Introduction
Heart failure (HF) is a long-standing public health
issue, causing high morbidity and mortality worldwide (1). More than 30 million individuals are
currently suffering from HF, and this number is expected to
continue rising owing to the global aging population (2). HF is characterized by the inability
of the heart to pump sufficient blood for the metabolic demands of
the body, resulting in dyspnea, fatigue, poor exercise tolerance
and fluid retention (3,4). Therefore, HF poses a considerable
economic burden to global healthcare systems (1,5-7).
The development of HF is usually accompanied by the synchronous
reprogramming of gene expression, associated with modifications in
multiple genes and signaling pathways involved in HF pathogenesis
(8-11).
Dilated cardiomyopathy (DCM), one of the leading factors associated
with HF, is characterized by left ventricular dilation combined
with systolic dysfunction (12-15).
However, the overall picture of myocardial gene co-expression
signatures in DCM-induced HF remains unclear. Therefore, detecting
key genes and pathways in the pathogenesis of DCM-induced HF is
necessary and meaningful, and may perform an important role in the
prevention, diagnosis and treatment of DCM-induced HF.
With the development of second-generation sequencing
technologies, several gene chip techniques have been used in both
research and clinical settings for the treatment of cardiovascular
diseases (16-19).
Clarke et al (16) found
that two lipoprotein variants were associated with an increased
risk of coronary disease using a novel gene chip containing
single-nucleotide polymorphisms. Kuehl et al (17) identified distinctive microRNAs to
assess the risk of virus persistence and progressive clinical
deterioration in the course of enterovirus cardiomyopathy using
microRNAs gene chips. Weighted gene co-expression network analysis
(WGCNA) has been introduced as a novel and powerful systems biology
method designed for constructing a co-expression network among
identified genes (20). WGCNA
divides genes into separate modules according to gene expression
patterns that connect with the complex changes of the clinical
phenotype. Compared with other bioinformatics analysis methods,
this technique offers a more convenient and effective way to obtain
key modules that are more closely related to clinical traits;
subsequently, hub genes and key pathways can be identified within
these modules (20,21). WGCNA has been widely used to
identify novel biomarkers and pathways in a number of
cardiovascular diseases, including human atrial fibrillation,
ischemic cardiomyopathy and acute myocardial infarction (22-24).
In the present study, WGCNA was performed to
identify key genes and biological pathways associated with
DCM-induced HF. The potential function of the key module was
explored using Gene Ontology (GO) annotation, Gene Set Enrichment
Analysis (GSEA) and the Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway enrichment analyses with the clusterProfiler package
in R. The diagnostic efficacy of key genes was evaluated using a
receiver operating characteristic (ROC) curve. The key genes were
also verified in the GSE116250 dataset and in vitro
experiments. Furthermore, the analysis of infiltrated immune cells
in heart tissues with DCM-induced HF was performed using the
CIBERSORT method.
Materials and methods
Data collection and processing
A flow chart of the present study is presented in
Fig. 1. Microarray datasets
GSE79962 and GSE116250 were extracted from the GEO database
(https://www.ncbi.nlm.nih.gov/geo). The
GSE79962 dataset, comprised of 9 patients with DCM-induced HF and
11 non-failing donors, was used to explore novel biomarkers
(25). The GSE116250 dataset
comprised 37 patients with DCM-induced HF and 14 control patients
and was served as the independent testing set to validate the
expressions and diagnostic efficacy of key genes (26,27).
The selection criteria for these datasets were: i) Microarray
dataset had to be available for both patients with DCM-induced HF
and healthy controls; and ii) an annotation file for the platform
was required for each dataset. All data analyses and processing
were conducted using R software.
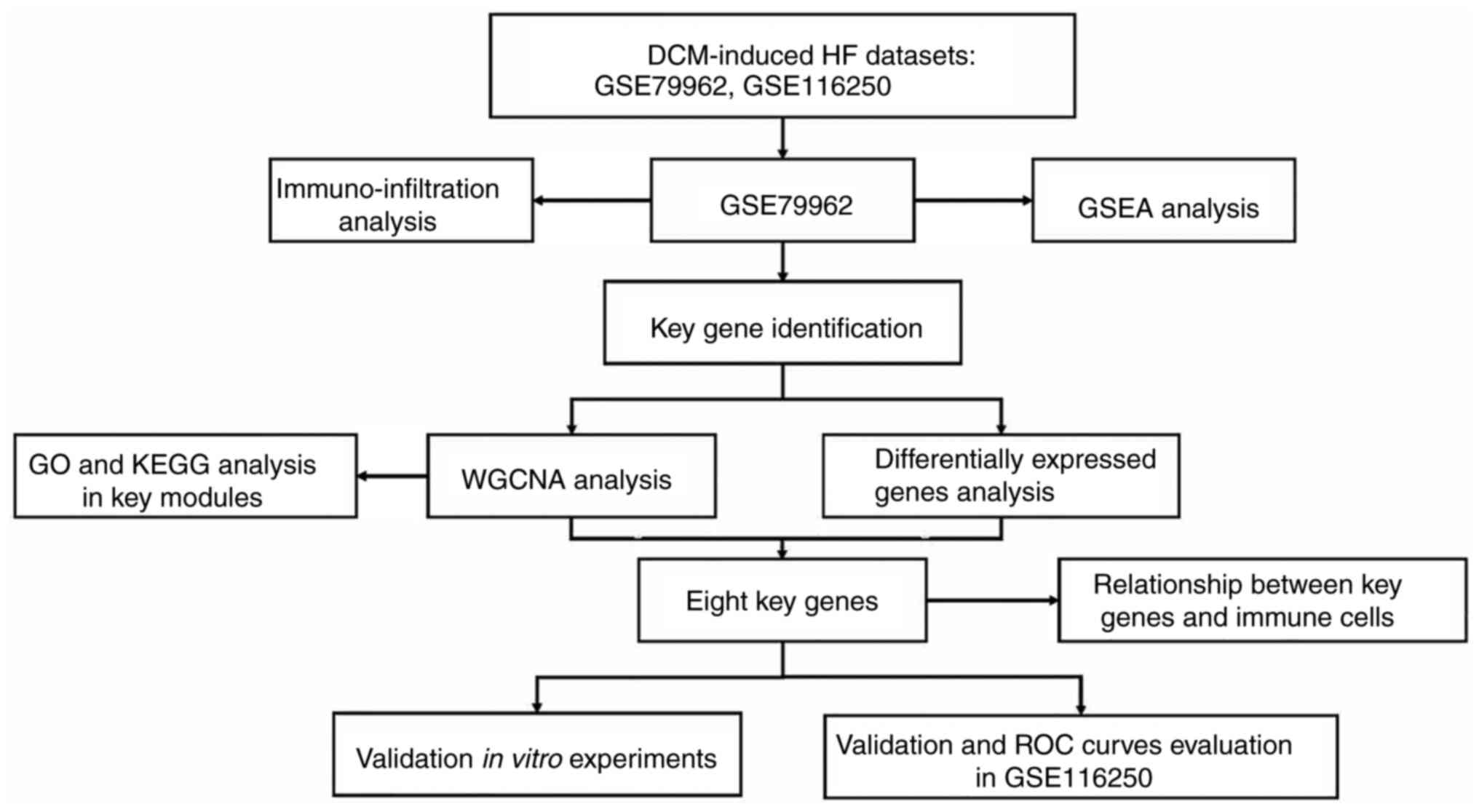 | Figure 1Flow chart of the study. In this
study, two DCM-induced HF datasets were collected. WGCNA and
differentially expressed genes analysis were used to identify key
genes in the GSE79962 dataset. The relative fractions of
infiltrated immune cells were evaluated the CIBERSORT algorithm.
