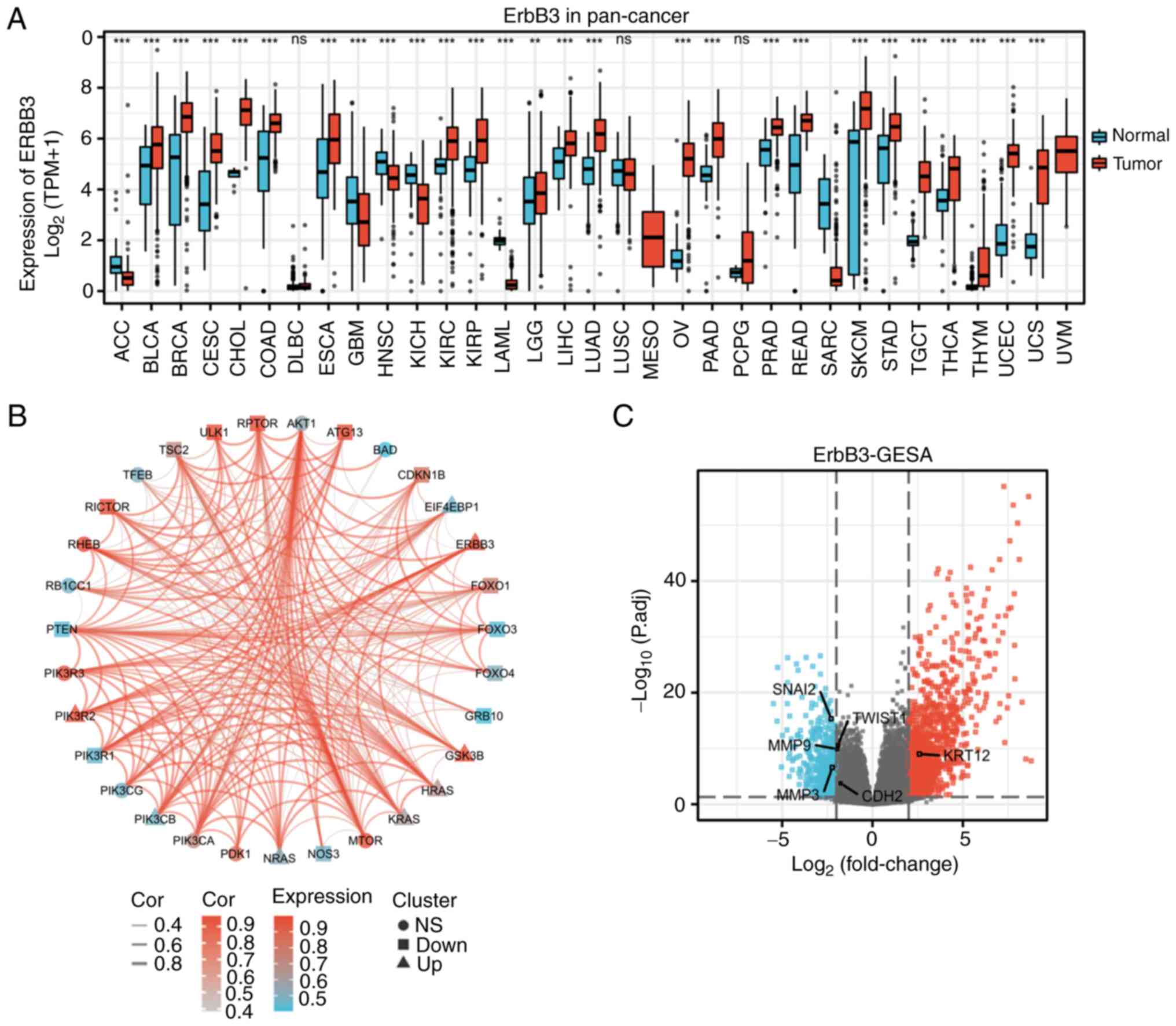
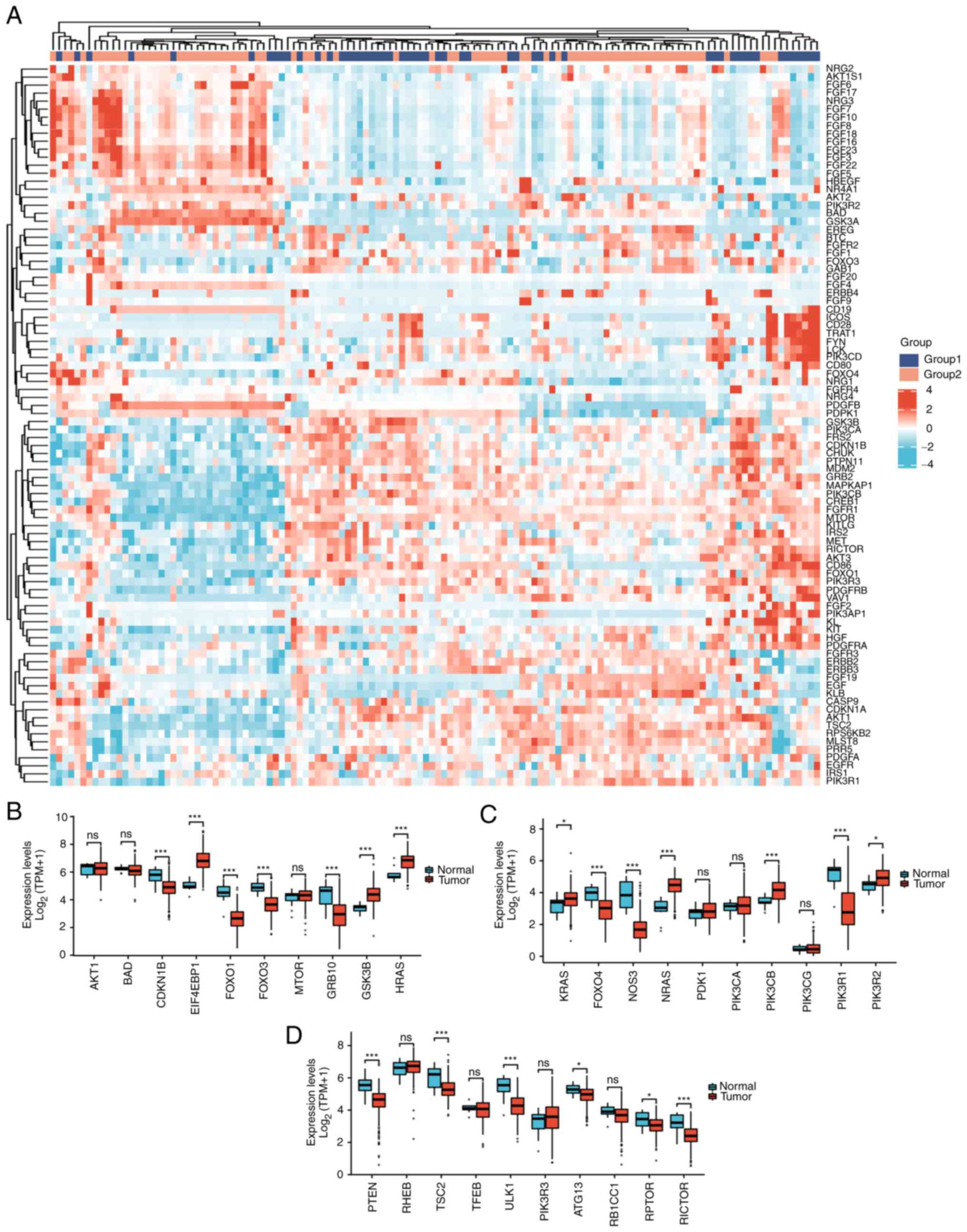
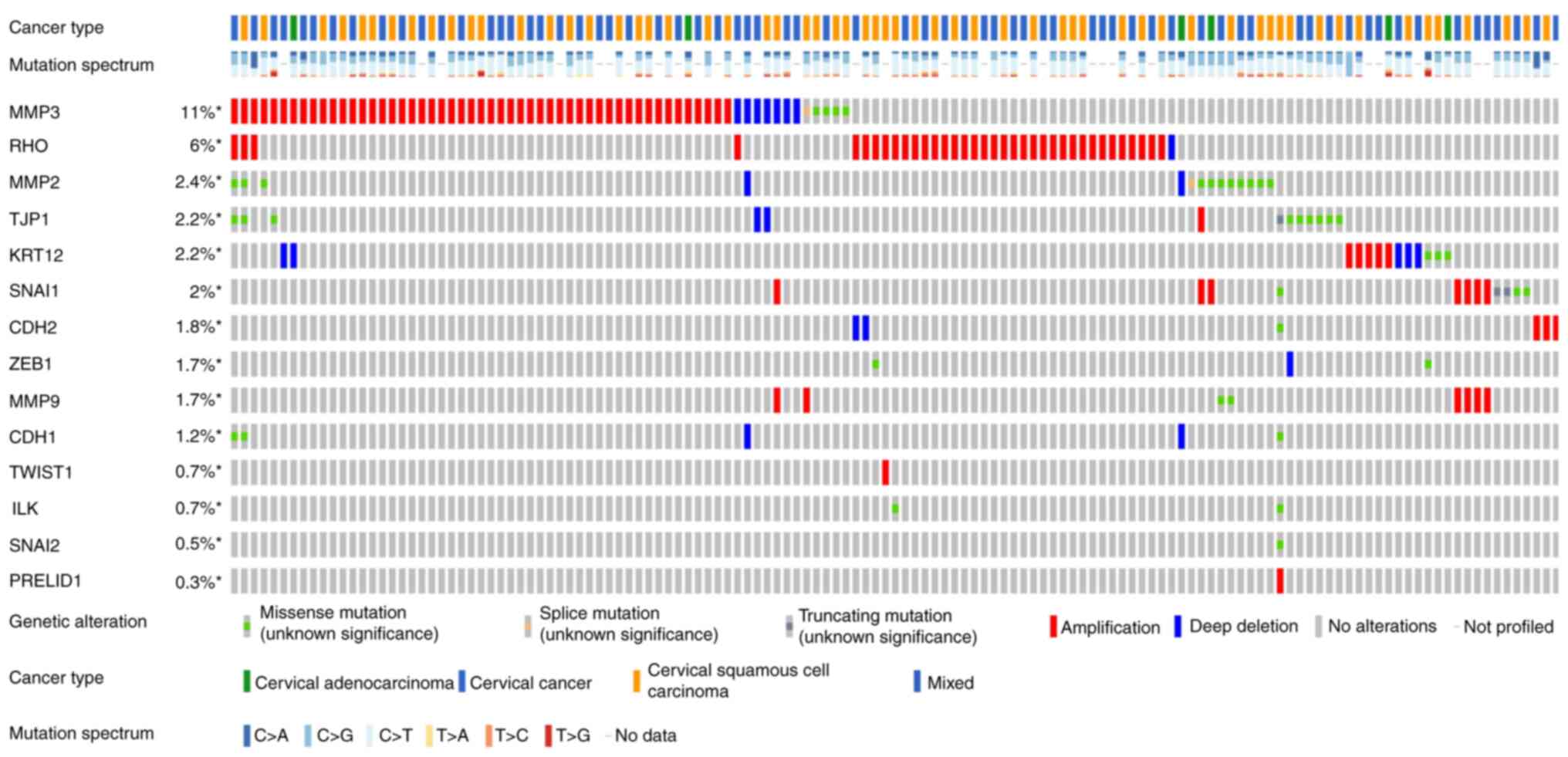
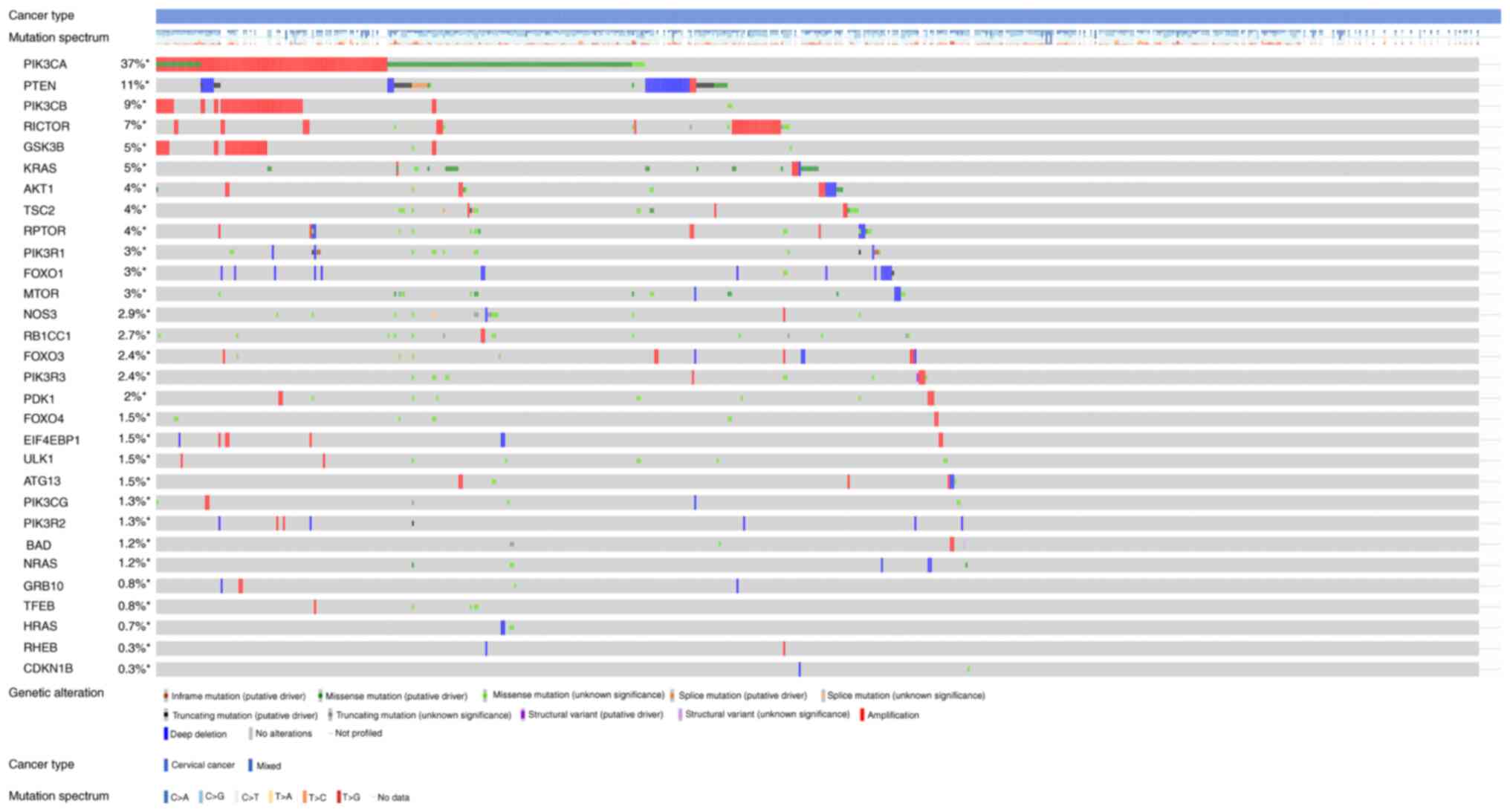
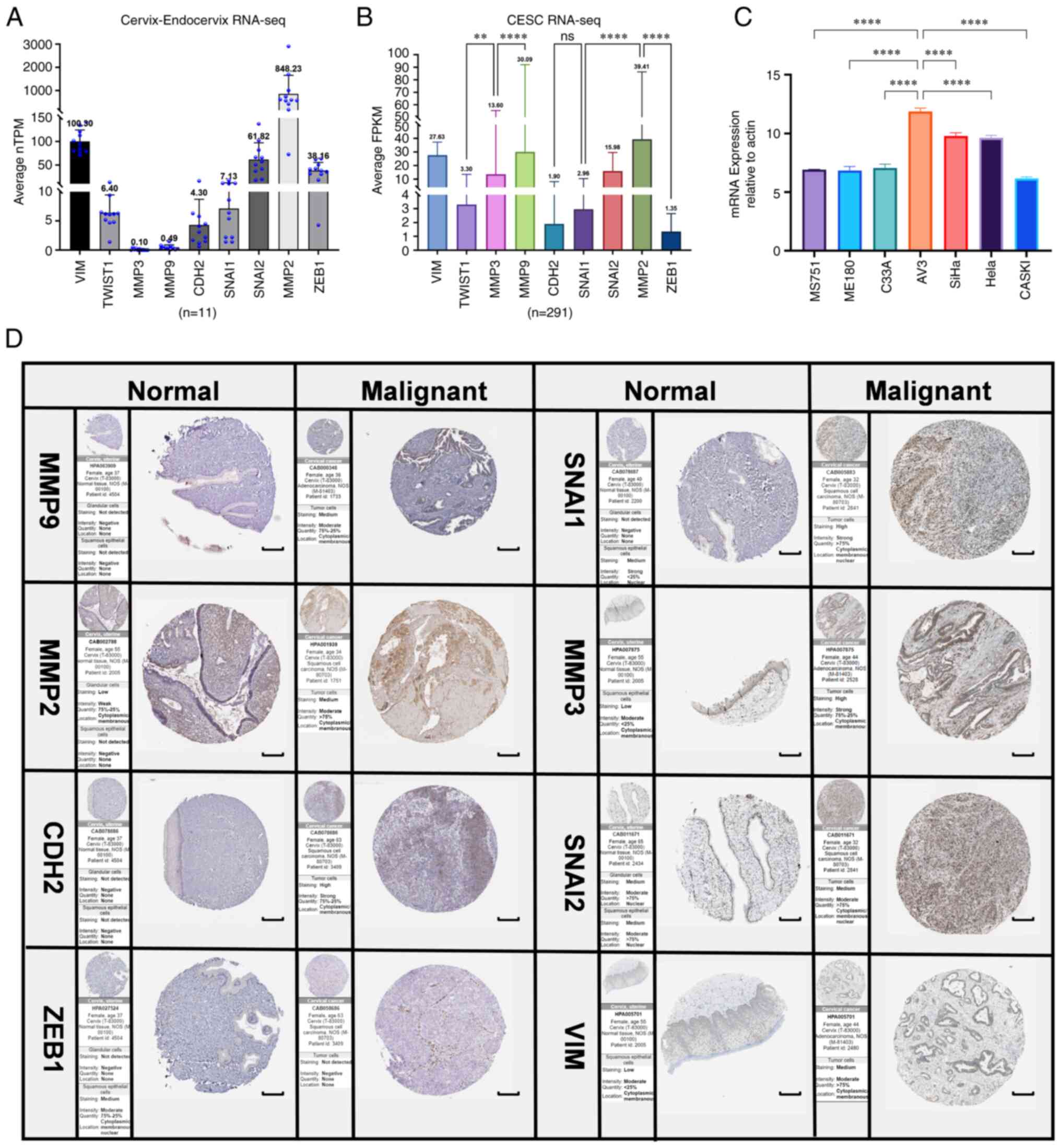
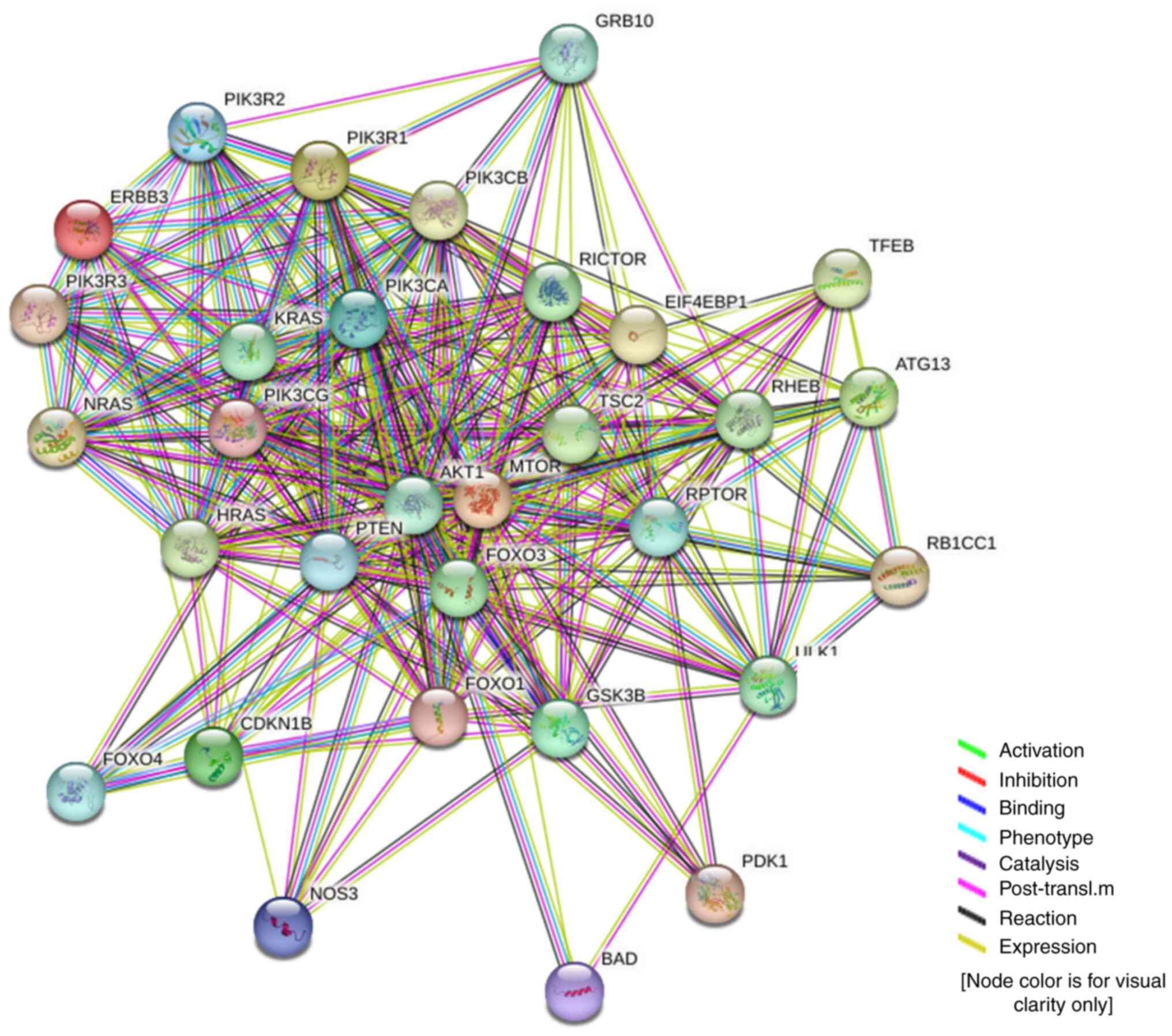
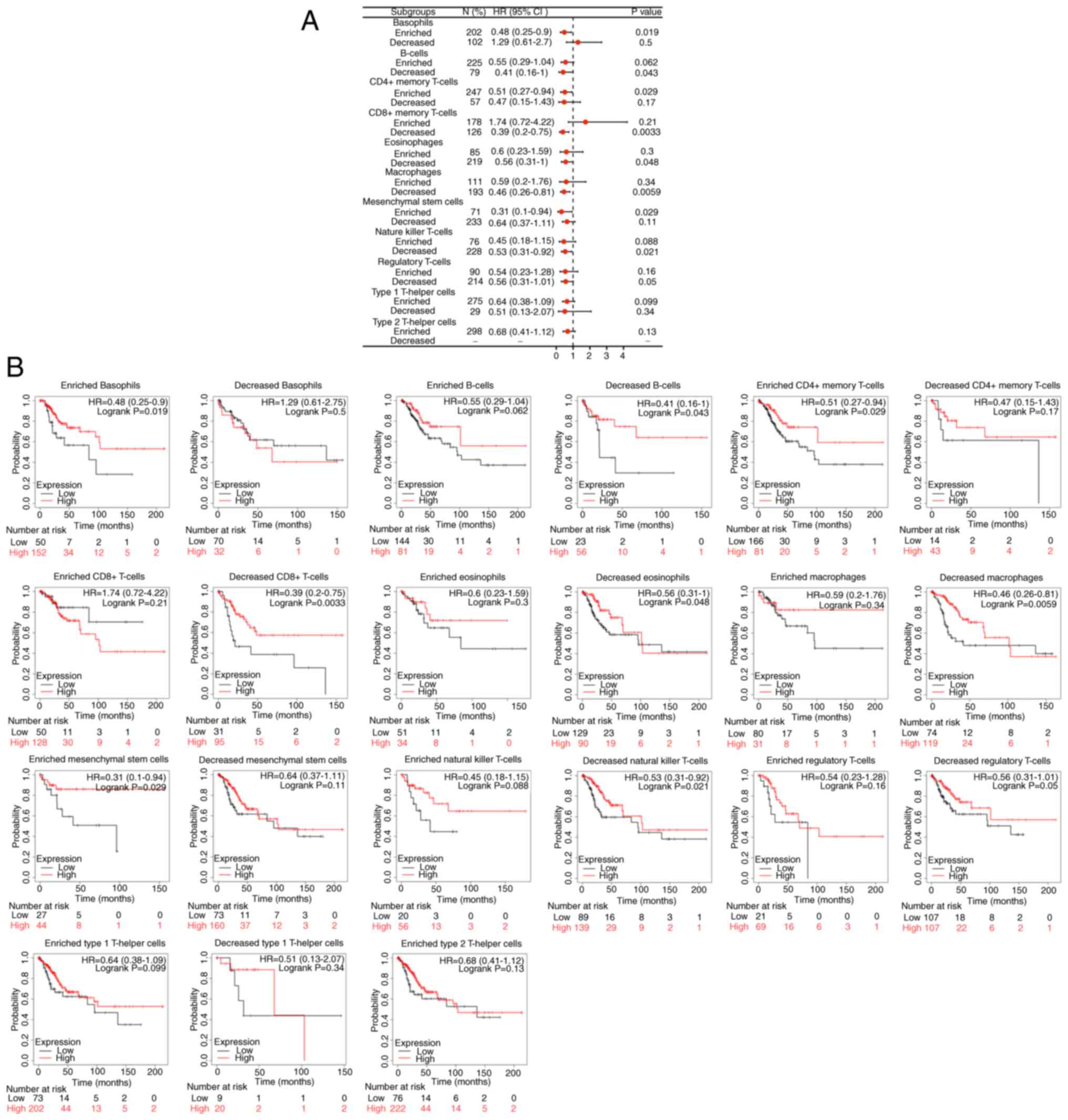
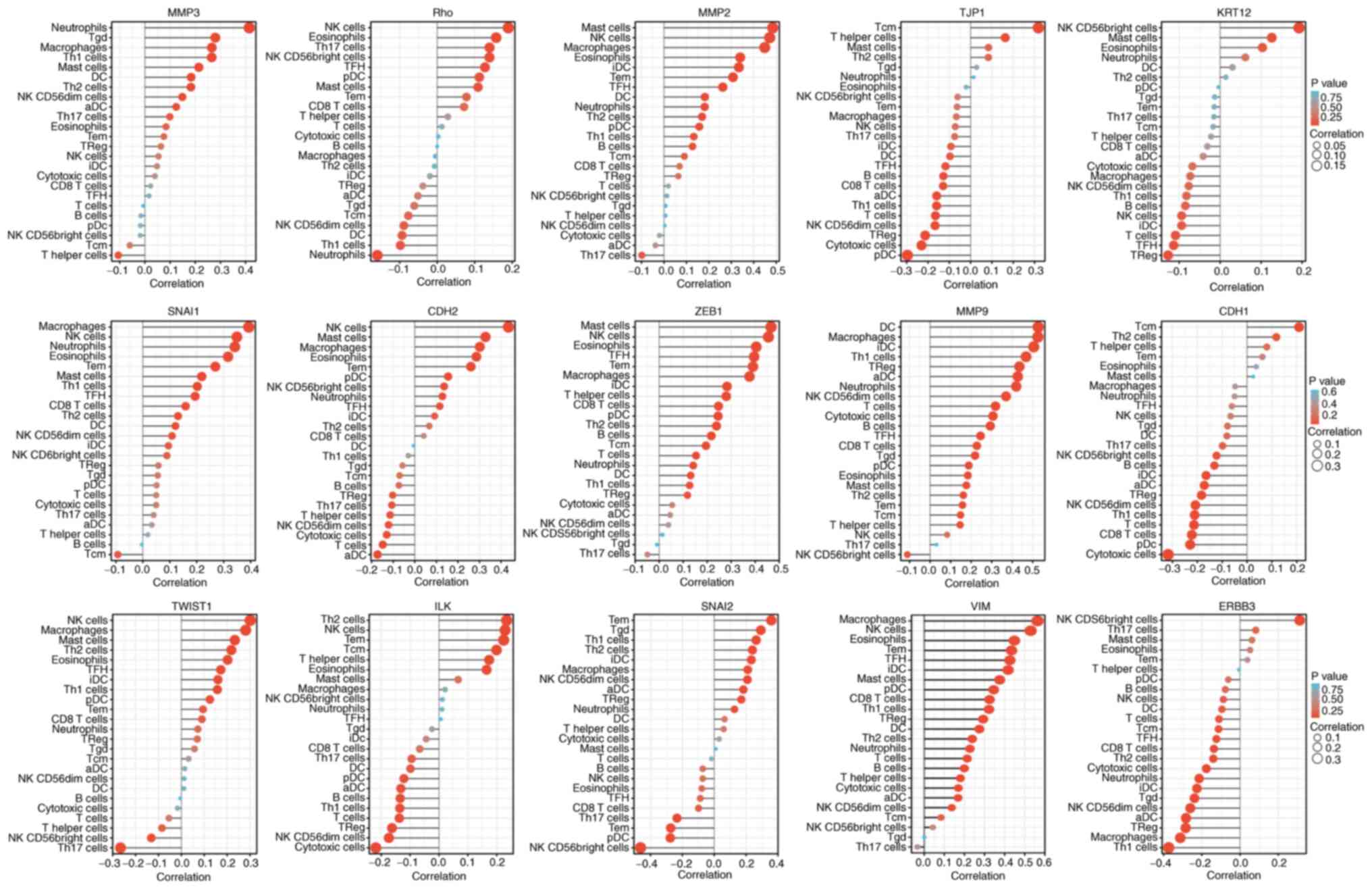
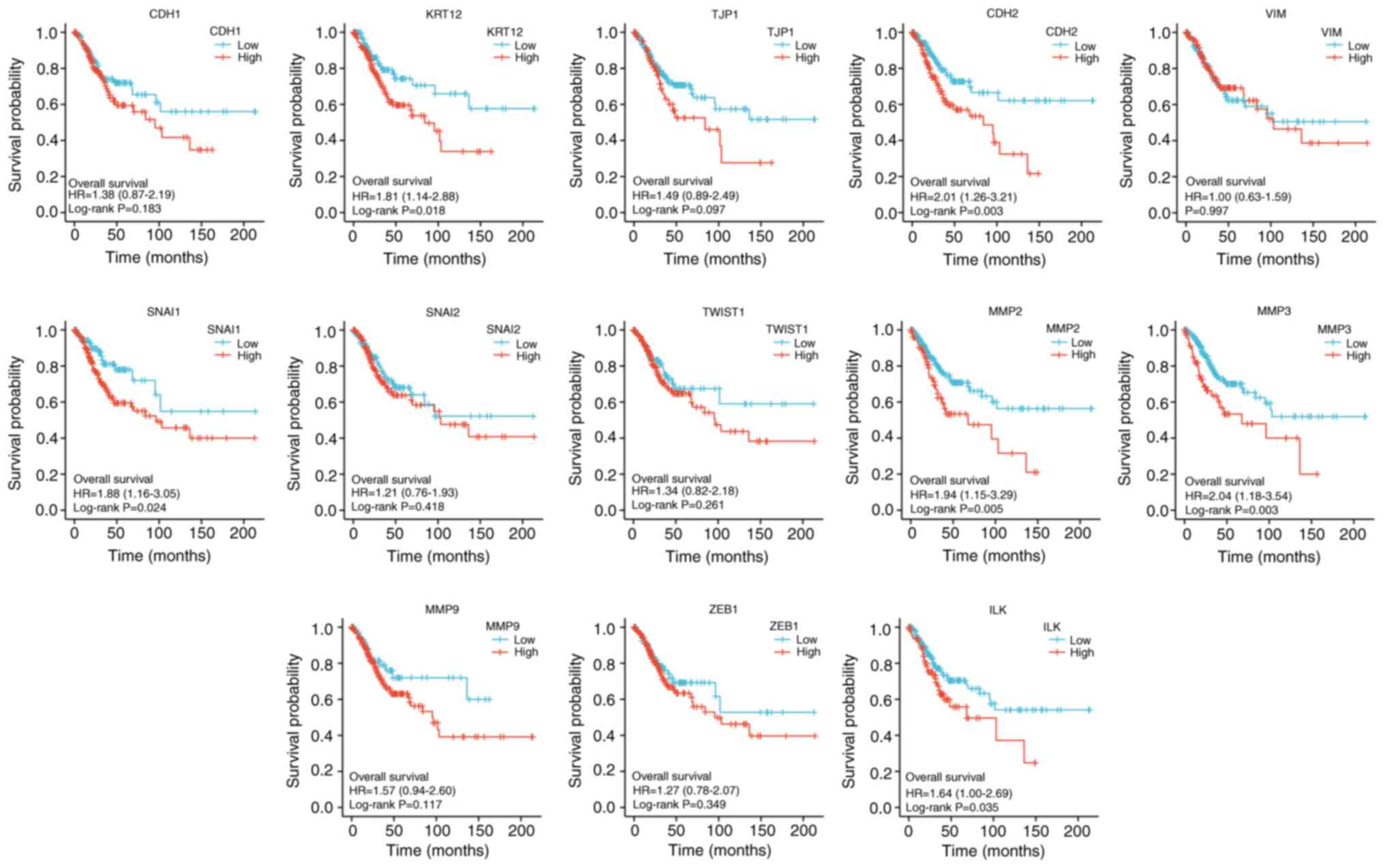
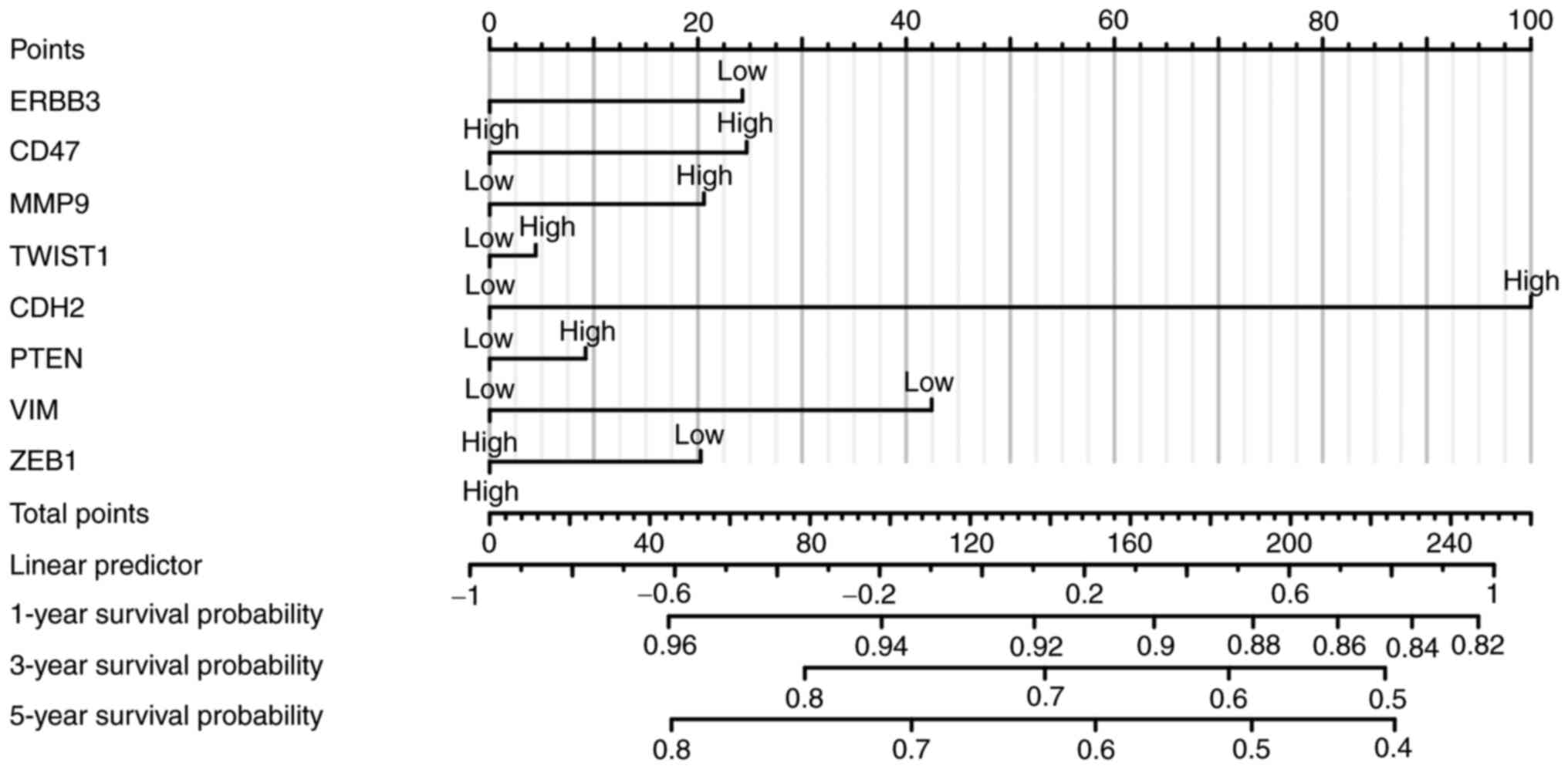
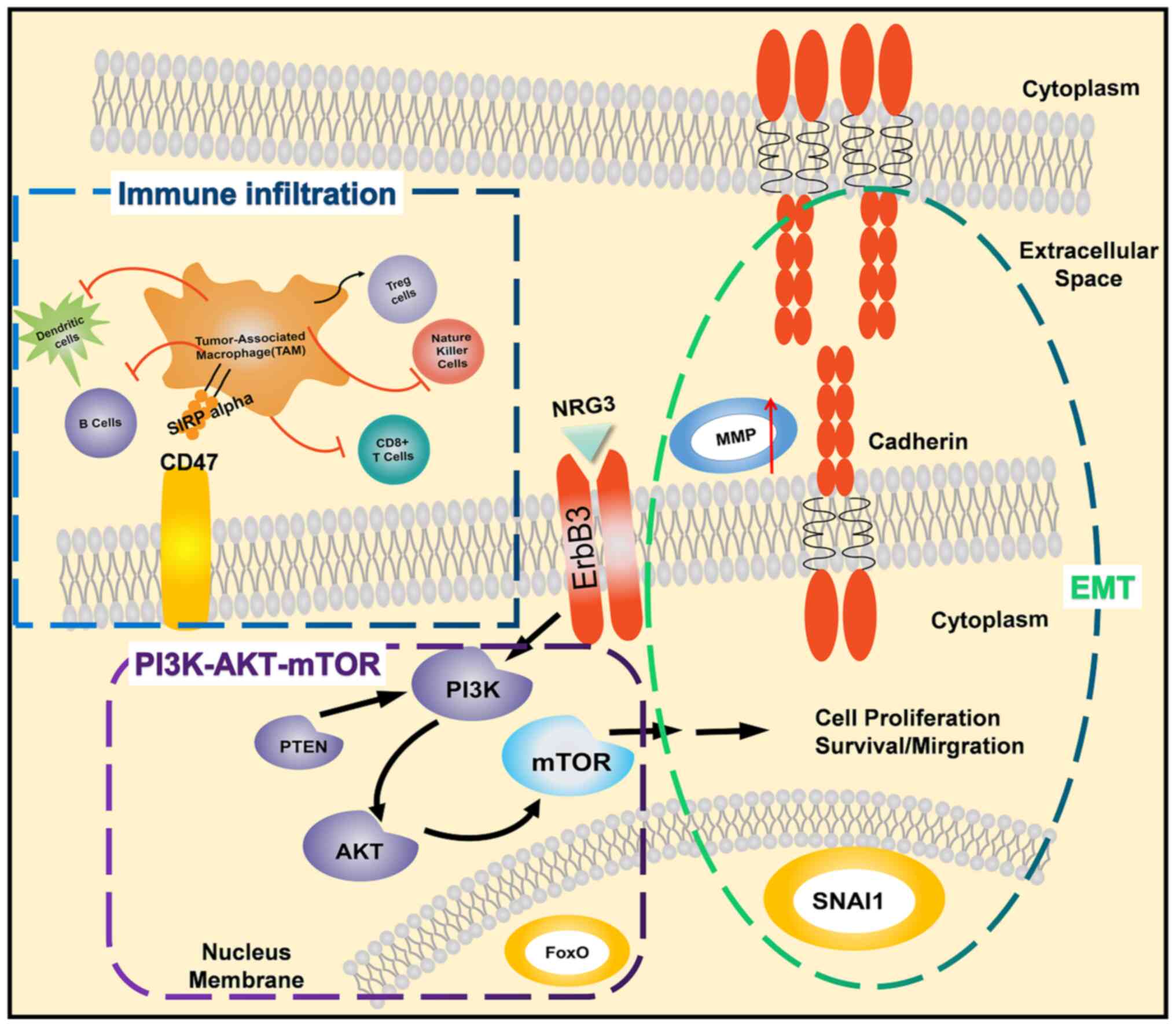
|
1
|
Cohen PA, Jhingran A, Oaknin A and Denny
L: Cervical cancer. Lancet. 393:169–182. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yugawa T and Kiyono T: Molecular
