIST1 (R=-0.407; P<0.001), TJP1
(R=0.323; P<0.001), KRT12 (R=0.307; P<0.001), MMP9 (R=-0.306;
P<0.001). Statistical method, Spearman's correlation;
*P<0.05, **P<0.01 and
***P<0.001. ns, not significant; CDH1, E-cadherin;
KRT12, cytokeratin 12; TJP1, tight junction protein 1; CDH2,
N-cadherin; VIM, vimentin; ZEB1, zinc finger E-box-binding homeobox
1; ILK, integrin-linked protein kinase. |
Transcriptomics
The key regulators in the PI3K/AKT/mTOR pathway are
as follows: AKT1, BAD, CDKN1B, FOXO1, FOXO3, FOXO4, GSK3B, HRAS,
KRAS, NOS3, NRAS, PDK1, PIK3CA, PIK3R3, PTEN, RPTOR, TSC2 and ULK1.
It was revealed that nine transcription factors (TSC22D3, TP53, AR,
RELA, NFKB1, STAT3, PPARG, SP1 and JUN) were associated with the
regulation of the PI3K/AKT/mTOR pathway (Table I).
 | Table IKey transcription factors of
PI3K/AKT/mTOR biomarkers in cervical squamous cell carcinoma and
adenocarcinoma. |
Table I
Key transcription factors of
PI3K/AKT/mTOR biomarkers in cervical squamous cell carcinoma and
adenocarcinoma.
| Key transcription
factor | Description | Regulated gene | P-value | FDR |
|---|
| TSC22D3 | TSC22 domain
family, member 3 | FOXO4, FOXO3,
FOXO1 |
3.62x10-8 |
3.26x10-7 |
| TP53 | Tumor protein
p53 | HRAS, PTEN, CDKN1B,
AKT1, FOXO3 |
5.57x10-6 |
2.51x10-5 |
| AR | Androgen
receptor | AKT1, TSC2, NRAS,
PTEN |
1.37x10-5 |
4.12x10-5 |
| RELA | V-rel
reticuloendotheliosis viral oncogene homolog A (avian) | PTEN, BAD, FOXO3,
NOS3, AKT1 |
1.02x10-4 |
1.90x10-4 |
| NFKB1 | Nuclear factor of
kappa light polypeptide gene enhancer in B-cells 1 | PTEN, AKT1, GSK3B,
FOXO3, NOS3 |
1.06x10-4 |
1.90x10-4 |
| STAT3 | Signal transducer
and activator of transcription 3 (acute-phase response factor) | PTEN, CDKN1B,
AKT1 |
1.46x10-3 |
2.19x10-3 |
| PPARG | Peroxisome
proliferator-activated receptor gamma | BAD, PTEN |
4.92x10-3 |
6.33x10-3 |
| SP1 | Sp1 transcription
factor | CDKN1B, PTEN, NOS3,
HRAS |
6.32x10-3 |
7.11x10-3 |
| JUN | Jun
proto-oncogene | PDK1, NOS3 |
2.33x10-2 |
2.33x10-2 |
Proteomics
By comparing with the RNA level in normal cervical
epithelium (Fig. 9A), it was
revealed that the expression level of MMP9 in cervical cancer was
significantly different. Through IHC analysis of the mRNA-protein
expression of the EMT-related genes in cervical cancer tissues, it
was revealed that the expression level of MMP family was relatively
higher compared with those of other EMT-related genes (Fig. 9B). EMT of cervical cancer is
associated with upregulation of MMP9. According to our previous
findings of ERBB3 sequencing of cervical cancer cell lines
(43), the mRNA levels of
different primary cervical cancer cell lines indicated that ERBB3
is highly expressed in cervical malignant cell lines dominated by
SiHa and HeLa (Fig 9C). After
further research on pathological types and HPV typing, it was
revealed that in the three types of cells with higher expression of
ERBB3, there was no significant difference between SiHa and HeLa,
while there was a significant difference between SiHa and C33A
(P<0.0001). These data show that ERBB3 is closely associated
with adenocarcinoma and HPV-positive cervical carcinoma. Fig. 10 shows the protein interactions of
the 30 key factors in the PI3K/AKT/mTOR signaling pathway with
ERBB3. The color of the lines in the figure shows that ERBB3 may
play a stimulating role in the PI3K/AKT/mTOR pathway.
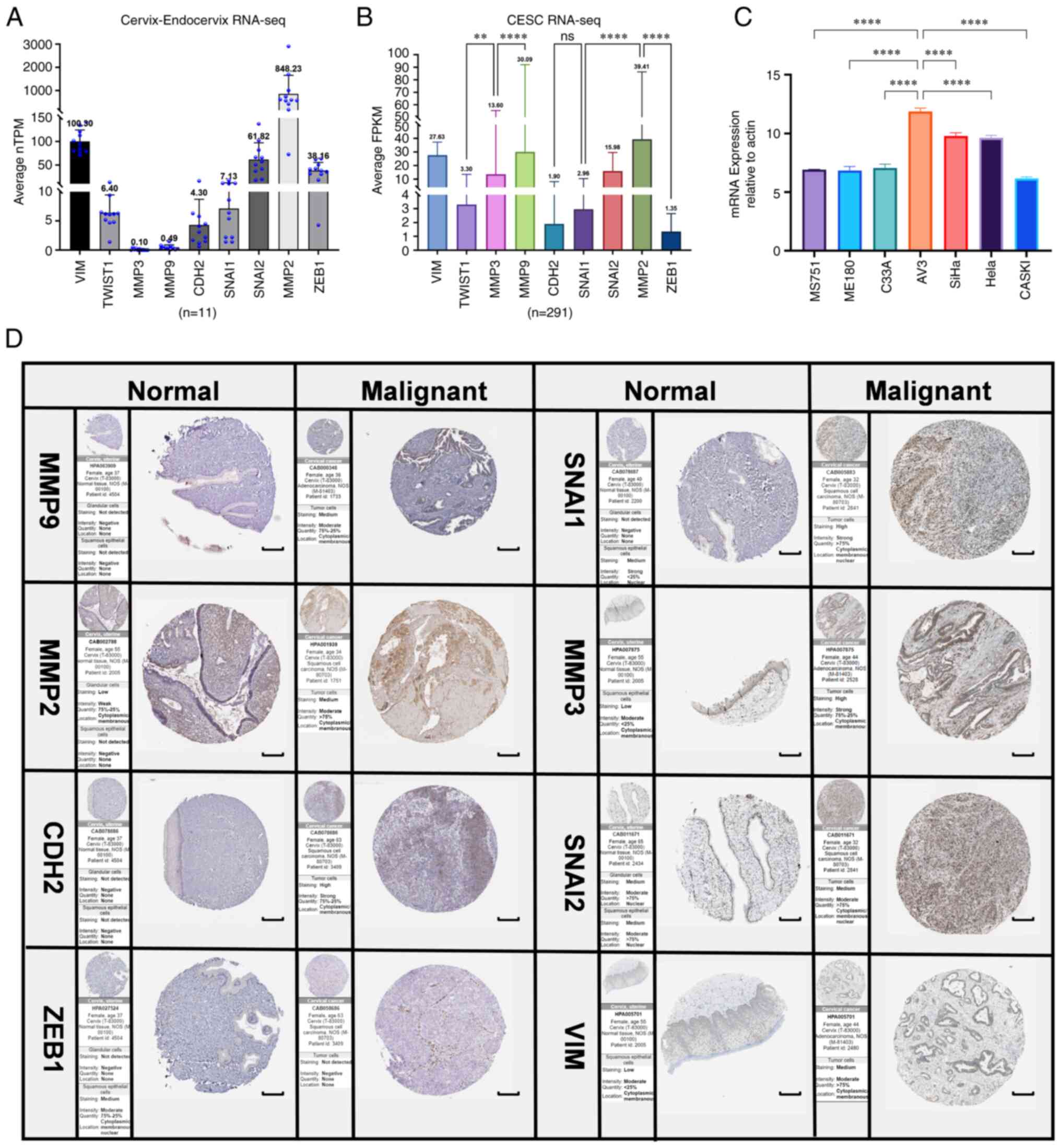 | Figure 9RNA-sequencing and protein level
analysis of EMT upregulated factors (PRELI domain-containing
protein 1, CDH2, TWIST1, MMP2, MMP3, MMP9, SNAI1, SNAI2 and ZEB1)
in normal cervical epithelium and tumor tissues by
immunohistochemistry for cervical cancer invasiveness assessment.
