Introduction
Coronavirus disease 2019 (COVID-19), caused by
severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a
major health threat all over the world. As of 8 April 2022, a
cumulative total of 494,587,638 COVID-19 cases were reported
globally, including 6,170,283 deaths, according to the World Health
Organization (1).
The spectrum of SARS-CoV-2 infection ranges from
asymptomatic to clinical symptoms, including fever, dry cough,
dyspnea, fatigue, muscle pain, headache, diarrhea, loss of taste
and/or smell, nasal obstruction and runny nose (2). The majority of patients with COVID-19
have a mild or moderate form. However, infection in certain
patients becomes severe, presenting with respiratory failure. In
the most severe cases, acute respiratory distress and multiple
organ dysfunction syndrome, coagulation abnormalities and shock
have been observed (2).
Older age (>60 years), unvaccinated, associated
comorbidities (such as cardiovascular disease, hypertension,
chronic pulmonary disease, diabetes, chronic liver and kidney
disease and malignancy), immunodeficiency, obesity and heavy
smoking are key host risk factors for severe COVID-19(2). Apart from these factors, studies have
shown that host genetics may also be key in the development of
severe COVID-19 and should be considered in COVID-19 prognosis
(3,4).
Interferon-induced transmembrane proteins (IFITM1, 2
and 3) are stimulated by interferon and serve a critical role in
the immune defense against viral infection (5-7).
Among them, IFITM3 has the strongest antiviral effect. It blocks
fusion with the host cellular membranes in a broad spectrum of
enveloped viruses, such as SARS-CoV, influenza, human
immunodeficiency virus (HIV), Ebola and Zika (5-7).
The human IFITM3, located on chromosome 11p15.5, is composed
of two exons and one intron. The single nucleotide polymorphism
(SNP) rs12252-C allele of IFITM3 truncates the protein,
leading to decreased restriction of virus replication in
vitro (8). A meta-analysis
indicated that this SNP may be associated with severe influenza
infection (9).
Recently, studies have investigated the association
of the IFITM3 rs12252 polymorphism with COVID-19 severity
(10-14).
However, the findings are inconsistent. Such inconsistency may be
due partly to population differences in genotype distribution,
insufficient power, a small effect of the IFITM3 rs12252
polymorphism on COVID-19 severity and false-positive results.
Therefore, a meta-analysis of published studies was performed to
investigate whether the IFITM3 rs12252 polymorphism is
associated with COVID-19 severity.
Materials and methods
Searching
Major databases and preprint servers, including
PubMed (pubmed.ncbi.nlm.nih.gov), EMBASE (https://www.embase.com), China National Knowledge
Infrastructure (https://www.cnki.net), Wanfang
(https://www.wanfangdata.com.cn), MedRxiv
(https://www.medrxiv.org) and BioRxiv (https://www.biorxiv.org), were searched for studies
concerning IFITM3 and COVID-19. The last search update was
performed on March 30, 2022. The search strategy is supplied in
Appendix S1. No language
restrictions were applied. Manual searching was conducted for
references of relevant reviews and included articles.
Study selection
Studies that were included met the following
criteria: i) Evaluated the association between IFITM3
rs12252 polymorphism and COVID-19 severity; ii) cohort or
case-control study; iii) contained sufficient data to calculate
odds ratio (OR) and iv) COVID-19 infection was clearly defined.
COVID-19 was diagnosed based on symptoms and laboratory tests.
Laboratory confirmation was defined as a positive result using
reverse transcription PCR, serological tests (anti-SARS-CoV-2 IgM)
or both. Studies were excluded when they were: i) Review articles,
