At present, the incidence of non-specific neck and
lower back pain in the global population is high and the etiology
is complex (1,2); this seriously affects the quality of
life or even causes disability in affected individuals, reduces
life expectancy and increases the economic and social burden
(3,4). The total annual expenses associated
with lower back pain were ~£12 billion in the UK in 1998(5), and US$7.4 billion in the USA from
1997 to 2007(6). Moreover, the
total annual costs associated with neck pain were US$686 million in
The Netherlands in 1996(7).
Efforts have been made by researchers and clinicians to elucidate
the pathogenesis of neck and lower back pain and promote treatment
strategies (8,9). One of the main pathogenic factors of
these afflictions is intervertebral disc degeneration (IDD)
(10-13).
Currently, various intervention measures are available for chronic
musculoskeletal pain, including psychological based therapies
(14-20),
pharmacological treatments (21-24),
and physical-based therapies (20,25-27).
However, satisfactory results have not been achieved in terms of
pain relief and functional improvement using these methods.
In recent years, various novel interventions have
been used to explore the treatment of IDD, such as stem cell
transplantation (28), and
nanoparticles (29). At present,
stem cell therapy for IDD includes hematopoietic precursor stem
cells (30), mesenchymal stem
cells (MSCs) and adipose-derived stem cells (31). Among them, MSCs have been well
studied, including autologous and allogeneic MSCs. In 2011, Orozco
et al (32) used autologous
MSCs transplantation to treat IDD and showed some pain relief. In
2017, they further used allogeneic MSCs to treat IDD, confirming
the feasibility and safety of this method and having some pain
relief (33). However, the
treatment of IDD with stem cells has yet to achieve satisfactory
outcomes, possibly due to unclear treatment mechanisms, low
survival rates of stem cells, different sources and injection
methods of stem cells and a lack of large-scale clinical studies.
Recently, nanoparticles have been increasingly used to treat IDD.
Prussian blue nanoparticles relieve intracellular oxidative stress
and increases the activity of intracellular antioxidant enzymes to
rescue IDD (34); polydopamine
nanoparticles alleviate IDD by reactive oxygen species consumption,
iron chelation and glutathione peroxidase-4 ubiquitination
inhibition (29). However, these
treatments remain in vitro or have been tested in animal
models and not in appropriate IDD models. Bioreactors are culture
systems that can mimic the physiological environment of the IDD and
provide an accurate nutritional and mechanical environment for the
culture of intervertebral discs (IVD) organs (35). However, current cultivation
techniques do not allow for extended reactor cultivation time.
According to the research conducted by Šećerović et al, the
survival rate of fibrous rings decreases by >30% within 3 weeks
(35). Additionally, there are
still significant differences between the simulated physiological
state of the IVD in the current IVD bioreactor and the actual human
IVD. Therefore, there are still significant limitations in the
research and treatment of IDD using bioreactors.
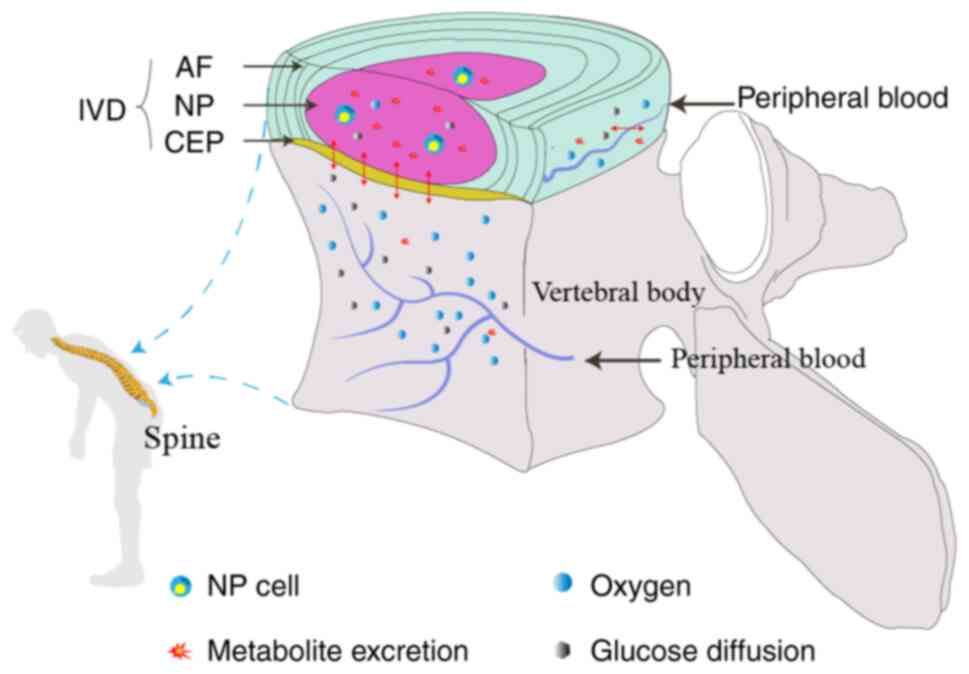
IVDs are often referred to as the largest avascular
structures of the human body (36), which consist of gelatinous nucleus
pulposus (NP) as the central structure, surrounded by lamellar
annulus fibrosus (AF) and sandwiched by the superior and inferior
cartilage endplate (CEP) (37,38).
Due to the avascular nature of the IVD, small molecules (such as
nutrients) have to reach the cells through the extracellular matrix
(ECM) mainly by diffusion (39),
from the blood vessels at disc margins via two pathways: The CEP-NP
pathway and the AF periphery pathway. Several researchers have
reported that the CEP-NP nutritional pathway is primarily
responsible for nurturing cells in the NP and inner AF regions,
while the AF periphery is mainly for cells in the outer AF region
(40-42)
(Fig. 1). CEP degeneration can
hinder the transport of nutrients and causes the dysfunctions of NP
and CEP cells (43-45),
including senescence, apoptosis and aberrant cell proliferation.
Thus, CEP degeneration is considered one of the major causes of
IDD, which causes neck or lower back pain (46-48).
At present, there are numerous clinical treatments available for
neck and lower back pain; however, these treatments can only
partially improve some symptoms of patients and cannot
fundamentally delay or reverse the pathological process of IDD
(49,50). Therefore, restoring the biological
function of the CEP and the nutrient supply of IVD, and preventing
or even reversing CEP degeneration at the molecular level are the
new aims of treatment for neck and lower back pain.
Non-coding RNAs (ncRNAs) are present in the majority
of tissues of different species and account for 99% of the total
RNA content (51-54).
In addition, ncRNAs, DNA methylation and histone modifications are
the main mechanisms in epigenetics (55,56).
They have been defined as a class of RNA molecules transcribed from
the genome, but not encoding proteins, such as long ncRNAs
(lncRNAs) (57,58), microRNAs (miRNAs/miRs) (59,60)
and circular RNAs (circRNAs) (61-63),
with known biological functions, as well as unknown functions
(64). ncRNAs have been found to
be involved in the development of various diseases, including
cancer, heart failure and even nervous system diseases (65-67).
Notably, an increasing number of studies have demonstrated that
ncRNAs are involved in chondrocyte degeneration through multiple
mechanisms (68,69) (Fig.
2).