(A) Normalized TPM was quantified, as shown. The dot plot depicts
the means and standard deviation of 11 images of normal
cervix-endocervix tissues. (B) FPKM was quantified of 291 images of
cervical cancer tissues. (C) ERBB3 expression in seven different
cell lines were examined using reverse transcription-quantitative
PCR. All PCR data were calculated relative to β-actin and represent
the average ± SD of triplicate samples. (D) Immunohistochemical
validation of the most significant EMT-related genes in cervical
cancer and normal cervix tissues by The Human Protein Atlas
database (scale bars, 200 µm). The translational expression level
(mRNA and protein) of the eight EMT-related genes was positively
correlated with disease status as they were upregulated in cervical
squamous cell carcinoma and adenocarcinoma samples. ANOVA followed
by Tukey's multiple-comparison tests; **P<0.01 and
****P<0.0001. EMT, epithelial-mesenchymal transition;
CDH1, E-cadherin; KRT12, cytokeratin 12; TJP1, tight junction
protein 1; CDH2, N-cadherin; VIM, vimentin; ZEB1, zinc finger
E-box-binding homeobox 1; ILK, integrin-linked protein kinase;
FPKM, fragments per kilobase per million; ERBB3, Erb-B2 receptor
tyrosine kinase 3; TPM, transcripts per million. |
EMT status and immuno-oncology
insights from the RNA-seq-based analyses. Immune microenvironment
characteristics of CESC
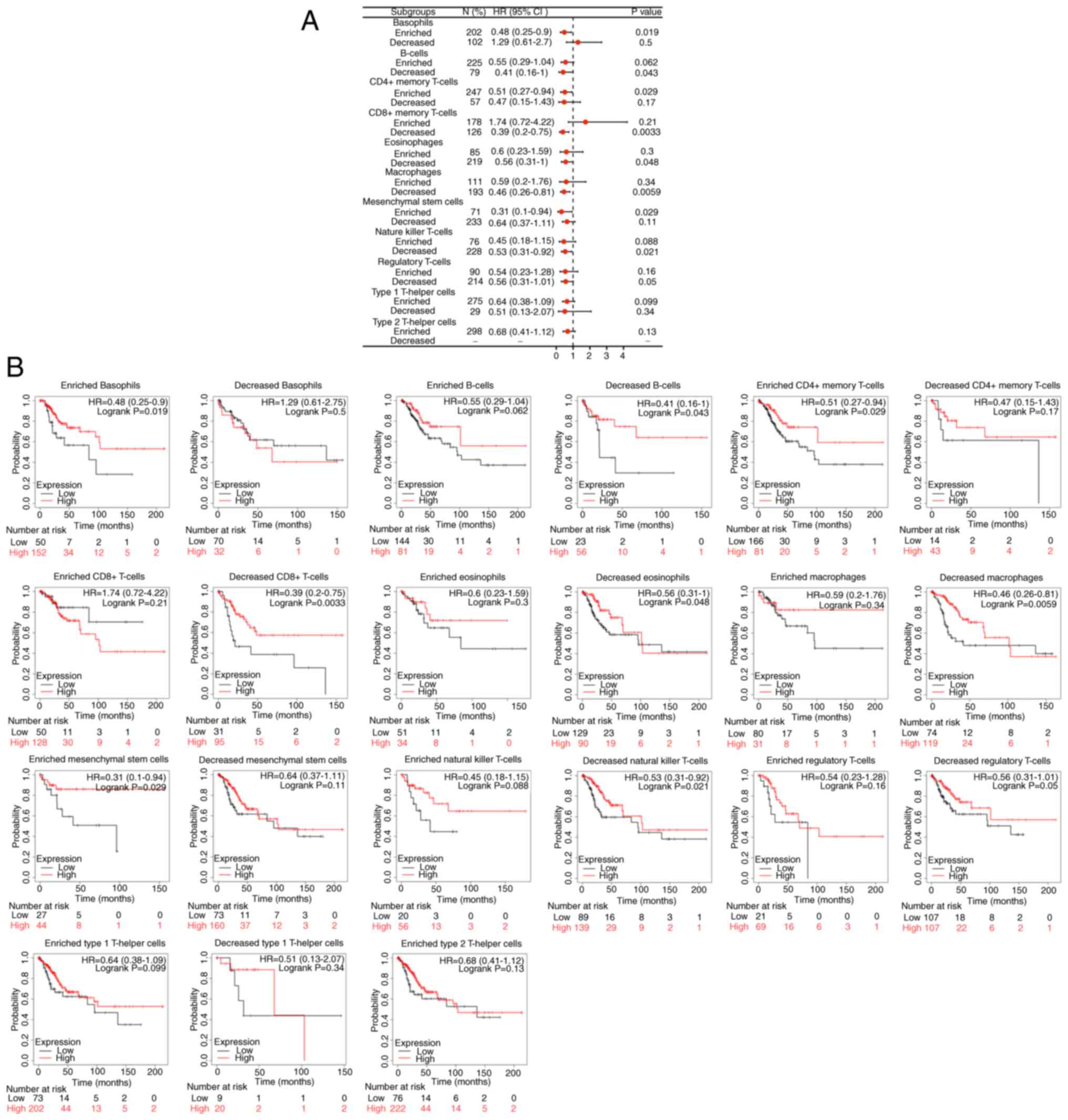
ERBB3 influenced the survival time of patients with
CESC, partially through immune cell infiltration. Enriched
basophils (P<0.05), decreased B cells (P<0.05), enriched
CD4+ memory T cells (P<0.05), enriched mesenchymal
stem cells (P<0.05), decreased eosinophils (P<0.05),
decreased natural-killer T cells (P<0.05), decreased
CD8+ memory T cells (P<0.01) and decreased
macrophages (P<0.01). Among the abovementioned eight types of
immune cell infiltration, the prognosis of the group with a higher
ERBB3 mRNA level was decreased (Fig.
11).
EMT-related genes change the immune
cell infiltration
If the absolute value of R is below 0.3, there is no
straight phase off; R of ≥0.3 denotes linear correlations; R values
between 0.3 and 0.5 indicate low-degree correlations; R values
between 0.5 and 0.8 refer to significant correlations; and R values
of 0.8 and above are high-degree correlations. As shown in Table II, correlation between the
EMT-related factors and immune cell infiltration was not high.
Among the EMT-related factors, MMP9, MMP2, and ZEB1 were closely
associated with the immune system. The EMT status (Fig. 12) may be related to MMP9 changing
the tumor immune microenvironment through dendritic cells and
macrophages (10).