comments, responses or case reports; ii) not associated with
IFITM3 rs12252 polymorphism and COVID-19 severity or iii)
non-human studies. When there were multiple studies involving the
same or overlapping population, only the most recent study with the
largest sample size was included. A total of two authors (KY and
JW) assessed each study independently. The titles and abstracts of
all citations were screened. Then, full texts of relevant citations
were examined for inclusion. Disagreements between reviewers were
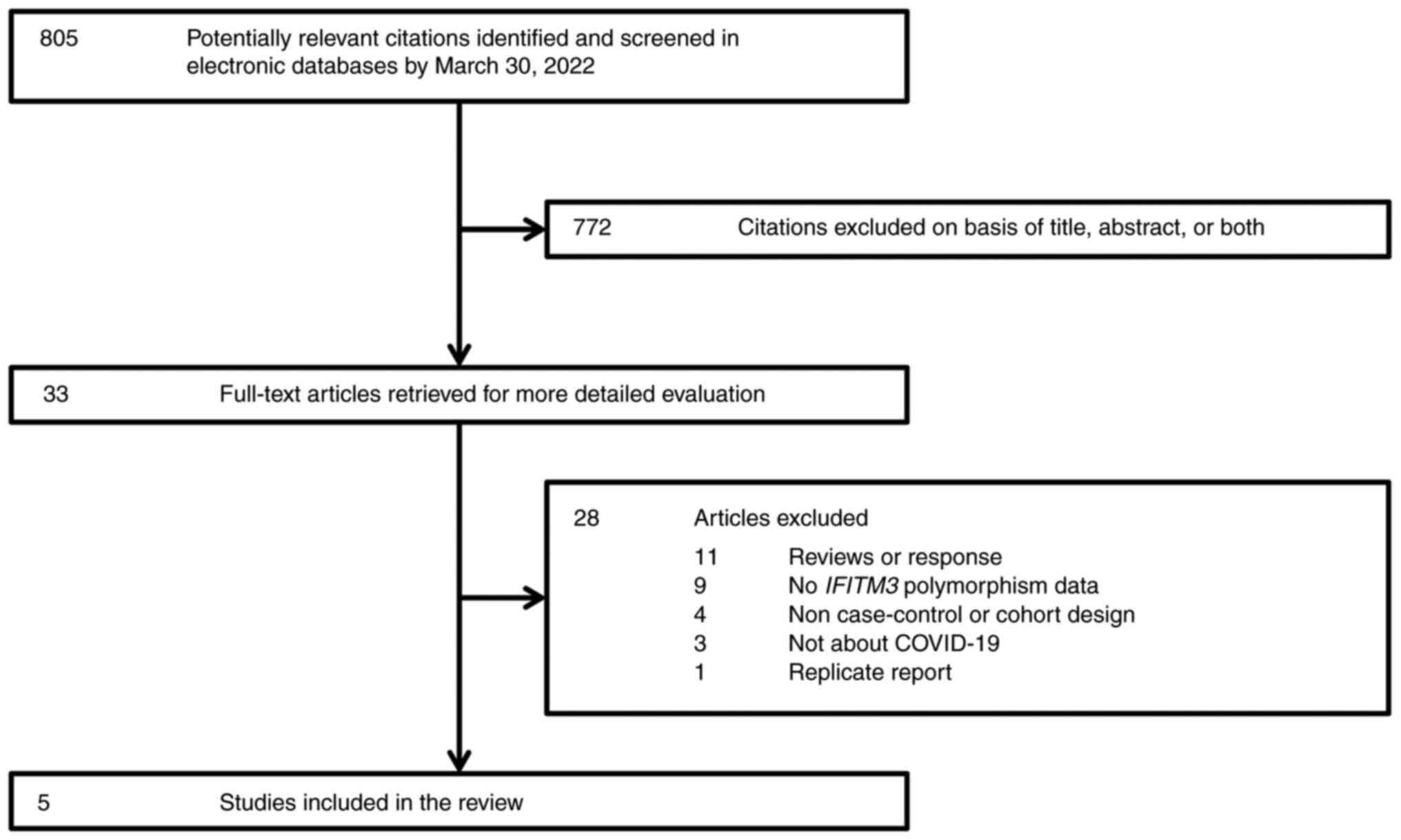
resolved through discussion with a third author (WW). Fig. 1 outlines the study selection
process that led to the final five studies in the present
meta-analysis.
Data extraction and quality
assessment
A total of two authors (KY and JW) independently
extracted the following information from each included study: Study
design, ethnicity, definition and number of mild-to-moderate and
severe cases, age, sex, confounding factors by matching or
adjustment, genotyping method, frequency of genotype and
consistency of genotype frequencies with Hardy-Weinberg equilibrium
(HWE) in control subjects without COVID-19 or mild-to-moderate
COVID-19 cases.
A total of two authors (KY and JW) independently
assessed the methodological quality of each included case-control
study using the Newcastle Ottawa Scale (NOS) for selection,
comparability and exposure. The NOS scores ranged from 0 to 9 and a
score ≥7 indicated high quality (15). Disagreements were resolved as
aforementioned.
Statistical analysis
ORs with their corresponding 95% confidence
intervals (CIs) were calculated to assess the strength of the
association between the IFITM3 rs12252 polymorphism and
COVID-19 severity. When mortality data was available, the
association between IFITM3 rs12252 polymorphism and COVID-19
mortality was also assessed. The associations were examined under
three genetic models: Additive (CC vs. TT), dominant (CC/CT vs. TT)
and recessive (CC vs. CT/TT). HWE was evaluated using χ2
test and P<0.05 was considered to indicate a statistically
significant difference. Sensitivity analysis was performed using
the one-study remove approach to assess the impact of each study on
the combined effect.
Between-study heterogeneity was evaluated using
Cochran's Q test and I2 statistic. If the P-value
for the Q test was <0.10 or if the I2
statistic was ≥50%, significant heterogeneity was considered and
the random-effect model was used. Otherwise, the fixed-effect model
was used. Egger's test and Begg's funnel plot were not used to
provide a diagnosis of the potential publication bias as there were
only five studies included in the meta-analysis (16). All statistical analyses were
performed with Stata 15.0 (StataCorp LP). A two-sided P<0.05 was
considered to indicate a statistically significant difference. The
performance and reporting of the present meta-analysis complied
with the Meta-analyses Of Observational Studies in Epidemiology
statement (Appendix S2) (17).
Results
Characteristics of included
studies
A total of five independent studies were identified
regarding IFITM3 rs12252 polymorphism and COVID-19 severity
(Fig. 1). These five studies were
published from 2020 to 2021, with four in the English language and
one in Chinese. A total of two studies were conducted in the
Chinese population (10,11), one in the Saudi population
(12), one in the Spanish
population (13) and one in the
German population (14). In total,
2,110 patients with COVID-19 were included. Of these, 1,443 were
mild-to-moderate cases and 667 were severe cases, including 121
deaths. The definition of severity was not identical across studies
(Appendix S3). All studies but
one showed that patients with severe cases had older ages compared
with mild-to-moderate cases (14).