The relationship between key genes and immune cells was calculated
by Spearman's correlation analysis. The potential function of the
key module was explored using GO, GSEA and KEGG analysis. Moreover,
key genes were validated in the GSE116250 dataset and in
vitro experiments. The diagnostic efficacy of key genes was
evaluated using a ROC curve in the GSE116250 dataset. DCM, dilated
cardiomyopathy; GO, gene ontology; GSEA, Gene Set Enrichment
Analysis; HF, heart failure; KEGG, Kyoto encyclopedia of genes and
genomes; ROC, receiver operating characteristic; WGCNA, weighted
gene co-expression network analysis. |
Co-expression network
construction
Co-expression networks were established for 4,343
genes (the top 25% of rank genes with the largest variance) in the
GSE79962 dataset using the WGCNA package v1.70-3 (https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA).
The soft threshold (β) was set to 9 to construct a co-expression
network that conformed to the scale-free distribution when the
degree of independence was 0.9. Next, an adjacency matrix was
constructed by raising the correlation matrix to the power of 9,
and then a topological overlap matrix (TOM) was used to measure
similarity based on the adjacency matrix. Genes were hierarchically
clustered and visualized in a dendrogram according to the
dissimilarity TOM. Hierarchical clustering was performed to obtain
modules using a dynamic tree cutting algorithm with a minimum
module size of 30 for the gene dendrogram. The modules with similar
expression profile were merged, and 24 modules were subsequently
obtained.
Functional enrichment of the key
modules
Gene significance (GS) is defined as the absolute
value of the correlation between between the gene expression and
clinical traits. Module significance (MS) is the average GS in a
specific module, which represents the correlation between the
module and clinical traits (20).
The module with the highest MS value was considered the key module
most relevant to DCM-induced HF. To explore the potential
mechanisms and functions of the target modules, GO functional term
and KEGG pathway enrichment analyses were performed using the R
package ‘clusterprofiler’ (v 4.0.5) (28).
GSEA
The related biological pathways in DCM-induced HF
from the GSE79962 dataset were further analyzed using GSEA in the R
package ‘clusterprofiler’ (v 4.0.5).
Differentially expressed genes
(DEGs)
The ‘limma’ R package was used to identify DEGs
between healthy individuals and patients with DCM-induced HF in the
GSE79962 dataset. An adjusted P<0.05 and log(fold change)
>1.5 were set as the cut-off values for DEG screening.
Identification of key genes
Key genes were mined according to the threshold
value of module membership (MM), GS and DEGs analysis. MM refers to
the Pearson's correlation coefficient between genes and the module
eigengene, where MM reflects the module connectivity of each gene.
GS refers to the correlation coefficient between genes and clinical
traits, representing the correlation between each gene and
DCM-induced HF (20). Hence, genes
with larger absolute GS and MM values were associated with
DCM-induced HF. Genes with |MM| >0.8 and |GS|>0.8 in the
clinically relevant gene modules were defined as hub genes.
Finally, the overlapping parts of hub genes in the crucial modules
and DEGs were considered as key genes and visualized using a Venn
diagram.
Validation of key genes and ROC curve
analyses
The GSE116250 dataset was used to verify the
expressions of the key genes. In addition, the ROC curves and the
area under the curve (AUC) were performed to calculate the
diagnostic value of key genes identified using ‘pROC’ (v 1.18.0) R
package.
Immune infiltration analysis
To estimate the relative fractions of infiltrating
immune cells in DCM-induced HF, the CIBERSORT algorithm was used to
determine the characteristics of immune cell infiltrations
(29).
Correlation analysis between
diagnostic markers and immune cells
Spearman's correlation analysis was performed to
examine the correlation between immune cells and diagnostic
markers.
Validation of the identified key genes
in vitro
AC16 is a proliferating human cardiomyocyte cell
line from human ventricular tissue. They can be differentiated
in vitro and used to study molecular mechanism of
cardiomyocytes in physiological and pathological settings (30). The AC16 cells were purchased from
the Shanghai EK-Bioscience Biotechnology Co., Ltd. The cells were
cultivated in a humidified atmosphere with 5% CO2 at
37˚C. The cells were cultured in six-well plates in DMEM
(MilliporeSigma) containing 8% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin-streptomycin solution
(MilliporeSigma). Doxorubicin (DOX; 2 µM; MedChemExpress) was used
to stimulate AC16 cells at 37˚C in a humidified atmosphere of 5%
CO2 for 24 h.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was isolated from AC16 cells using the RNA
Rapid Extraction Kit (ShangHai YiShan Biotechnology Co., Ltd).
PrimeScript™ RT Master Mix (cat. no. RR036A; Takara Bio, Inc.) was
used to synthesize cDNA according to the manufacturer's protocol.
SYBR Premix (cat. no. RR420A; Takara Bio, Inc.) was used for qPCR
(reaction conditions: 95˚C pre-denaturation for 30 sec, 95˚C
denaturation for 5 sec and 60˚C annealing for 31 sec, for 40
cycles). The relative expression levels of genes were assessed
using the 2-ΔΔCq method; GAPDH was used as an endogenous
loading control (31). The primers
used are listed in Table SI.
Western blotting
AC16 cells were cultured in 6-well plates to 80%
density and then incubated with 2 µM DOX at 37˚C for 24 h.
Subsequently, the total cell proteins were extracted from AC16
cells by RIPA Lysate (Beyotime Institute of Biotechnology).
SDS-PAGE gel preparation kit (cat. no. P0012A; Beyotime Institute
of Biotechnology) was used to prepare the gel (5% stacking gel, 12%
separating gel concentration), and then 20 µg protein per lane was
added for electrophoresis on 0.22-µM PVDF membranes
(MilliporeSigma). The membranes were blocked with 5% skimmed milk
for 2 h at room temperature, and then incubated in anti-BCL2 (cat.
no. 26593; 1:2,500; Proteintech Group, Inc.), anti-BAX (cat. no.
50599; 1:1,000; Proteintech Group, Inc.), anti-atrial natriuretic
peptide (ANP; cat. no. 27426; 1:1,000; Proteintech Group, Inc.) and
HRP-conjugated β-actin (cat. no. HRP-60008; 1:1,000; Proteintech
Group, Inc.) overnight at 4˚C. HRP-conjugated Affinipure Goat
Anti-Rabbit IgG (cat. no. SA00001-2; 1:5,000; Proteintech Group,
Inc.) was added at room temperature for 2 h. After washing,
proteins were visualized using an ECL luminescence kit (cat. no.
WBKLS0500; MilliporSigma).
TUNEL assay
AC16 cells were cultured in 24-well plates to 80%
density and then incubated with 2 µM DOX for 24 h. Apoptosis was
also evaluated with a fluorescence microscope (five fields of view)
using One Step TUNEL Apoptosis Detection Kit (cat. no. C1086;
Beyotime Institute of Biotechnology) according to the
manufacturer's instructions.
Cell Counting Kit-8 (CCK-8) assay
AC16 cells were inoculated into 96-well plates at a
density of 1x104 cells/well (100 µl/well). The cells
were cultured with various concentrations of DOX (0, 0.5, 1, 2 and
4 µM/ml) at 37˚C for 24 h. Next, the cells were incubated with 10
µl CCK-8 reagent (Dojindo Laboratories, Inc.) at 37˚C for 1 h in a
humidified CO2 incubator. The absorbance (optical
density) value was analyzed at 450 nm, according to the
manufacturer's instructions.