Previously, it has been determined that miRNAs play
a critical role in complex gene regulatory networks (70). According to statistics, >1,500
miRNAs have been found in the human genome, and each miRNA can
target multiple mRNAs; in addition, each mRNA can also be regulated
by several miRNAs (71-73).
Short RNA molecules of 19-25 nucleotides in size are a class of
ncRNAs that regulate the post-transcriptional silencing of target
genes by directly binding to the 3'-untranslated region (UTR),
5'-UTR and coding sequence regions of their target mRNAs (74). The majority of miRNA sequences are
conserved across species (75).
However, miRNA expression varies depending on the time and period
examined and tissue type, which indicates that changes in miRNA
expression may reflect different cellular composition or activation
states (76,77). There is evidence to indicate that
miRNAs participate in diverse chondrocyte processes, such as cell
proliferation (78), apoptosis
(79) and differentiation
(80). They are therefore involved
in a wider range of processes, such as cartilaginous development,
degeneration (81) and
regeneration (82). Consequently,
CEP degeneration is the primary factor leading to IDD and
maintaining the physiological function of CEP is essential for
prevention and treatment of IDD (46).
Previous studies have demonstrated that intermittent
cyclic mechanical tension (ICMT) can lead to CEP degeneration
(83,84). However, the role of miRNAs in
regulating chondrocyte responses to ICMT needs to be elucidated. In
the study by Feng et al (85), CEP chondrocytes from patients
without ICMT stimulation were used as controls and specimens were
obtained from patients who underwent posterior discectomy and a
fusion procedure for IDD. They identified a total of 21
significantly upregulated and 62 downregulated identified compared
with the control.
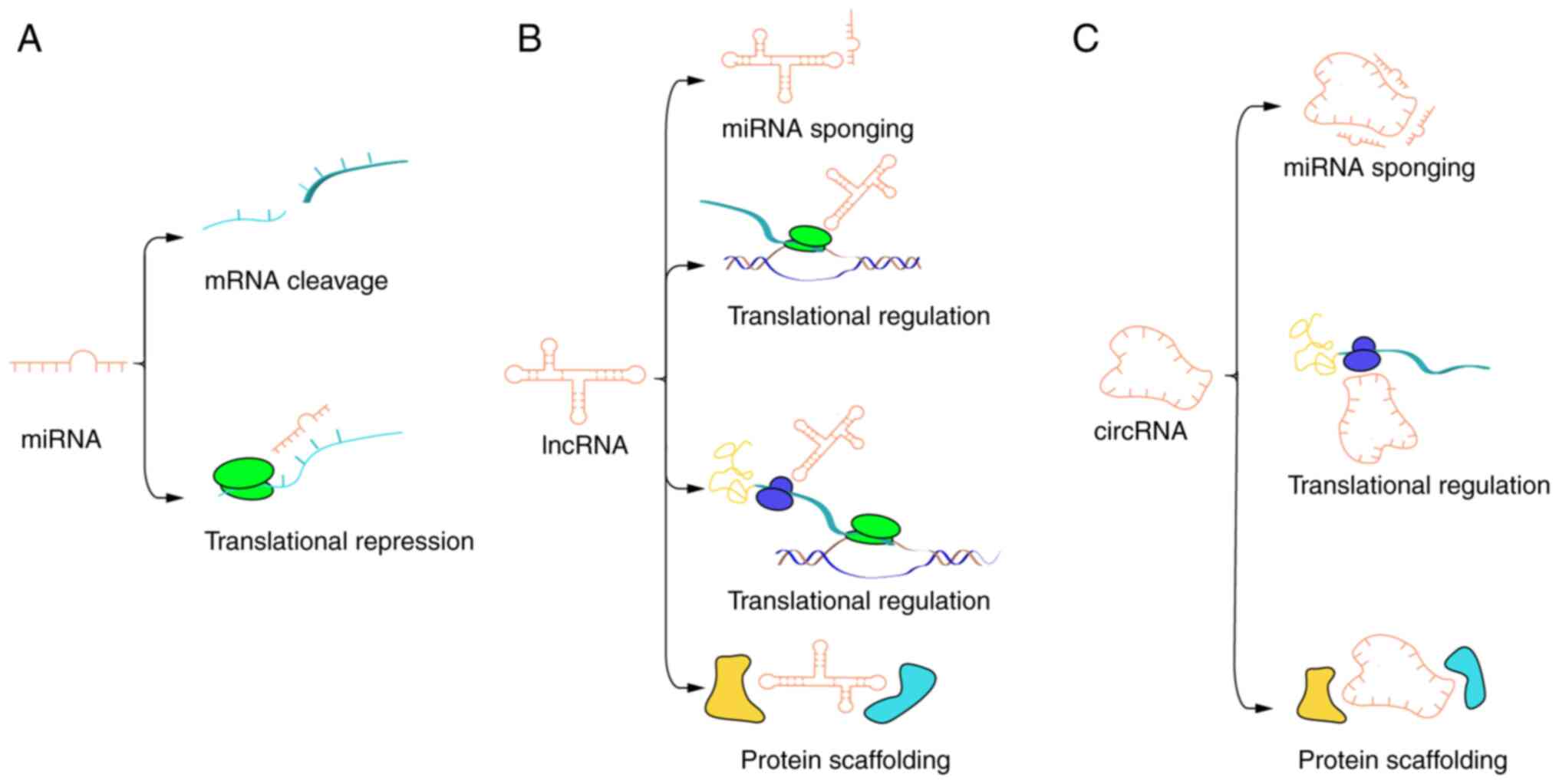
The biological potency of miRNAs has been
well-established, with their regulatory effects primarily exerted
through sponge target genes, as depicted in Fig. 2, which illustrates the underlying
molecular mechanisms.
miRNAs are involved in the regulation of multiple
mechanisms as a novel subtype ncRNAs. There is evidence to indicate
that the apoptosis of chondrocytes in the CEP is implicated in the
pathogenesis IDD. Chen et al (86) demonstrated that the expression of
miR-34a is markedly elevated in human degenerated CEP chondrocytes
compared with normal CEP chondrocytes. Furthermore, luciferase
assays from the same study indicated that Bcl-2 is a target of
miR-34a, while miR-34a represses the expression of Bcl-2.
Functionally, the inhibition of miR-34a rescues the fas-induced
apoptosis of CEP chondrocytes by releasing Bcl-2, which plays an
important role in the development of IDD.
It has been shown that miRNAs are involved in the
tension-induced degeneration of endplate chondrocytes by regulating
the miR-455-5p/runt-related transcription factor 2 (RUNX2) axis. In
the majority of cases, more tension is borne by endplate
chondrocytes compared with other cells in human body (87), which is responsible for chondrocyte
degeneration (88,89). In chondrocytes, the aberrantly
low-expression of miR-455-5p increases the degeneration level of
chondrocytes by upregulating RUNX2 expression using ICMT.
Furthermore, Xiao et al (90) revealed that the up- or
downregulation of miR-455-5p does not affect the proliferation or
apoptosis of endplate chondrocytes, while RUNX2 expression also
exhibits a down- and upregulation, respectively. Therefore, these
findings result indicate that miR-455-5p is a therapeutic target
for tension-induced degeneration. There is previous evidence to
indicate that miRNAs are involved in the calcification of CEP
chondrocytes induced by matrix stiffness. For example, it has been
shown that the inhibition of miR-20a attenuates calcium deposition
and calcification-related gene expression, whereas the
overexpression of miR-20a enhances the calcification of CEP
chondrocytes on a stiff matrix, which is positively associated with
the degree of IDD (91).