A total of three studies genotyped healthy controls or controls
without COVID-19 (11,13,14).
The IFITM3 rs12252 C allele frequency was higher in the
Chinese population compared with Caucasian population (49.2-55.4%
vs. 2.8-9.6%, respectively; Table
I). All studies were consistent with HWE. Detailed
characteristics of the five studies are described in Table I. NOS scores of all included
studies ranged from 8 to 9, which indicated good quality (Appendix S4).
 | Table ICharacteristics of studies included
in the meta-analysis. |
Table I
Characteristics of studies included
in the meta-analysis.
| |
Mild-to-moderate | Severe | |
|---|
| Study | Population | Genotype
method | Male, % | Age, years | CC/CT/TT | Male (%) | Age, years | CC/CT/TT | Died CC/CT/TT | Control
CC/CT/TT | Factors
adjusted | C allele, % | HWE P-value |
|---|
| Zhang et al,
2020(10) | Chinese | Sequencing | 42.9 | 43.5
(34.0-56.5) | 16/30/10 | 37.5% | 67.5
(57.8-74.3) | 12/7/5 | 2/1/0 | NA | Age | 55.4 | >0.5 |
| Pan et al,
2021(11) | Chinese | Sequencing | 54.2 | 39.5 | 126/203/105 | 50.0% | 51.8 | 7/2/3 | 3/0/0 | 15/34/16 | Age | 49.2 | >0.7 |
| Alghamdi et
al, 2021a
(12) | Saudi | Taqman | 64.0 | 30.0 | 3/82/372 | 56.0% | 59.0 | 1/73/330 | 0/21/56 | NA | Age, sex | 9.6 | >0.5 |
| Cuesta-Llavona
et al, 2021(13) | Spanish | Taqman | 59.0 | 64.0±16.0 | 2/30/300 | 74.0% | 67.0±16.0 | 2/17/133 | 1/5/32 | 0/10/172 | Age, sex | 2.8 | >0.7 |
| Schönfelder et
al, 2021(14) | German | Sequencing | 52.4 | 57.0
(18.0-94.0) | 2/15/147 | 73.3% | 64.0
(26.0-99.0) | 0/7/68 | NA | 0/19/234 | Age, sex | 3.8 | >0.5 |
Association between IFITM3 rs12252
polymorphism and COVID-19 severity
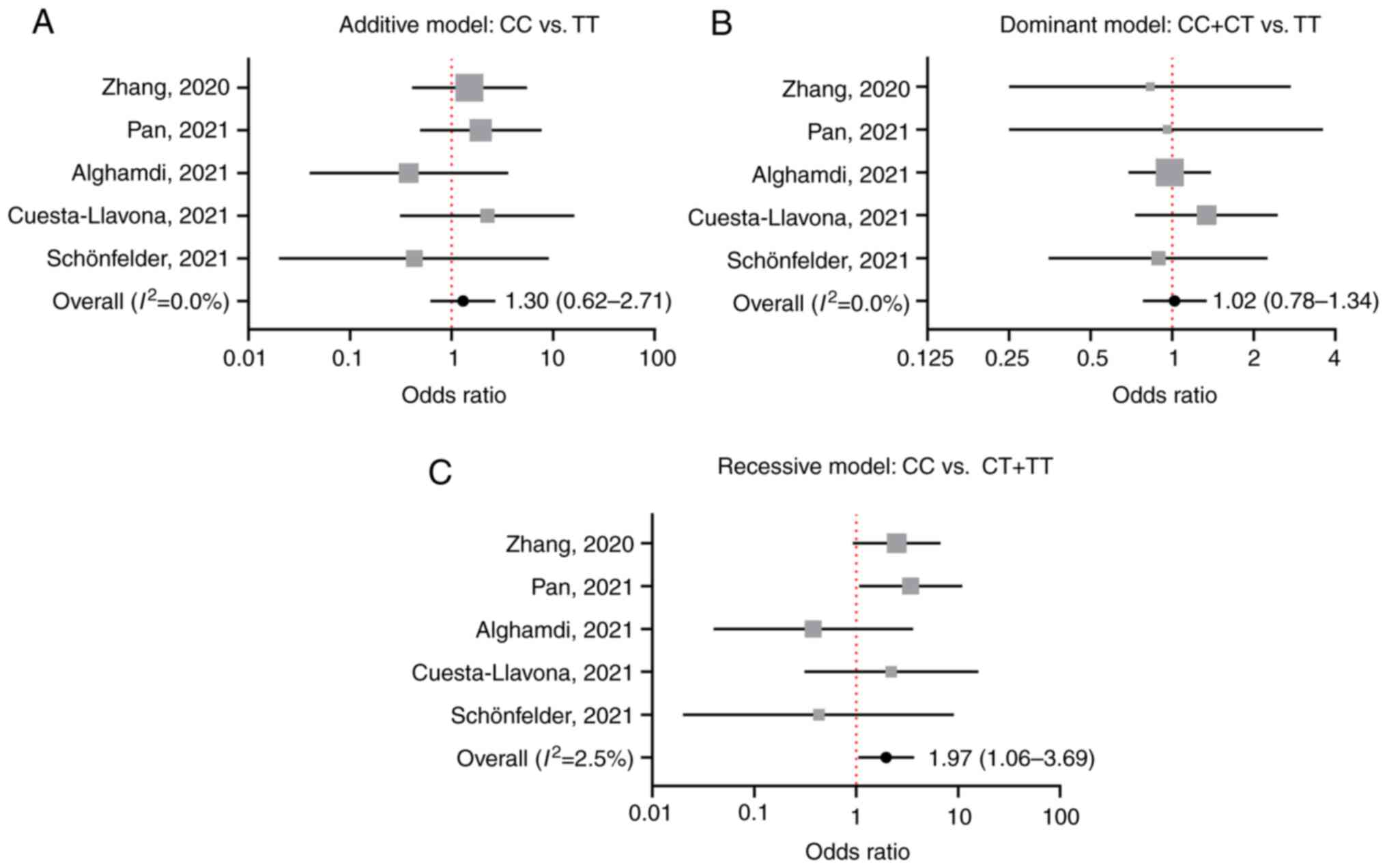
Under each genetic model, no significant
heterogeneity was detected by the Q test and I2
statistic (Fig. 2A-C). Thus, in
the meta-analysis, the fixed-effect model was used for each genetic
model. Only under the recessive model (CC vs. CT/TT) was a
