Introduction
Esophageal cancer (ESCA) is one of the most
prevalent malignant cancers worldwide and ranks as the sixth
commonest cause of death (1).
Esophageal squamous cell carcinomas (ESCC) and adenocarcinoma are
two major subtypes of ESCA with broad pathological heterogeneity
and together comprise the majority of diagnosed ESCA cases
(2). ESCC is the primary
histologic type and accounts for >90% of ESCA (3). Currently, the primary clinical
therapies have focused on the improvement of patient outcomes,
including chemotherapy and surgical resection (4). Extensive tumor cell metastasis
remains the primary cause of high mortality and poor prognosis of
ESCC (5). Previous reports
demonstrate that cancer immunotherapies have been considered a
central tool against cancer in the past few decades (6,7).
Therefore, it is urgent to elucidate the more specific detection
indicators and the underlying pathological mechanism of ESCC.
Split hand and foot malformation 1 (SHFM1) is a key
transcription factor that regulates various genes important for
embryonic morphogenesis and tumor progression (8). Notably, SHFM1 has been considered an
oncogene that is highly expressed in a number of types of human
cancer, such as lung cancer, oral squamous cell carcinoma,
osteosarcoma and ovarian cancer (9-12).
The tumor promotion effect of SHFM1 has attracted great attention.
Previous reports indicate that SHFM1 promoted osteosarcoma
progression and ablation of SHFM1 inhibited cell proliferation and
promoted cell apoptosis both in vitro and in vivo
(10). In addition, SHFM1 exerts
tumor promotion effects on ovarian cancer progression and knockdown
of SHFM1 causes an inhibition of cell growth and cell cycle
progression (12). These emerging
findings suggest that SHFM1 expression is closely associated with
tumor progression and considered an oncogene in cancerous
development. However, the functional roles of SHFM1 in ESCC
progression have yet to be determined. In addition, emerging
evidence indicates that SHFM1 is associated with the activation of
multiple signaling pathways that participated in tumorigeneses such
as the Akt, Notch and Wnt signaling pathways (11,12).
It is well established that the NF-κB signaling is aberrantly
dysregulated in numerous types of cancer cells and is associated
with tumorigenesis (13). However,
the role of SHFM1 in the regulation of NF-κB signaling in ESCC
progression remains to be elucidated.
The immunotherapy of cancer has been well-documented
and the regulation of immune responses serves an important role in
tumor progression (14). Notably,
natural killer (NK) cell-mediated cellular cytotoxicity is valuable
for cancer immunotherapy (15).
c-Myc is a proto-oncogene in the majority of types of human cancer
(16,17) and promising research indicates that
c-Myc expression regulates cytotoxicity-induced apoptosis, which is
one of the mechanisms of NK cell-mediated immune response in tumors
(18,19). c-Myc acts as a transcriptional
target of SHFM1 and SHFM1 overexpression promotes cell
proliferation by regulating the transcriptional expression of c-Myc
in lung cancer (20). c-Myc could
regulate tumor immune response by mediating an immune checkpoint
programmed death-ligand 1 (PD-L1) in a number of types of tumors
(21-23).
Notably, c-Myc can bind to the promoter of PD-L1, thereby
positively regulating the expression of PD-L1 in ESCC progression
(24). Blockade of PD-L1 by immune
checkpoint inhibitors exhibits promising clinical results for
antitumor activity via enhancing NK cell responses (25). Hence, the present study
hypothesized that SHFM1 also regulated immune response in ESCC
progression.
In the present study SHFM1 was identified as a
potential biomarker in ESCC through bioinformatics analysis. SHFM1
expression was frequently upregulated in ESCC patient tissues and
was significantly associated with clinicopathologic features and
overall survival of patients. In addition, the present study
focused on the effects of SHFM1 on the malignant phenotypes of ESCC
cell lines. Functional studies demonstrated that SHFM1 promoted
cell viability, cell cycle progression and migration in ESCC cells
and accelerated tumor formation in a xenograft mouse model.
Furthermore, the present study specifically assessed whether SHFM1
expression was involved in tumor immunity response in ESCC cells.
These findings confirmed the oncogenic role of SHFM1 in the
progression of ESCC and highlighted its potential role as a target
for ESCC treatment.
Materials and methods
Bioinformatics and database
analysis
A total of two gene expression profiles GSE17351 and
GSE33810 were selected from the Gene Expression Omnibus (GEO;
http://www.ncbi.nlm.nih.gov/geo)
database. The GSE17351 dataset included five ESCC and normal
tissues. The GSE33810 dataset consisted of one normal and two ESCC
samples. The differential expressed genes between ESCC and normal
samples were screened by the GEO2R database (http://www.ncbi.nlm.nih.gov/geo/geo2r/). The P-value
<0.05 and |log2FC| value >2 were set as significance. The
differential genes in ESCC were presented using volcano plots. Venn
diagram analysis was performed to explore the overlapped
upregulated genes according to these two profiles. Gene Ontology
(GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathways assays were performed using DAVID (https://david.ncifcrf.gov/) to annotate overlapped
genes. Protein-protein interaction (PPI) network was constructed to
analyze the interactions among selected proteins. Kaplan-Meier
plots (http://www.kmplot.com/analysis/index.php?p) database
was performed to explore the correlation between gene expression
and overall survival. The Gene Set Cancer Analysis database (GSCA;
http://bioinfo.life.hust.edu.cn/GSCA)
was performed to analyze SHFM1 expression across The Cancer Genome
Atlas (TCGA) cancer types. The expression level of SHFM1 in
clinical cases was validated using the Gene Expression Profiling
Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) database and the UALCAN
database (http://ualcan.path.uab.edu). UALCAN
dataset was also performed to assess the association between SHFM1
expression and clinical parameters of ESCC.
Human tissue samples
A total of 58 patients with ESCC (male, n=41 and
female, n=17; age range, 56-87 years; with a median age of 68
years) were harvested from the Xingtai People's Hospital. All
procedures were approved by the Ethics Committee of Xingtai
People's Hospital (approval no. 2022-021) and conducted according
to the guidelines of the Declaration of Helsinki; all patients
provided written informed consent prior to entering the study. ESCC
and adjacent non-tumor tissues were collected and stored at
-80˚C.
Cell culture and transfection
Human ESCC cell lines TE-1 and KYSE-410 were
obtained from Zhong Qiao Xin Zhou Biotechnology (cat. no. ZQ0235)
and Procell (cat. no. CL-0586), respectively and were cultured at
37˚C with 5% CO2 in RPMI-1640 medium (cat. no. 31800;
Beijing Solarbio Science & Technology Co., Ltd.) supplemented
with 10% fetal bovine serum (FBS). Small interfering RNA (siRNA)
sequences were designed and synthesized by General Biology (Anhui)
Co., Ltd., comprising two siRNA sequences against SHFM1 (siSHFM1-1
and siSHFM1-2) or nonspecific control siRNA (siControl). In brief,
20 µM siRNAs (3.75 µl) and Lipofectamine® 3000 regent
(7.5 µl; cat. no. L3000015; Invitrogen; Thermo Fisher Scientific,
Inc.) were added to Opti-MEM (125 µl; cat. no. 31985070,
Invitrogen; Thermo Fisher Scientific, Inc.) and incubated for 15
min at room temperature. The sequences used in the present study
were as follows: siSHFM1-1, 5'-GAUCAAGAAGAUCAUGAAATT-3'; siSHFM1-2,
5'-AGAUCAAGAAGAUCAUGAATT-3'; and siControl,
5'-UUCUCCGAACGUGUCACGUTT-3'. For SHFM1 overexpression, the coding
sequence of SHFM1 was constructed to the pcDNA3.1 vector (cat. no.
G109090; Youbao Biology). pcDNA3.1 empty vector (cat. no. V79020;
Invitrogen; Thermo Fisher Scientific, Inc.) served as a control.
Briefly, SHFM1 expression plasmid or empty vector (4 µg) was
complexed with 7.5 µl Lipofectamine® 3000 (cat. no.
L3000015; Invitrogen; Thermo Fisher Scientific, Inc.) and then
incubated for 15 min at room temperature for transfection into TE-1
and KYSE-410 cells. All experiments were performed 48 h after the
transfection.
Western blotting analysis
ESCC tissues and cells were lysed using the RIPA
buffer (cat. no. P0013B; Beyotime Institute of Biotechnology) with
phenylmethylsulfonyl fluoride (cat. no. ST506; Beyotime Institute
of Biotechnology) for the isolation of proteins. A BCA protein
assay kit (cat. no. P0009; Beyotime Institute of Biotechnology) was
used for protein concentration quantification. Equal amounts of
proteins (15-30 µg) were loaded into a 10% SDS-PAGE gel (cat. no.
P0015; Beyotime Institute of Biotechnology) and blotted to
polyvinylidene fluoride (PVDF) membranes (cat. no. LC2005; Thermo
Fisher Scientific, Inc.). Following blocking with bovine albumin
(5%; BSA; cat. no. BS043; Biosharp Life Sciences) for 1 h at room
temperature, PVDF membranes were immunoblotted with antibody
against SHFM1 (1:500; cat. no. 10592-1-AP, Wuhan Sanying
Biotechnology), c-Myc (1:500; cat. no. 10828-1-AP; Wuhan Sanying
Biotechnology), PD-L1 (1:1,000; cat. no. 28076-1-AP, Wuhan Sanying
Biotechnology), phosphorylated (p-)P65 (Ser536; 1:1,000; cat. no.
AF2006; Affinity Biosciences), P65 (1:1,000; cat. no. AF5006;
Affinity Biosciences), matrix metalloproteinase 9 (MMP9) (1:500;
cat. no. 10375-2-AP; Wuhan Sanying Biotechnology), or MMP2
(1:1,000; cat. no. 10373-2-AP; Wuhan Sanying Biotechnology),
respectively, overnight at 4˚C, followed by the incubation of
horseradish peroxidase (HRP)-conjugated goat anti-rabbit (1:10,000;
IgG, cat. no. SA00001-2; Wuhan Sanying Biotechnology) at 37˚C for
40 min. The blots were visualized with an ECL detection reagent
(cat. no. E003; Seven Sea biotech, China) and protein expression
was normalized to β-actin. The blots were detected using the
Gel-Pro-Analyzer (cat. no. WD-9413B; Beijing Liuyi Biotechnology
Co., Ltd.).
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was isolated from ESCC cells (6-well
plates at a density of 1x106 cells per well) with
TRIpure solution (cat. no. RP1001; BioTeke Corporation) according
to the manufacturer's instructions. The synthesis and
quantification of cDNA were performed with the Exicycler 96 SYBR
Green PCR system (Bioneer Corporation) according to the
manufacturer's instructions. qPCR was conducted using the SYBR
Green Master Mix (cat. no. SY1020; Beijing Solarbio Science &
Technology Co., Ltd.). The PCR program consisted of: 94˚C for 5
min; 94˚C for 20 sec and 60˚C for 30 sec, for 40 cycles. Gene
expression was conducted by the method of 2-ΔΔCq
(26) and β-actin was used for
normalization. Three biological replicates were analyzed for each
experiment. The primer sequences were: SHFM1 forward,
5'-ACCTCGGCTTCCTATGGC-3' and reverse, 5'-CTGGGTTTACGAACTTTCTTTG-3';
c-Myc forward, 5'-ACACCCTTCTCCCTTCG-3' and reverse,
5'-CCGCTCCACATACAGTCC-3'; PD-L1 forward, 5'-AACTACCTCTGGCACATC-3'
and reverse, 5'-ATCCATCATTCTCCCTTT-3'; β-actin forward,
5'-GGCACCCAGCACAATGAA-3' and reverse, 5'-TAGAAGCATTTGCGGTGG-3'.