The role of CEP chondrocytes in ECM synthesis and
catabolism, such as collagens and proteoglycans, plays an important
role in maintaining the structural stability of the IVD and in
resisting mechanical loads (92,93).
In patients with IDD, an imbalance in matrix synthesis and
breakdown in the CEP is observed, as shown by the increased
expression of breakdown proteins, such as MMP-3 and MMP-9, and a
corresponding reduction in the expression of synthetic proteins.
Sheng et al (94) found
that the overexpression of miR-221 in degenerative CEP tissue
accelerates apoptosis by downregulating the level of estrogen
receptor α. Furthermore, the increased level of miR-221
deteriorates the degradation of the ECM by disrupting the balance
in the expression of ECM-degrading and anti-ECM-degrading
genes.
Recent studies have demonstrated that cartilage
endplate stem cells (CESCs) can maintain the normal function of the
NP and CEP through miRNAs. Chen et al (95) revealed that miR-637 is expressed in
low levels in the degenerative CEP, and the inhibition of miR-637
promotes the osteogenic differentiation ability of degenerative
CESCs. However, the upregulation of Wnt family member 5A partially
annuls the inhibitory effects of miR-637 overexpression on the
osteogenic differentiation of degenerative CESCs. In addition, Chen
and Jiang (96) examined the
effects of normal CESC-derived exosomes on autophagy, apoptosis and
ECM metabolism in the NP. Bioinformatics analysis was used to
analyze differences in miRNA expression, and dual-luciferase
reporter assays were used to detect target associations. They
confirmed that exosomes-derived miR-125-5p from CESCs regulate
autophagy and ECM metabolism in the NP by targeting SUV38H1.
There is evidence to indicate that the reduction of
the proliferation of CEP chondrocytes is implicated in the
pathogenesis of IDD. Using a double luciferase assay, Wang et
al (97) indicated that the
target gene of miR-142-3p is high mobility group box 1 (HMGB1), the
expression of which is significantly increased during the process
of IDD. Functionally, the inhibition of chondrocytes proliferation
ability follows the addition of a HMGB1 inhibitor.
In conclusion, the aberrant expression of seven
miRNAs has been discovered to be involved in various cellular
processes, such as proliferation, apoptosis and
calcification-induced apoptosis, with their specific regulatory
mechanisms and expressions documented in Table I.
It has been shown that there is a clear difference
in lncRNA expression between degenerated endplate chondrocytes and
normal endplate chondrocytes (116). In 2020, the expression profile of
lncRNAs was reported for the first time in the endplate of
degenerated cartilage. In degenerated chondrocytes, 369 lncRNAs
exhibited a differential expression, including 316 upregulated and
53 downregulated lncRNAs, contrasting with the non-degenerated CEP
of cervical fractures. In addition, Li et al (117) identified the highly selective
expression of 34 lncRNAs in human fetal growth plate chondrocytes
by employing RNA sequencing. A total of eight lncRNAs were adjacent
to the loci of protein coding genes that participate in skeletal
development, suggesting that cartilage-selective lncRNAs may be
involved in chondrogenesis is through the regulation of protein
coding genes.
In summary, the biological functions of lncRNA in
chondrocytes can be mediated by various mechanisms, including miRNA
sponging, protein scaffolding and translational regulation.
Additionally, specific expression patterns of lncRNAs in
degenerated endplate chondrocytes have been demonstrated.
Evidence suggests that diabetes causes CEP
degeneration by altering endplate thickening and reducing porosity
(118-121).
Furthermore, chondrocyte apoptosis, characterized by various
signaling molecules, is involved in the degeneration of CEPs
(122,123). Based on these findings, Jiang
et al (124) induced CEP
cell degeneration with high-glucose medium and revealed that the
knockdown of lncRNA MALAT1 reduces the apoptosis of chondrocytes.
Furthermore, they demonstrated that lncRNA MALAT1 promotes high
glucose-induced rat CEP apoptosis via the p38/MAPK signaling
pathway. lncRNAs, as gene expression modulators, are expected to be
a novel target for the treatment of disc degeneration; however, to
the best of our knowledge, studies on lncRNAs in CEP degeneration
are limited and, thus, further studies on their mechanism of action
in CEP degeneration are warranted.
circRNAs, which are single-stranded and covalently
closed, were first reported as viroids (125), which are pathogens of certain
plants in 1976 and were first detected in human HeLa cells by
electron microscopy in 1979(126). Later on, with the development of
high-throughput RNA sequencing and bioinformatics tools, circRNAs
began to be considered as a general feature of the human
transcriptome and are ubiquitous in numerous other metazoans,
including mammals (127),
unicellular eukaryotes (128),
prokaryotes (129) and viruses
(130). Previous studies have
identified multiple functions of circRNAs, including serving as
protein scaffolds or miRNA sponges and being translated into
polypeptides (131,132). In addition, the unique covalently
closed structure of circRNAs that provides them with a longer
half-life and greater resistance to RNase R compared with linear
RNAs (133), renders them as
potential candidates for use as diagnostic biomarkers and
therapeutic targets.
We previously conducted a study to compare
degenerative CEP to healthy CEP using a human ceRNA microarray
(134). It was revealed that 578
circRNAs were differentially regulated in degenerative CEP samples
compared with healthy tissues. Of these, 435 circRNAs were highly
expressed, while 143 were significantly repressed. In addition, it
has been indicated that biomechanical stimulation is essential for
the growth and maintenance of endplate cartilage function (102). Excessive mechanical loading, on
the other hand, alters the distribution of the ECM in the CEP,
ultimately leading to the destruction of normal cartilage structure
and the interruption of nutrient supply (135). By applying an ICMT of 0.5 Hz and
an extension of 10% to primitive human endplate chondrocytes, Xiao
et al (136) verified
upregulated expression levels of 17circRNAs and the downregulated
expression of another 12 with fold changes 1.5 by using a circRNA
microarray technique.
Compared with miRNAs and lncRNAs, circRNAs exhibit
an enhanced richness, stability and specific expression (137). Furthermore, various mechanisms
such as miRNA sponging, protein scaffolding and translational
regulation can be utilized to regulate the chondrocyte process by
circRNAs (138) (Fig. 2).
Specific circRNAs regulate the ECM and proliferation
via a ceRNA mechanism, which contributes to the development of IDD
(139). Specifically,
circRNA_0058097 and circ small nucleolar RNA host gene 5 (SNHG5)
are involved in ECM regulation (136,140). circSNHG5 is related to CEP cell
proliferation. miR-495-3p stimulates ECM degradation and inhibits
chondrocyte cell proliferation by inhibiting Cbp/P300-interacting
transactivator with glu/asp rich carboxy-terminal domain 2, whereas
circSNHG5 alleviates the negative effects by sponging miR-495-3p.
However, in IDD tissues, the expression of circSNHG5 is repressed,
resulting in an aberrantly higher level of miR-495-3p and IDD. The
upregulation of circRNA_0058097 expression was observed in the
loading group that was subjected to an ICMT of 0.5 Hz and 10%
elongation degeneration. Furthermore, circRNA_0058097 can sponge
miR-365a-5p and overexpression of miR-365a-5p alleviates
tension-induced chondrocyte degeneration (130). These results suggest that the
fate of CEP cells in IDD can be modulated by circRNAs, which have
the potential to serve as therapeutic targets.