significant association between the IFITM3 rs12252
polymorphism and COVID-19 severity observed. Overall, the OR of
developing severe COVID-19 was 1.97 (95% CI, 1.06-3.69) in patients
carrying the CC genotype compared with those with other genotypes
(CT/TT; Fig. 2C). The association
was not observed under the additive (CC vs. TT) or the dominant
model (CC/CT vs. TT; Fig. 2A and
B).
The Chinese population had a significantly higher
frequency of IFITM3 rs12252 C allele compared with the
Caucasian population. Stratified analysis by ethnicity showed that
only in the Chinese population was the CC genotype associated with
severe COVID-19 (CC vs. CT/TT; OR=2.84; 95% CI, 1.34-6.04). This
association was not observed in the Caucasian population (Table II).
 | Table IIMeta-analysis of studies on
associations of the IFITM3 rs12252 polymorphism with
COVID-19 severity and mortality. |
Table II
Meta-analysis of studies on
associations of the IFITM3 rs12252 polymorphism with
COVID-19 severity and mortality.
| | | | Additive model (CC
vs. TT) | Dominant model
(CC/CT vs. TT) | Recessive model (CC
vs. CT/TT) |
|---|
| Overall/stratified
analysis | No. of studies | No. of
patients | OR (95% CI) | Z-value | P-value | OR (95% CI) | Z-value | P-value | OR (95% CI) | Z-value | P-value |
|---|
| Severity (severe
vs. mild/moderate) | | | | | | | | | | | |
|
Overall | 5 | 2,110 | 1.30
(0.62-2.71) | 0.70 | 0.487 | 1.02
(0.78-1.34) | 0.17 | 0.862 | 1.97
(1.06-3.69) | 2.13 | 0.033 |
|
Chinese
population | 2 | 526 | 1.70
(0.66-4.39) | 1.10 | 0.270 | 0.88
(0.36-2.14) | 0.27 | 0.784 | 2.84
(1.34-6.04) | 2.72 | 0.007 |
|
Caucasian
population | 3 | 1,584 | 0.80
(0.23-2.83) | 0.35 | 0.730 | 1.04
(0.78-1.38) | 0.27 | 0.788 | 0.79
(0.23-2.80) | 0.36 | 0.720 |
| Mortality (died vs.
survived) | | | | | | | | | | | |
|
Overall | 4 | 1,871 | 3.33
(0.85-12.96) | 1.73 | 0.083 | 1.75
(1.11-2.75) | 2.43 | 0.015 | 4.61