Cell Counting Kit-8 (CCK-8) assay
ESCC cells (3x103 cells) were seeded onto
96-well plates and cultured in RPMI-1640 medium. Following
transfection, cell viability was assessed at 0, 24, 48 and 72 h,
respectively. The cells were treated with CCK-8 solution (10 µl;
cat. no. KGA317; Nanjing KeyGen Biotech Co., Ltd.) and were
cultured for another 2 h. Cell proliferation ability was
represented by detecting the absorbance value at 450 nm under a
microplate reader (800Ts; BioTek Instruments, Inc.).
Cell cycle assay
Cell cycle analysis was performed using the Cell
Cycle Analysis kit (cat. no. KGA512; Nanjing KeyGen Biotech Co.,
Ltd.) according to the manufacturer's protocol. Briefly, the
transfected ESCC cells were collected and fixed in 70% cold ethanol
at 4˚C overnight. Subsequently, the fixed cells were washed with 1X
phosphate-buffered saline (PBS; pH 7.4; Sangon Biotech Co., Ltd.),
followed by incubation with the prepared propidium iodide (PI)
staining solution in darkness for 30 min. The DNA content was
detected using a flow cytometer (NovoCyte; Agilent Technologies,
Inc.). Flow Plus software (version 1.5.6; Agilent Technologies,
Inc.) was used to analyze the results. The percentage of cells at
the G1, S and G2 phases were calculated.
Migration and invasion assays
The effect of SHFM1 on the migratory and invasive
capabilities of ESCC cells was performed by Transwell assay. For
invasion assay, the Transwell chambers were pre-coated with 40 µl
of diluted (1:3) Matrigel (cat. no. 3422; Corning, Inc.) at 37˚C
for 2 h. In brief, the transfected TE-1 and KYSE-410 cells
(6x103 cells) were suspended and seeded in 200 µl
serum-free medium in the top chamber. RPMI-1640 medium (800 µl)
containing 10% FBS was added to the bottom chambers. Following
cultivation at 37˚C with 5% CO2 for 24 h, the cells were
washed and fixed with 4% paraformaldehyde for 15 min at 37˚C,
followed by staining with 0.4% crystal violet solution (cat. no.
0528; Amresco, LLC) at 37˚C for 5 min. The migrated and invaded
cells from five random fields were quantified and captured under an
inverted light microscope (IX53; Olympus Corporation).
Immunofluorescence assay
P65 localization was developed by the
immunofluorescence assay. The transfected ESCC cells were fixed
with 4% paraformaldehyde at 37˚C and reacted with 0.1% Triton X-100
(cat. no. ST795; Beyotime Institute of Biotechnology). Following
blocking with 1% BSA for 15 min at 37˚C, cells were incubated with
an antibody against P65 (1:200; cat. no. A11201; ABclonal Biotech
Co., Ltd.) overnight at 4˚C. Cy3-labeled goat anti-rabbit IgG
(1:200; cat. no. A27039; Invitrogen; Thermo Fisher Scientific,
Inc.) were used as secondary antibodies. Cells were counterstained
with DAPI staining solution (cat. no. D106471-5mg, Aladdin, China)
for 5 min at 37˚C and the immunofluorescent images were captured
using a fluorescence microscope (BX53; Olympus Corporation).
Cell cytotoxicity assay
For cell cytotoxicity assay, NK-92 cells were
obtained from Procell Life Science & Technology Co., Ltd. and
grown in a specific medium (cat. no. CM-0530; Procell Life Science
& Technology Co., Ltd.) at 37˚C with 5% CO2. The
transfected TE-1 and KYSE-410 cells were stained with
5-6-carboxyfluorescein diacetate succinimidyl ester (CFSE; cat. no.
S19285; Shanghai Yuanye Biotechnology Co., Ltd.) for 10 min at
37˚C. Following incubation for 24 h, NK-92 cells were co-incubated
with TE-1 or KYSE-410 cells (5:1) for 4 h, respectively. TE-1 and
KYSE-410 cells were also incubated separately to detect basal
levels. The collected cells were then stained with PI (100 µg/ml)
for 5 min at 4˚C. Cells were then examined by flow cytometry
(NovoCyte; Agilent Technologies, Inc.). The specific lysis rate was
analyzed by Flow Plus software (version 1.5.6; Agilent
Technologies, Inc.) according to the following formula: Specific
lysis%=(sample ratio-basal ratio) x100%, where ratio=% CFSE+PI+/%
CFSE+ (27).
ELISA assay
To assess granzyme B and perforin expression levels,
ESCC cells were harvested following transfection for 48 h. NK-92
cells were co-incubated with ESCC cell at 5:1 radio for a further
12 h at 37˚C. The levels of granzyme B (cat. no. EH0157) and
perforin (cat. no. EH1487) from culture media supernatants were
detected using the human ELISA kits (Wuhan Fine Biotech Co., Ltd.)
according to the manufacturer's instructions.
Animal experiments
A total of 24 BALB/c male nude mice (aged 4-5 weeks;
weight 15-16 g) purchased from Changzhou Cavince Laboratory Animal
Co., Ltd. were used for the xenograft growth assay. Mice were
maintained under standard conditions (temperature, 22±1˚C; 45-55%
humidity; 12 h light/dark cycle). The mice were randomly divided
into four groups (6 mice in each group): The shControl group, the
shSHFM1 group, the Vector group and the SHFM1 group; none of nude
mice succumbed during the study. For xenograft models,
KYSE-410/vector cells or SHFM1-overexpression KYSE-410 cells
(1x106 cells) were subcutaneously injected into the
right flank of mice. In addition, a total of 1x106 cells
with SHFM1 knockdown or without gene intervention were injected
into mice. Tumor volumes (LxW2)/2 were examined every 4
days. Loss of weight >20% of the body weight of mice and tumor
position severely impairing usual body function were applied as
humane endpoints. After 28 days, the animals were sacrificed using
30% volume/min CO2. Tumor tissues were dissected and
fixed in 4% paraformaldehyde at room temperature for 24 h for
further analysis. Mortality was confirmed by observation of
cessation of heartbeat, respiratory arrest and dilated pupils. All
animal experiments were approved by the Institutional Animal Care
and Use Committee of the Xingtai People's Hospital (approval no.
2022-026) and the ARRIVE Guidelines 2.0 for Reporting Animal
Research (28) were followed.
Immunohistochemistry (IHC)
For IHC staining analysis, the samples were fixed in
4% paraformaldehyde at room temperature overnight. The sections
were embedded in paraffin and sliced into 5 µm sections. Tumor
sections were deparaffinized with xylene, followed by rehydration
with ethanol. The sections were performed by heat-induced epitope
retrieval with antigen retrieval buffer and endogenous peroxidase
activity was quenched with 3% H2O2 for 15
min. The samples were blocked with 1% BSA at 37˚C for 15 min and
incubated with antibodies against SHFM1 (1:100; cat. no. DF3220;
Affinity Biosciences) or Ki-67 (1:100; cat. no. AF0198; Affinity
Biosciences) overnight at 4˚C. Slides were then cultivated with
goat anti-rabbit HRP-conjugated secondary antibody (1:500; cat. no.
31460; Thermo Fisher Scientific, Inc.) for 1 h at 37˚C. The
sections were counterstained with DAB solution (cat. no. DAB-1031;
Fuzhou Maixin Biotech Co., Ltd.). The images were obtained under a
light microscope (BX53; Olympus Corporation).
Statistical analysis
GraphPad Prism 8.0 software (Dotmatics) was used for
statistical analysis. Statistical tests for data analysis were
calculated with Student's t-test or one-way analysis of variance
(ANOVA) with Tukey's multiple comparisons test. The data were
expressed as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
Identification of differentially
expressed genes in ESCC and analysis
The present study first screened ESCC-related
differential genes in two GSE data sets (GSE17351 and GSE33810).
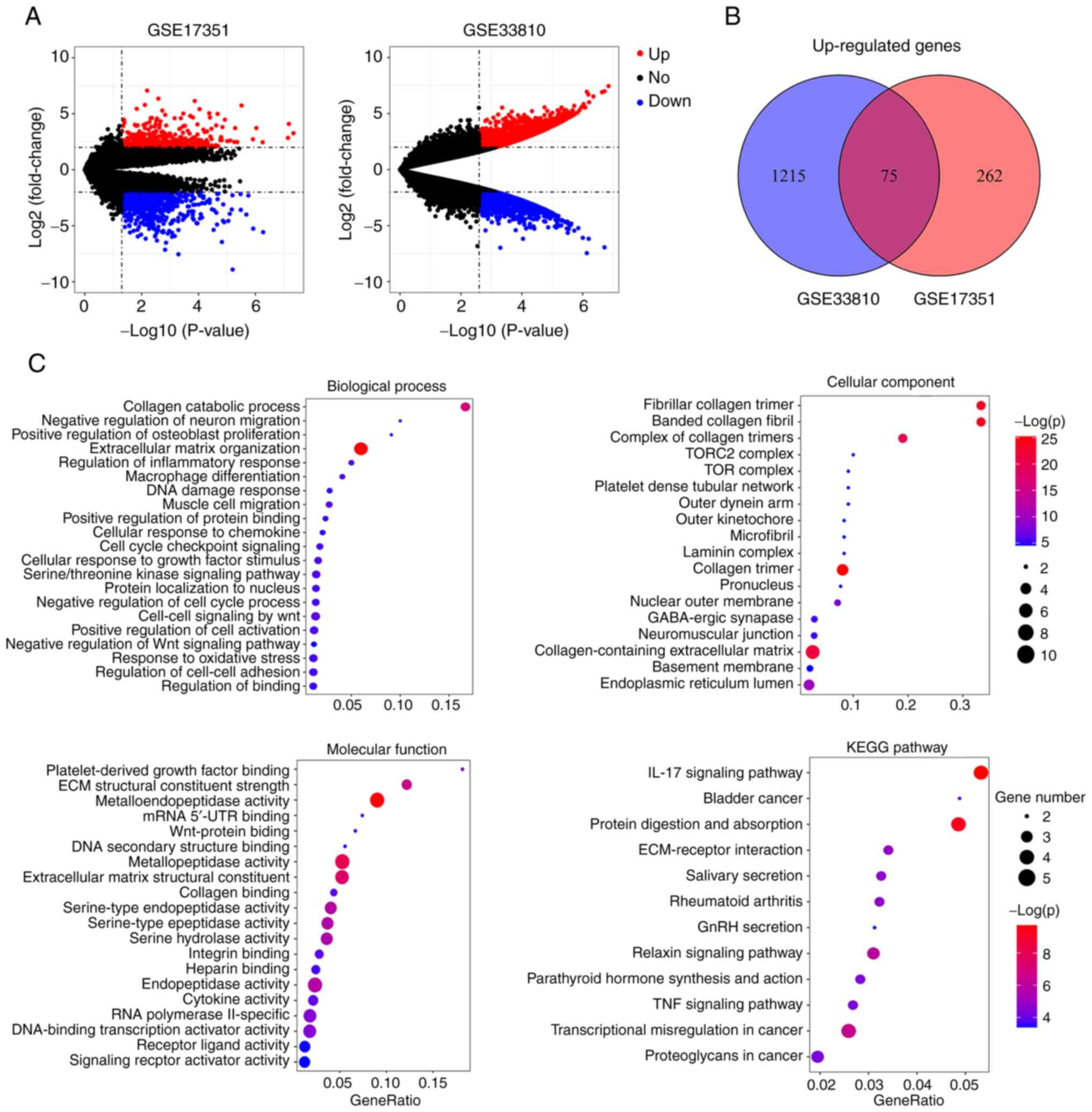
The regulated genes were shown in the volcano plot (Fig. 1A). The Venn diagram showed the
overlap of two profiles and 75 upregulated genes were screened
(Fig. 1B).