mechanisms of cervical carcinogenesis by high-risk human
papillomaviruses: Novel functions of E6 and E7 oncoproteins. Rev
Med Virol. 19:97–113. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Zhang L, Wu J, Ling MT, Zhao L and Zhao
KN: The role of the PI3K/Akt/mTOR signalling pathway in human
cancers induced by infection with human papillomaviruses. Mol
Cancer. 14(87)2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bossler F, Hoppe-Seyler K and Hoppe-Seyler
F: PI3K/AKT/mTOR signaling regulates the virus/host cell crosstalk
in HPV-positive cervical cancer cells. Int J Mol Sci.
20(2188)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Shi X, Wang J, Lei Y, Cong C, Tan D and
Zhou X: Research progress on the PI3K/AKT signaling pathway in
gynecological cancer. Mol Med Rep. 19:4529–4535. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Senga SS and Grose RP: Hallmarks of
cancer-the new testament. Open Biol. 11(200358)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
O'Donnell JS, Teng MWL and Smyth MJ:
Cancer immunoediting and resistance to T cell-based immunotherapy.
Nat Rev Clin Oncol. 16:151–167. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Fukumura D, Kloepper J, Amoozgar Z, Duda
DG and Jain RK: Enhancing cancer immunotherapy using
antiangiogenics: Opportunities and challenges. Nat Rev Clin Oncol.
15:325–340. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Aiello NM and Kang Y: Context-dependent
EMT programs in cancer metastasis. J Exp Med. 216:1016–1026.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Taki M, Abiko K, Ukita M, Murakami R,
Yamanoi K, Yamaguchi K, Hamanishi J, Baba T, Matsumura N and Mandai
M: Tumor immune microenvironment during epithelial-mesenchymal
transitionthe review of the loop between EMT and immunosuppression.
Clin Cancer Res. 27:4669–4679. 2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Dongre A, Rashidian M, Reinhardt F,