(1.44-14.75) | 2.58 | 0.010 |
|
Chinese
population | 2 | 526 | 4.31
(0.51-36.34) | 1.34 | 0.180 | 2.00
(0.24-16.55) | 0.64 | 0.521 | 7.91
(1.29-48.44) | 2.24 | 0.025 |
|
Caucasian
population | 2 | 1,345 | 2.37
(0.41-13.87) | 0.96 | 0.337 | 1.73
(1.09-2.75) | 2.33 | 0.020 | 2.16
(0.37-12.55) | 0.86 | 0.391 |
Association between IFITM3 rs12252
polymorphism and COVID-19 mortality
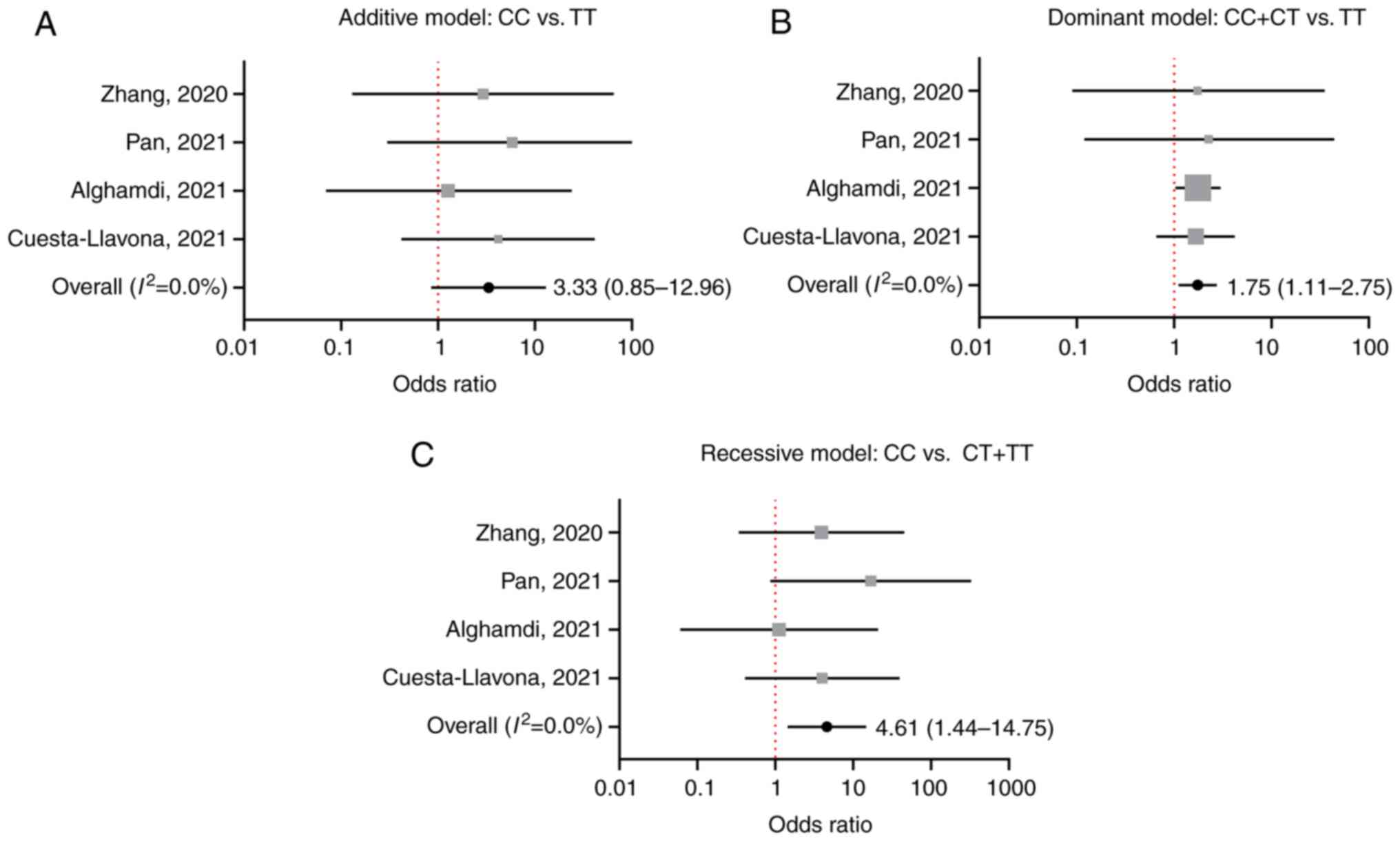
A total of four studies included IFITM3
rs12252 polymorphism data for patients who died with COVID-19. In
total, 121 patients who died and 1,989 who survived were included
to assess its association with COVID-19 mortality. Under each
genetic model, no significant heterogeneity was detected and the
fixed model was used (Fig. 3A-C).
Overall, the CC genotype of IFITM3 rs12252 was strongly
associated with increased COVID-19 mortality risk compared with the
CT/TT genotype [recessive model (CC vs. CT/TT); OR=4.61; 95% CI,
1.44-14.75; Fig. 3C]. Compared
with the TT genotype, the CC/CT genotype was associated with
increased COVID-19 mortality risk [dominant model (CC/CT vs. TT);
OR=1.75; 95% CI, 1.11-2.75; Fig.
3B]. This trend was observed when comparing the CC and TT
genotype but it was not statistically significant [the additive
model (CC vs. TT); OR=3.33; 95% CI, 0.85-12.96; Fig. 3A].
Stratified analysis by ethnicity showed that in the
Chinese population the CC genotype of IFITM3 rs12252 was
strongly associated with increased COVID-19 mortality risk compared
with the CT/TT genotype (CC vs. CT/TT; OR=7.91; 95% CI,
1.29-48.44). In the Caucasian population, the CC/CT genotype was
associated with increased COVID-19 mortality risk compared with the
TT genotype (CC/CT vs. TT; OR=1.73; 95% CI, 1.09-2.75; Table II). Using other genetic models,
the association between IFITM3 rs12252 polymorphism and
COVID-19 mortality was not observed in either the Chinese or
Caucasian population (Table
II).