Enriched GO and the KEGG pathway analysis was
performed with DAVID. As indicated in Fig. 1C, the GO terms showed that 75
upregulated genes were mainly enriched in the extracellular matrix
organization and collagen catabolic process on biological process
group; collagen-containing extracellular matrix and collagen trimer
on cellular component; metallopeptidase activity and extracellular
matrix structural constituent on molecular function. In addition,
in the KEGG pathway analysis, 75 upregulated genes were mostly
enriched on the IL-17 signaling pathway and transcriptional
misregulation in cancer.
PPI network analysis and key genes
selection
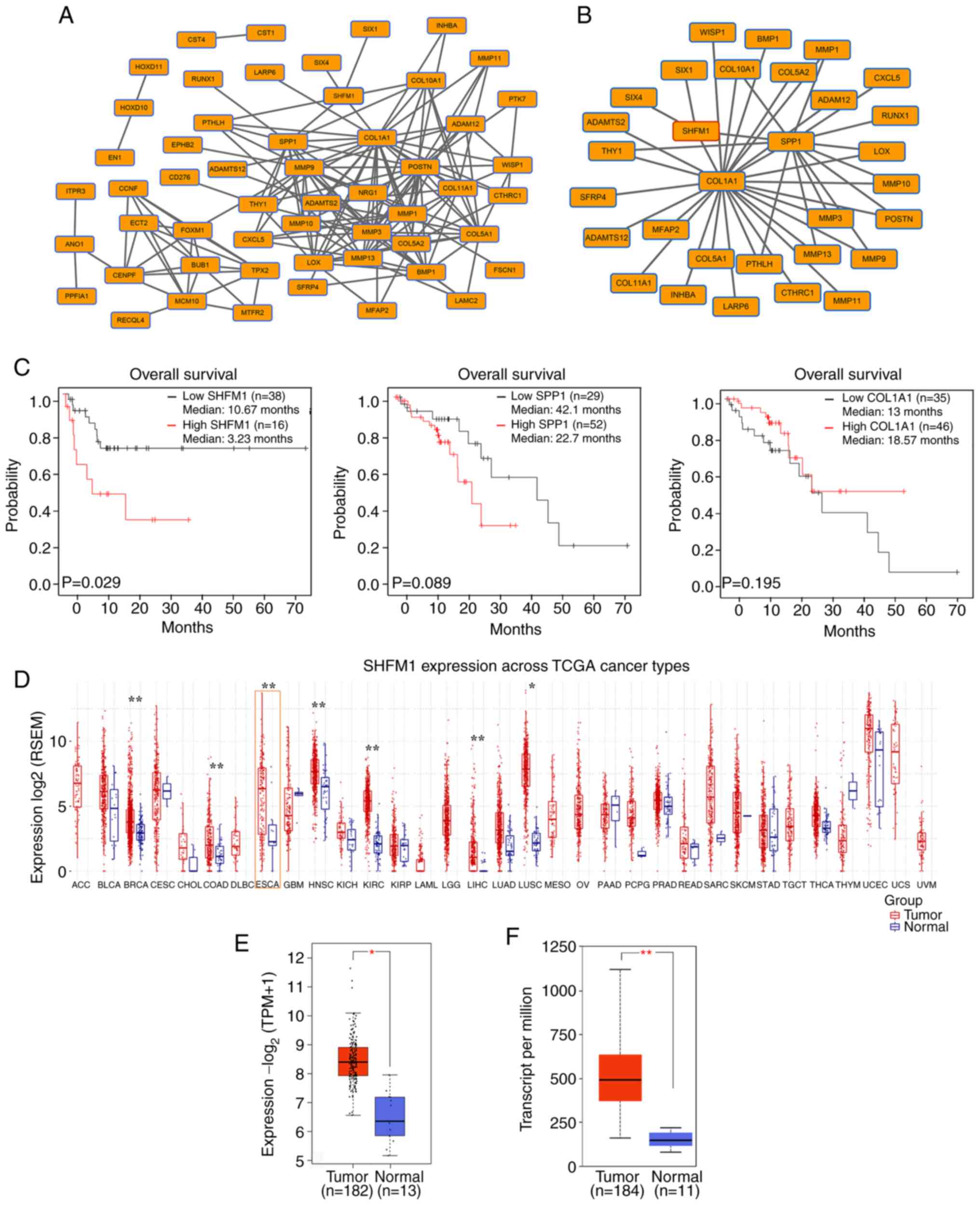
As shown in Fig.
2A, the PPI network revealed a correlation among these
significantly co-expressed genes. A significant module was
identified and the most significant three node genes were collagen
type I alpha 1 (COL1A1), secreted phosphoprotein 1 (SPP1) and SHFM1
(Fig. 2B). Survival analysis was
further performed to evaluate prognostic value of these significant
genes. Survival analysis data from TCGA indicated a negative
association between SHFM1 expression and overall survival rate of
patients (P<0.05; Fig. 2C). In
addition, there was no significant association between COL1A1 or
SPP1 expression and survival rate of patients with ESCC. Among
these key genes, SHFM1 was identified as a candidate biomarker
based on the prognostic value and SHFM1 expression in different
types of cancer was performed across TCGA database. Analysis of
data from the GSCA database revealed that SHFM1 expression was
highly expressed in various types of cancer, including ESCC
(Fig. 2D). Furthermore, the SHFM1
expression pattern focused on ESCC in clinical cases was explored
based on the GEPIA and UALCAN databases (P<0.05; Fig. 2E and F). Consistently, the expression of SHFM1
was markedly higher in ESCC tissues compared with normal esophageal
tissues.
SHFM1 expression and
clinical-pathological parameters of patients with ESCC
Given that SHFM1 was related to the prognosis of
ESCC, the present study further explored the association between
SHFM1 expression and clinical parameters based on TCGA datasets.
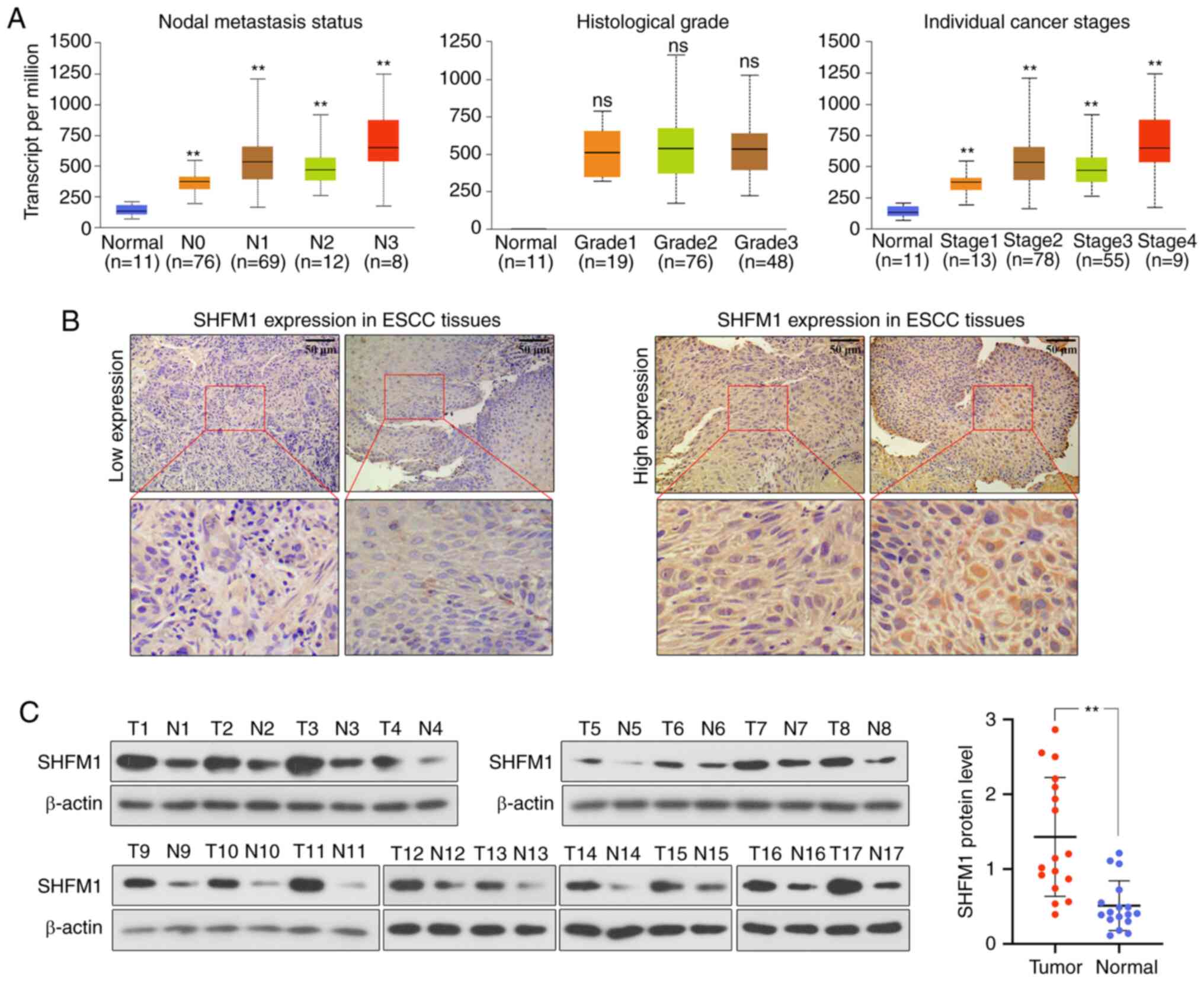
SHFM1 expression in ESCC patients was increased and positively
associated with different clinical features, including nodal
metastasis status and pathologic stages in ESCC and was not
associated with histological grade (Fig. 3A). The present study revealed that
SHFM1 expression in the primary tumors with nodal metastasis
(N1-N3) was markedly increased compared with those without lymph
node metastasis (N0) in ESCC (P<0.01) and SHFM1 expression
increased in stages 1-4 compared with normal samples, suggesting
that SHFM1 expression was significantly related to the prognosis of
patients with ESCC (Fig. 3A). In
addition, SHFM1 expression was gradually upregulated with an
increase in tumor stages and lymphatic invasion in ESCC. To further
confirm the expression of SHFM1 in ESCC progression, 58 ESCC
tissues were collected and the correlation among SHFM1 and
clinicopathological variables was detected. Notably, SHFM high
expression was positively associated with TNM stage (P=0.048) and
lymph node metastasis (P=0.006; Table
I), this was consistent with database results, suggesting that
SHFM might be independent prognostic factor for the patients with
ESCC. IHC staining showed that SHFM1 expression was divided into
the low expression and high expression groups (Fig. 3B). As shown in Fig. 3C, the protein expression of SHFM1
in ESCC tissues was significantly increased in cancerous tissues
compared with adjacent normal tissues.