Bagnato A, Keckesova Z, Ploegh HL and Weinberg RA:
Epithelial-to-mesenchymal transition contributes to
immunosuppression in breast carcinomas. Cancer Res. 77:3982–3989.
2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hoxhaj G and Manning BD: The PI3K-AKT
network at the interface of oncogenic signalling and cancer
metabolism. Nat Rev Cancer. 20:74–88. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Coussy F, El Botty R, Lavigne M, Gu C,
Fuhrmann L, Briaux A, de Koning L, Dahmani A, Montaudon E, Morisset
L, et al: Combination of PI3K and MEK inhibitors yields durable
remission in PDX models of PIK3CA-mutated metaplastic breast
cancers. J Hematol Oncol. 13(13)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li M, Liu F, Zhang F, Zhou W, Jiang X,
Yang Y, Qu K, Wang Y, Ma Q, Wang T, et al: Genomic ERBB2/ERBB3
mutations promote PD-L1-mediated immune escape in gallbladder
cancer: a whole-exome sequencing analysis. Gut. 68:1024–1033.
2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ross JS, Fakih M, Ali SM, Elvin JA,
Schrock AB, Suh J, Vergilio JA, Ramkissoon S, Severson E, Daniel S,
et al: Targeting HER2 in colorectal cancer: The landscape of
amplification and short variant mutations in ERBB2 and ERBB3.