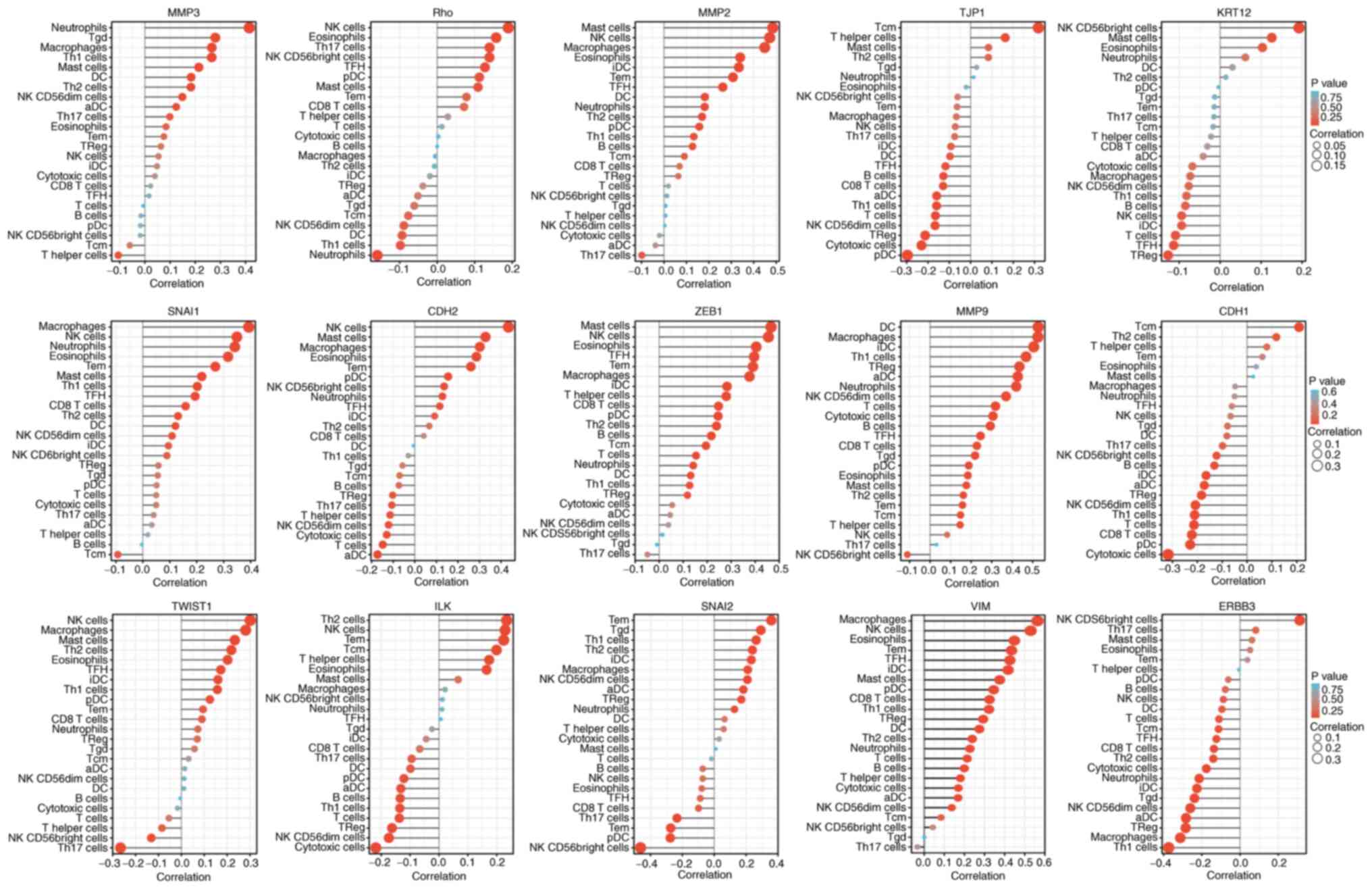 | Figure 12Single sample GSEA enrichment set the
score to infer the infiltration by immune cells in each sample. The
size of the circle represents the degree of relevance. The greater
the height of the bar (the distance from 0), the higher the degree
of correlation (a positive number represents a positive
correlation, and a negative number represents a negative
correlation). The depth of the circle represents the P-value
obtained by the correlation analysis, the legend on the right is
the color scale value, and the range of the color scale is
automatically generated according to the range of the P-value
obtained in the figure. CDH1, E-cadherin; KRT12, cytokeratin 12;
TJP1, tight junction protein 1; CDH2, N-cadherin; VIM, vimentin;
ZEB1, zinc finger E-box-binding homeobox 1; ILK, integrin-linked
protein kinase. |
 | Table IIERBB3 methylation and immune
infiltration in tumor microenvironment of cervical cancer. |
Table II
ERBB3 methylation and immune
infiltration in tumor microenvironment of cervical cancer.
| Cell type | CDH1 | CDH2 | ERBB3 | ILK | KRT12 | MMP2 | MMP3 | MMP9 | PRELID1 | RHO | SNAI2 | SNAI1 | TJP1 | TWIST1 | ZEB1 |
|---|
| aDC | -0.172a | -0.173a | -0.283b | -0.132 | -0.041 | -0.038 | 0.124c | 0.427b,d | 0.028 | -0.051 | 0.183a | 0.033 | -0.159a | 0.015 | 0.046 |
| B cells | -0.131c | -0.074 | -0.078 | -0.134 | -0.085 | 0.127c | -0.016 | 0.293b | 0.014 | -0.000 | -0.072 | -0.004 | -0.128c | -0.005 | 0.217b |
| CD8 T cells | -0.222c | 0.042 | -0.137c | -0.067 | -0.031 | 0.069 | 0.022 | 0.228b | 0.056 | 0.072 | -0.099 | 0.159a | -0.129c | 0.090 | 0.247b |
| Cytotoxic
cells | -0.318c,d | -0.130c | -0.177a | -0.219b | -0.068 | -0.021 | 0.040 | 0.306b,d | 0.077 | 0.003 | 0.031 | 0.049 | -0.231b | -0.017 | 0.054 |
| DC | -0.081 | -0.005 | -0.095 | -0.099 | 0.030 | 0.182a | 0.184a | 0.527c,e | 0.064 | -0.094 | 0.065 | 0.121c | -0.097 | 0.011 | 0.131c |
| Eosinophils | 0.037 | 0.285b | 0.052 | 0.165a | 0.103 | 0.339b,d | 0.084 | 0.184a | 0.009 | 0.157a | -0.078 | 0.316b,d | -0.022 | 0.202b | 0.404b,d |
| iDC | -0.165a | 0.092 | -0.226b | -0.045 | -0.094 | 0.334b,d | 0.048 | 0.505b,e | 0.006 | -0.019 | 0.233b | 0.095 | -0.092 | 0.160a | 0.283b |
| Macrophages | -0.048 | 0.301b | -0.313b,d | 0.021 | -0.073 | 0.449b,d | 0.266b | 0.526c,e | 0.142c | -0.005 | 0.210b | 0.393b,d | -0.067 | 0.281b | 0.376b,d |
| Mast cells | 0.025 | 0.328b,d | 0.062 | 0.066 | 0.126c | 0.485b,d | 0.214b | 0.177a | -0.108 | 0.109 | 0.011 | 0.219b | 0.084 | 0.234b | 0.466b,d |
| Neutrophils | -0.050 | 0.128c | -0.214b | 0.011 | 0.062 | 0.181a | 0.415b,d | 0.420b,d | 0.112 | -0.160a | 0.127a | 0.341b | 0.015 | 0.072 | 0.142c |
| NK CD56 bright
cells | -0.123a | 0.136c | 0.312b,d | 0.012 | 0.193b | 0.013 | -0.019 | -0.111 | -0.081 | 0.140c | -0.463b,d | 0.089 | -0.061 | -0.131c | 0.014 |
| NK CD56dim
cells | -0.208b | -0.122c | -0.261b | -0.174a | -0.076 | 0.004 | 0.149a | 0.370c,d | 0.028 | -0.089 | 0.208b | 0.108 | -0.167a | 0.012 | 0.039 |
| NK cells | -0.066 | 0.434b,d | -0.088 | 0.229b | -0.094 | 0.471b,d | 0.054 | 0.083 | 0.043 | 0.190b | -0.072 | 0.349b | -0.072 | 0.301b | 0.455b,d |
| pDC | -0.229c | 0.155a | -0.062 | -0.121 | -0.004 | 0.157a | -0.018 | 0.188b | 0.070 | 0.113c | -0.275b | 0.051 | -0.296b | 0.124c | 0.245b |
| T cells | -0.213c | -0.148a | -0.111 | -0.137 | -0.109 | 0.020 | -0.007 | 0.319b,d | 0.052a | 0.012 | -0.018 | 0.049 | -0.165a | -0.055 | 0.153a |
| T helper cells | 0.079 | -0.114c | -0.006 | 0.174a | -0.022 | 0.006 | -0.106 | 0.146c | -0.015 | 0.029 | 0.059 | 0.019 | 0.162a | -0.085 | 0.279b |
| TCM | 0.209b | -0.070 | -0.113c | 0.199b | -0.017 | 0.091 | -0.060 | 0.148a | -0.182 | -0.077 | 0.358b,d | -0.093 | 0.318b,d | 0.032 | 0.195b |
| TEM | 0.062 | 0.260b | 0.038 | 0.224b | -0.014 | 0.307b,d | 0.075 | 0.157a | 0.101 | 0.078 | -0.273b | 0.269b | -0.064 | 0.095 | 0.391b,d |
| TFH | -0.061 | 0.116c | -0.124c | 0.007 | -0.113c | 0.262b | 0.017 | 0.246b | 0.032 | 0.128c | -0.088 | 0.194b | -0.118c | 0.172a | 0.396b,d |
| TGD | -0.079 | -0.057 | -0.239b | -0.025 | -0.013 | 0.007 | 0.280b | 0.219b | -0.011 | -0.061 | 0.292b | 0.055 | 0.028 | 0.057 | -0.010 |
| Th1 cells | -0.211b | -0.030 | -0.374b,d | -0.136 | -0.082 | 0.133c | 0.265b | 0.468b,d | -0.014 | -0.098 | 0.263b | 0.202b | -0.160a | 0.157a | 0.127c |
| Th17 cells | -0.099 | -0.106 | 0.082 | -0.095 | -0.015 | -0.099 | 0.099 | 0.031 | 0.136c | 0.140c | -0.234b | 0.040 | -0.075 | -0.266b | -0.049 |
| Th2 cells | 0.118c | 0.067 | -0.140c | 0.235b | 0.014 | 0.170a | 0.183a | 0.162a | -0.043 | -0.007 | 0.240b | 0.130c | 0.083 | 0.218b | 0.239b |
| T Reg | -0.183a | -0.103 | -0.285b | -0.163a | -0.127c | 0.064 | 0.063 | 0.435 | 0.033 | -0.038 | 0.170a | 0.058 | -0.213b | 0.070 | 0.118c |
Prognostic analysis of
microenvironment phenotypes
Statistically significant EMT-related genes that can
predict the OS index of CESC included KRT12 (P<0.05), VIM
(P<0.05), SNAI1 (P<0.05), ILK (P<0.05), CDH2 (P<0.01),
MMP2 (P<0.01) and MMP3 (P<0.01). All of the seven factors
were the risk factors for CESC (Fig.