 | Table ICorrelation between SHFM1 expression
and clinicopathologic parameters in ESCC patients. |
Table I
Correlation between SHFM1 expression
and clinicopathologic parameters in ESCC patients.
| | SHFM1
expression | |
|---|
| Parameters | n | High (n=33) | Low (n=25) | P-value |
|---|
| Age (years) | | | | 0.114 |
|
≥65 | 40 | 20 (50%) | 20 (50%) | |
|
<65 | 18 | 13 (72.2) | 5 (27.8%) | |
| Sex | | | | 0.942 |
|
Male | 41 | 23 (56.1%) | 18 (43.9%) | |
|
Female | 17 | 10 (58.9%) | 7 (41.1%) | |
| TNM stage | | | |
0.048* |
|
T1 | 1 | 0 (0) | 1(1) | |
|
T2 | 21 | 7 (33.3%) | 14 (66.7%) | |
|
T3 | 24 | 17 (70.8%) | 7 (29.2%) | |
|
T4 | 12 | 9 (75%) | 3 (25%) | |
| Histological
grade | | | | 0.359 |
|
Low | 27 | 18 (66.7%) | 9 (33.3%) | |
|
Middle | 22 | 11 (50%) | 11 (50%) | |
|
High | 9 | 4 (44.4%) | 5 (55.6%) | |
| Lymph node
metastasis | | | |
0.006* |
|
Positive | 26 | 20 (76.9%) | 6 (23.1%) | |
|
Negative | 32 | 13 (40.6%) | 19 (59.4%) | |
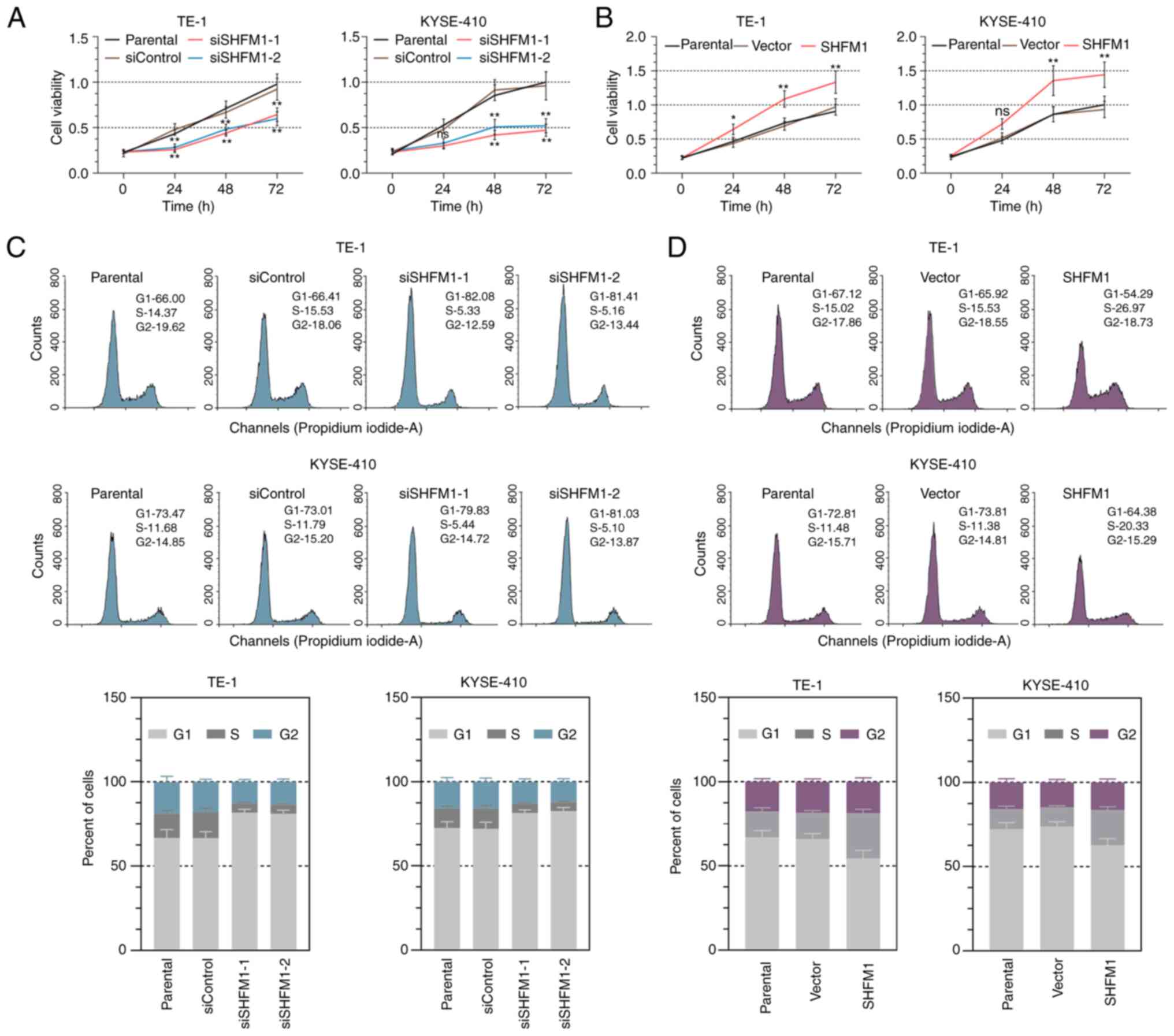
SHFM1 promotes cell growth and cell
cycle progression in ESCC cells
The biological function of SHFM1 in ESCC cell lines
was further explored. SHFM1 was silenced or overexpressed in ESCC
cells by loss-of or gain-of-function approaches. RT-qPCR and
western blotting analysis were performed to confirm the expression
of SHFM1 following transfection in TE-1 and KYSE-410 cells
(Fig. S1A-1D). Subsequently, cell
proliferation ability was monitored in ESCC cells by CCK-8
analysis. The results revealed that knockdown of SHFM1
significantly reduced cell proliferation and the cell viability was
increased following SHFM1 overexpression (Fig. 4A and B). To further confirm these results, flow
cytometry analysis was conducted to evaluate the role of SHFM1 in
cell cycle progression. As shown in Fig. 4C, in the absence of SHFM1, the cell
proportion of the G1 phase was remarkably increased,
while the number of cells was decreased in the S phase, suggesting
SHFM1 deficiency caused cell cycle arrest in the G1
phase. by contrast, SHFM1 overexpression caused a decrease in the
cell counts of the G1 phase and was accompanied by
increased cells in the S phase (Fig.
4D).
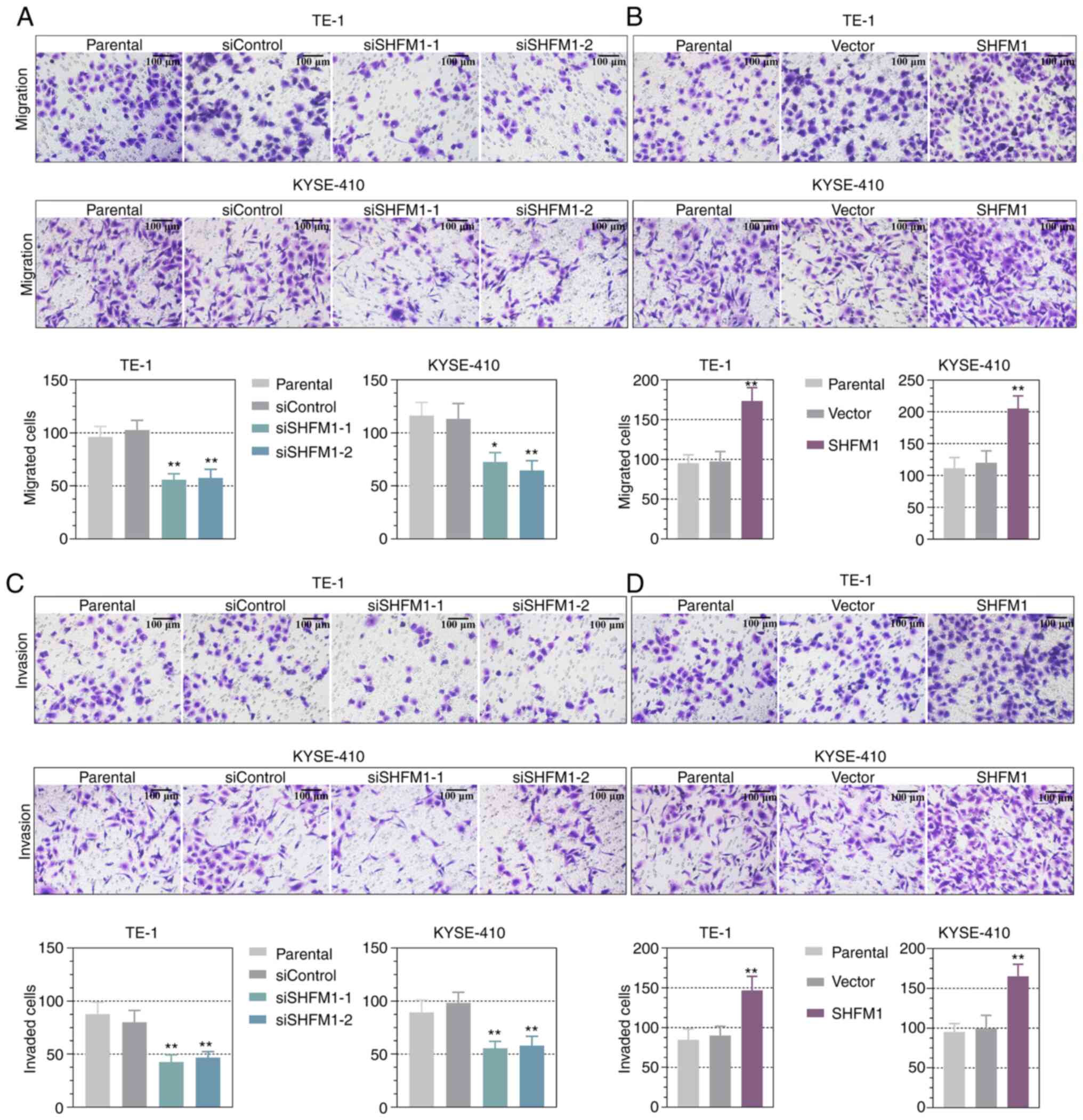
SHFM1 promotes migration and invasion
of ESCC cells
Furthermore, the effect of SHFM1 on cell
aggressiveness in SHFM1-overexpressed or -silenced cells was
determined by Transwell assay. Migration analysis revealed that the
downregulation of SHFM1 markedly inhibited the migration capability
of TE-1 and KYSE-410 cells and overexpression of SHFM1 markedly
increased migration ability and the number of the migrated cells
(Fig. 5A and B). In addition, the Matrigel invasion
assay indicated that SHFM1 depletion caused a significant decrease
in cell invasiveness compared with the corresponding control group
and the invasive ability was increased in the SHFM1-overexpressed
TE-1 and KYSE-410 cells (Fig. 5C
and D).