Statistical analysis
Data are presented as the mean ± SD; Microsoft Excel
software 2019 (Microsoft Corporation) and GraphPad Prism 8 software
(GraphPad Software; Dotmatics) were used for data analysis. The
diagnostic value of key genes was assessed by ROC curve. The
differences between two groups were assessed by unpaired Student's
t-test, and one-way ANOVA with Bonferroni's multiple comparison
post hoc test was used to compare multiple groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Gene co-expression module construction
using WGCNA
From the 20 samples (9 patients with DCM-induced HF
and 11 control patients) in the GSE79962 dataset, the top 4,343
genes were identified and used to construct a co-expression
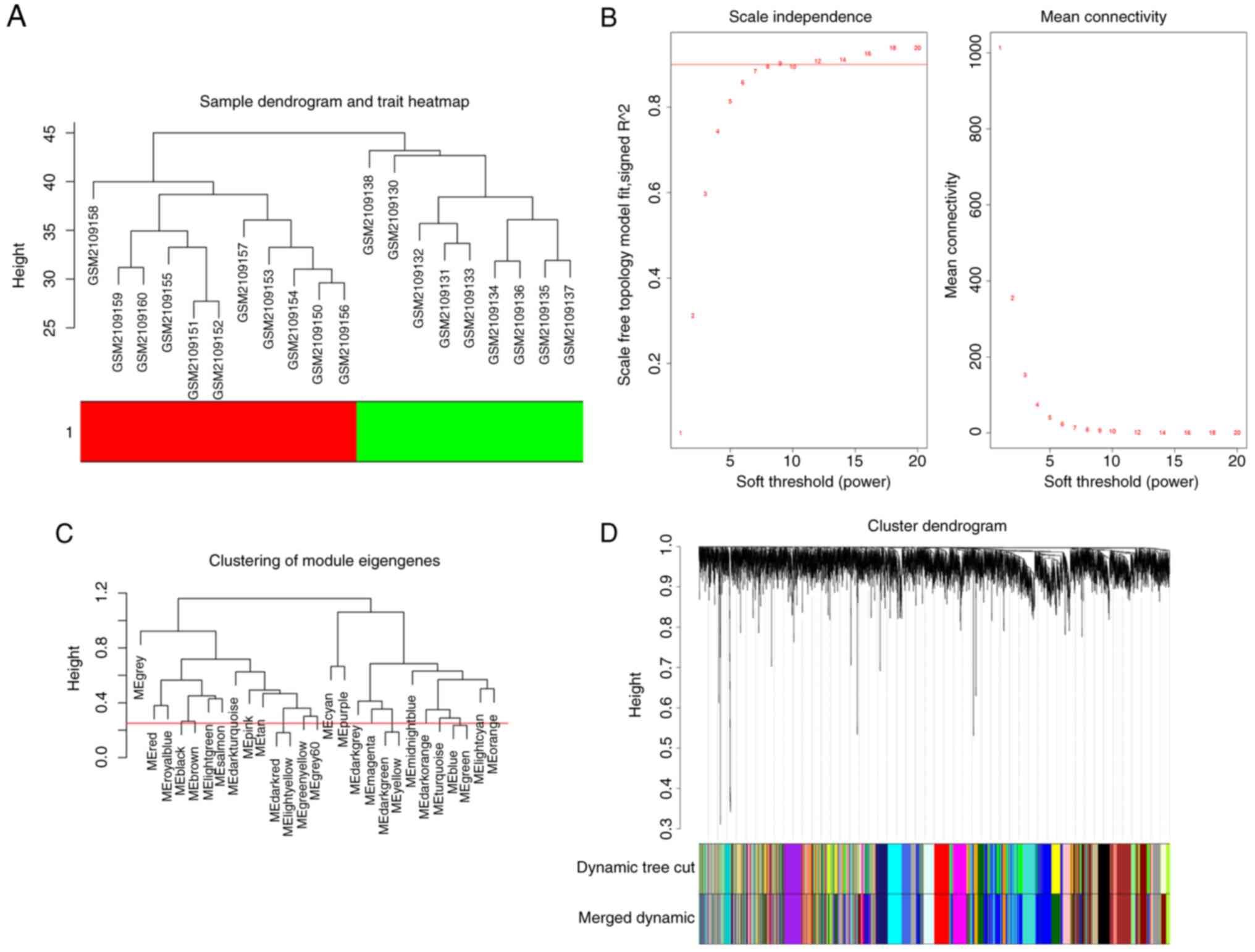
network. The results of the cluster analysis of the samples are
presented in Fig. 2A, which shows
a clear distinction between the samples from patients with
DCM-induced HF and the control group. An appropriate
softthresholding was screened out by analysis of scale independence
and mean connectivity for various soft-threshold powers (β). The
soft-thresholding power (β) was set as 9 (R2=0.9) to
construct a scale-free co-expression network (Fig. 2B). MergeCutHeight was used as the
dendrogram cut height for module merging. Modules with similar
expression patterns were merged. By setting the mergeCutHeight as
0.25, 24 modules were obtained (Fig.