13).
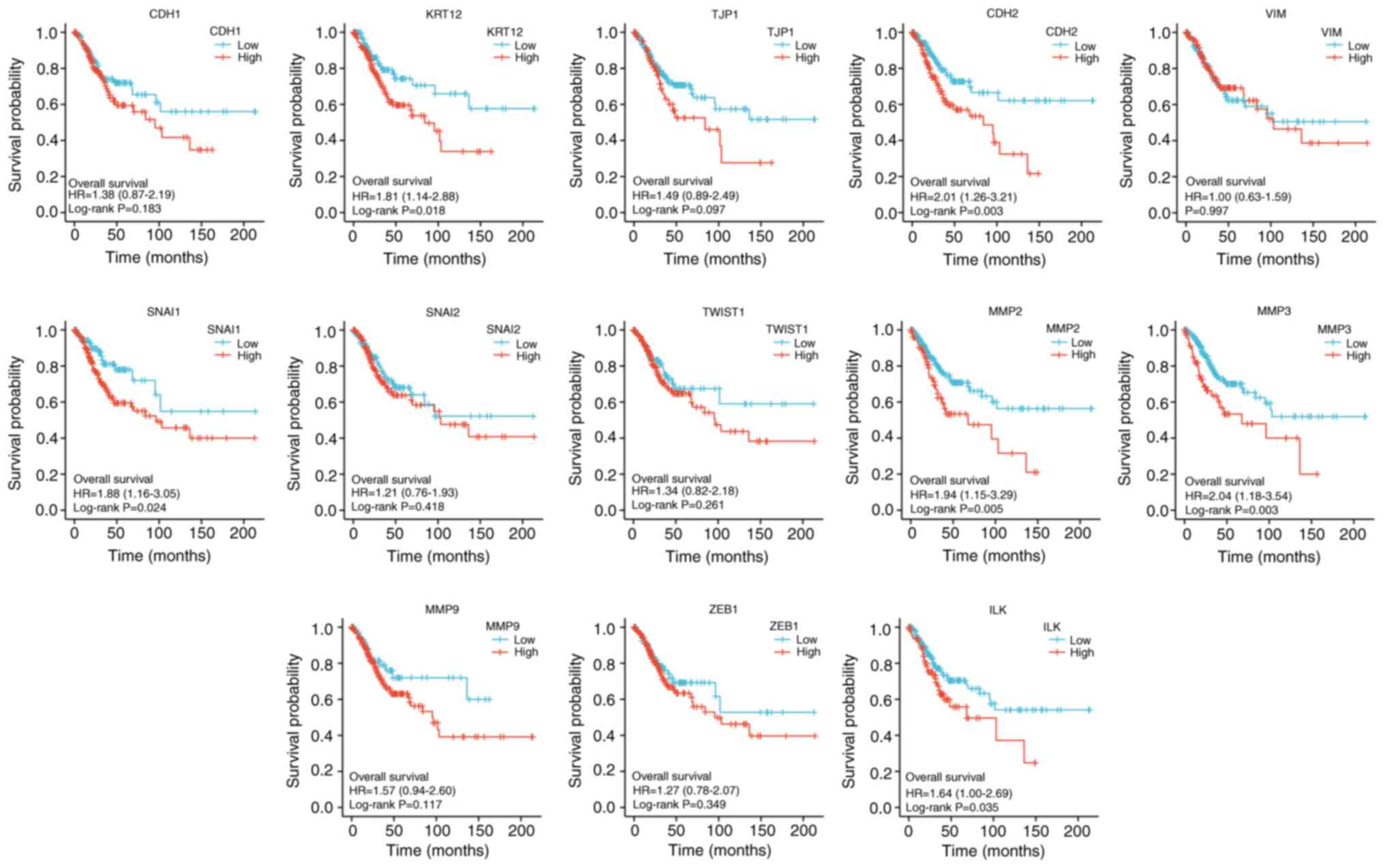 | Figure 13Kaplan-Meier overall survival curves
according to high and low expression of CDH1, KRT12, TJP1,
CDH2,VIM, SNAI1, SNAI2, TWIST1, MMP2, MMP3, MMP9, ZEB1, and ILK in
cervical squamous cell carcinoma and adenocarcinoma samples tumor
immune microenvironment. CDH1, E-cadherin; KRT12, cytokeratin 12;
TJP1, tight junction protein 1; CDH2, N-cadherin; VIM, vimentin;
ZEB1, zinc finger E-box-binding homeobox 1; ILK, integrin-linked
protein kinase; HR, hazard ratio. |
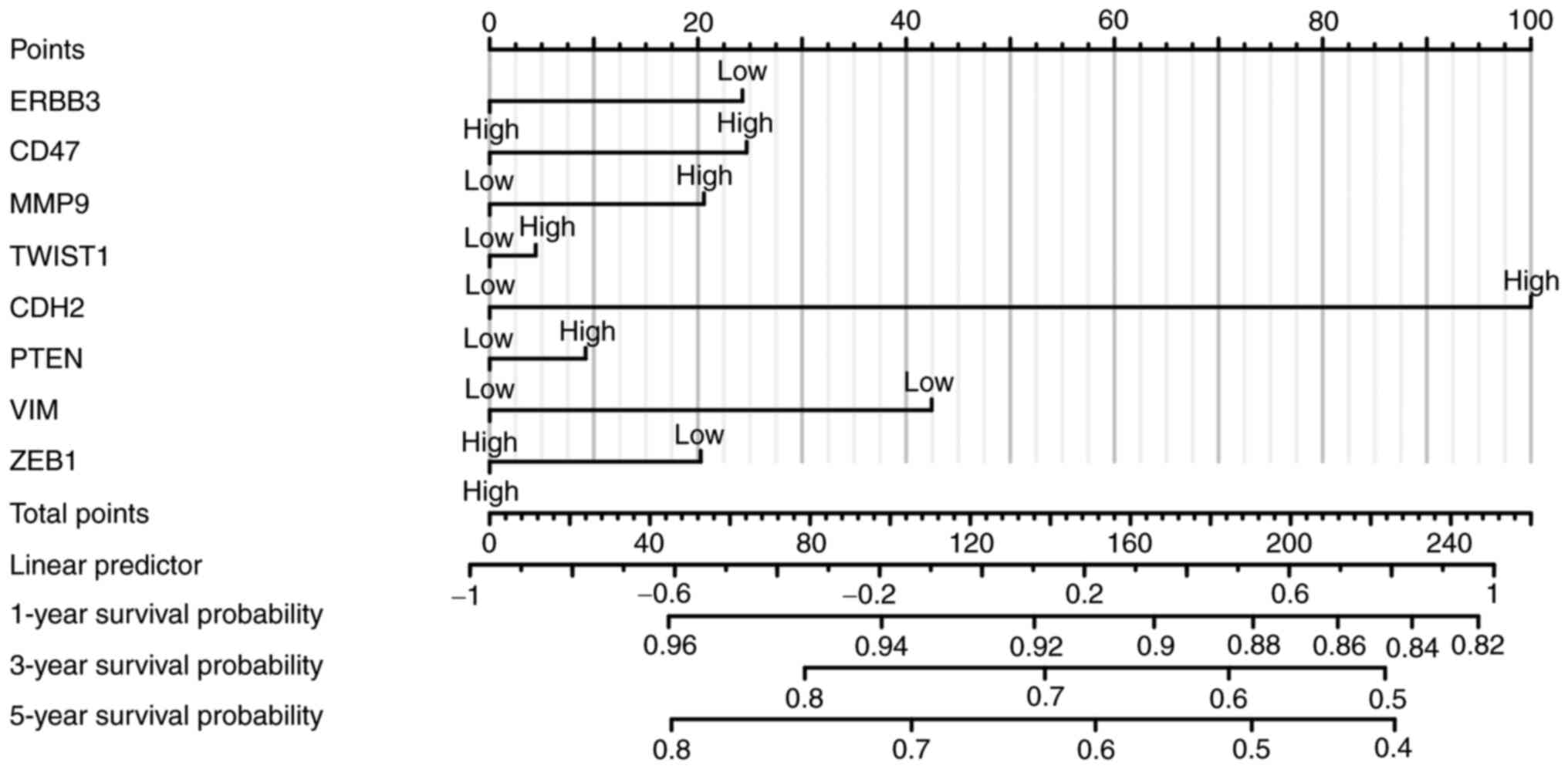
Risk score was constructed using eight selected
genes through the multifactor analysis of the disease prognosis
model, and three prognostic types of OS, disease-specific survival
and progression-free interval (PFI) were analyzed. ERBB3, CD47,
MMP9, TWIST1, CDH2, PTEN, VIM and ZEB1 were included. Multivariate
analysis revealed that CDH2, MMP9, and VIM were significant factors
in the assessment of PFI. Gene signatures of cervical cancer cell
immune-oncology microenvironment positively correlated with the
patients' survival. These analyses (Fig. 14) indicated that the selected
genes and constructed risk prognostic models had good prognostic
value.