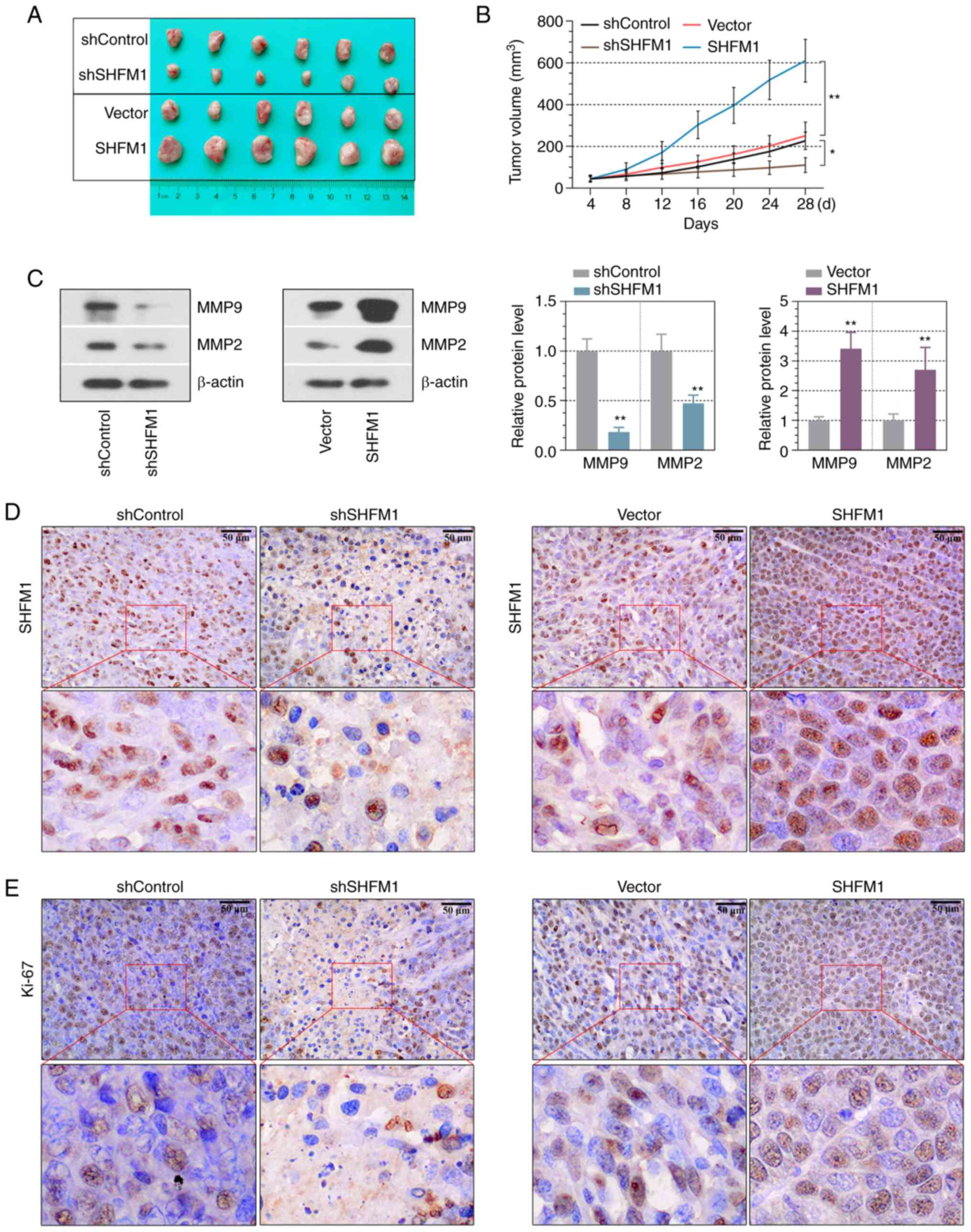
SHFM1 promotes ESCC tumor growth in
vivo
Given that SHFM1 associated with ESCC cell growth
in vitro, a xenograft tumor model was established to explore
whether SHFM1 promoted ESCC progression in vivo. As
indicated in Fig. 6A and B, the tumors formed by SHFM1-silenced
ESCC cells demonstrated a decreased growth rate and reduced tumor
volumes compared with those of the control group, while SHFM1
overexpression clearly accelerated ESCC tumor growth in
vivo. In addition, the contribution of SHFM1 in tumor
progression in vivo was explored. MMP9 and MMP2 expression
in ESCC tissues was significantly upregulated in
SHFM1-overexpression tumors and downregulated following SHFM1
depletion (Fig. 6C). IHC staining
of the xenograft sections demonstrated that overexpression of SHFM1
resulted in increased expression of SHFM1 and Ki-67 levels and
SHFM1 and Ki-67 expression was decreased following knockdown of
SHFM1 (Fig. 6D and E). These results further suggested that
SHFM1 promoted ESCC progression in vivo.
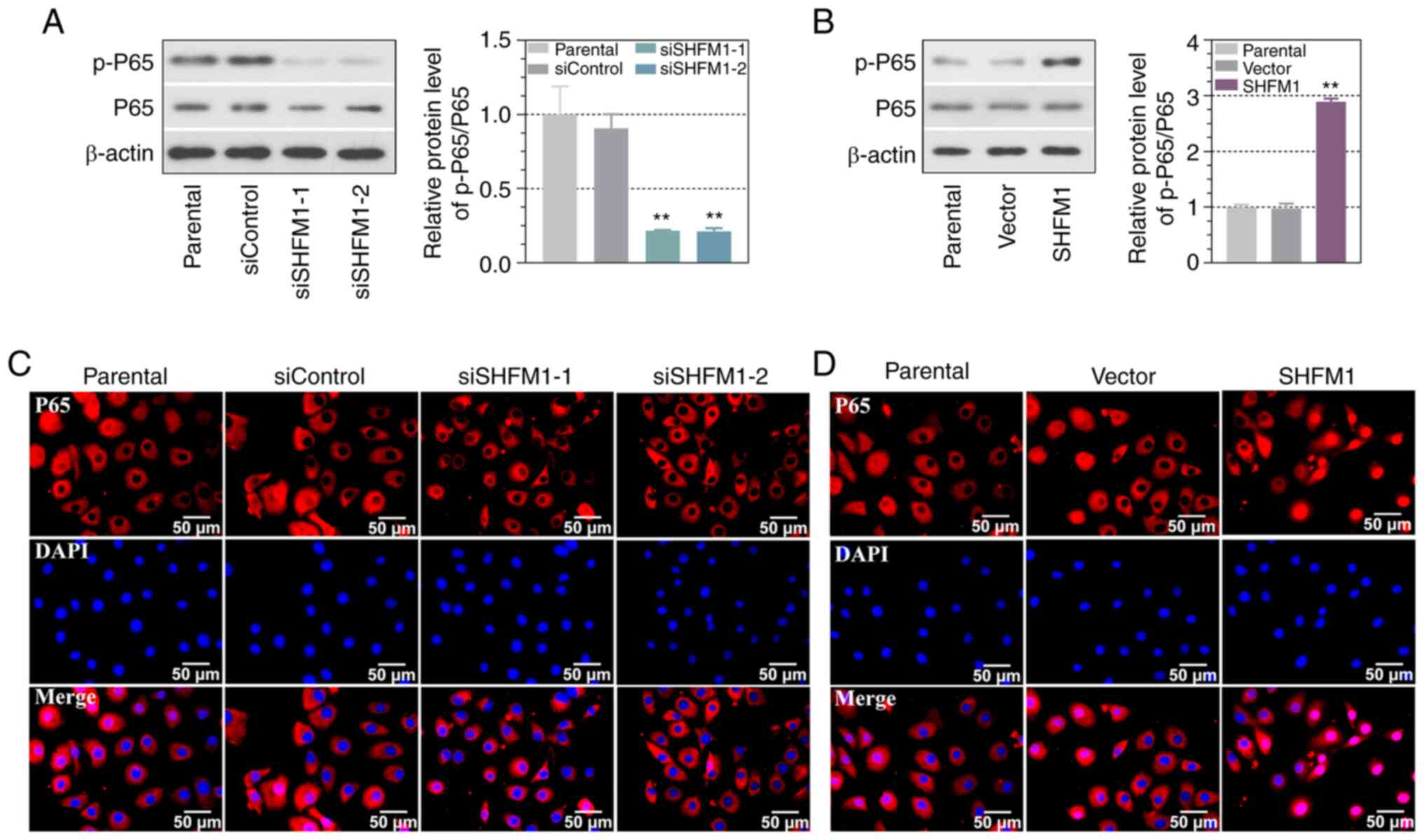
Effects of SHFM1 on the NF-κB
signaling pathway
The present study further investigated the effect of
SHFM1 on the NF-κB signaling pathway. Following transfection for 48
h, ESCC cells were collected to detect the expression of total and
phosphorylated P65 content. As indicated in Fig. 7A and B, the knockdown of SHFM1 in ESCC cells
markedly decreased the expression levels of phosphorylation of P65,
while SHFM1 overexpression strongly promoted P65 phosphorylation
and the expression of total P65 protein was not significantly
changed. These data suggested that SHFM1 might exert the role in
activating the NF-κB signaling pathway. In addition, the location
of P65 was further detected by immunofluorescence assay and the
results revealed that P65 was located predominantly in the
cytoplasm of control cells. Notably, SHFM1 overexpression showed an
increase in nuclear translocation of P65, while P65 was mainly in
the cytoplasm of SHFM1-silencing cells (Fig. 7C and D).