Cancer. 124:1358–1373. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Van Lengerich B, Agnew C, Puchner EM,
Huang B and Jura N: EGF and NRG induce phosphorylation of
HER3/ERBB3 by EGFR using distinct oligomeric mechanisms. Proc Natl
Acad Sci USA. 114:E2836–E2845. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sithanandam G and Anderson LM: The ERBB3
receptor in cancer and cancer gene therapy. Cancer Gene Ther.
15:413–448. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Moghbeli M, Makhdoumi Y, Delgosha MS,
Aarabi A, Dadkhah E, Memar B, Abdollahi A and Abbaszadegan MR:
ErbB1 and ErbB3 co-over expression as a prognostic factor in
gastric cancer. Biol Res. 52(2)2008.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shi DM, Li LX, Bian XY, Shi XJ, Lu LL,
Zhou HX, Pan TJ, Zhou J, Fan J and Wu WZ: miR-296-5p suppresses EMT
of hepatocellular carcinoma via attenuating NRG1/ERBB2/ERBB3
signaling. J Exp Clin Cancer Res. 37(294)2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Vivian J, Rao AA, Nothaft FA, Ketchum C,
Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD,
Musselman-Brown A, et al: Toil enables reproducible, open source,
big biomedical data analyses. Nat Biotechnol. 35:314–316.
2017.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Yang J, Antin P, Berx G, Blanpain C,
Brabletz T, Bronner M, Campbell K, Cano A, Casanova J, Christofori
G, et al: EMT International Association (TEMTIA). Guidelines and
definitions for research on epithelial-mesenchymal transition. Nat
Rev Mol Cell Biol. 21:341–352. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Den Boon JA, Pyeon D, Wang SS, Horswill M,
Schiffman M, Sherman M, Zuna RE, Wang Z, Hewitt SM, Pearson R, et
al: Molecular transitions from papillomavirus infection to cervical
precancer and cancer: Role of stromal estrogen receptor signaling.
Proc Natl Acad Sci USA. 112:E3255–E3264. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M and Murty VV: Identification of copy
number gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995.
2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Smyth GK: Limma: Linear models for
microarray data. Bioinformatics and computational biology solutions
using R and Bioconductor. Springer, New York, NY, 2005.
397-420.
|
|
27
|
Gu Z, Eils R and Schlesner M: Complex
heatmaps reveal patterns and correlations in multidimensional
genomic data. Bioinformatics. 32:2847–2849. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bindea G, Mlecnik B, Tosolini M,
Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T,
Lafontaine L, Berger A, et al: Spatiotemporal dynamics of
intratumoral immune cells reveal the immune landscape in human
cancer. Immunity. 39:782–795. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Walter W, Sánchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucleic Acids Res. 36(Database issue):D480–D484.
2007.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lanczky A and Gyorffy B: Web-based
survival analysis tool tailored for medical research (KMplot):
Development and implementation. J Med Internet Res.
23(e27633)2021.PubMed/NCBI View
Article : Google Scholar
|
|
34
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Goel AL and Wu SM: Determination of ARL
and a contour nomogram for CUSUM charts to control normal mean.
Technometrics. 13:221–230. 1971.
|
|
36
|
Han H, Cho JW, Lee S, Yun A, Kim H, Bae D,
Yang S, Kim CY, Lee M, Kim E, et al: TRRUST v2: An expanded
reference database of human and mouse transcriptional regulatory
interactions. Nucleic Acids Res. 46:D380–D386. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
R Core Team: A language and environment
for statistical computing. R Foundation for Statistical Computing,
Vienna, 2013. http://www.R-project.org/.
|
|
38
|
Jeppson H and Hofmann H: Generalized
Mosaic Plots in the ggplot2 Framework: Exploratory analysis of
high-dimensional data with visual tools 17, 2021. https://rdrr.io/cran/ggmosaic/.
|
|
39
|
Csardi G and Nepusz T: The igraph software
package for complex network research. InterJournal Complex Systems.
1695:1–9. 2006.
|
|
40
|
Gustavsson EK, Zhang D, Reynolds RH, et
al: ggtranscript: an R package for the visualization and
interpretation of transcript isoforms using ggplot2.
Bioinformatics. 38(15):3844–3846. 2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kassambara A, Kosinski M and Biecek P:
Package ‘survminer’. Drawing Survival Curves using ‘ggplot2’(R
package version 03 1), 2017.
|
|
42
|
Cancer Genome Atlas Research Network.
Integrated genomic and molecular characterization of cervical
cancer. Nature. 543:378–384. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yang X, Chen Y, Li M and Zhu W: ERBB3
methylation and immune infiltration in tumor microenvironment of
cervical cancer. Sci Rep. 12(8112)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Soto D, Song C and McLaughlin-Drubin ME:
Epigenetic alterations in human papillomavirus-associated cancers.
Viruses. 9(248)2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Senapati R, Senapati NN and Dwibedi B:
Molecular mechanisms of HPV mediated neoplastic progression. Infect
Agent Cancer. 11(59)2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Narisawa-Saito M and Kiyono T: Basic
mechanisms of high-risk human papillomavirus-induced
carcinogenesis: Roles of E6 and E7 proteins. Cancer Sci.
98:1505–1511. 2007.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Martinez-Zapien D, Ruiz FX, Poirson J,
Mitschler A, Ramirez J, Forster A, Cousido-Siah A, Masson M, Pol
SV, Podjarny A, et al: Structure of the E6/E6AP/p53 complex
required for HPV-mediated degradation of p53. Nature. 529:541–545.
2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Karamanou K, Franchi M, Vynios D and
Brézillon S: Epithelial-to-mesenchymal transition and invadopodia
markers in breast cancer: Lumican a key regulator. Semin Cancer
Biol. 62:125–133. 2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Yang HL, Thiyagarajan V, Shen PC, Mathew
DC, Lin KY, Liao JW and Hseu YC: Anti-EMT properties of CoQ0
attributed to PI3K/AKT/NFKB/MMP-9 signaling pathway through
ROS-mediated apoptosis. J Exp Clin Cancer Res.
38(186)2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Mao X, Xu J, Wang W, Liang C, Hua J, Liu
J, Zhang B, Meng Q, Yu X and Shi S: Crosstalk between
cancer-associated fibroblasts and immune cells in the tumor
microenvironment: New findings and future perspectives. Mol Cancer.
20(131)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Looi CK, Chung FFL, Leong CO, Wong SF,
Rosli R and Mai SW: Therapeutic challenges and current
immunomodulatory strategies in targeting the immunosuppressive
pancreatic tumor microenvironment. J Exp Clin Cancer Res.
38(162)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Cheng YQ, Wang SB, Liu JH, Jin L, Liu Y,
Li CY, Su YR, Liu YR, Sang X, Wan Q, et al: Modifying the tumour
microenvironment and reverting tumour cells: New strategies for
treating malignant tumours. Cell Prolif. 53(e12865)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Li D and Wu M: Pattern recognition
receptors in health and diseases. Signal Transduct Target Ther.
6(291)2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Mendes F, Domingues C, Rodrigues-Santos P,
Abrantes AM, Gonçalves AC, Estrela J, Encarnação J, Pires AS,
Laranjo M, Alves V, et al: The role of immune system exhaustion on
cancer cell escape and anti-tumor immune induction after
irradiation. Biochim Biophys Acta. 1865:168–175. 2016.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Liu Y and Cao X: Immunosuppressive cells
in tumor immune escape and metastasis. J Mol Med (Berl).
94:509–522. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Che Y, Yang Y, Suo J, An Y and Wang X:
Induction of systemic immune responses and reversion of
immunosuppression in the tumor microenvironment by a therapeutic
vaccine for cervical cancer. Cancer Immunol Immunother.
69:2651–2664. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Jayshree RS: The immune microenvironment
in human papilloma virus-induced cervical lesions-evidence for
estrogen as an immunomodulator. Front Cell Infect Microbiol.