Sensitivity analysis
Sensitivity analysis was performed to assess the
impact of each study on the pooled results by omitting individual
studies in turn (Table III). The
pooled OR of developing severe COVID-19 was sensitive to the two
Chinese studies (10,11) and for COVID-19 mortality was
sensitive to Pan et al (11) and Alghamdi et al (12).
 | Table IIISensitivity analysis using the
one-study remove approach. |
Table III
Sensitivity analysis using the
one-study remove approach.
| | Additive model (CC
vs. TT) | Dominant model
(CC/CT vs. TT) | Recessive model (CC
vs. CT/TT) |
|---|
| Study omitted | OR (95% CI) | Z-value | P-value | OR (95% CI) | Z-value | P-value | OR (95% CI) | Z-value | P-value |
|---|
| Severity | | | | | | | | | |
|
Overall (5
studies) | 1.30
(0.62-2.71) | 0.70 | 0.487 | 1.02
(0.78-1.34) | 0.17 | 0.862 | 1.97
(1.06-3.69) | 2.13 | 0.033 |
|
Zhang et
al, 2020(10) | 1.21
(0.50-2.96) | 0.42 | 0.673 | 1.04
(0.78-1.37) | 0.25 | 0.803 | 1.67
(0.74-3.79) | 1.24 | 0.217 |
|
Pan et
al, 2021(11) | 1.08
(0.44-2.63) | 0.17 | 0.865 | 1.03
(0.78-1.36) | 0.19 | 0.849 | 1.58
(0.75-3.35) | 1.19 | 0.233 |
|
Alghamdi
et al, 2021(12) | 1.57
(0.70-3.49) | 1.10 | 0.272 | 1.10
(0.71-1.71) | 0.42 | 0.674 | 2.40
(1.23-4.69) | 2.57 | 0.010 |
|
Cuesta-Llavona
et al, 2021(13) | 1.19
(0.54-2.64) | 0.44 | 0.660 | 0.96
(0.71-1.30) | 0.27 | 0.788 | 1.95
(1.01-3.78) | 1.98 | 0.048 |
|
Schönfelder
et al, 2021(14) | 1.42
(0.66-3.08) | 0.90 | 0.371 | 1.04
(0.78-1.38) | 0.26 | 0.795 | 2.18
(1.14-4.19) | 2.34 | 0.019 |
| Mortality | | | | | | | | | |
|
Overall (4
studies) | 3.33
(0.85-12.96) | 1.73 | 0.083 | 1.75
(1.11-2.75) | 2.43 | 0.015 | 4.61
(1.44-14.75) | 2.58 | 0.010 |
|
Zhang et
al, 2020(10) | 3.47
(0.79-15.28) | 1.64 | 0.101 | 1.75
(1.11-2.77) | 2.40 | 0.016 | 4.90
(1.34-17.96) | 2.40 | 0.017 |
|
Pan et
al, 2021(11) | 2.56
(0.54-12.21) | 1.18 | 0.238 | 1.73
(1.10-2.74) | 2.36 | 0.018 | 2.76
(0.68-11.11) | 1.43 | 0.154 |
|
Alghamdi
et al, 2021(12) | 4.27
(0.81-22.55) | 1.71 | 0.087 | 1.74
(0.74-4.06) | 1.28 | 0.202 | 6.63
(1.61-27.38) | 2.61 | 0.009 |
|
Cuesta-Llavona
et al, 2021(13) | 3.12
(0.63-15.48) | 1.39 | 0.163 | 1.77
(1.06-2.98) | 2.17 | 0.030 | 4.77
(1.25-18.21) | 2.29 | 0.022 |
Discussion
Recently, the association between the IFITM3
rs12252 polymorphism and COVID-19 severity has been investigated in
several studies but the results were inconsistent (10-14).
To the best of our knowledge, the present study is the first
meta-analysis conducted on the aforementioned association. The
current meta-analysis, which pooled five studies with 2,110
patients, showed that the CC genotype of IFITM3 rs12252 was
associated with increased risk of severe COVID-19 and mortality.
Subgroup analyses revealed that this association was strong in the
Chinese population. Larger-scale studies are required to determine
whether genotyping for rs12252 SNP of IFITM3 in Chinese and
other Asian patients infected with SARS-Cov-2 predicts those who
might progress to severe disease. Useful tools combining this SNP
with other risk factors may be developed to improve prognosis of
the patients by early targeted intervention.
Notably, the CC genotype of IFITM3 rs12252 is
rare in Europeans (0.3%) and common in Asians (25-44%) (8,18).
Thus, it is possible that the power of the present meta-analysis
may have been insufficient to detect an effect of the CC genotype
in the Caucasian population. This was indicated by a lower but
statistically significant OR regarding COVID-19 mortality when
patients with CC/CT and TT genotypes were compared in the Caucasian
population (OR=1.73; 95% CI, 1.09-2.75). Similar results have been
reported for Caucasian patients with influenza, in which studies
found no or weak association between IFITM3 rs12252
polymorphism and disease severity (19,20).
IFITM3 is a virus restriction factor mediating
cellular resistance to multiple classes of enveloped viral
pathogens that enter cells via the acidic endosome (7). IFITM3 inhibits human coronaviruses,
including SARS-CoV-1 and Middle East respiratory syndrome
coronavirus, as well as SARS-CoV-2 (21,22).
Previous studies showed an increased homozygosity of the minor C
allele of SNP rs12252 in IFITM3 in patients with severe
viral infection, such as influenza (18), cytomegalovirus (23), enterovirus (24), Hantaan virus (25) and HIV (26). SNP rs12252-C allele encodes a
splice acceptor site of the human IFITM3 gene. Everitt et
al (8) found that the
rs12252-C allele may be associated with a truncated protein with an
N-terminal 21 amino acid deletion, which may lead to decreased
restriction of virus replication in vitro. However, other
evidence does not support the hypothesis that CC genotype carriers
express truncated IFITM3 (27); full-length transcript/protein is
dominant in all rs12252 genotypes (27). It is hypothesized that rs12252 may
be in linkage disequilibrium with a causative SNP near the
IFITM3 locus.