Neck and lower back pain is the most prevalent of
all musculoskeletal conditions, and it places a major strain on
individuals, health systems and social care systems (141). CEP degeneration is one of the
primary causes of IDD that leads to neck and lower back pain
(46). However, the mechanisms
involved have not yet been fully elucidated. Recently, it has been
shown that ncRNAs are involved in the degeneration of chondrocytes,
including endplate chondrocytes (142).
The present review summarizes the latest evidence
concerning the regulation of endplate chondrocytes in IDD based on
miRNAs, lncRNAs and circRNAs. In addition, the present review
summarizes the mechanisms through which proliferation,
calcification, apoptosis and ECM degradation of the CEP can be
regulated by regulating downstream target genes (Table I). The data presented herein
provides novel insights into the etiology of endplate chondrocyte
degeneration and identify ncRNAs as potential novel targets for the
treatment of IDD. However, effective therapeutic approaches, such
as bone/cartilage targeted hydrogel (143), and exosome-based bone-targeting
(144), are hampered by an
incomplete understanding of the mechanisms of CEP homeostasis and
degeneration. Recently, the advent of novel materials like lipid
nanoparticles and cationic polymers has enhanced the targeting
specificity of therapy, while also mitigating toxicity and
immunogenicity concerns. Furthermore, technological breakthroughs
such as CRISPR/Cas gene editing have lowered off-target effects and
boosted RNA interference levels. Therefore, injectable hydrogels or
nanoparticles (145,146), recombinant adeno-associated viral
vector-mediated gene delivery (147), and mesenchymal stem cell-based
therapies (148) interfere with
RNA expression in endplate chondrocytes to achieve the purpose of
treating disc degeneration. At the same time, interfering with the
central nodes in the regulatory network allows ncRNAs to provide a
future for IDD treatment.
Not applicable.
Funding: The present study was supported by the National Natural
Science Foundation of China (grant nos. 8166090137 and 8186090165),
the National Natural Science Foundation of Jiangxi Province (grant
no. 20202ACBL206012) and the Graduate Innovative Special Fund
Projects of Jiangxi Province, China (grant no. YC2022-s212).
Not applicable.
XKZ, JHY, JYJ, JZ, JHL, QC, TL, ZWW, HW, XXM, TLW,
BL and XGC contributed to the conception and design of the study.
JHL, QC, TL, ZWW and HW examined the relevant literature, and XKZ
wrote the manuscript. JHY, JYJ, JZ, XXM TLW and BL provided advice
and are responsible for revising the manuscript. All authors read
and approved the final manuscript. Data sharing is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
GBD 2015 Disease and Injury Incidence and
Prevalence Collaborators. Global, regional, and national incidence,
prevalence, and years lived with disability for 310 diseases and
injuries, 1990-2015: A systematic analysis for the Global Burden of
Disease Study 2015. Lancet. 388:1545–1602. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Minghelli B: Musculoskeletal spine pain in
adolescents: Epidemiology of non-specific neck and low back pain
and risk factors. J Orthop Sci. 25:776–780. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Frymoyer JW and Cats-Baril WL: An overview
of the incidences and costs of low back pain. Orthop Clin North Am.
22:263–271. 1991.PubMed/NCBI
|
|
4
|
Steenstra IA, Verbeek JH, Prinsze FJ and
Knol DL: Changes in the incidence of occupational disability as a
result of back and neck pain in the Netherlands. BMC Public Health.
6(190)2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Maniadakis N and Gray A: The economic
burden of back pain in the UK. Pain. 84:95–103. 2000.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Dagenais S, Caro J and Haldeman S: A
systematic review of low back pain cost of illness studies in the
United States and internationally. Spine J. 8:8–20. 2008.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Borghouts JAJ, Koes BW, Vondeling H and
Bouter LM: Cost-of-illness of neck pain in The Netherlands in 1996.
Pain. 80:629–636. 1999.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Brockow T, Dillner A, Franke A and Resch
KL: Analgesic effectiveness of subcutaneous carbon-dioxide
insufflations as an adjunct treatment in patients with non-specific
neck or low back pain. Complement Ther Med. 9:68–76.
2001.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Miyamoto GC, Lin CC, Cabral CMN, van
Dongen JM and van Tulder MW: Cost-effectiveness of exercise therapy
in the treatment of non-specific neck pain and low back pain: A
systematic review with meta-analysis. Br J Sports Med. 53:172–181.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Nakamura M, Nishiwaki Y, Ushida T and
Toyama Y: Prevalence and characteristics of chronic musculoskeletal
pain in Japan. J Orthop Sci. 16:424–432. 2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Nakamura M, Toyama Y, Nishiwaki Y and
Ushida T: Prevalence and characteristics of chronic musculoskeletal
pain in Japan: A second survey of people with or without chronic
pain. J Orthop Sci. 19:339–350. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Samartzis D, Karppinen J, Mok F, Fong DY,
Luk KD and Cheung KM: A population-based study of juvenile disc
degeneration and its association with overweight and obesity, low
back pain, and diminished functional status. J Bone Joint Surg Am.
93:662–670. 2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gibson J, Nouri A, Krueger B, Lakomkin N,
Nasser R, Gimbel D and Cheng J: Degenerative cervical myelopathy: A
clinical review. Yale J Biol Med. 91:43–48. 2018.PubMed/NCBI
|
|
14
|
Slade SC and Keating JL: Unloaded movement
facilitation exercise compared to no exercise or alternative
therapy on outcomes for people with nonspecific chronic low back
pain: A systematic review. J Manipulative Physiol Ther. 30:301–311.
2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Furlan AD, Imamura M, Dryden T and Irvin
E: Massage for low-back pain. Cochrane Database Syst Rev.
(4)(Cd001929)2008.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hall J, Swinkels A, Briddon J and McCabe
CS: Does aquatic exercise relieve pain in adults with neurologic or
musculoskeletal disease? A systematic review and meta-analysis of
randomized controlled trials. Arch Phys Med Rehabil. 89:873–883.
2008.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hendrick P, Te Wake AM, Tikkisetty AS,
Wulff L, Yap C and Milosavljevic S: The effectiveness of walking as
an intervention for low back pain: A systematic review. Eur Spine
J. 19:1613–1620. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Miller J, Gross A, D'Sylva J, Burnie SJ,
Goldsmith CH, Graham N, Haines T, Brønfort G and Hoving J: Manual
therapy and exercise for neck pain: A systematic review. Man Ther.
15:334–354. 2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Rubinstein SM, van Middelkoop M,
Assendelft WJ, de Boer MR and van Tulder MW: Spinal manipulative
therapy for chronic low-back pain: An update of a Cochrane review.
Spine (Phila Pa 1976). 36:E825–E846. 2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
van Middelkoop M, Rubinstein SM, Kuijpers
T, Verhagen AP, Ostelo R, Koes BW and van Tulder MW: A systematic
review on the effectiveness of physical and rehabilitation
interventions for chronic non-specific low back pain. Eur Spine J.
20:19–39. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Noble M, Treadwell JR, Tregear SJ, Coates
VH, Wiffen PJ, Akafomo C and Schoelles KM: Long-term opioid
management for chronic noncancer pain. Cochrane Database Syst Rev.