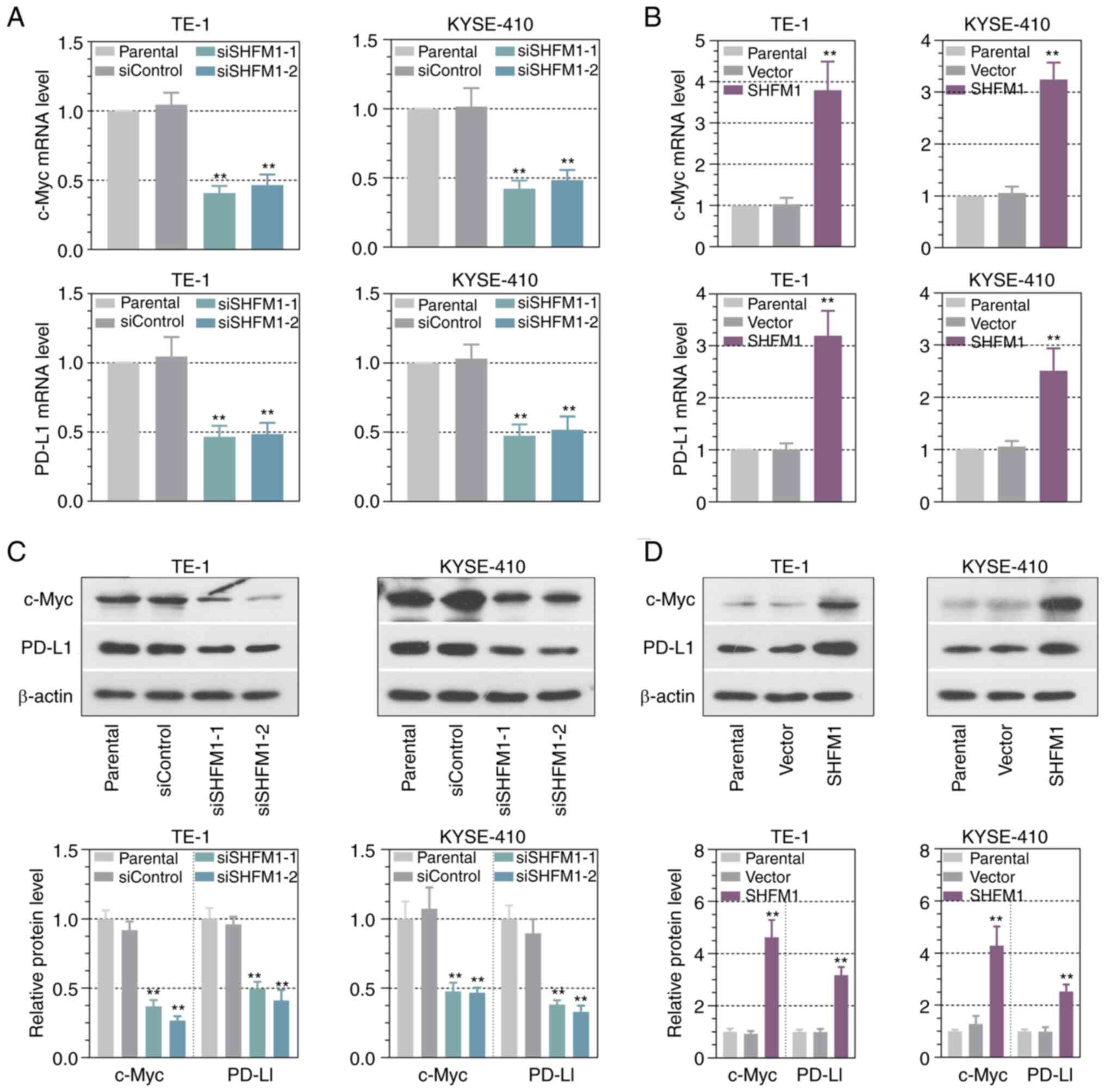
Effects of SHFM1 on NK cell-mediated
immune response
It is well-known that c-Myc and PD-L1 expression is
involved in the immune response in ESCC progression (24,29).
Thus, the effect of SHFM1 on the expression level of c-Myc and
PD-L1 in ESCC cells was investigated. The results revealed that
downregulation of SHFM1 significantly reduced c-Myc and PD-L1
levels, while upregulation of SHFM1 enhanced c-Myc and PD-L1
expression in ESCC cells (Fig.
8A-D). It has been reported that human leukocyte antigen (HLA)
class-I molecules and PD-L1 on cancer cell surfaces are pivotal for
tumor immunity (30). The present
study further verified HLA class-I expression in ESCC cells.
Results indicated that HLA class-I expression was significantly
upregulated following SHFM1 knockdown (Fig. S2). Having shown that SHFM1
knockdown inhibited c-Myc and PD-L1 expression, the present study
next explored the effect of SHFM1 on the NK cell-mediated immune
cell killing and co-incubated NK cells with ESCC cells. ESCC cells
were used as the target cells and NK-92 cells as the effector cells
and a CFSE/PI flow cytometry assay was conducted. As shown in
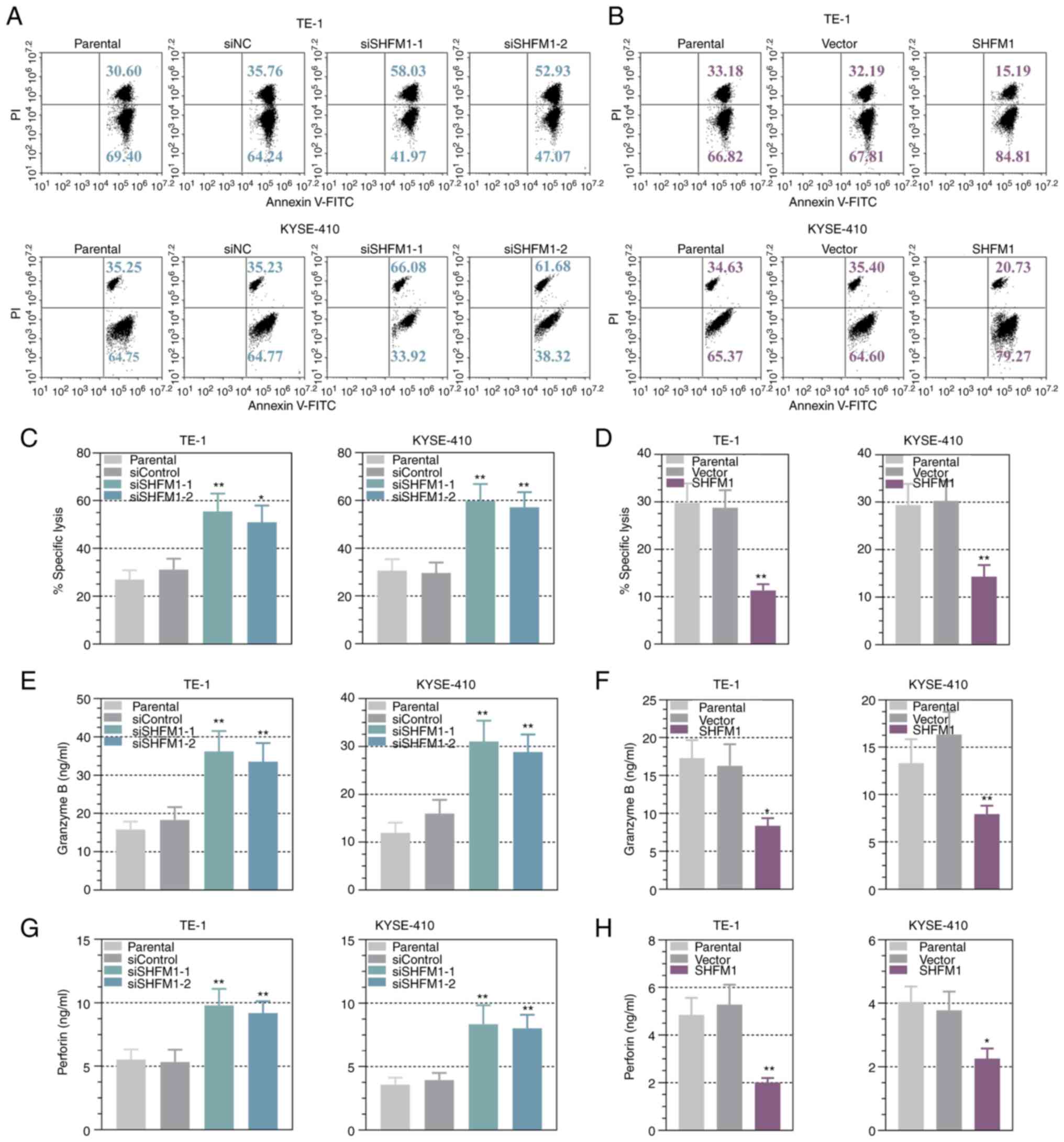
Fig. 9A and B, SHFM1 silencing significantly increased
the dead target cells, while ESCC cells overexpressing SHFM1
exhibited a decreased number of dead cells, suggesting that
knockdown of SHFM1 enhanced the capability of immune cell killing.
Additionally, NK-92 cell-mediated specific cell lysis results
showed that SHFM1 deficiency increased the percent specific lysis
of the ESCC cells, whereas overexpression of SHFM1 yielded the
reverse results (Fig. 9C and
D). It is generally known that
granzyme B and perforin synergize to mediate target cell apoptosis
in NK-mediated killing (15,31).
The present study detected the expression levels of granzyme B and
perforin in culture medium from SHFM1-silenced or -overexpressed
ESCC cells that co-cultured with NK-92 cells. The results of ELISA
indicated that the knockdown of SHFM1 significantly increased the
expression of granzyme B and perforin and upregulation of SHFM1
decreased levels of these cytolytic agents (Fig. 9E-H). These findings suggested that
SHFM1 expression partly blocked the susceptibility of cancer cells
toward immune attack.
Discussion
In the present study, bioinformatics methods were
performed to identify differential genes based on the cohorts
profile datasets. GO and KEGG pathway analysis was performed for
annotating these genes and PPI network was also developed to
identify central node genes. Through bioinformatical analysis,
three significant genes were screened, including COL1A1, SPP1 and
SHFM1. The clinical significance and function of COL1A1 in ESCC
have been well documented in previous studies (32,33).
In addition, SPP1 has been identified based on four GEO databases
from ESCC samples (34). Thus,
SHFM1 was selected as a biological marker in ESCC for further
study. Accumulating research indicate that SHFM1 is involved in
tumorigenesis in a number of types of human cancer (8,35,36).
Consistently, the present study observed a significantly higher
level of SHFM1 in ESCC progression based on clinic patient tissues
and aberrantly increased SHFM1 expression in cancer patients
predicted poor survival. Furthermore, the prognosis value of SHFM1
was investigated based on online databases and clinical cases. It
was found that SHFM1 expression was higher in tumors with lymph
node metastasis compared with non-metastatic focus. Highly
expressed SHFM1 was positively associated with lymph node
metastasis and TNM stage, indicating a significant association of
high SHFM1 expression with tumor growth and metastasis in ESCC. A
previous study reported the functional significance of SHFM1 in
ESCC progression in that it promotes cell growth, invasiveness
ability, colony formation and xenograft growth (37). These findings demonstrated the
important role of SHFM1 in cancer progression. Indeed, the present
study presented the oncogenic role of SHFM1 in ESCC progression and
SHFM1 was associated with the aggressiveness of ESCC cells both
in vitro and in vivo. Suppression of SHFM1 inhibited
the proliferation ability and the migrated and invasive capacity of
ESCC cells and overexpression of SHFM1 promoted ESCC
progression.
Abnormal activation of NF-κB signaling serves a
vital role in progressions in various types of cancer, including
ESCC (38). It has been indicated
that the homeoprotein of SHFM1 is associated with enhanced NF-κB
activity in ovarian cancer (39).