The primary limitation of the present meta-analysis
is that it only included five studies with a relatively small size.
Thus, the findings in the current meta-analysis are not robust.
Sensitivity analysis in the present study also indicated this.
Larger cohort studies are needed to confirm the genetic association
with COVID-19 severity. As with most meta-analyses, another
limitation of the present study is that it is based on unadjusted
estimates. Individual information was not available to adjust for
confounding factors, such as age, sex and underlying diseases.
Finally, all patients included in the present meta-analysis were
infected with early strains of SARS-CoV-2 and vaccination coverage
was relatively low at that time. Whether and to what extent
IFITM3 rs12252 polymorphism has an effect on COVID-19 needs
to be investigated as the virus mutates and herd immunity develops
(28-30).
In conclusion, the present meta-analysis suggested
that IFTM3-rs12252 CC genotype was significantly associated
with increased risks of severe COVID-19 and mortality in the
Chinese population and that the IFTM3-rs12252 C allele may
be associated with increased risk of COVID-19 mortality in the
Caucasian population. Large-scale studies are needed to confirm the
genetic association with COVID-19 severity in different global
populations.
Supplementary Material
Search strategies
Checklist for Meta-analyses of
Observational Studies.
Definition of severe COVID-19 of
included studies
Quality of included studies assessed
using the Newcastle Ottawa Scale (8).
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
KY, JW, HL and WW conceived and designed the study,
collected, analyzed and interpretated data and wrote and revised
the manuscript. KY and WW confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization: WHO Coronavirus
(COVID-19) Dashboard. https://covid19.who.int/. Accessed April 9, 2022.
|
|
2
|
The National Health Commission of China:
The diagnosis and treatment of COVID-19. http://www.nhc.gov.cn/yzygj/s7653p/202203/b74ade1ba4494583805a3d2e40093d88.shtml.
Accessed 1 April 2022, 2022.
|
|
3
|
Ferreira de Araújo JL, Menezes D, Saraiva
Duarte JM, de Lima Ferreira L, Santana de Aguiar R and Pedra de
Souza R: Systematic review of host genetic association with
Covid-19 prognosis and susceptibility: What have we learned in
2020? Rev Med Virol. 32(e2283)2022.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Suh S, Lee S, Gym H, Yoon S, Park S, Cha
J, Kwon DH, Yang Y and Jee SH: A systematic review on papers that
study on single nucleotide polymorphism that affects coronavirus
2019 severity. BMC Infect Dis. 22(47)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Brass AL, Huang IC, Benita Y, John SP,
Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig
E, et al: The IFITM proteins mediate cellular resistance to
influenza A H1N1 virus, West Nile virus, and dengue virus. Cell.
139:1243–1254. 2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Perreira JM, Chin CR, Feeley EM and Brass
AL: IFITMs restrict the replication of multiple pathogenic viruses.
J Mol Biol. 425:4937–4955. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ren L, Du S, Xu W, Li T, Wu S, Jin N and
Li C: Current progress on host antiviral factor IFITMs. Front
Immunol. 11(543444)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Everitt AR, Clare S, Pertel T, John SP,
Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, et al:
IFITM3 restricts the morbidity and mortality associated with
influenza. Nature. 484:519–523. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Prabhu SS, Chakraborty TT, Kumar N and
Banerjee I: Association between IFITM3 rs12252 polymorphism and
influenza susceptibility and severity: A meta-analysis. Gene.
674:70–79. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang Y, Qin L, Zhao Y, Zhang P, Xu B, Li
K, Liang L, Zhang C, Dai Y, Feng Y, et al: Interferon-induced
transmembrane protein 3 genetic variant rs12252-C associated with
disease severity in coronavirus disease 2019. J Infect Dis.
222:34–37. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Pan Y, Li F, Wang X, Liang Z, Cui S, Peng
X, Lu G, Zhao J, Liu Y, Wang Q and Zhang D: Association between
rs12252 polymorphism in IFITM3 gene and COVID-19. Int J Virology.
28:192–195. 2021.
|
|
12
|
Alghamdi J, Alaamery M, Barhoumi T, Rashid
M, Alajmi H, Aljasser N, Alhendi Y, Alkhalaf H, Alqahtani H,
Algablan O, et al: Interferon-induced transmembrane protein-3
genetic variant rs12252 is associated with COVID-19 mortality.
Genomics. 113:1733–1741. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cuesta-Llavona E, Albaiceta GM,
García-Clemente M, Duarte-Herrera ID, Amado-Rodríguez L,
Hermida-Valverde T, Enríquez-Rodriguez AI, Hernández-González C,
Melón S, Alvarez-Argüelles ME, et al: Association between the
interferon-induced transmembrane protein 3 gene (IFITM3) rs34481144
/ rs12252 haplotypes and COVID-19. Curr Res Virol Sci.
2(100016)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Schönfelder K, Breuckmann K, Elsner C,
Dittmer U, Fistera D, Herbstreit F, Risse J, Schmidt K, Sutharsan
S, Taube C, et al: The influence of IFITM3 polymorphisms on
susceptibility to SARS-CoV-2 infection and severity of COVID-19.