2010(Cd006605)2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chou R and Huffman LH: American Pain
Society; American College of Physicians. Medications for acute and
chronic low back pain: A review of the evidence for an American
Pain Society/American College of Physicians clinical practice
guideline. Ann Intern Med. 147:505–514. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Roelofs PD, Deyo RA, Koes BW, Scholten RJ
and van Tulder MW: Non-steroidal anti-inflammatory drugs for low
back pain. Cochrane Database Syst Rev. (1)(Cd000396)2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Mason L, Moore RA, Edwards JE, Derry S and
McQuay HJ: Topical NSAIDs for chronic musculoskeletal pain:
Systematic review and meta-analysis. BMC Musculoskelet Disord.
5(28)2004.PubMed/NCBI View Article : Google Scholar
|
|
25
|
van Geen JW, Edelaar MJ, Janssen M and van
Eijk JT: The long-term effect of multidisciplinary back training: A
systematic review. Spine (Phila Pa 1976). 32:249–255.
2007.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Scascighini L, Toma V, Dober-Spielmann S
and Sprott H: Multidisciplinary treatment for chronic pain: A
systematic review of interventions and outcomes. Rheumatology
(Oxford). 47:670–678. 2008.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ravenek MJ, Hughes ID, Ivanovich N, Tyrer
K, Desrochers C, Klinger L and Shaw L: A systematic review of
multidisciplinary outcomes in the management of chronic low back
pain. Work. 35:349–367. 2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sakai D and Andersson GB: Stem cell
therapy for intervertebral disc regeneration: Obstacles and
solutions. Nat Rev Rheumatol. 11:243–256. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yang X, Chen Y, Guo J, Li J, Zhang P, Yang
H, Rong K, Zhou T, Fu J and Zhao J: Polydopamine nanoparticles
targeting ferroptosis mitigate intervertebral disc degeneration via
reactive oxygen species depletion, iron ions chelation, and GPX4
ubiquitination suppression. Adv Sci (Weinh).
10(e2207216)2023.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Haufe SM and Mork AR: Intradiscal
injection of hematopoietic stem cells in an attempt to rejuvenate
the intervertebral discs. Stem Cells Dev. 15:136–137.
2006.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hoogendoorn RJ, Lu ZF, Kroeze RJ, Bank RA,
Wuisman PI and Helder MN: Adipose stem cells for intervertebral
disc regeneration: Current status and concepts for the future. J
Cell Mol Med. 12:2205–2216. 2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Orozco L, Soler R, Morera C, Alberca M,
Sanchez A and Garcia-Sancho J: Intervertebral disc repair by
autologous mesenchymal bone marrow cells: A pilot study.
Transplantation. 92:822–828. 2011.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Noriega DC, Ardura F, Hernandez-Ramajo R,
Martín-Ferrero MÁ, Sánchez-Lite I, Toribio B, Alberca M, García V,
Moraleda JM, Sánchez A and García-Sancho J: Intervertebral disc
repair by allogeneic mesenchymal bone marrow cells: A randomized
controlled trial. Transplantation. 101:1945–1951. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhou T, Yang X, Chen Z, Yang Y, Wang X,
Cao X, Chen C, Han C, Tian H, Qin A, et al: Prussian blue
nanoparticles stabilize SOD1 from ubiquitination-proteasome
degradation to rescue intervertebral disc degeneration. Adv Sci
(Weinh). 9(e2105466)2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Šećerović A, Ristaniemi A, Cui S, Li Z,
Soubrier A, Alini M, Ferguson SJ, Weder G, Heub S, Ledroit D and
Grad S: Toward the next generation of spine bioreactors: Validation
of an ex vivo intervertebral disc organ model and customized
specimen holder for multiaxial loading. ACS Biomater Sci Eng.
8:3969–3976. 2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Grunhagen T, Wilde G, Soukane DM,
Shirazi-Adl SA and Urban JP: Nutrient supply and intervertebral
disc metabolism. J Bone Joint Surg Am. 88 (Suppl 2):S30–S35.
2006.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Song Y, Lu S, Geng W, Feng X, Luo R, Li G
and Yang C: Mitochondrial quality control in intervertebral disc
degeneration. Exp Mol Med. 53:1124–1133. 2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Roughley PJ: Biology of intervertebral
disc aging and degeneration: Involvement of the extracellular
matrix. Spine (Phila Pa 1976). 29:2691–2699. 2004.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Urban JP, Holm S and Maroudas A: Diffusion
of small solutes into the intervertebral disc: As in vivo study.
Biorheology. 15:203–221. 1978.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ogata K and Whiteside LA: 1980 Volvo award
winner in basic science. Nutritional pathways of the intervertebral
disc. An experimental study using hydrogen washout technique. Spine
(Phila Pa 1976). 6:211–216. 1981.PubMed/NCBI
|
|
41
|
van der Werf M, Lezuo P, Maissen O, van
Donkelaar CC and Ito K: Inhibition of vertebral endplate perfusion
results in decreased intervertebral disc intranuclear diffusive
transport. J Anat. 211:769–774. 2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Rajasekaran S, Babu JN, Arun R, Armstrong
BR, Shetty AP and Murugan S: ISSLS prize winner: A study of
diffusion in human lumbar discs: A serial magnetic resonance
imaging study documenting the influence of the endplate on
diffusion in normal and degenerate discs. Spine (Phila Pa 1976).
29:2654–2667. 2004.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kang R, Li H, Ringgaard S, Rickers K, Sun
H, Chen M, Xie L and Bünger C: Interference in the endplate
nutritional pathway causes intervertebral disc degeneration in an
immature porcine model. Int Orthop. 38:1011–1017. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Yin S, Du H, Zhao W, Ma S, Zhang M, Guan M
and Liu M: Inhibition of both endplate nutritional pathways results
in intervertebral disc degeneration in a goat model. J Orthop Surg
Res. 14(138)2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Hutton WC, Murakami H, Li J, Elmer WA,
Yoon ST, Minamide A, Akamaru T and Tomita K: The effect of blocking
a nutritional pathway to the intervertebral disc in the dog model.
J Spinal Disord Tech. 17:53–63. 2004.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Jiang C, Guo Q, Jin Y, Xu JJ, Sun ZM, Zhu
DC, Lin JH, Tian NF, Sun LJ, Zhang XL and Wu YS: Inhibition of EZH2
ameliorates cartilage endplate degeneration and attenuates the
progression of intervertebral disc degeneration via demethylation
of Sox-9. EBioMedicine. 48:619–629. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Määttä JH, Kraatari M, Wolber L, Niinimäki
J, Wadge S, Karppinen J and Williams FM: Vertebral endplate change
as a feature of intervertebral disc degeneration: A heritability
study. Eur Spine J. 23:1856–1862. 2014.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wang Y, Videman T and Battié MC: ISSLS
prize winner: Lumbar vertebral endplate lesions: Associations with
disc degeneration and back pain history. Spine (Phila Pa 1976).
37:1490–1496. 2012.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Livshits G, Popham M, Malkin I, Sambrook
PN, Macgregor AJ, Spector T and Williams FM: Lumbar disc
degeneration and genetic factors are the main risk factors for low
back pain in women: The UK Twin Spine Study. Ann Rheum Dis.