The present study hypothesized that SHFM1 might exert the
functional role in ESCC progression through the regulation of the
NF-κB signaling pathway. The data demonstrated that overexpressing
SHFM1 increased the expression level of p-P65 and P65 nuclear
translocation, suggesting that SHFM1 might regulate the NF-κB
pathway by promoting NF-κB nuclear translocation. Furthermore,
nuclear translocation of the NF-κB P65 was decreased in
SHFM1-silenced ESCC cells. This finding suggested that SHFM1
depletion might be a valuable approach for the treatment of cancers
by regulating the NF-κB signaling pathway in ESCC.
In addition, the present study studied the effect of
SHFM1 on the NK cell-mediated immune response in ESCC cells. SHFM1
exerts functional roles through regulating c-Myc expression in lung
cancer (11,20). Evidence indicates that the
expression of PD-L1 exerts important roles in the malignant
progression of various types of human tumors based on the immune
response (40,41). PD-L1 is highly expressed in various
tumors and elevated PD-L1 expression has been indicated to inhibit
the immune response (42). The
expression PD-L1 level is commonly regulated by c-Myc in the
antitumor immune response (22,43).
Thus, the inhibition of PD-L1 and c-Myc could regulate immune
responses that benefit tumor immunotherapy (44,45).
The present study demonstrated a positive association between SHFM1
expression and c-Myc and PD-L1 levels. SHFM1 knockdown
significantly decreased the expression of c-Myc and PD-L1 in ESCC
cells. It has been demonstrated that c-Myc and PD-L1 expression is
associated with cytotoxicity-induced apoptosis and NK cell
responses against tumors (22,46).
Therefore, the present study further explored the effect of SHFM1
on cellular cytotoxicity-mediated immune response in ESCC.
Consistently, immune killing assays were performed to explore the
role of SHFM1 in the capacity of cellular cytotoxicity-mediated
cell death. Cytotoxic T lymphocytes and NK cells are prevalent
players in the innate immune response that exert an antitumor role
through recognizing and eradicating cancer cells (47). NK cell-mediated cytotoxicity assay
was performed to investigate the cellular and molecular that
regulates NK cell anticancer function (48). In the present study, the effect of
SHFM1 on NK cell-mediated killing activity was investigated and the
percent ESCC cell death was quantified and it was found that
suppression of SHFM1 enhanced the ability of immune cell killing of
these NK cells on ESCC cells. Notably, ESCC cells overexpressing
SHFM1 showed clear resistance to NK cell-mediated cell death.
Cellular cytotoxicity is mediated by the secretion of lytic
granules, including pore-forming protein perforin and granzymes
(15). Perforin-mediated escape of
granzymes initiating cell apoptosis is one of the major mechanisms
of the ability of NK cells to kill tumor cells (49,50).
Thus, the release of perforin and granzymes is vital in cellular
cytotoxicity. Inhibition of SHFM1 expression significantly
increased the expression levels of perforin and granzyme B. In
brief, the increased cell death and lytic granules secretion
provided promising evidence of the effect of SHFM1 on immune
response during ESCC progression, suggesting that SHFM1 might
consider a potential target for ESCC immunotherapy.
The primary mediators of cellular cytotoxicity are
CD8+ T cells and NK cells (51).
HLA in tumors is another major escape mechanism in cellular
cytotoxicity-mediated immune response that is based on triggering
tumor specific CD8+ T lymphocyte-mediated responses (52). It has been demonstrated that tumor
cells tend to lose expression of HLA class I molecules and reduced
expression of HLA class I in tumor cells is associated with
mechanisms of tumor escape from immune recognition by
tumor-specific cytotoxic T lymphocytes (53) and recovery of HLA class I in
cancers is important for T-cell mediated cancer immunotherapy
(54). Therefore, HLA class I
expression was investigated and it was found that SHFM1 depletion
increased HLA class I expression, suggesting that SHFM1 might be
involved in T cell-mediated immunity through the regulation of HLA
class I. However, information about epigenetic factors for the
regulation of HLA-class I expression is limited. The molecular
mechanisms underlying SHFM1-dependent HLA-A regulation were not
elucidated in the current study. Future work will remedy these
deficiencies.
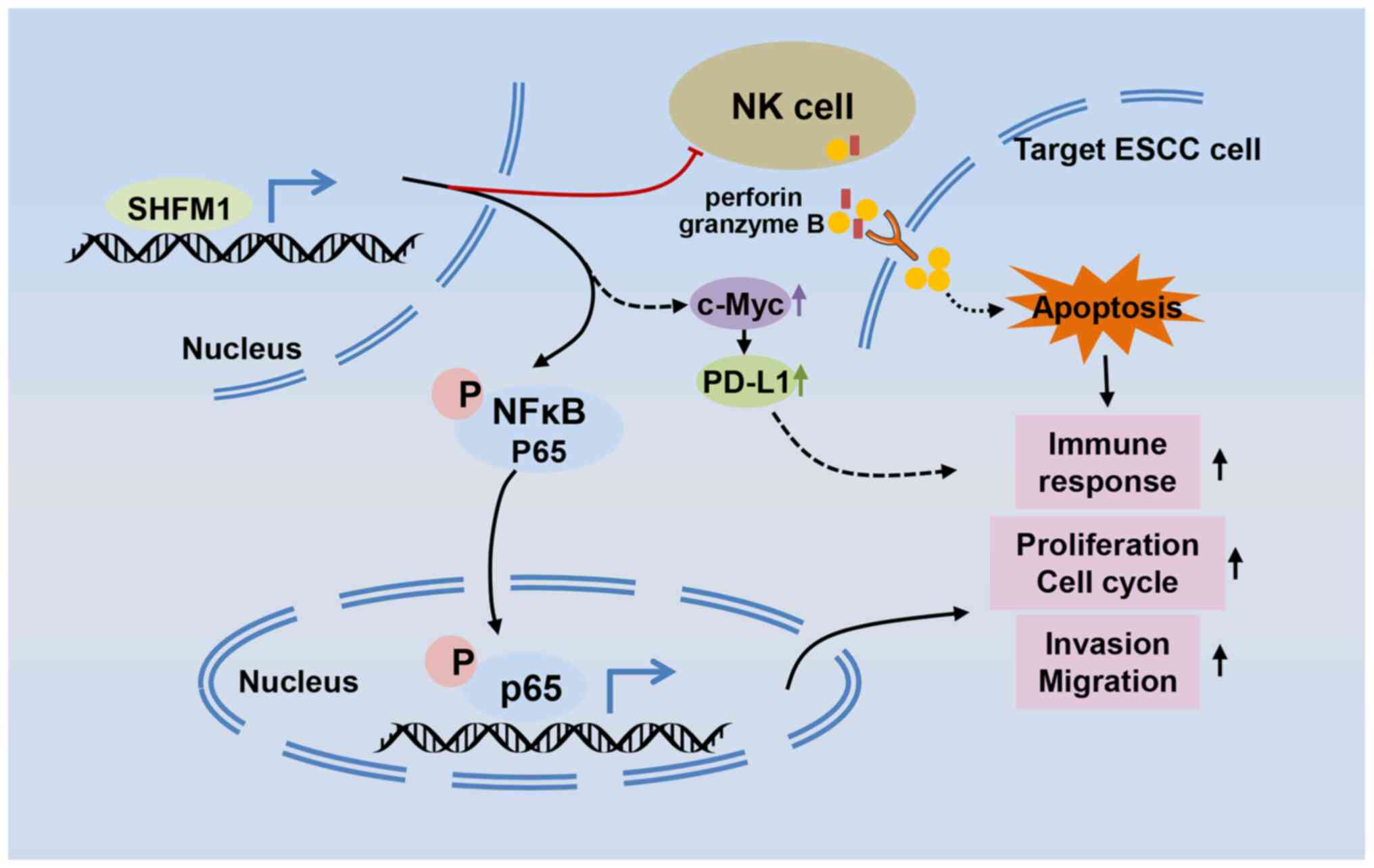
In conclusion, the findings of the current study
highlighted the functional role of SHFM1 in ESCC progression. In
addition, it further demonstrated that the effects of SHFM1 on the
progression of ESCC might be through the regulation of the NF-κB
signaling and the effect of SHFM1 on the NK cell-mediated immune
response was ascertained (Fig.
10). In summary, the findings of the present study implicated
the multi-faceted role of SHFM1 and it might be an attractive
diagnostic marker for the therapy of ESCC.
Supplementary Material
SHFM1 was silenced or overexpressed in
TE-1 and KYSE-410 cells; (A and B) SHFM1 mRNA levels in ESCC cells
were detected by reverse transcription-quantitative PCR. (C and D)
SHFM1 protein levels in ESCC cells were detected by Western
blotting. The relative expression was analyzed. Data are presented
as the mean ± standard deviation. **P<0.01 vs.
siControl or Vector group. SHFM1, split hand and foot malformation
1; ESCC, esophageal squamous cell carcinoma; si, short
interfering.
The protein expression of HLA class-I
in SHFM1-silenced TE-1 cells. The relative expression was analyzed.
Data are presented as the mean ± standard deviation.
**P<0.01 vs. siControl group. HLA, human leukocyte
antigen; SHFM1, split hand and foot malformation 1; si, short
interfering.
The list of siRNAs sequences used in
the present study
The list of primer sequences used in
the reverse transcription-quantitative PCR.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used in the current study are available
from the corresponding author on reasonable request.
Authors' contributions
YW and ZW were responsible for the conception and
design of the present study. YW and ZW confirm the authenticity of
all the raw data. YW, SL and XC obtained the study materials. YW,
SL, XC and SZ were responsible for data acquisition and data
analysis. YW wrote the manuscript. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
All clinical and animal experiments were approved by
the Ethics Committee of Xingtai People's Hospital (approval no.
2022-021). Written informed consent was provided by all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhou X, Ren T, Zan H, Hua C and Guo X:
Novel immune checkpoints in esophageal cancer: From biomarkers to
therapeutic targets. Front Immunol. 13(864202)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Jun Y, Tang Z, Luo C, Jiang B, Li X, Tao
M, Gu H, Liu L, Zhang Z, Sun S, et al: Leukocyte-mediated combined
targeted chemo and gene therapy for esophageal cancer. ACS Appl
Mater Interfaces. 12:47330–47341. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Yang YM, Hong P, Xu WW, He QY and Li B:
Advances in targeted therapy for esophageal cancer. Signal
Transduct Target Ther. 5(229)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lambert AW, Pattabiraman DR and Weinberg
RA: Emerging biological principles of metastasis. Cell.
168:670–691. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
He S, Xu J, Liu X and Zhen Y: Advances and
challenges in the treatment of esophageal cancer. Acta Pharm Sin B.
11:3379–3392. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sharma P, Wagner K, Wolchok JD and Allison
JP: Novel cancer immunotherapy agents with survival benefit: Recent
successes and next steps. Nat Rev Cancer. 11:805–812.
2011.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Tan Y and Testa JR: DLX genes: Roles in
development and cancer. Cancers. 13(3005)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang J, Wu J, Chen Y and Zhang W: Dlx5
promotes cancer progression through regulation of CCND1 in oral
squamous cell carcinoma (OSCC). Biochem Cell Biol. 99:424–434.