11(649815)2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Welters MJ, van der Sluis TC, van Meir H,
Loof NM, van Ham VJ, van Duikeren S, Santegoets SJ, Arens R, de Kam
ML, Cohen AF, et al: Vaccination during myeloid cell depletion by
cancer chemotherapy fosters robust T cell responses. Sci Transl
Med. 8(334ra52)2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Todd RW, Roberts S, Mann CH, Luesley DM,
Gallimore PH and Steele JC: Human papillomavirus (HPV) type
16-specific CD8+ T cell responses in women with high grade vulvar
intraepithelial neoplasia. Int J Cancer. 108:857–862.
2004.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Torres-Poveda K, Bahena-Román M,
Madrid-González C, Burguete-García AI, Bermúdez-Morales VH,
Peralta-Zaragoza O and Madrid-Marina V: Role of IL-10 and TGF-β1 in
local immunosuppression in HPV-associated cervical neoplasia. World
J Clin Oncol. 5:753–763. 2014.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Kalathil SG and Thanavala Y: High
immunosuppressive burden in cancer patients: A major hurdle for
cancer immunotherapy. Cancer Immunol Immunother. 65:813–819.
2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Huang YK, Wang M, Sun Y, Di Costanzo N,
Mitchell C, Achuthan A, Hamilton JA, Busuttil RA and Boussioutas A:
Macrophage spatial heterogeneity in gastric cancer defined by
multiplex immunohistochemistry. Nat Commun. 10(3928)2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Zhang Y and Zhang Z: The history and
advances in cancer immunotherapy: Understanding the characteristics
of tumor-infiltrating immune cells and their therapeutic
implications. Cell Mol Immunol. 17:807–821. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Chao MP, Weissman IL and Majeti R: The
CD47-SIRPα pathway in cancer immune evasion and potential
therapeutic implications. Curr Opin Immunol. 24:225–232.
2012.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Feng M, Jiang W, Kim BYS, Zhang CC, Fu YX
and Weissman IL: Phagocytosis checkpoints as new targets for cancer
immunotherapy. Nat Rev Cancer. 19:568–586. 2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Oronsky B, Carter C, Reid T, Brinkhaus F
and Knox SJ: Just eat it: A review of CD47 and SIRP-α antagonism.
Semin Oncol. 47:117–124. 2020.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Li Z, Li Y, Gao J, Fu Y, Hua P, Jing Y,
Cai M, Wang H and Tong T: The role of CD47-SIRPα immune checkpoint
in tumor immune evasion and innate immunotherapy. Life Sci.
273(119150)2021.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Huang CY, Ye ZH, Huang MY and Lu JJ:
Regulation of CD47 expression in cancer cells. Transl Oncol.
13(100862)2020.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Yang H, Shao R, Huang H, Wang X, Rong Z
and Lin Y: Engineering macrophages to phagocytose cancer cells by
blocking the CD47/SIRPα axis. Cancer Med. 8:4245–4253.
2019.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Braicu EI, Gasimli K, Richter R, Nassir M,
Kümmel S, Blohmer JU, Yalcinkaya I, Chekerov R, Ignat I, Ionescu A,
et al: Role of serum VEGFA, TIMP2, MMP2 and MMP9 in monitoring
response to adjuvant radiochemotherapy in patients with primary
cervical cancer-results of a companion protocol of the randomized
NOGGO-AGO phase III clinical trial. Anticancer Res. 34:385–391.
2014.PubMed/NCBI
|
|
71
|
Riani M, Le Jan S, Plée J, Durlach A, Le
Naour R, Haegeman G, Bernard P and Antonicelli F: Bullous
pemphigoid outcome is associated with CXCL10-induced matrix
metalloproteinase 9 secretion from monocytes and neutrophils but
not lymphocytes. J Allergy Clin Immunol. 139:863–872.e3.
2017.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Yi W, Tu MJ, Liu Z, Zhang C, Batra N, Yu
AX and Yu AM: Bioengineered miR-328-3p modulates GLUT1-mediated
glucose uptake and metabolism to exert synergistic
antiproliferative effects with chemotherapeutics. Acta Pharm Sin B.
10:159–170. 2020.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Vasconcelos RC, de Oliveira Moura JM,
Junior VLB, da Silveira ÉJ and de Souza LB: Immunohistochemical
expression of GLUT-1, GLUT-3, and carbonic anhydrase IX in benign
odontogenic lesions. J Oral Pathol Med. 45:712–717. 2016.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Cheng SC, Quintin J, Cramer RA, Shepardson
KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao
NA, Aghajanirefah A, et al: mTOR- and HIF-1α-mediated aerobic
glycolysis as metabolic basis for trained immunity. Science.
345(1250684)2014.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Giovanelli P, Sandoval TA and
Cubillos-Ruiz JR: Dendritic cell metabolism and function in tumors.
Trends Immunol. 40:699–718. 2019.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Kim IS and Zhang XHF: One microenvironment
does not fit all: Heterogeneity beyond cancer cells. Cancer
Metastasis Rev. 35:601–629. 2016.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Sobral-Leite M, Salomon I, Opdam M, Kruger
DT, Beelen KJ, van der Noort V, van Vlierberghe RLP, Blok EJ,
Giardiello D, Sanders J, et al: Cancer-immune interactions in
ER-positive breast cancers: PI3K pathway alterations and
tumor-infiltrating lymphocytes. Breast Cancer Res.
21(90)2019.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Rivera LB, Meyronet D, Hervieu V,
Frederick MJ, Bergsland E and Bergers G: Intratumoral myeloid cells
regulate responsiveness and resistance to antiangiogenic therapy.
Cell Rep. 11:577–591. 2015.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Lambertz U, Silverman JM, Nandan D,
McMaster WR, Clos J, Foster LJ and Reiner NE: Secreted virulence
factors and immune evasion in visceral leishmaniasis. J Leukoc
Biol. 91:887–899. 2012.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Crane CA, Panner A, Murray JC, Wilson SP,
Xu H, Chen L, Simko JP, Waldman FM, Pieper RO and Parsa AT: PI (3)
kinase is associated with a mechanism of immunoresistance in breast
and prostate cancer. Oncogene. 28:306–312. 2009.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Bahrami A, Hasanzadeh M, Hassanian SM,
ShahidSales S, Ghayour-Mobarhan M, Ferns GA and Avan A: The
potential value of the PI3K/Akt/mTOR signaling pathway for
assessing prognosis in cervical cancer and as a target for therapy.
J Cell Biochem. 118:4163–4169. 2017.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Chai J, Modak C, Mouazzen W, Narvaez R and
Pham J: Epithelial or mesenchymal: Where to draw the line? Biosci
Trends. 4:130–142. 2010.PubMed/NCBI
|
|
83
|
Gerber TS, Ridder DA, Schindeldecker M,
Weinmann A, Duret D, Breuhahn K, Galle PR, Schirmacher P, Roth W,
Lang H and Straub BK: Constitutive occurrence of E: N-cadherin
heterodimers in adherens junctions of hepatocytes and derived
tumors. Cells. 11(2507)2022.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Deshmukh AP, Vasaikar SV, Tomczak K,
Tripathi S, den Hollander P, Arslan E, Chakraborty P, Soundararajan
R, Jolly MK, Rai K, et al: Identification of EMT signaling
cross-talk and gene regulatory networks by single-cell RNA
sequencing. Proc Natl Acad Sci USA. 118(e2102050118)2021.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Suarez-Carmona M, Lesage J, Cataldo D and
Gilles C: EMT and inflammation: Inseparable actors of cancer
progression. Mol Oncol. 11:805–823. 2017.PubMed/NCBI View Article : Google Scholar
|