Cytokine. 142(155492)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wells GA, Shea B, O'Connell D, Peterson J,
Welch V, Losos M and Tugwell P: The Newcastle-Ottawa scale (NOS)
for assessing the quality of nonrandomised studies in
meta-analyses. Ottawa Hospital Research Institute, Ottawa, ON,
2000. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
|
|
16
|
Harbord RM, Harris RJ and Sterne JAC:
Updated tests for small-study effects in meta-analyses. Stata J.
9:197–210. 2009.
|
|
17
|
Stroup DF, Berlin JA, Morton SC, Olkin I,
Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA and Thacker
SB: Meta-analysis of observational studies in epidemiology: A
proposal for reporting. Meta-analysis Of observational studies in
epidemiology (MOOSE) group. JAMA. 283:2008–2012. 2000.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang YH, Zhao Y, Li N, Peng YC,
Giannoulatou E, Jin RH, Yan HP, Wu H, Liu JH, Liu N, et al:
Interferon-induced transmembrane protein-3 genetic variant
rs12252-C is associated with severe influenza in Chinese
individuals. Nat Commun. 4(1418)2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Randolph AG, Yip WK, Allen EK, Rosenberger
CM, Agan AA, Ash SA, Zhang Y, Bhangale TR, Finkelstein D,
Cvijanovich NZ, et al: Evaluation of IFITM3 rs12252 association
with severe pediatric influenza infection. J Infect Dis. 216:14–21.
2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
López-Rodríguez M, Herrera-Ramos E,
Solé-Violán J, Ruíz-Hernández JJ, Borderías L, Horcajada JP,
Lerma-Chippirraz E, Rajas O, Briones M, Pérez-González MC, et al:
IFITM3 and severe influenza virus infection. No evidence of genetic
association. Eur J Clin Microbiol Infect Dis. 35:1811–1817.
2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Prelli Bozzo C, Nchioua R, Volcic M,
Koepke L, Krüger J, Schütz D, Heller S, Stürzel CM, Kmiec D,
Conzelmann C, et al: IFITM proteins promote SARS-CoV-2 infection
and are targets for virus inhibition in vitro. Nat Commun.
12(4584)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shi G, Kenney AD, Kudryashova E, Zani A,
Zhang L, Lai KK, Hall-Stoodley L, Robinson RT, Kudryashov DS,
Compton AA and Yount JS: Opposing activities of IFITM proteins in
SARS-CoV-2 infection. EMBO J. 40(e106501)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang YS, Luo QL, Guan YG, Fan DY, Luan GM
and Jing A: HCMV infection and IFITM3 rs12252 are associated with
Rasmussen's encephalitis disease progression. Ann Clin Transl
Neurol. 8:558–570. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li M, Li YP, Deng HL, Wang MQ, Chen Y,
Zhang YF, Wang J and Dang SS: DNA methylation and SNP in IFITM3 are
correlated with hand, foot and mouth disease caused by enterovirus
71. Int J Infect Dis. 105:199–208. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Xu-Yang Z, Pei-Yu B, Chuan-Tao Y, Wei Y,
Hong-Wei M, Kang T, Chun-Mei Z, Ying-Feng L, Xin W, Ping-Zhong W,
et al: Interferon-induced transmembrane protein 3 inhibits Hantaan
virus infection, and its single nucleotide polymorphism rs12252
influences the severity of hemorrhagic fever with renal syndrome.
Front Immunol. 7(535)2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang Y, Makvandi-Nejad S, Qin L, Zhao Y,
Zhang T, Wang L, Repapi E, Taylor S, McMichael A, Li N, et al:
Interferon-induced transmembrane protein-3 rs12252-C is associated
with rapid progression of acute HIV-1 infection in Chinese MSM
cohort. AIDS. 29:889–894. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Makvandi-Nejad S, Laurenson-Schafer H,
Wang L, Wellington D, Zhao Y, Jin B, Qin L, Kite K, Moghadam HK,
Song C, et al: Lack of truncated IFITM3 transcripts in cells
homozygous for the rs12252-C variant that is associated with severe
influenza infection. J Infect Dis. 217:257–262. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Neagu M, Calina D, Docea AO, Constantin C,
Filippini T, Vinceti M, Drakoulis N, Poulas K, Nikolouzakis TK,
Spandidos DA and Tsatsakis A: Back to basics in COVID-19: Antigens
and antibodies-completing the puzzle. J Cell Mol Med. 25:4523–4533.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Petrakis D, Nikolouzakis TK, Karzi V,
Vardavas AI, Vardavas CI and Tsatsakis A: The growing anthropogenic
immune deficit and the COVID-19 pandemic. Public Health Toxicol.
1:1–5. 2021.
|
|
30
|
Tsatsakis A, Vakonaki E, Tzatzarakis M,
Flamourakis M, Nikolouzakis TK, Poulas K, Papazoglou G, Hatzidaki
E, Papanikolaou NC, Drakoulis N, et al: Immune response (IgG)
following full inoculation with BNT162b2 COVID-19 mRNA among
healthcare professionals. Int J Mol Med. 48(200)2021.PubMed/NCBI View Article : Google Scholar
|