70:1740–1745. 2011.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Pennicooke B, Moriguchi Y, Hussain I,
Bonssar L and Härtl R: Biological treatment approaches for
degenerative disc disease: A review of clinical trials and future
directions. Cureus. 8(e892)2016.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Mattick JS: Non-coding RNAs: The
architects of eukaryotic complexity. EMBO Rep. 2:986–991.
2001.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Patrushev LI and Kovalenko TF: Functions
of noncoding sequences in mammalian genomes. Biochemistry (Mosc).
79:1442–1469. 2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Palazzo AF and Lee ES: Non-coding RNA:
What is functional and what is junk? Front Genet.
6(2)2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Watson CN, Belli A and Di Pietro V: Small
Non-coding RNAs: New class of biomarkers and potential therapeutic
targets in neurodegenerative disease. Front Genet.
10(364)2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Braicu C, Calin GA and Berindan-Neagoe I:
MicroRNAs and cancer therapy - from bystanders to major players.
Curr Med Chem. 20:3561–3573. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Cătană CS, Pichler M, Giannelli G, Mader
RM and Berindan-Neagoe I: Non-coding RNAs, the Trojan horse in
two-way communication between tumor and stroma in colorectal and
hepatocellular carcinoma. Oncotarget. 8:29519–29534.
2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Guo TF, Zhou MW, Li SH, Ye BL, Chen W and
Fu ZB: Long non-coding RNA for metabolism of bone tissue. Zhongguo
Gu Shang. 31:286–291. 2018.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
58
|
Wang J, Sun Y, Liu J, Yang B, Wang T,
Zhang Z, Jiang X, Guo Y and Zhang Y: Roles of long non-coding RNA
in osteoarthritis (Review). Int J Mol Med. 48(133)2021.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Liu Q, Peng F and Chen J: The role of
exosomal MicroRNAs in the tumor microenvironment of breast cancer.
Int J Mol Sci. 20(3884)2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Lan T, Shiyu-Hu Shen Z, Yan B and Chen J:
New insights into the interplay between miRNAs and autophagy in the
aging of intervertebral discs. Ageing Res Rev.
65(101227)2021.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Guo HY, Guo MK, Wan ZY, Song F and Wang
HQ: Emerging evidence on noncoding-RNA regulatory machinery in
intervertebral disc degeneration: A narrative review. Arthritis Res
Ther. 22(270)2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Wang T, Hao Z, Liu C, Yuan L, Li L, Yin M,
Li Q, Qi Z and Wang Z: LEF1 mediates osteoarthritis progression
through circRNF121/miR-665/MYD88 axis via NF-кB signaling pathway.
Cell Death Dis. 11(598)2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Zhao R, Fu J, Zhu L, Chen Y and Liu B:
Designing strategies of small-molecule compounds for modulating
non-coding RNAs in cancer therapy. J Hematol Oncol.
15(14)2022.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Matsui M and Corey DR: Non-coding RNAs as
drug targets. Nat Rev Drug Discov. 16:167–179. 2017.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Huang W, Li H, Yu Q, Xiao W and Wang DO:
LncRNA-mediated DNA methylation: An emerging mechanism in cancer
and beyond. J Exp Clin Cancer Res. 41(100)2022.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Zhao Y, Ling S, Li J, Zhong G, Du R, Li Y,
Wang Y, Liu C, Jin X, Liu W, et al: 3' untranslated region of
Ckip-1 inhibits cardiac hypertrophy independently of its cognate
protein. Eur Heart J. 42:3786–3799. 2021.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Wu AC, Yang WB, Chang KY, Lee JS, Liou JP,
Su RY, Cheng SM, Hwang DY, Kikkawa U, Hsu TI, et al: HDAC6 involves
in regulating the lncRNA-microRNA-mRNA network to promote the
proliferation of glioblastoma cells. J Exp Clin Cancer Res.
41(47)2022.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Chen X, Gong W, Shao X, Shi T, Zhang L,
Dong J, Shi Y, Shen S, Qin J, Jiang Q and Guo B: METTL3-mediated
m(6)A modification of ATG7 regulates autophagy-GATA4 axis to
promote cellular senescence and osteoarthritis progression. Ann
Rheum Dis. 81:87–99. 2022.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Chen J, Huang T, Liu R, Wang C, Jiang H
and Sun H: Congenital microtia patients: The genetically engineered
exosomes released from porous gelatin methacryloyl hydrogel for
downstream small RNA profiling, functional modulation of microtia
chondrocytes and tissue-engineered ear cartilage regeneration. J
Nanobiotechnology. 20(164)2022.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Li Z, Yu X, Shen J, Chan MT and Wu WK:
MicroRNA in intervertebral disc degeneration. Cell Prolif.
48:278–283. 2015.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Ambros V and Chen X: The regulation of
genes and genomes by small RNAs. Development. 134:1635–1641.
2007.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Zhu Y, Li K, Yan L, He Y, Wang L and Sheng
L: miR-223-3p promotes cell proliferation and invasion by targeting
Arid1a in gastric cancer. Acta Biochim Biophys Sin (Shanghai).
52:150–159. 2020.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Rau CS, Yang JC, Wu SC, Chen YC, Lu TH,
Lin MW, Wu YC, Tzeng SL, Wu CJ and Hsieh CH: Profiling circulating
microRNA expression in a mouse model of nerve allotransplantation.
J Biomed Sci. 20(64)2013.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Guerau-de-Arellano M, Smith KM, Godlewski
J, Liu Y, Winger R, Lawler SE, Whitacre CC, Racke MK and
Lovett-Racke AE: Micro-RNA dysregulation in multiple sclerosis
favours pro-inflammatory T-cell-mediated autoimmunity. Brain.
134(Pt 12):3578–3589. 2011.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Nie H, Zhang K, Xu J, Liao K, Zhou W and
Fu Z: Combining bioinformatics techniques to study diabetes
biomarkers and related molecular mechanisms. Front Genet.
11(367)2020.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Shen P, Yang Y, Liu G, Chen W, Chen J,
Wang Q, Gao H, Fan S, Shen S and Zhao X: CircCDK14 protects against
Osteoarthritis by sponging miR-125a-5p and promoting the expression
of Smad2. Theranostics. 10:9113–9131. 2020.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Li S, Liu J, Liu S, Jiao W and Wang X:
Mesenchymal stem cell-derived extracellular vesicles prevent the
development of osteoarthritis via the circHIPK3/miR-124-3p/MYH9
axis. J Nanobiotechnology. 9(194)2021.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Wu Z, Qiu X, Gao B, Lian C, Peng Y, Liang
A, Xu C, Gao W, Zhang L, Su P, et al: Melatonin-mediated
miR-526b-3p and miR-590-5p upregulation promotes chondrogenic
differentiation of human mesenchymal stem cells. J Pineal Res.
65(e12483)2018.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Chen L, Li Q, Wang J, Jin S, Zheng H, Lin
J, He F, Zhang H, Ma S, Mei J and Yu J: MiR-29b-3p promotes
chondrocyte apoptosis and facilitates the occurrence and
development of osteoarthritis by targeting PGRN. J Cell Mol Med.
21:3347–3359. 2017.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Razmara E, Bitaraf A, Yousefi H, Nguyen
TH, Garshasbi M, Cho WC and Babashah S: Non-Coding RNAs in
cartilage development: An updated review. Int J Mol Sci.