2C and D).
Identification and functional
annotation of the blue module corresponding to DCM-induced HF
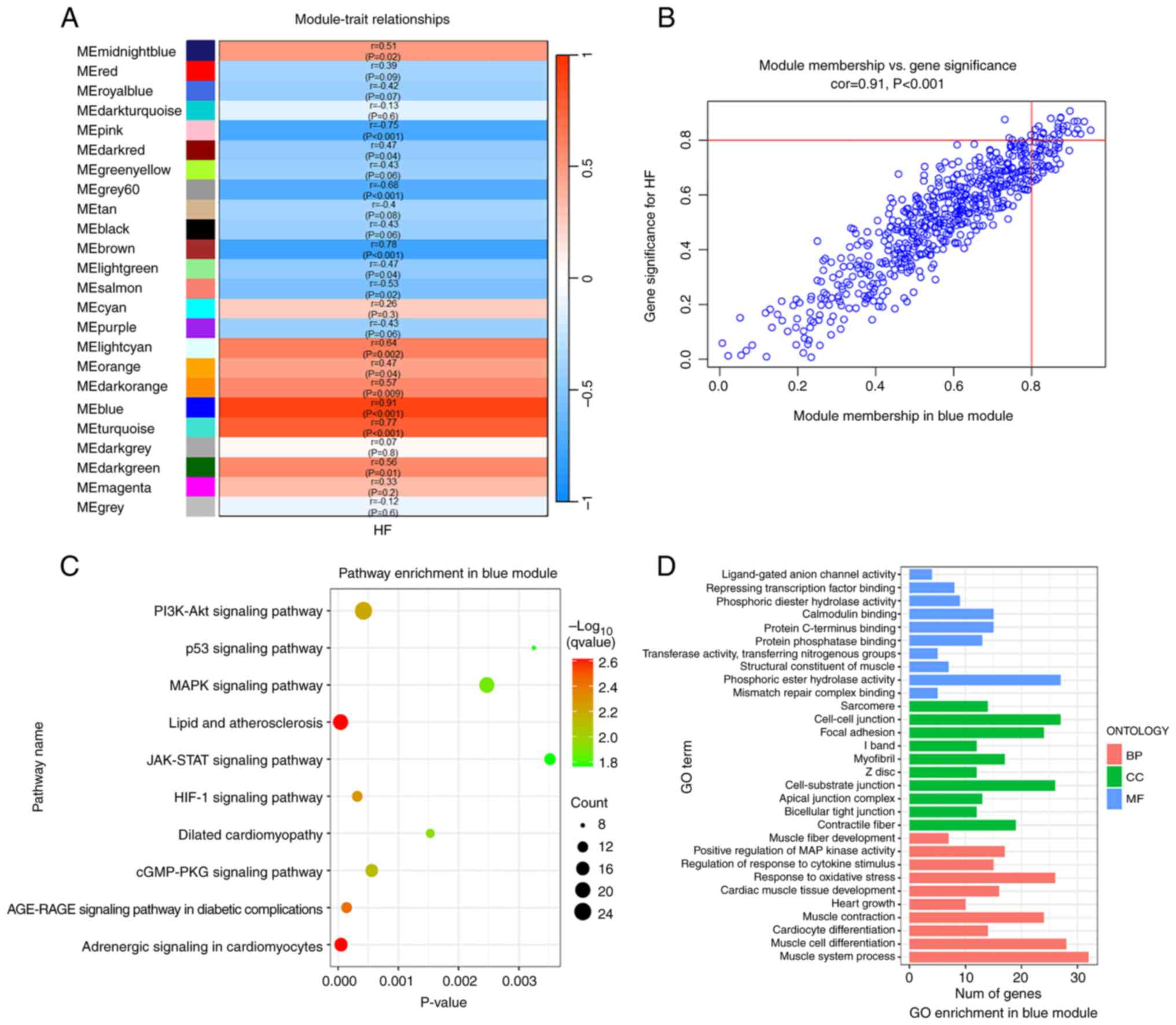
The aforementioned data indicated that the blue
module showed the strongest correlation with DCM-induced HF
(r=0.91; P<0.001; Fig. 3A and
B). Therefore, 602 genes in the
blue module were extracted to perform functional annotation
analysis. KEGG pathway analysis revealed that that the blue module
was enriched in the ‘p53 signaling pathway’, ‘MAPK signaling
pathway’, ‘AGE-RAGE signaling pathway in diabetic complications’,
‘Adrenergic signaling in cardiomyocytes’, ‘JAK/STAT signaling
pathway’ and ‘cGMP/PKG signaling pathway’ (Fig. 3C). In addition, GO enrichment
analysis indicated that the most significant functional terms in
the blue module were mainly ‘cardiac muscle tissue development’,
‘response to oxidative stress’, ‘muscle contraction’, ‘focal
adhesion’ and ‘phosphoric ester hydrolase activity’ (Fig. 3D).
GSEA
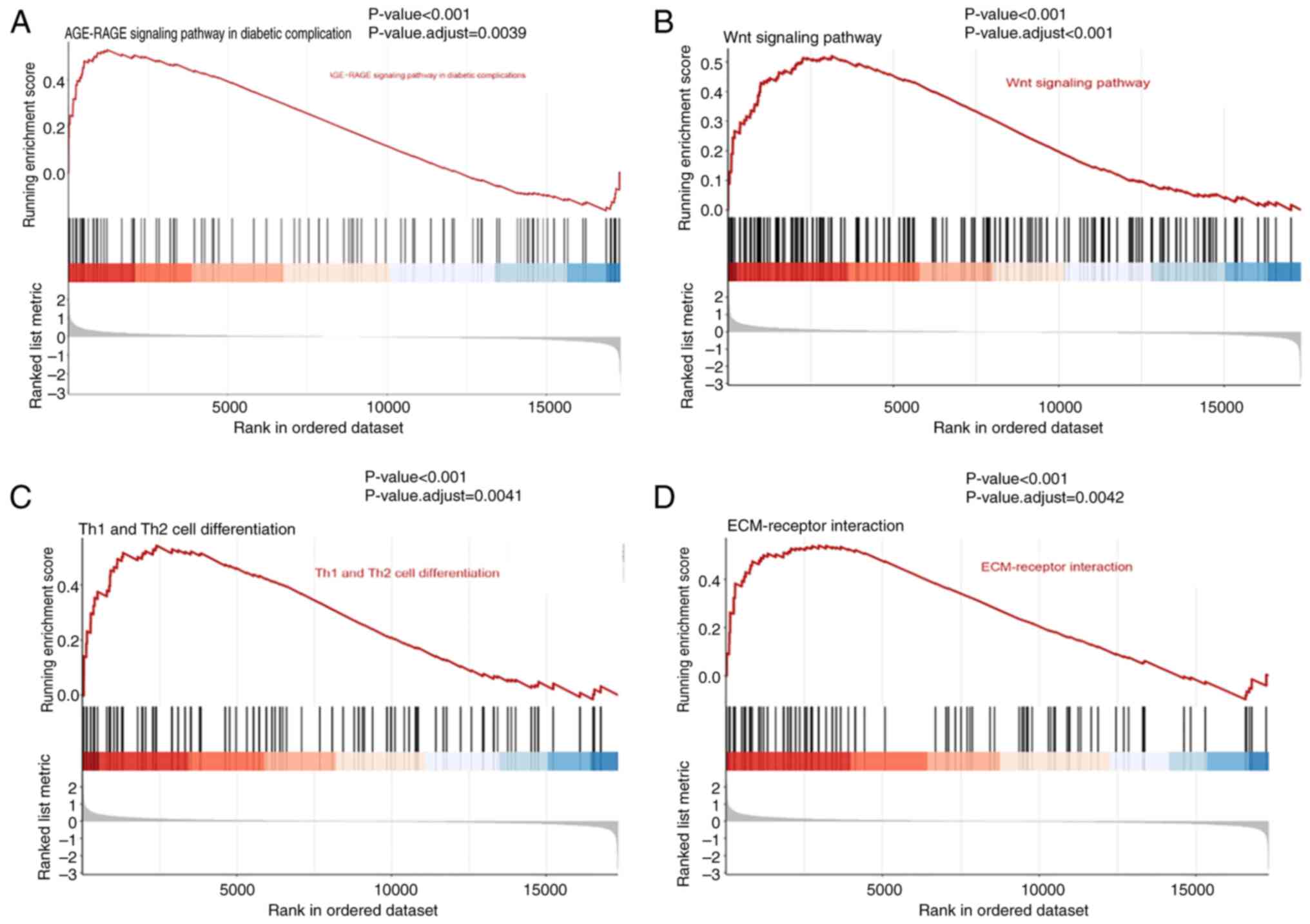
To further explore the key biological pathways
underlying the development of DCM-induced HF, GSEA was performed on
the GSE79962 dataset. The results showed that the pathways of
‘AGE-RAGE signaling pathway in diabetic complications’, ‘Wnt
signaling pathway’, ‘Th1 and Th2 cell differentiation’ and
‘ECM-receptor interaction’ were enriched in patients with
DCM-induced HF (Fig. 4).