Discussion
High-risk HPV16 DNA is integrated into the host cell
genome (HPV16: q21-q31 of chromosome 2.13; HPV18: chromosome 24.8)
(44), disrupts the open reading
frame, and causes overexpression of E6 and E7 genes (45). It has been demonstrated that E6 and
E7 exert carcinogenic effects by combining with cell cycle
regulators, such as p53 (a transcription factor related to the PI3K
pathway as shown in Table I) and
retinoblastoma (46). E6 can
interact with E6-related protein E6AP to form a complex and bind to
p53(47), hydrolyze p53, and cause
the loss of negative regulation of cell proliferation induced by
p53, thereby leading to uncontrolled cell proliferation and
malignant transformation. The present study also revealed that
ERBB3 was highly expressed in HPV-infected cell lines and was
associated with adenocarcinoma. Therefore, it can be speculated
that HPV-positive cervical cancer cells and adenocarcinoma are the
carcinogenic factors or prognostic factors of cervical cancer.
To investigate the association between the actual
activation of the PI3K pathway and immune infiltration, the DEGs of
ERBB3 in CESC was assessed (GSE63514, GSE9750 and GSE44001 datasets
from the Gene Expression Omnibus database were analyzed) for hub
genes of the PI3K/AKT/mTOR pathway and cancer progression. Pathway
enrichment, protein-protein interaction and pathway crosstalk
analyses were performed to identify key genes and pathways. The
current study illustrated that cancer-immune interactions might
differ depending on specific alterations in the PI3K pathway,
demonstrating that genetic aberrations in malignant cells influence
the immune landscape of tumors.
The diversity of EMT creates a wide range of
heterogeneity in tumors, and may provide tumor cells with increased
adaptability and resistance, enabling them to survive and
proliferate in a complex TME, and metastasize and invade lymph and
blood vessels. The present study demonstrated that MMPs, especially
MMP9 as a prominent representative, are highly relevant for TME and
immune cells. MMPs are a family of zinc-dependent endopeptidases
(48,49). The biological function of MMPs is
to degrade various molecules used for cell adhesion and regulate
the interaction between cells and the extracellular matrix. Recent
studies (50-52)
have shown that MMPs are highly associated with the
microenvironment of tumors and immune cells, and targeting MMPs may
overcome the barriers of immunosuppression. However, the present
study revealed that the expression of MMP9 was not a significant
predictor of OS in patients with cervical cancer. MMP9 was
significantly associated with ERBB3.
In the complex TME, the same anti-infection immune
cells can be destroyed by tumor cells (53). As a result, the antitumor immune
cells not only do not destroy the transformed cells, but they even
change to immune cells that promote tumor growth and metastasis
(54,55). These immune cells secrete factors
that promote survival, promote migration, and resist detection.
Hence, the present study discussed the mechanism of accelerating
cervical cancer tumor progression from the perspective of
EMT-associated immune evasion.
Cellular immunity is necessary for clearing
HPV-infected and HPV-transformed tumor cells. HPV-specific CD8
cytotoxic T lymphocytes (CTLs) are needed for the immune defense
against cervical cancer. However, the function of CTLs may be
blunted by systemic and local immunosuppressive environments
associated with tumor growth (56,57).
A series of clinical trials (58-60)
have shown that the immune system is unable to completely eradicate
the tumor despite the presence of HPV-specific T cells in
HPV-associated neoplastic tissue, which suggests the possible
existence of systemic immunosuppression and an immunosuppressive
TME that significantly influence the efficacy of therapeutic
vaccines.
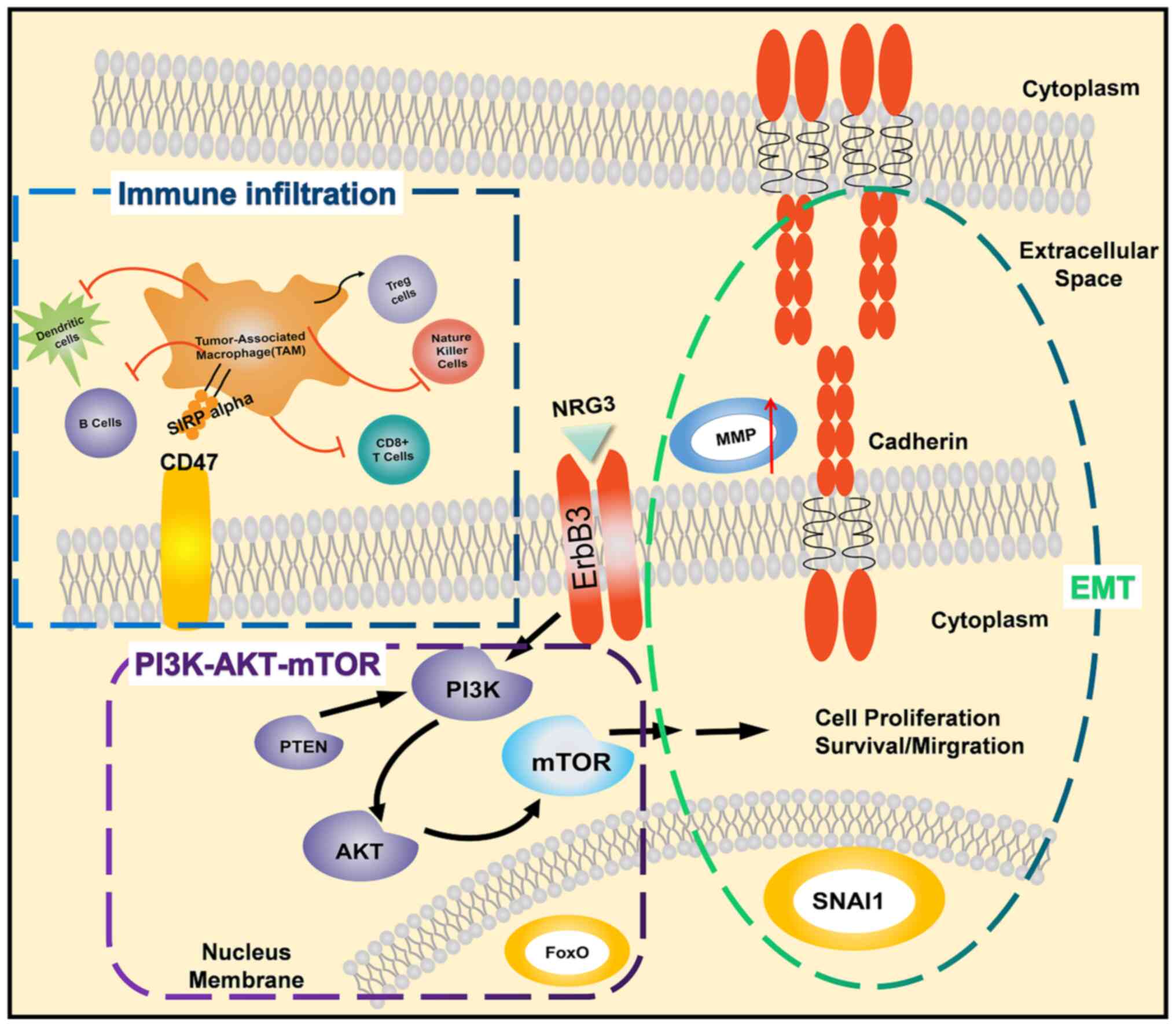
Tumor-associated macrophages (TAMs) may cause the
disruptive change of antitumor immunity in TME and promote tumor
growth and metastasis (61). TAMs
are a heterogeneous population of cells that display a range of
phenotypes depending on the type of tumor and their location in TME
(62,63). TAMs are commonly the most abundant
infiltrating leukocytes in most tumors and are predominantly
thought to have pro-tumor effects. These include both
immunosuppressive effects in addition to pro-antigenic and
metastatic effects. TAMs also promote tumor immune evasion through
expression of signal regulatory protein α (SIRPα) (64,65).