20(4475)2019.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Peng B, Hou S, Shi Q and Jia L: The
relationship between cartilage end-plate calcification and disc
degeneration: An experimental study. Chin Med J (Engl).
114:308–312. 2001.PubMed/NCBI
|
|
84
|
Bian Q, Liang QQ, Wan C, Hou W, Li CG,
Zhao YJ, Lu S, Shi Q and Wang YJ: Prolonged upright posture induces
calcified hypertrophy in the cartilage end plate in rat lumbar
spine. Spine (Phila Pa 1976). 36:2011–2020. 2011.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Feng C, Liu M, Fan X, Yang M, Liu H and
Zhou Y: Intermittent cyclic mechanical tension altered the microRNA
expression profile of human cartilage endplate chondrocytes. Mol
Med Rep. 17:5238–5246. 2018.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Chen H, Wang J, Hu B, Wu X, Chen Y, Li R
and Yuan W: MiR-34a promotes Fas-mediated cartilage endplate
chondrocyte apoptosis by targeting Bcl-2. Mol Cell Biochem.
406:21–30. 2015.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Onodera K, Takahashi I, Sasano Y, Bae JW
and Mitani H, Kagayama M and Mitani H: Stepwise mechanical
stretching inhibits chondrogenesis through cell-matrix adhesion
mediated by integrins in embryonic rat limb-bud mesenchymal cells.
Eur J Cell Biol. 84:45–58. 2005.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Bleuel J, Zaucke F, Brüggemann GP and
Niehoff A: Effects of cyclic tensile strain on chondrocyte
metabolism: A systematic review. PLoS One.
10(e0119816)2015.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Yuan W, Che W, Jiang YQ, Yuan FL, Wang HR,
Zheng GL, Li XL and Dong J: Establishment of intervertebral disc
degeneration model induced by ischemic sub-endplate in rat tail.
Spine J. 15:1050–1059. 2015.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Xiao L, Xu S, Xu Y, Liu C, Yang B, Wang J
and Xu H: TGF-β/SMAD signaling inhibits intermittent cyclic
mechanical tension-induced degeneration of endplate chondrocytes by
regulating the miR-455-5p/RUNX2 axis. J Cell Biochem.
119:10415–10425. 2018.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Liu MH, Sun C, Yao Y, Fan X, Liu H, Cui
YH, Bian XW, Huang B and Zhou Y: Matrix stiffness promotes
cartilage endplate chondrocyte calcification in disc degeneration
via miR-20a targeting ANKH expression. Sci Rep.
6(25401)2016.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Zhang F, Zhao X, Shen H and Zhang C:
Molecular mechanisms of cell death in intervertebral disc
degeneration (Review). Int J Mol Med. 37:1439–1448. 2016.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Chen WK, Yu XH, Yang W, Wang C, He WS, Yan
YG, Zhang J and Wang WJ: lncRNAs: Novel players in intervertebral
disc degeneration and osteoarthritis. Cell Prolif.
50(e12313)2017.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Sheng B, Yuan Y, Liu X, Zhang Y, Liu H,
Shen X, Liu B and Chang L: Protective effect of estrogen against
intervertebral disc degeneration is attenuated by miR-221 through
targeting estrogen receptor α. Acta Biochim Biophys Sin (Shanghai).
50:345–354. 2018.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Chen Y, Chen Q, Zhong M, Xu C, Wu Y and
Chen R: miR-637 inhibits osteogenic differentiation of human
intervertebral disc cartilage endplate stem cells by targeting
WNT5A. J Invest Surg. 35:1313–1321. 2022.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Chen D and Jiang X: Exosomes-derived
miR-125-5p from cartilage endplate stem cells regulates autophagy
and ECM metabolism in nucleus pulposus by targeting SUV38H1. Exp
Cell Res. 414(113066)2022.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Wang B, Ji D, Xing W, Li F, Huang Z, Zheng
W, Xue J, Zhu Y and Yang X: miR-142-3p and HMGB1 are negatively
regulated in proliferation, apoptosis, migration, and autophagy of
cartilage endplate cells. Cartilage. 13 (2_suppl):592S–603S.
2021.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Qian X, Zhao J, Yeung PY, Zhang QC and
Kwok CK: Revealing lncRNA structures and interactions by
sequencing-based approaches. Trends Biochem Sci. 44:33–52.
2019.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Khan S, Masood M, Gaur H, Ahmad S and Syed
MA: Long non-coding RNA: An immune cells perspective. Life Sci.
271(119152)2021.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Bridges MC, Daulagala AC and Kourtidis A:
LNCcation: lncRNA localization and function. J Cell Biol.
220(e202009045)2021.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Huang H, Xing D, Zhang Q, Li H and Lin J,
He Z and Lin J: LncRNAs as a new regulator of chronic
musculoskeletal disorder. Cell Prolif. 54(e13113)2021.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Liu X, Li W, Jiang L, Lü Z, Liu M, Gong L,
Liu B, Liu L and Yin X: Immunity-associated long non-coding RNA and
expression in response to bacterial infection in large yellow
croaker (Larimichthys crocea). Fish Shellfish Immunol. 94:634–642.
2019.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Li Z, Li X, Chen C, Li S, Shen J, Tse G,
Chan MTV and Wu WKK: Long non-coding RNAs in nucleus pulposus cell
function and intervertebral disc degeneration. Cell Prolif.
51(e12483)2018.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Zhu J, Zhang X, Gao W, Hu H, Wang X and
Hao D: lncRNA/circRNA-miRNA-mRNA ceRNA network in lumbar
intervertebral disc degeneration. Mol Med Rep. 20:3160–3174.
2019.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Wan ZY, Song F, Sun Z, Chen YF, Zhang WL,
Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al: Aberrantly
expressed long noncoding RNAs in human intervertebral disc
degeneration: A microarray related study. Arthritis Res Ther.
16(465)2014.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Kitagawa M, Kitagawa K, Kotake Y, Niida H
and Ohhata T: Cell cycle regulation by long non-coding RNAs. Cell
Mol Life Sci. 70:4785–4794. 2013.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Solé C, Nadal-Ribelles M, de Nadal E and
Posas F: A novel role for lncRNAs in cell cycle control during
stress adaptation. Curr Genet. 61:299–308. 2015.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Guiducci G and Stojic L: Long Noncoding
RNAs at the crossroads of cell cycle and genome integrity. Trends
Genet. 37:528–546. 2021.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Ballarino M, Morlando M, Fatica A and
Bozzoni I: Non-coding RNAs in muscle differentiation and
musculoskeletal disease. J Clin Invest. 126:2021–2030.
2016.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Delás MJ, Sabin LR, Dolzhenko E, Knott SR,
Munera Maravilla E, Jackson BT, Wild SA, Kovacevic T, Stork EM,
Zhou M, et al: lncRNA requirements for mouse acute myeloid leukemia
and normal differentiation. Elife. 6(e25607)2017.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Deniz E and Erman B: Long noncoding RNA
(lincRNA), a new paradigm in gene expression control. Funct Integr
Genomics. 17:135–143. 2017.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Mondal T, Subhash S, Vaid R, Enroth S,
Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, et al:
MEG3 long noncoding RNA regulates the TGF-β pathway genes through
formation of RNA-DNA triplex structures. Nat Commun.