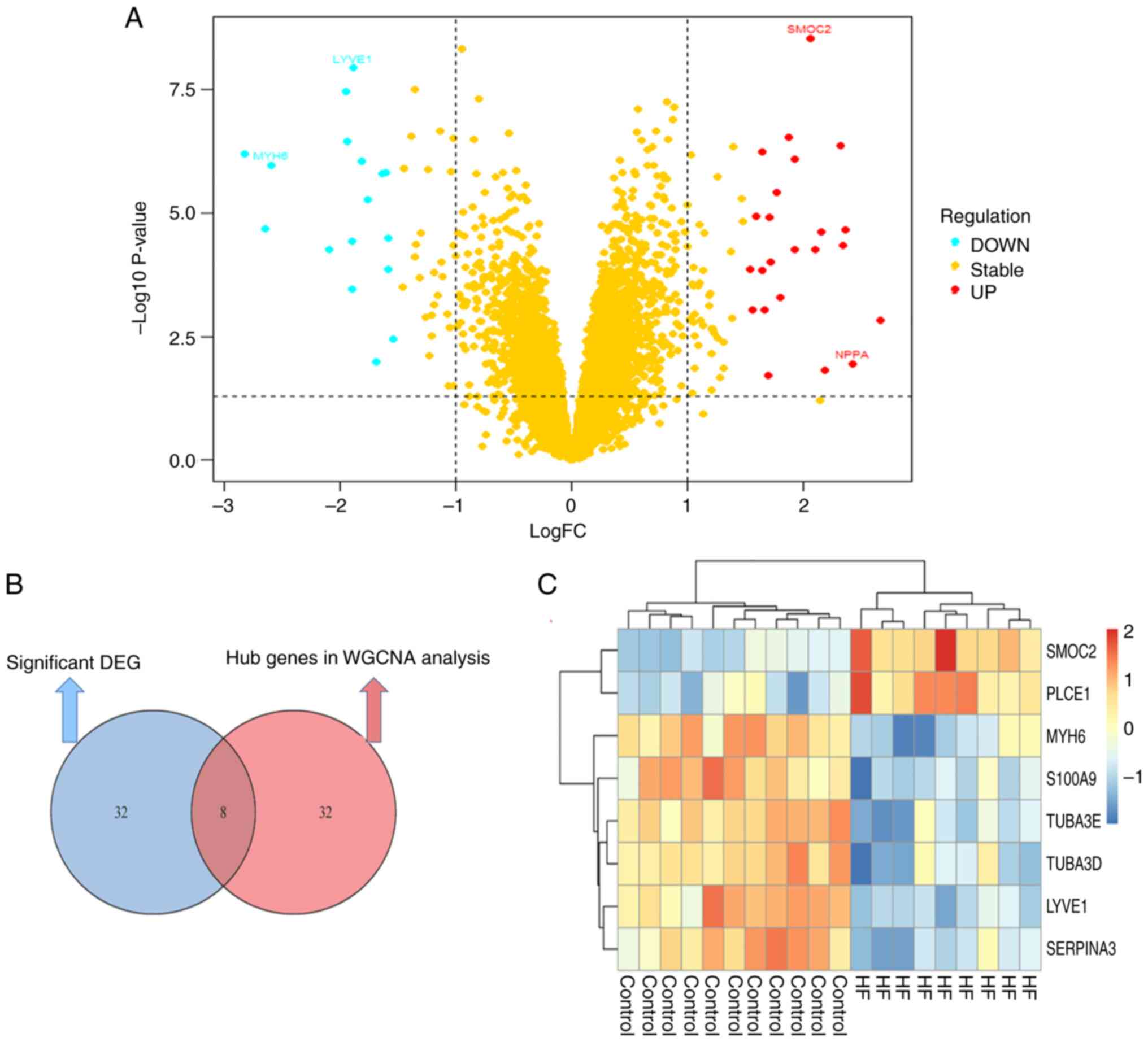
Key genes identification
The blue module, which contained 602 genes, was
regarded as the most relevant to DCM-induced HF compared with the
other modules. Using |MM|>0.8 and |GS|>0.8 as cut-off values,
40 genes were identified as hub genes (Table SII). The DEGs in patients with
DCM-induced HF were then screened with 40 genes being defined as
significant DEGs (Table SIII);
volcano plots show the significant DEGs (Fig. 5A). Subsequently, A Venn diagram
identified eight overlapping hub genes from the WGCNA analysis and
the DEGs analysis, which were considered to be key genes (Fig. 5B). The mutual genes included
secreted protein acidic and rich in cysteine (SPARC)-related
modular calcium-binding protein 2 (SMOC2), serpin family A member 3
(SERPINA3), myosin heavy chain 6 (MYH6), S100 calcium binding
protein A9 (S100A9), tubulin α3 (TUBA3)E, TUBA3D, lymphatic vessel
endothelial hyaluronic acid receptor 1 (LYVE1) and phospholipase C
ε1 (PLCE1). SMOC2, and PLCE1 were upregulated in patients with
DCM-induced HF compared with normal healthy controls in the
GSE79962 dataset, whereas SERPINA3, MYH6, S100A9, TUBA3E, TUBA3D,
and LYVE1 were downregulated (Fig.
5C).
Validation of key candidate genes
The mRNA expression levels of the eight key genes
were verified in the GSE116250 dataset. Patients with DCM-induced
HF showed increased expression levels of SMOC2 (Fig. 6A) and PLCE1 (Fig. 6B), whereas the expressions of