SIRPα is a receptor for CD47(66),
a cell surface protein that typically protects normal cells from
phagocytosis by macrophages or dendritic cells. CD47 is frequently
overexpressed on tumor cells and plays a key role in tumor escape
by binding to SIRPα and sending macrophages a ‘don't eat me’ signal
(67,68). Blockade of the CD47-SIRPα signal
has been shown to stimulate phagocytosis, leading to tumor cell
elimination (69).
In the present study, TAMs were revealed to drive
tumor angiogenesis and progression in a spontaneous model of
cervical cancer through the production of MMP-9. Previous studies
(60,70) showed that MMP9 alone did not
significantly affect the survival rate of cervical cancer; however,
in the present study, when TME macrophages decreased, there was an
impact on OS, and the OS with high ERBB3 was significantly reduced.
The GO and KEGG enrichment of MMP9 demonstrated that it
participated in the biological process of the IL-17 signaling
pathway. In patients with bullous pemphigoid, monocyte-derived
macrophages, but not lymphocytes, respond to CXCL10 (upregulated by
IL-17) by increasing MMP-9 release, potentially creating an
inflammatory loop associated with disease outcome (71).
Late after pattern recognition receptors
stimulation, bone-marrow-derived dendritic cells (BMDCs) induce the
glucose transporter GLUT1 (72,73)
and commit to aerobic glycolysis via the mTOR-HIF-1α/iNOS axis
(74), which generates NO,
inhibiting the electron transport chain. This process might
decrease expression of MHC and co-stimulatory molecules by
activated BMDCs (75).
Intratumoral immune cells are more often present in
tumors with increasing PI3K downstream phosphorylation (76-78).
This is most pronounced for MMP9-positive cells. Research data
shows that disturbances in the PI3K pathway may help immune escape
(79). A prospective trial in
cervical cancer suggested that PI3K pathway alterations may be
associated with the composition of TME (80,81).
Previous studies (82-84)
have suggested that when cells undergo EMT and shift progressively
from an epithelial to a mesenchymal state, genetic alterations
include decreased expression levels of CDH1, cytokeratin 12 and
TJP1, increased expression levels of CDH2, VIM, SNAI1, SNAI2,
TWIST1, MMP2, MMP3, MMP-9 and ZEB1 and increased activity of ILK
and RHO.
It can be inferred from the present study that tumor
infiltration by CD8-positive lymphocytes was associated with PIK3CA
mutations and worse clinical outcome. At the molecular level, EMT
transcriptional factors, including SNAl, ZEB1 and TWIST1, regulated
immunosuppressive cells or enhanced the expression of
immunosuppressive checkpoint molecules through the production of
chemokines, thereby resulting in immunosuppression of TME.
Immunosuppressive factors can also induce EMT in tumor cells. This
mutual feedback between EMT and immunosuppression promotes tumor
progression (85).
In conclusion, integrating the characteristics of
biomarkers in multiple dimensions can ensure the most efficient
management choice for each patient with cancer (Fig. 15). The EMT status cannot be
assessed based on one or a few molecular markers, but should be
assessed in conjunction with changes in cell characteristics to
assess the current ability of cell metastasis and distant invasion.
Immuno-oncology research can generate the discriminating power and
richness of data required for these features. In particular, simple
MMP changes are not a prognostic factor in cervical cancer. When
ERBB3 activates the PI3K pathway to change immune cell
infiltration, the cervical cancer prognosis model is
meaningful.
Acknowledgements
Not applicable.
Funding
Funding: This research was funded by ‘Research on Early
Diagnosis of Cervical Cancer Based on Terahertz Technology’ in
Zhenjiang Social Development Project (grant no. SH2020051);
‘Observation and study on the clinical efficacy of a vaginal gel
for the treatment of Female Genital HPV Infection and Related
Diseases’ by 2019 Key Laboratory of Pharmacodynamics and Safety
Evaluation of Traditional Chinese Medicine in Jiangsu Province
(grant no. JKLPSE201906); ‘Study on the Innovation of Distance
Teaching Mode of Gynecological Laparoscopic Surgery Under the
Background of New Medicine’ Project of Cooperative Education
Between Industry and Education in 2020 (grant no. 202002144004);
and The Open Project of State Key Laboratory of Radiology and
Radiation Protection ‘LINC00460/miR-361-3p/Gli1 Pathway and
Radiosensitivity of Cervical Cancer Cells’ (grant no.
GZK1202106).
Availability of data and materials
The datasets generated and analyzed during the
current study are available in the cervical squamous cell carcinoma
and endocervical adenocarcinoma (CESC) data in TCGA (https://portal.gdc.cancer.gov/). The datasets
used and/or analyzed during the current study are available from
the corresponding author on reasonable request.
Authors' contributions
XY performed the data analysis work and aided in
writing the manuscript. WZ designed the study and edited the
manuscript. XY and WZ confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cohen PA, Jhingran A, Oaknin A and Denny
L: Cervical cancer. Lancet. 393:169–182. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yugawa T and Kiyono T: Molecular
mechanisms of cervical carcinogenesis by high-risk human
papillomaviruses: Novel functions of E6 and E7 oncoproteins. Rev
Med Virol. 19:97–113. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Zhang L, Wu J, Ling MT, Zhao L and Zhao
KN: The role of the PI3K/Akt/mTOR signalling pathway in human
cancers induced by infection with human papillomaviruses. Mol
Cancer. 14(87)2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bossler F, Hoppe-Seyler K and Hoppe-Seyler
F: PI3K/AKT/mTOR signaling regulates the virus/host cell crosstalk
in HPV-positive cervical cancer cells. Int J Mol Sci.
20(2188)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Shi X, Wang J, Lei Y, Cong C, Tan D and
Zhou X: Research progress on the PI3K/AKT signaling pathway in
gynecological cancer. Mol Med Rep. 19:4529–4535. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Senga SS and Grose RP: Hallmarks of
cancer-the new testament. Open Biol. 11(200358)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
O'Donnell JS, Teng MWL and Smyth MJ:
Cancer immunoediting and resistance to T cell-based immunotherapy.
Nat Rev Clin Oncol. 16:151–167. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Fukumura D, Kloepper J, Amoozgar Z, Duda
DG and Jain RK: Enhancing cancer immunotherapy using
antiangiogenics: Opportunities and challenges. Nat Rev Clin Oncol.
15:325–340. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Aiello NM and Kang Y: Context-dependent
EMT programs in cancer metastasis. J Exp Med. 216:1016–1026.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Taki M, Abiko K, Ukita M, Murakami R,
Yamanoi K, Yamaguchi K, Hamanishi J, Baba T, Matsumura N and Mandai
M: Tumor immune microenvironment during epithelial-mesenchymal
transitionthe review of the loop between EMT and immunosuppression.
Clin Cancer Res. 27:4669–4679. 2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Dongre A, Rashidian M, Reinhardt F,
Bagnato A, Keckesova Z, Ploegh HL and Weinberg RA:
Epithelial-to-mesenchymal transition contributes to
immunosuppression in breast carcinomas. Cancer Res. 77:3982–3989.