2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang X, Bian H, Wei W, Wang Q, Chen J,
Hei R, Chen C, Wu X, Yuan H, Gu J, et al: DLX5 promotes
osteosarcoma progression via activation of the NOTCH signaling
pathway. Am J Cancer Res. 11:3354–3374. 2021.PubMed/NCBI
|
|
11
|
Sun S, Yang F, Zhu Y and Zhang S: KDM4A
promotes the growth of non-small cell lung cancer by mediating the
expression of Myc via DLX5 through the Wnt/β-catenin signaling
pathway. Life Sci. 262(118508)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tan Y, Cheung M, Pei J, Menges CW, Godwin
AK and Testa JR: Upregulation of DLX5 promotes ovarian cancer cell
proliferation by enhancing IRS-2-AKT signaling. Cancer Res.
70:9197–9206. 2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang L, Zhou W, Zhong Y, Huo Y, Fan P,
Zhan S, Xiao J, Jin X, Gou S, Yin T, et al: Overexpression of G
protein-coupled receptor GPR87 promotes pancreatic cancer
aggressiveness and activates NF-κB signaling pathway. Mol Cancer.
16(61)2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang Y and Zhang Z: The history and
advances in cancer immunotherapy: Understanding the characteristics
of tumor-infiltrating immune cells and their therapeutic
implications. Cell Mol Immunol. 17:807–821. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Prager I, Liesche C, van Ooijen H, Urlaub
D, Verron Q, Sandström N, Fasbender F, Claus M, Eils R, Beaudouin
J, et al: NK cells switch from granzyme B to death
receptor-mediated cytotoxicity during serial killing. J Exp Med.
216:2113–2127. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lourenco C, Resetca D, Redel C, Lin P,
MacDonald AS, Ciaccio R, Kenney TMG, Wei Y, Andrews DW, Sunnerhagen
M, et al: MYC protein interactors in gene transcription and cancer.
Nat Rev Cancer. 21:579–591. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wu X, Nelson M, Basu M, Srinivasan P,
Lazarski C, Zhang P, Zheng P and Sandler AD: MYC oncogene is
associated with suppression of tumor immunity and targeting Myc
induces tumor cell immunogenicity for therapeutic whole cell
vaccination. J Immunother Cancer. 9(e001388)2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang P, Wu X, Basu M, Dong C, Zheng P,
Liu Y and Sandler AD: MYCN amplification is associated with
repressed cellular immunity in neuroblastoma: An in ailico
immunological analysis of TARGET satabase. Front Immunol.
8(1473)2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Casey SC, Baylot V and Felsher DW: The MYC
oncogene is a global regulator of the immune response. Blood.
131:2007–2015. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Xu J and Testa JR: DLX5 (distal-less
homeobox 5) promotes tumor cell proliferation by transcriptionally
regulating MYC. J Biol Chem. 284:20593–20601. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Casey SC, Baylot V and Felsher DW: MYC:
Master regulator of immune privilege. Trends Immunol. 38:298–305.
2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Casey SC, Tong L, Li Y, Do R, Walz S,
Fitzgerald KN, Gouw AM, Baylot V, Gütgemann I, Eilers M and Felsher
DW: MYC regulates the antitumor immune response through CD47 and
PD-L1. Science. 352:227–231. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kim EY, Kim A, Kim SK and Chang YS: MYC
expression correlates with PD-L1 expression in non-small cell lung
cancer. Lung Cancer. 110:63–67. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Liang MQ, Yu FQ and Chen C: C-Myc
regulates PD-L1 expression in esophageal squamous cell carcinoma.
Am J Transl Res. 12:379–388. 2020.PubMed/NCBI
|
|
25
|
Chiossone L, Vienne M, Kerdiles YM and
Vivier E: Natural killer cell immunotherapies against cancer:
Checkpoint inhibitors and more. Semin Immunol. 31:55–63.
2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sun X, Zhang J, Hou Z, Han Q, Zhang C and
Tian Z: MiR-146a is directly regulated by STAT3 in human
hepatocellular carcinoma cells and involved in anti-tumor immune
suppression. Cell Cycle. 14:243–252. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Percie N, Hurst V, Ahluwalia A, Alam S,
Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U, et al:
The ARRIVE guidelines 2.0: Updated guidelines for reporting animal
research. PLoS Biol. 18(e3000410)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lian Y, Niu X, Cai H, Yang X, Ma H, Ma S,
Zhang Y and Chen Y: Clinicopathological significance of c-MYC in
esophageal squamous cell carcinoma. Tumour Biol.
39(1010428317715804)2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ito S, Okano S, Morita M, Saeki H,
Tsutsumi S, Tsukihara H, Nakashima Y, Ando K, Imamura Y, Ohgaki K,
et al: Expression of PD-L1 and HLA class I in esophageal squamous
cell carcinoma: Prognostic factors for patient outcome. Ann Surg
Oncol. 23:508–515. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Sefid F, Payandeh Z, Azamirad G, Baradaran
B, Afjadi MN, Islami M, Darvish M, Kalantar SM, Kahroba H and
Ardakani MA: Atezolizumab and granzyme B as immunotoxin against
PD-L1 antigen; an insilico study. In Silico Pharmacol.
9(20)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Li Y, Wang X, Shi L, Xu J and Sun B:
Predictions for high and expression resulting in a poor prognosis
in esophageal squamous cell carcinoma by bioinformatics analyses.
Transl Cancer Res. 9:85–94. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Fang S, Dai Y, Mei Y, Yang M, Hu L, Yang
H, Guan X and Li J: Clinical significance and biological role of
cancer-derived type I collagen in lung and esophageal cancers.
Thorac Cancer. 10:277–288. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li M, Wang K, Pang Y, Zhang H, Peng H, Shi
Q, Zhang Z, Cui X and Li F: Secreted phosphoprotein 1 (SPP1) and
fibronectin 1 (FN1) are associated with progression and prognosis
of esophageal cancer as identified by integrated expression
profiles analysis. Med Sci Monit. 26(e920355)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Karsli-Ceppioglu S, Dagdemir A, Judes G,
Lebert A, Penault-Llorca F, Bignon YJ and Bernard-Gallon D: The
epigenetic landscape of promoter genome-wide analysis in breast
cancer. Sci Rep. 7(6597)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Tamilzhalagan S, Muthuswami M, Periasamy
J, Lee MH, Rha SY, Tan P and Ganesan K: Upregulated, 7q21-22
amplicon candidate gene SHFM1 confers oncogenic advantage by
suppressing p53 function in gastric cancer. Cell Signal.
27:1075–1086. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Huang Y, Yang Q, Zheng Y, Lin L, Xu X, Xu
XE, Silva TC, Hazawa M, Peng L, Cao H, et al: Activation of
bivalent factor DLX5 cooperates with master regulator TP63 to
promote squamous cell carcinoma. Nucleic Acids Res. 49:9246–9263.
2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhao Y, Wei L, Shao M, Huang X, Chang J,
Zheng J, Chu J, Cui Q, Peng L, Luo Y, et al: BRCA1-associated
protein increases invasiveness of esophageal squamous cell
carcinoma. Gastroenterology. 153:1304–1319.e1305. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Haria D, Trinh BQ, Ko SY, Barengo N, Liu J
and Naora H: The homeoprotein DLX4 stimulates NF-κB activation and
CD44-mediated tumor-mesothelial cell interactions in ovarian
cancer. Am J Pathol. 185:2298–2308. 2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chen L and Han X: Anti-PD-1/PD-L1 therapy
of human cancer: Past, present, and future. J Clin Invest.
125:3384–3391. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Doi T, Piha-Paul SA, Jalal SI, Saraf S,
Lunceford J, Koshiji M and Bennouna J: Safety and antitumor
activity of the anti-programmed death-1 antibody pembrolizumab in
patients with advanced esophageal carcinoma. J Clin Oncol.
36:61–67. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wang X, Teng F, Kong L and Yu J: PD-L1
expression in human cancers and its association with clinical
outcomes. Onco Targets Ther. 9:5023–5039. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhou C, Che G, Zheng X, Qiu J, Xie Z, Cong
Y, Pei X, Zhang H, Sun H and Ma H: Expression and clinical
significance of PD-L1 and c-Myc in non-small cell lung cancer. J
Cancer Res Clin Oncol. 145:2663–2674. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Qu S, Jiao Z, Lu G, Yao B, Wang T, Rong W,
Xu J, Fan T, Sun X, Yang R, et al: PD-L1 lncRNA splice isoform
promotes lung adenocarcinoma progression via enhancing c-Myc
activity. Genome Biol. 22(104)2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Han H, Jain AD, Truica MI,
Izquierdo-Ferrer J, Anker JF, Lysy B, Sagar V, Luan Y, Chalmers ZR,
Unno K, et al: Small-molecule MYC inhibitors suppress tumor growth
and enhance immunotherapy. Cancer Cell. 36:483–497.e415.
2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wu Y, Xie J, Jin X, Lenchine RV, Wang X,
Fang DM, Nassar ZD, Butler LM, Li J and Proud CG: eEF2K enhances
expression of PD-L1 by promoting the translation of its mRNA.
Biochem J. 477:4367–4381. 2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Uppendahl LD, Felices M, Bendzick L, Ryan
C, Kodal B, Hinderlie P, Boylan KLM, Skubitz APN, Miller JS and
Geller MA: Cytokine-induced memory-like natural killer cells have
enhanced function, proliferation, and in vivo expansion against
ovarian cancer cells. Gynecol Oncol. 153:149–157. 2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lorenzo-Herrero S, Sordo-Bahamonde C,
González S and López-Soto A: A flow cytometric NK cell-mediated
cytotoxicity assay to evaluate anticancer immune responses in
vitro. Methods Mol Biol. 1884:131–139. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Prager I and Watzl C: Mechanisms of
natural killer cell-mediated cellular cytotoxicity. J Leukoc Biol.
105:1319–1329. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Zhou Z, He H, Wang K, Shi X, Wang Y, Su Y,
Wang Y, Li D, Liu W, Zhang Y, et al: Granzyme A from cytotoxic
lymphocytes cleaves GSDMB to trigger pyroptosis in target cells.
Science. 368(eaaz7548)2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Cerwenka A and Lanier LL: Natural killer
cell memory in infection, inflammation and cancer. Nat Rev Immunol.
16:112–123. 2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Aptsiauri N, Ruiz-Cabello F and Garrido F:
The transition from HLA-I positive to HLA-I negative primary
tumors: The road to escape from T-cell responses. Curr Opin
Immunol. 51:123–132. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ramia E, Chiaravalli AM, Eddine FBN,
Tedeschi A, Sessa F, Accolla RS and Forlani G: CIITA-related block
of HLA class II expression, upregulation of HLA class I, and
heterogeneous expression of immune checkpoints in hepatocarcinomas:
Implications for new therapeutic approaches. Oncoimmunology.
8(1548243)2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Garrido F, Aptsiauri N, Doorduijn EM, Lora
AM and van Hall T: The urgent need to recover MHC class I in
cancers for effective immunotherapy. Curr Opin Immunol. 39:44–51.
2016.PubMed/NCBI View Article : Google Scholar
|