SERPINA3 (Fig. 6C), MYH6 (Fig. 6D), S100A9 (Fig. 6E), LYVE1 (Fig. 6F), TUBA3D (Fig. 6G) and TUBA3E (Fig. 6H) were decreased. The results were
similar to those exhibited in the GSE79962 dataset.
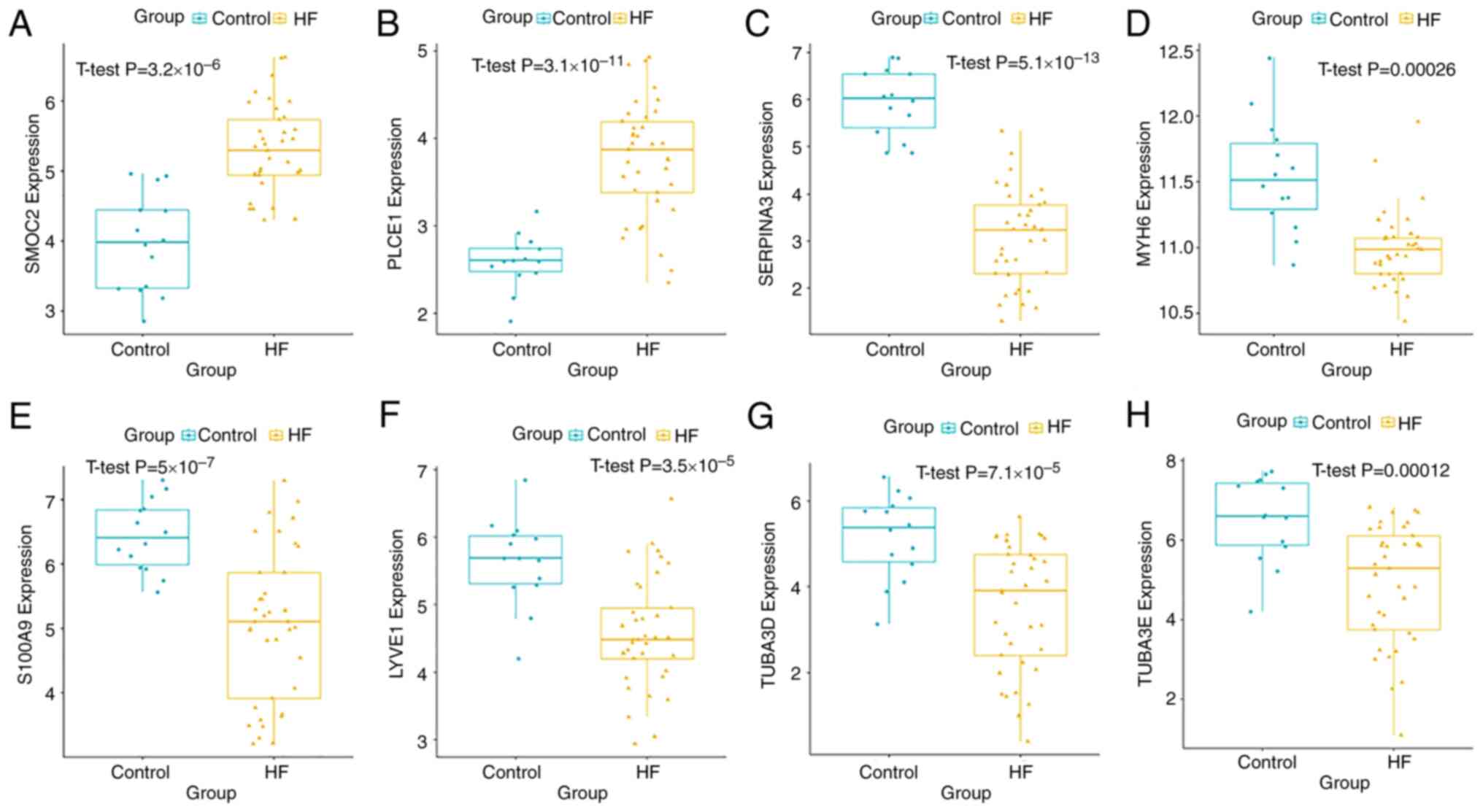 | Figure 6Key gene validation in the Gene
Expression Omnibus dataset GSE116250. mRNA expression levels of (A)
SMOC2 and (B) PLCE1 were significantly increased in patients with
DCM-induced HF compared with the control group. Expression levels
of (C) SERPINA3, (D) MYH6, (E) S100A9, (F) LYVE1, (G) TUBA3D and
(H) TUBA3E were significantly downregulated in patients with
DCM-induced HF. DCM, dilated cardiomyopathy; HF, heart failure;
LYVE1, lymphatic vessel endothelial hyaluronic acid receptor 1;
MYH6, myosin heavy chain 6; PLCE1, phospholipase C ε1; S100A9, S100
calcium binding protein A9; SERPINA3, serpin family A member 3;
SMOC2, cysteine-related modular calcium-binding protein 2; TUBA3,
tubulin α3. |
ROC analysis
To evaluate the ability of key genes to serve as
potential diagnostic biomarkers of DCM-induced HF, ROC curves were
performed in the GSE116250 dataset. In the GSE116250 dataset, the
eight key genes exhibited high predictive accuracy for diagnosing
DCM-induced HF (Fig. 7). The AUC
values of SMOC2, PLCE1 and SERPINA3 were >0.9, indicating that
these three key genes carried the highest accuracies. The other
five key genes (MYH6, S100A9, LYVE1, TUBA3D and TUBA3E) also showed
high specificity with AUCs >0.8.
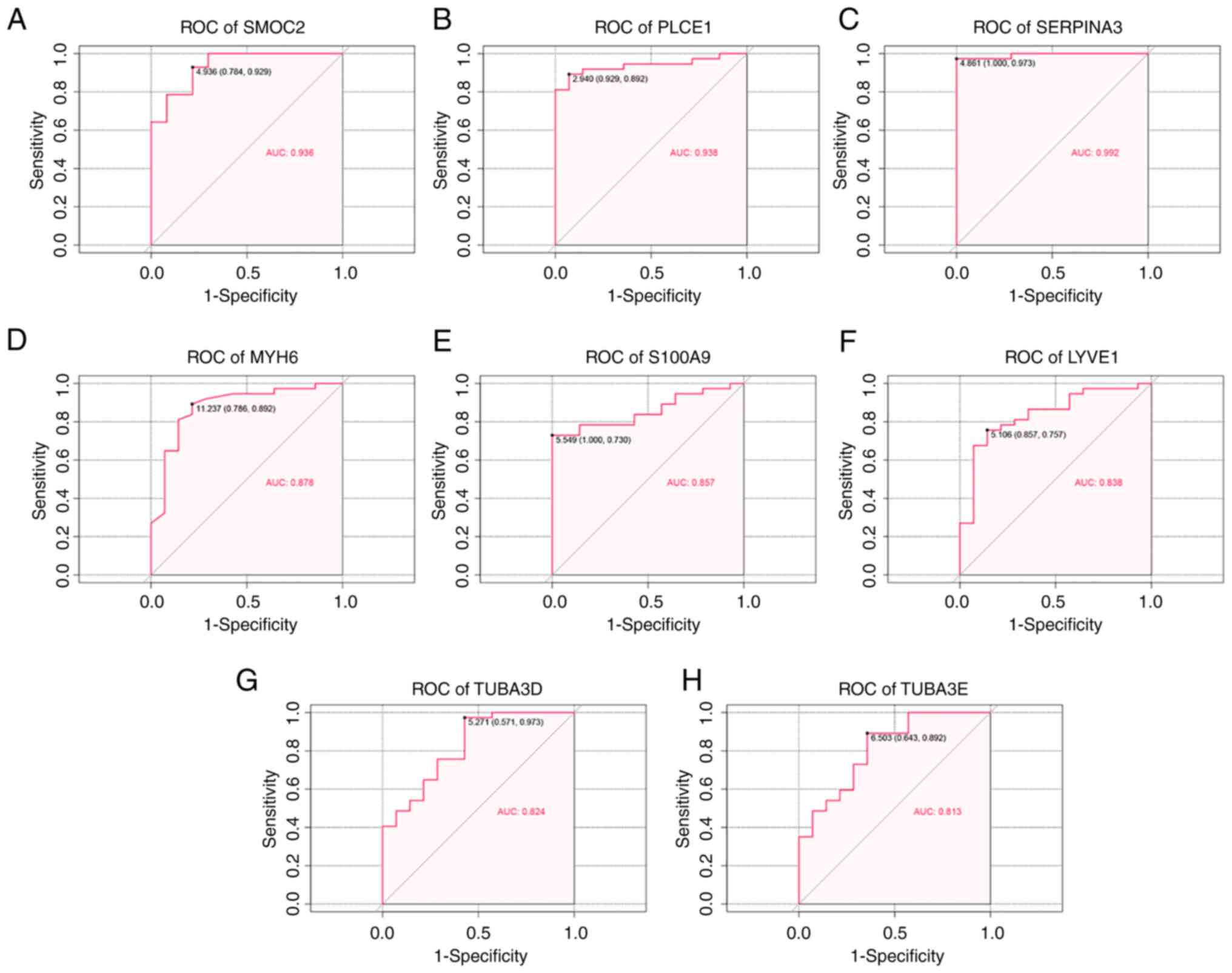 | Figure 7ROC curve analysis. The eight genes