2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hoxhaj G and Manning BD: The PI3K-AKT
network at the interface of oncogenic signalling and cancer
metabolism. Nat Rev Cancer. 20:74–88. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Coussy F, El Botty R, Lavigne M, Gu C,
Fuhrmann L, Briaux A, de Koning L, Dahmani A, Montaudon E, Morisset
L, et al: Combination of PI3K and MEK inhibitors yields durable
remission in PDX models of PIK3CA-mutated metaplastic breast
cancers. J Hematol Oncol. 13(13)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li M, Liu F, Zhang F, Zhou W, Jiang X,
Yang Y, Qu K, Wang Y, Ma Q, Wang T, et al: Genomic ERBB2/ERBB3
mutations promote PD-L1-mediated immune escape in gallbladder
cancer: a whole-exome sequencing analysis. Gut. 68:1024–1033.
2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ross JS, Fakih M, Ali SM, Elvin JA,
Schrock AB, Suh J, Vergilio JA, Ramkissoon S, Severson E, Daniel S,
et al: Targeting HER2 in colorectal cancer: The landscape of
amplification and short variant mutations in ERBB2 and ERBB3.
Cancer. 124:1358–1373. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Van Lengerich B, Agnew C, Puchner EM,
Huang B and Jura N: EGF and NRG induce phosphorylation of
HER3/ERBB3 by EGFR using distinct oligomeric mechanisms. Proc Natl
Acad Sci USA. 114:E2836–E2845. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sithanandam G and Anderson LM: The ERBB3
receptor in cancer and cancer gene therapy. Cancer Gene Ther.
15:413–448. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Moghbeli M, Makhdoumi Y, Delgosha MS,
Aarabi A, Dadkhah E, Memar B, Abdollahi A and Abbaszadegan MR:
ErbB1 and ErbB3 co-over expression as a prognostic factor in
gastric cancer. Biol Res. 52(2)2008.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shi DM, Li LX, Bian XY, Shi XJ, Lu LL,
Zhou HX, Pan TJ, Zhou J, Fan J and Wu WZ: miR-296-5p suppresses EMT
of hepatocellular carcinoma via attenuating NRG1/ERBB2/ERBB3
signaling. J Exp Clin Cancer Res. 37(294)2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Vivian J, Rao AA, Nothaft FA, Ketchum C,
Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD,
Musselman-Brown A, et al: Toil enables reproducible, open source,
big biomedical data analyses. Nat Biotechnol. 35:314–316.
2017.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Yang J, Antin P, Berx G, Blanpain C,
Brabletz T, Bronner M, Campbell K, Cano A, Casanova J, Christofori
G, et al: EMT International Association (TEMTIA). Guidelines and
definitions for research on epithelial-mesenchymal transition. Nat
Rev Mol Cell Biol. 21:341–352. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Den Boon JA, Pyeon D, Wang SS, Horswill M,
Schiffman M, Sherman M, Zuna RE, Wang Z, Hewitt SM, Pearson R, et
al: Molecular transitions from papillomavirus infection to cervical
precancer and cancer: Role of stromal estrogen receptor signaling.
Proc Natl Acad Sci USA. 112:E3255–E3264. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M and Murty VV: Identification of copy
number gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995.
2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Smyth GK: Limma: Linear models for
microarray data. Bioinformatics and computational biology solutions
using R and Bioconductor. Springer, New York, NY, 2005.
397-420.
|
|
27
|
Gu Z, Eils R and Schlesner M: Complex
heatmaps reveal patterns and correlations in multidimensional
genomic data. Bioinformatics. 32:2847–2849. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bindea G, Mlecnik B, Tosolini M,
Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T,
Lafontaine L, Berger A, et al: Spatiotemporal dynamics of
intratumoral immune cells reveal the immune landscape in human
cancer. Immunity. 39:782–795. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Walter W, Sánchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucleic Acids Res. 36(Database issue):D480–D484.
2007.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lanczky A and Gyorffy B: Web-based
survival analysis tool tailored for medical research (KMplot):
Development and implementation. J Med Internet Res.
23(e27633)2021.PubMed/NCBI View
Article : Google Scholar
|
|
34
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Goel AL and Wu SM: Determination of ARL
and a contour nomogram for CUSUM charts to control normal mean.
Technometrics. 13:221–230. 1971.
|
|
36
|
Han H, Cho JW, Lee S, Yun A, Kim H, Bae D,
Yang S, Kim CY, Lee M, Kim E, et al: TRRUST v2: An expanded
reference database of human and mouse transcriptional regulatory
interactions. Nucleic Acids Res. 46:D380–D386. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
R Core Team: A language and environment
for statistical computing. R Foundation for Statistical Computing,
Vienna, 2013. http://www.R-project.org/.
|
|
38
|
Jeppson H and Hofmann H: Generalized
Mosaic Plots in the ggplot2 Framework: Exploratory analysis of
high-dimensional data with visual tools 17, 2021. https://rdrr.io/cran/ggmosaic/.
|
|
39
|
Csardi G and Nepusz T: The igraph software
package for complex network research. InterJournal Complex Systems.
1695:1–9. 2006.
|
|
40
|
Gustavsson EK, Zhang D, Reynolds RH, et
al: ggtranscript: an R package for the visualization and
interpretation of transcript isoforms using ggplot2.
Bioinformatics. 38(15):3844–3846. 2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kassambara A, Kosinski M and Biecek P:
Package ‘survminer’. Drawing Survival Curves using ‘ggplot2’(R
package version 03 1), 2017.
|
|
42
|
Cancer Genome Atlas Research Network.
Integrated genomic and molecular characterization of cervical
cancer. Nature. 543:378–384. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yang X, Chen Y, Li M and Zhu W: ERBB3
methylation and immune infiltration in tumor microenvironment of
cervical cancer. Sci Rep. 12(8112)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Soto D, Song C and McLaughlin-Drubin ME:
Epigenetic alterations in human papillomavirus-associated cancers.
Viruses. 9(248)2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Senapati R, Senapati NN and Dwibedi B:
Molecular mechanisms of HPV mediated neoplastic progression. Infect
Agent Cancer. 11(59)2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Narisawa-Saito M and Kiyono T: Basic
mechanisms of high-risk human papillomavirus-induced
carcinogenesis: Roles of E6 and E7 proteins. Cancer Sci.
98:1505–1511. 2007.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Martinez-Zapien D, Ruiz FX, Poirson J,
Mitschler A, Ramirez J, Forster A, Cousido-Siah A, Masson M, Pol
SV, Podjarny A, et al: Structure of the E6/E6AP/p53 complex
required for HPV-mediated degradation of p53. Nature. 529:541–545.
2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Karamanou K, Franchi M, Vynios D and
Brézillon S: Epithelial-to-mesenchymal transition and invadopodia
markers in breast cancer: Lumican a key regulator. Semin Cancer
Biol. 62:125–133. 2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Yang HL, Thiyagarajan V, Shen PC, Mathew
DC, Lin KY, Liao JW and Hseu YC: Anti-EMT properties of CoQ0
attributed to PI3K/AKT/NFKB/MMP-9 signaling pathway through
ROS-mediated apoptosis. J Exp Clin Cancer Res.
38(186)2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Mao X, Xu J, Wang W, Liang C, Hua J, Liu
J, Zhang B, Meng Q, Yu X and Shi S: Crosstalk between
cancer-associated fibroblasts and immune cells in the tumor
microenvironment: New findings and future perspectives. Mol Cancer.
20(131)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Looi CK, Chung FFL, Leong CO, Wong SF,
Rosli R and Mai SW: Therapeutic challenges and current
immunomodulatory strategies in targeting the immunosuppressive
pancreatic tumor microenvironment. J Exp Clin Cancer Res.
38(162)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Cheng YQ, Wang SB, Liu JH, Jin L, Liu Y,
Li CY, Su YR, Liu YR, Sang X, Wan Q, et al: Modifying the tumour
microenvironment and reverting tumour cells: New strategies for
treating malignant tumours. Cell Prolif. 53(e12865)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Li D and Wu M: Pattern recognition
receptors in health and diseases. Signal Transduct Target Ther.