6(7743)2015.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Mi D, Cai C, Zhou B, Liu X, Ma P, Shen S,
Lu W and Huang W: Long non-coding RNA FAF1 promotes intervertebral
disc degeneration by targeting the Erk signaling pathway. Mol Med
Rep. 17:3158–3163. 2018.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Yuan J, Jia J, Wu T, Liu X, Hu S, Zhang J,
Ding R, Pang C and Cheng X: Comprehensive evaluation of
differential long non-coding RNA and gene expression in patients
with cartilaginous endplate degeneration of cervical vertebra. Exp
Ther Med. 20(260)2020.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Li B, Balasubramanian K, Krakow D and Cohn
DH: Genes uniquely expressed in human growth plate chondrocytes
uncover a distinct regulatory network. BMC Genomics.
18(983)2017.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Gu W, Zhu Q, Gao X and Brown MD:
Simulation of the progression of intervertebral disc degeneration
due to decreased nutritional supply. Spine (Phila Pa 1976).
39:E1411–E1417. 2014.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Fields AJ, Berg-Johansen B, Metz LN,
Miller S, La B, Liebenberg EC, Coughlin DG, Graham JL, Stanhope KL,
Havel PJ and Lotz JC: Alterations in intervertebral disc
composition, matrix homeostasis and biomechanical behavior in the
UCD-T2DM rat model of type 2 diabetes. J Orthop Res. 33:738–746.
2015.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Agius R, Galea R and Fava S: Bone mineral
density and intervertebral disc height in type 2 diabetes. J
Diabetes Complications. 30:644–650. 2016.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Jiang Z, Lu W, Zeng Q, Li D, Ding L and Wu
J: High glucose-induced excessive reactive oxygen species promote
apoptosis through mitochondrial damage in rat cartilage endplate
cells. J Orthop Res. 36:2476–2483. 2018.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Li X, Wu FR, Xu RS, Hu W, Jiang DL, Ji C,
Chen FH and Yuan FL: Acid-sensing ion channel 1a-mediated calcium
influx regulates apoptosis of endplate chondrocytes in
intervertebral discs. Expert Opin Ther Targets. 18:1–14.
2014.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Yuan FL, Wang HR, Zhao MD, Yuan W, Cao L,
Duan PG, Jiang YQ, Li XL and Dong J: Ovarian cancer G
protein-coupled receptor 1 is involved in acid-induced apoptosis of
endplate chondrocytes in intervertebral discs. J Bone Miner Res.
29:67–77. 2014.PubMed/NCBI View Article : Google Scholar
|
|
124
|
Jiang Z, Zeng Q, Li D, Ding L, Lu W, Bian
M and Wu J: Long non-coding RNA MALAT1 promotes high
glucose-induced rat cartilage endplate cell apoptosis via the
p38/MAPK signalling pathway. Mol Med Rep. 21:2220–2226.
2020.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Grabowski PJ, Zaug AJ and Cech TR: The
intervening sequence of the ribosomal RNA precursor is converted to
a circular RNA in isolated nuclei of Tetrahymena. Cell. 23:467–476.
1981.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Ford E and Ares M Jr: Synthesis of
circular RNA in bacteria and yeast using RNA cyclase ribozymes
derived from a group I intron of phage T4. Proc Natl Acad Sci USA.
91:3117–3121. 1994.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Kos A, Dijkema R, Arnberg AC, van der
Meide PH and Schellekens H: The hepatitis delta (delta) virus
possesses a circular RNA. Nature. 323:558–560. 1986.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Xiao MS, Ai Y and Wilusz JE: Biogenesis
and functions of circular RNAs come into focus. Trends Cell Biol.
30:226–240. 2020.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461.
2014.PubMed/NCBI View Article : Google Scholar
|
|
134
|
O'Conor CJ, Case N and Guilak F:
Mechanical regulation of chondrogenesis. Stem Cell Res Ther.
4(61)2013.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Xia DD, Lin SL, Wang XY, Wang YL, Xu HM,
Zhou F and Tan J: Effects of shear force on intervertebral disc: An
in vivo rabbit study. Eur Spine J. 24:1711–1719. 2015.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Xiao L, Ding B, Xu S, Gao J, Yang B, Wang
J and Xu H: circRNA_0058097 promotes tension-induced degeneration
of endplate chondrocytes by regulating HDAC4 expression through
sponge adsorption of miR-365a-5p. J Cell Biochem. 121:418–429.
2020.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Ren S, Lin P, Wang J, Yu H, Lv T, Sun L
and Du G: Circular RNAs: Promising molecular biomarkers of human
aging-related diseases via functioning as an miRNA Sponge. Mol Ther
Methods Clin Dev. 18:215–229. 2020.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Xu D, Ma X, Sun C, Han J, Zhou C, Wong SH,
Chan MTV and Wu WKK: Circular RNAs in intervertebral disc
degeneration: An updated review. Front Mol Biosci.
8(781424)2022.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Zhang J, Hu S, Ding R, Yuan J, Jia J, Wu T
and Cheng X: CircSNHG5 Sponges Mir-495-3p and Modulates CITED2 to
protect cartilage endplate from degradation. Front Cell Dev Biol.
9(668715)2021.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Larsson ME and Nordholm LA: Responsibility
for managing musculoskeletal disorders-a cross-sectional postal
survey of attitudes. BMC Musculoskelet Disord.
9(110)2008.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Hu B, Xiao L, Wang C, Liu C, Zhang Y, Ding
B, Gao D, Lu Y and Xu H: Circ_0022382 ameliorated intervertebral
disc degeneration by regulating TGF-β3 expression through sponge
adsorption of miR-4726-5p. Bone. 154(116185)2022.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Zhang H, Wu S, Chen W, Hu Y, Geng Z and Su
J: Bone/cartilage targeted hydrogel: Strategies and applications.
Bioact Mater. 23:156–169. 2022.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Guo J, Wang F, Hu Y, Luo Y, Wei Y, Xu K,
Zhang H, Liu H, Bo L, Lv S, et al: Exosome-based bone-targeting
drug delivery alleviates impaired osteoblastic bone formation and
bone loss in inflammatory bowel diseases. Cell Rep Med.
4(100881)2023.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Ji ML, Jiang H, Zhang XJ, Shi PL, Li C, Wu
H, Wu XT, Wang YT, Wang C and Lu J: Preclinical development of a
microRNA-based therapy for intervertebral disc degeneration. Nat
Commun. 9(5051)2018.PubMed/NCBI View Article : Google Scholar
|
|
146
|
Hu Y, Li X, Zhang Q, Gu Z, Luo Y, Guo J,
Wang X, Jing Y, Chen X and Su J: Exosome-guided bone targeted
delivery of Antagomir-188 as an anabolic therapy for bone loss.
Bioact Mater. 6:2905–2913. 2021.PubMed/NCBI View Article : Google Scholar
|
|
147
|
Wang Y, Chu X and Wang B: Recombinant
adeno-associated virus-based gene therapy combined with tissue
engineering for musculoskeletal regenerative medicine. Biomater
Transl. 2:19–29. 2021.PubMed/NCBI View Article : Google Scholar
|
|
148
|
Ahn J, Park EM, Kim BJ, Kim JS, Choi B,
Lee SH and Han I: Transplantation of human Wharton's jelly-derived
mesenchymal stem cells highly expressing TGFβ receptors in a rabbit
model of disc degeneration. Stem Cell Res Ther.
6(190)2015.PubMed/NCBI View Article : Google Scholar
|