included (A) SMOC2, (B) PLCE1, (C) SERPINA3, (D) MYH6, (E) S100A9,
(F) LYVE1, (G) TUBA3D, and (H) TUBA3E. AUC, area under the curve;
LYVE1, lymphatic vessel endothelial hyaluronic acid receptor 1;
MYH6, myosin heavy chain 6; PLCE1, phospholipase C ε1; ROC,
receiver operating characteristic; S100A9, S100 calcium binding
protein A9; SERPINA3, serpin family A member 3; SMOC2,
cysteine-related modular calcium-binding protein 2; TUBA3, tubulin
α3. |
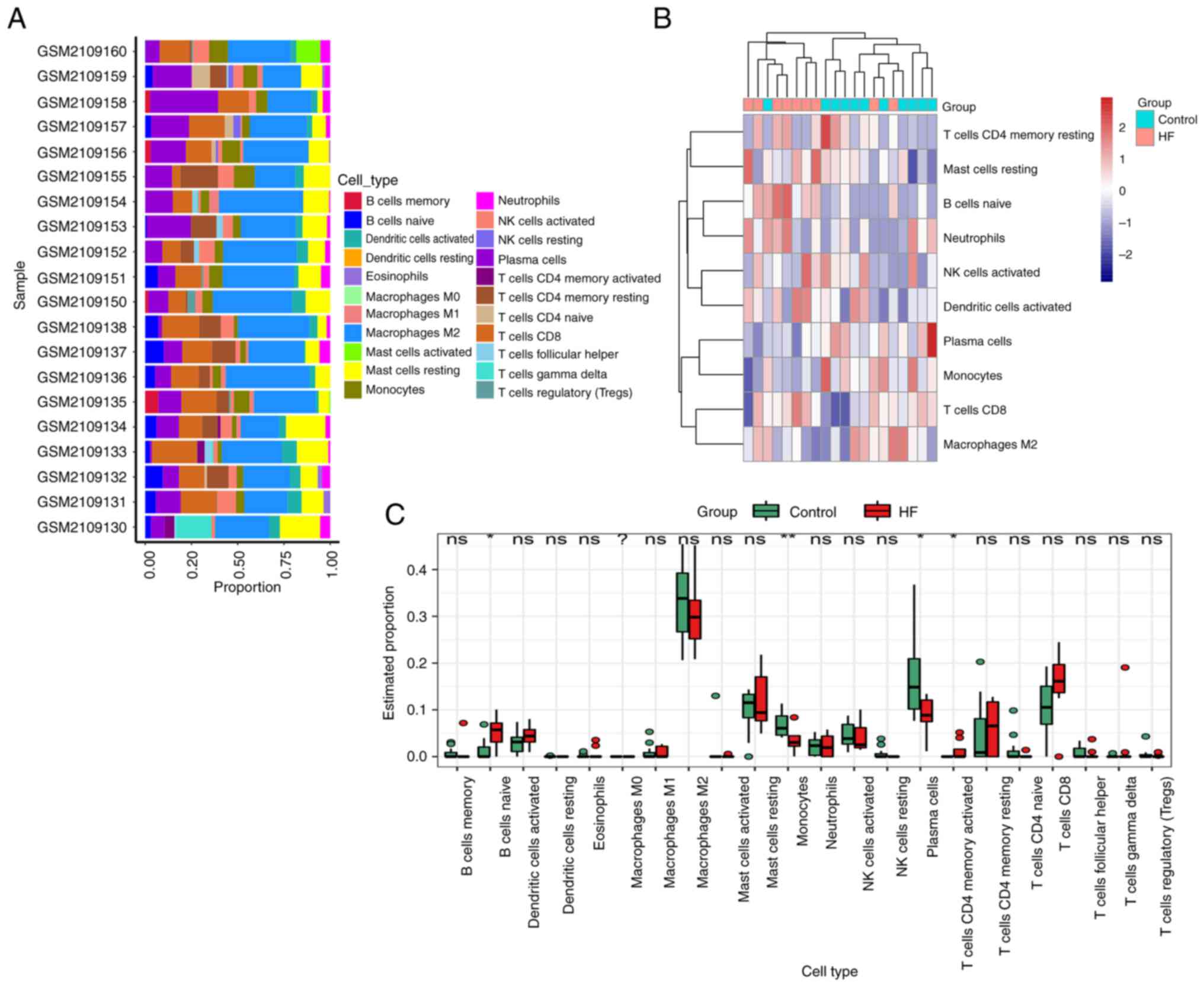
Immune cell infiltration analysis
Using the CIBERSORT model, 22 types of immune cell
were identified between DCM-induced HF patients and controls.
Fig. 8A demonstrates the immune
cell infiltration difference from 11 non-failing donors and 9
patients with DCM-induced HF. The difference in immune cell
infiltration showed that the fractions of naive B cells and
CD4-memory-activated T cells in DCM-induced HF groups were higher
compared with the control groups, whereas the infiltration of
monocytes and plasma cells was lower (Fig. 8B and C).
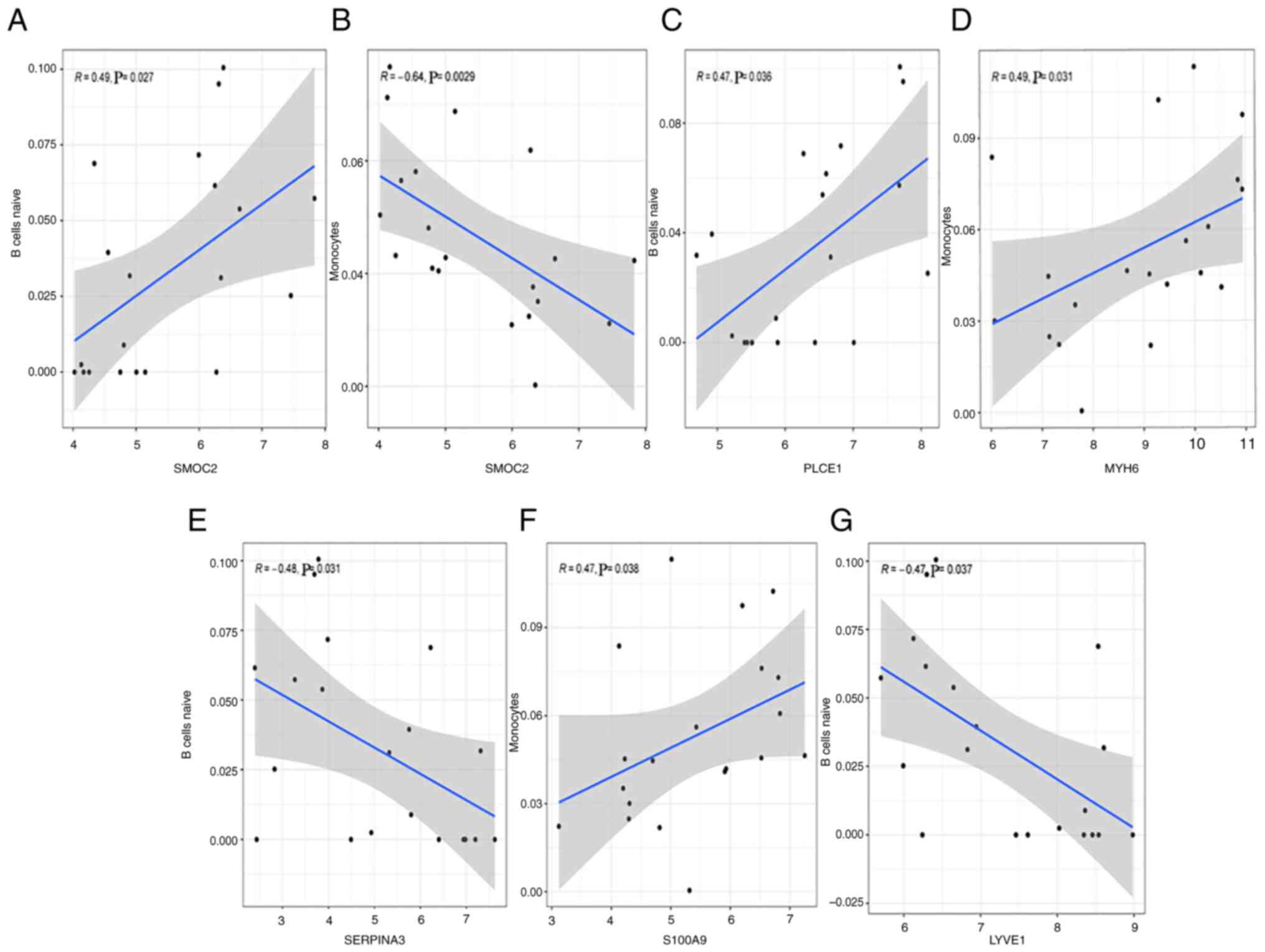
Analysis of key genes and immune
cells
A significant positive correlation was identified
between SMOC2 and naive B cells (r=0.49, P=0.027; Fig. 9A), and negative correlation was
found between SMOC2 and monocytes (r=-0.64, P=0.0029; Fig. 9B). PLCE1 was positively correlated
with naive B cells (r=0.47, P=0.036; Fig. 9C). MYH6 had a positive correlation
with monocytes (r=0.49, P=0.031; Fig.