6(291)2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Mendes F, Domingues C, Rodrigues-Santos P,
Abrantes AM, Gonçalves AC, Estrela J, Encarnação J, Pires AS,
Laranjo M, Alves V, et al: The role of immune system exhaustion on
cancer cell escape and anti-tumor immune induction after
irradiation. Biochim Biophys Acta. 1865:168–175. 2016.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Liu Y and Cao X: Immunosuppressive cells
in tumor immune escape and metastasis. J Mol Med (Berl).
94:509–522. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Che Y, Yang Y, Suo J, An Y and Wang X:
Induction of systemic immune responses and reversion of
immunosuppression in the tumor microenvironment by a therapeutic
vaccine for cervical cancer. Cancer Immunol Immunother.
69:2651–2664. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Jayshree RS: The immune microenvironment
in human papilloma virus-induced cervical lesions-evidence for
estrogen as an immunomodulator. Front Cell Infect Microbiol.
11(649815)2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Welters MJ, van der Sluis TC, van Meir H,
Loof NM, van Ham VJ, van Duikeren S, Santegoets SJ, Arens R, de Kam
ML, Cohen AF, et al: Vaccination during myeloid cell depletion by
cancer chemotherapy fosters robust T cell responses. Sci Transl
Med. 8(334ra52)2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Todd RW, Roberts S, Mann CH, Luesley DM,
Gallimore PH and Steele JC: Human papillomavirus (HPV) type
16-specific CD8+ T cell responses in women with high grade vulvar
intraepithelial neoplasia. Int J Cancer. 108:857–862.
2004.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Torres-Poveda K, Bahena-Román M,
Madrid-González C, Burguete-García AI, Bermúdez-Morales VH,
Peralta-Zaragoza O and Madrid-Marina V: Role of IL-10 and TGF-β1 in
local immunosuppression in HPV-associated cervical neoplasia. World
J Clin Oncol. 5:753–763. 2014.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Kalathil SG and Thanavala Y: High
immunosuppressive burden in cancer patients: A major hurdle for
cancer immunotherapy. Cancer Immunol Immunother. 65:813–819.
2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Huang YK, Wang M, Sun Y, Di Costanzo N,
Mitchell C, Achuthan A, Hamilton JA, Busuttil RA and Boussioutas A:
Macrophage spatial heterogeneity in gastric cancer defined by
multiplex immunohistochemistry. Nat Commun. 10(3928)2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Zhang Y and Zhang Z: The history and
advances in cancer immunotherapy: Understanding the characteristics
of tumor-infiltrating immune cells and their therapeutic
implications. Cell Mol Immunol. 17:807–821. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Chao MP, Weissman IL and Majeti R: The
CD47-SIRPα pathway in cancer immune evasion and potential
therapeutic implications. Curr Opin Immunol. 24:225–232.
2012.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Feng M, Jiang W, Kim BYS, Zhang CC, Fu YX
and Weissman IL: Phagocytosis checkpoints as new targets for cancer
immunotherapy. Nat Rev Cancer. 19:568–586. 2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Oronsky B, Carter C, Reid T, Brinkhaus F
and Knox SJ: Just eat it: A review of CD47 and SIRP-α antagonism.
Semin Oncol. 47:117–124. 2020.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Li Z, Li Y, Gao J, Fu Y, Hua P, Jing Y,
Cai M, Wang H and Tong T: The role of CD47-SIRPα immune checkpoint
in tumor immune evasion and innate immunotherapy. Life Sci.
273(119150)2021.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Huang CY, Ye ZH, Huang MY and Lu JJ:
Regulation of CD47 expression in cancer cells. Transl Oncol.
13(100862)2020.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Yang H, Shao R, Huang H, Wang X, Rong Z
and Lin Y: Engineering macrophages to phagocytose cancer cells by
blocking the CD47/SIRPα axis. Cancer Med. 8:4245–4253.
2019.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Braicu EI, Gasimli K, Richter R, Nassir M,
Kümmel S, Blohmer JU, Yalcinkaya I, Chekerov R, Ignat I, Ionescu A,
et al: Role of serum VEGFA, TIMP2, MMP2 and MMP9 in monitoring
response to adjuvant radiochemotherapy in patients with primary
cervical cancer-results of a companion protocol of the randomized
NOGGO-AGO phase III clinical trial. Anticancer Res. 34:385–391.
2014.PubMed/NCBI
|
|
71
|
Riani M, Le Jan S, Plée J, Durlach A, Le
Naour R, Haegeman G, Bernard P and Antonicelli F: Bullous
pemphigoid outcome is associated with CXCL10-induced matrix
metalloproteinase 9 secretion from monocytes and neutrophils but
not lymphocytes. J Allergy Clin Immunol. 139:863–872.e3.
2017.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Yi W, Tu MJ, Liu Z, Zhang C, Batra N, Yu
AX and Yu AM: Bioengineered miR-328-3p modulates GLUT1-mediated
glucose uptake and metabolism to exert synergistic
antiproliferative effects with chemotherapeutics. Acta Pharm Sin B.
10:159–170. 2020.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Vasconcelos RC, de Oliveira Moura JM,
Junior VLB, da Silveira ÉJ and de Souza LB: Immunohistochemical
expression of GLUT-1, GLUT-3, and carbonic anhydrase IX in benign
odontogenic lesions. J Oral Pathol Med. 45:712–717. 2016.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Cheng SC, Quintin J, Cramer RA, Shepardson
KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao
NA, Aghajanirefah A, et al: mTOR- and HIF-1α-mediated aerobic
glycolysis as metabolic basis for trained immunity. Science.
345(1250684)2014.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Giovanelli P, Sandoval TA and
Cubillos-Ruiz JR: Dendritic cell metabolism and function in tumors.
Trends Immunol. 40:699–718. 2019.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Kim IS and Zhang XHF: One microenvironment
does not fit all: Heterogeneity beyond cancer cells. Cancer
Metastasis Rev. 35:601–629. 2016.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Sobral-Leite M, Salomon I, Opdam M, Kruger
DT, Beelen KJ, van der Noort V, van Vlierberghe RLP, Blok EJ,
Giardiello D, Sanders J, et al: Cancer-immune interactions in
ER-positive breast cancers: PI3K pathway alterations and
tumor-infiltrating lymphocytes. Breast Cancer Res.
21(90)2019.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Rivera LB, Meyronet D, Hervieu V,
Frederick MJ, Bergsland E and Bergers G: Intratumoral myeloid cells
regulate responsiveness and resistance to antiangiogenic therapy.
Cell Rep. 11:577–591. 2015.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Lambertz U, Silverman JM, Nandan D,
McMaster WR, Clos J, Foster LJ and Reiner NE: Secreted virulence
factors and immune evasion in visceral leishmaniasis. J Leukoc
Biol. 91:887–899. 2012.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Crane CA, Panner A, Murray JC, Wilson SP,
Xu H, Chen L, Simko JP, Waldman FM, Pieper RO and Parsa AT: PI (3)
kinase is associated with a mechanism of immunoresistance in breast
and prostate cancer. Oncogene. 28:306–312. 2009.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Bahrami A, Hasanzadeh M, Hassanian SM,
ShahidSales S, Ghayour-Mobarhan M, Ferns GA and Avan A: The
potential value of the PI3K/Akt/mTOR signaling pathway for
assessing prognosis in cervical cancer and as a target for therapy.
J Cell Biochem. 118:4163–4169. 2017.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Chai J, Modak C, Mouazzen W, Narvaez R and
Pham J: Epithelial or mesenchymal: Where to draw the line? Biosci
Trends. 4:130–142. 2010.PubMed/NCBI
|
|
83
|
Gerber TS, Ridder DA, Schindeldecker M,
Weinmann A, Duret D, Breuhahn K, Galle PR, Schirmacher P, Roth W,
Lang H and Straub BK: Constitutive occurrence of E: N-cadherin
heterodimers in adherens junctions of hepatocytes and derived
tumors. Cells. 11(2507)2022.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Deshmukh AP, Vasaikar SV, Tomczak K,
Tripathi S, den Hollander P, Arslan E, Chakraborty P, Soundararajan
R, Jolly MK, Rai K, et al: Identification of EMT signaling
cross-talk and gene regulatory networks by single-cell RNA
sequencing. Proc Natl Acad Sci USA. 118(e2102050118)2021.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Suarez-Carmona M, Lesage J, Cataldo D and
Gilles C: EMT and inflammation: Inseparable actors of cancer
progression. Mol Oncol. 11:805–823. 2017.PubMed/NCBI View Article : Google Scholar
|