9D). SERPINA3 was positively correlated with naive B cells
(r=-0.48, P=0.031; Fig. 9E).
S100A9 showed a positive correlation with monocytes (r=0.47,
P=0.038; Fig. 9F). LYVE1 was
negatively correlated with naive B cells (r=-0.47, P=0.037;
Fig. 9G).
Validation of the identified key genes
in vitro
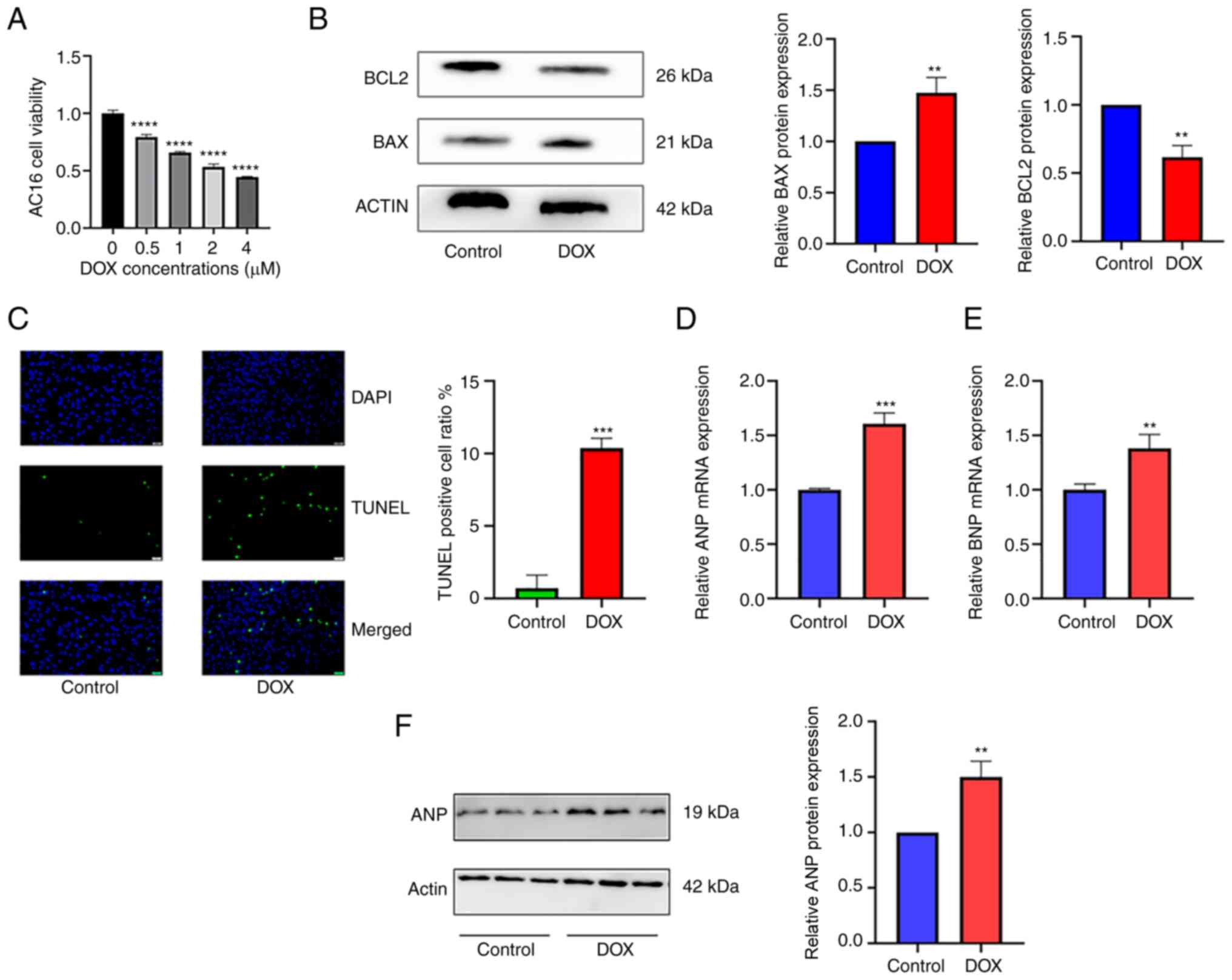
The mRNA expression levels of selected key genes
were examined by RT-qPCR in AC16 human cardiomyocyte cells treated
with DOX, which is known to induce cardiac injury can be used as an
in vitro model to mimic the mechanism of HF (32,33).
DOX treatment downregulated cardiomyocyte viability in a
concentration-dependent manner, with viability approaching 0.5 when
treated with 2 µM (Fig. 10A).
Therefore, 2 µM DOX was chosen for subsequent experiments.
Following treatment of AC16 cells with 2 µM DOX, the protein
expression level of BAX, a molecular marker of cell apoptosis, was
increased in the DOX group compared with the control group
(P<0.01; Fig. 10B). The
expression levels of BCl2, an anti-apoptotic protein, were
decreased in the DOX group compared with the control (P<0.01;
Fig. 10B). Additionally, the
apoptotic levels of AC16 cells were detected by TUNEL assay, which
showed that DOX stimulation significantly increased the number of
apoptotic AC16 cells to 10.37% (P<0.001; Fig. 10C). The mRNA expression levels of
ANP (P<0.001) and brain natriuretic peptide (BNP; P<0.01)
were also increased in the DOX groups compared with the controls
(Fig. 10D and E). The protein expression levels of ANP
were increased in the DOX group compared with the control
(P<0.01; Fig. 10F). These
results indicated that an in vitro model of DOX-induced
cardiac injury was successfully constructed, which was used to
validate the expression of hub genes.
As shown in Fig.
11, the gene expression for PLCE1, SERPINA3, MYH6, S100A9, and
LYVE1 was similar to that of the microarray analysis in the
GSE79962 and GSE116250 datasets. Specifically, PLCE1 (P<0.0001;
Fig. 11B) was upregulated in
DOX-induced cardiac injury, whereas SERPINA3 (P<0.0001; Fig. 11C), MYH6 (P<0.001; Fig. 11D), S100A9 (P<0.01; Fig. 11E) and LYVE1 (P<0.001: Fig. 11F) were downregulated compared
with the control group. However, the expression of SMOC2, TUBA3D
and TUBA3E were inconsistent with the results of microarray
datasets. Specifically, the expression of SMOC2 was decreased in
DOX-induced cardiac injury compared with the controls, but the
difference was not statistically significant (Fig. 11A). The expression of TUBA3E was
increased in DOX-induced cardiac injury compared with that in
untreated cells (Fig. 11H),
whereas there was no difference in the levels of TUBA3D (Fig. 11G).
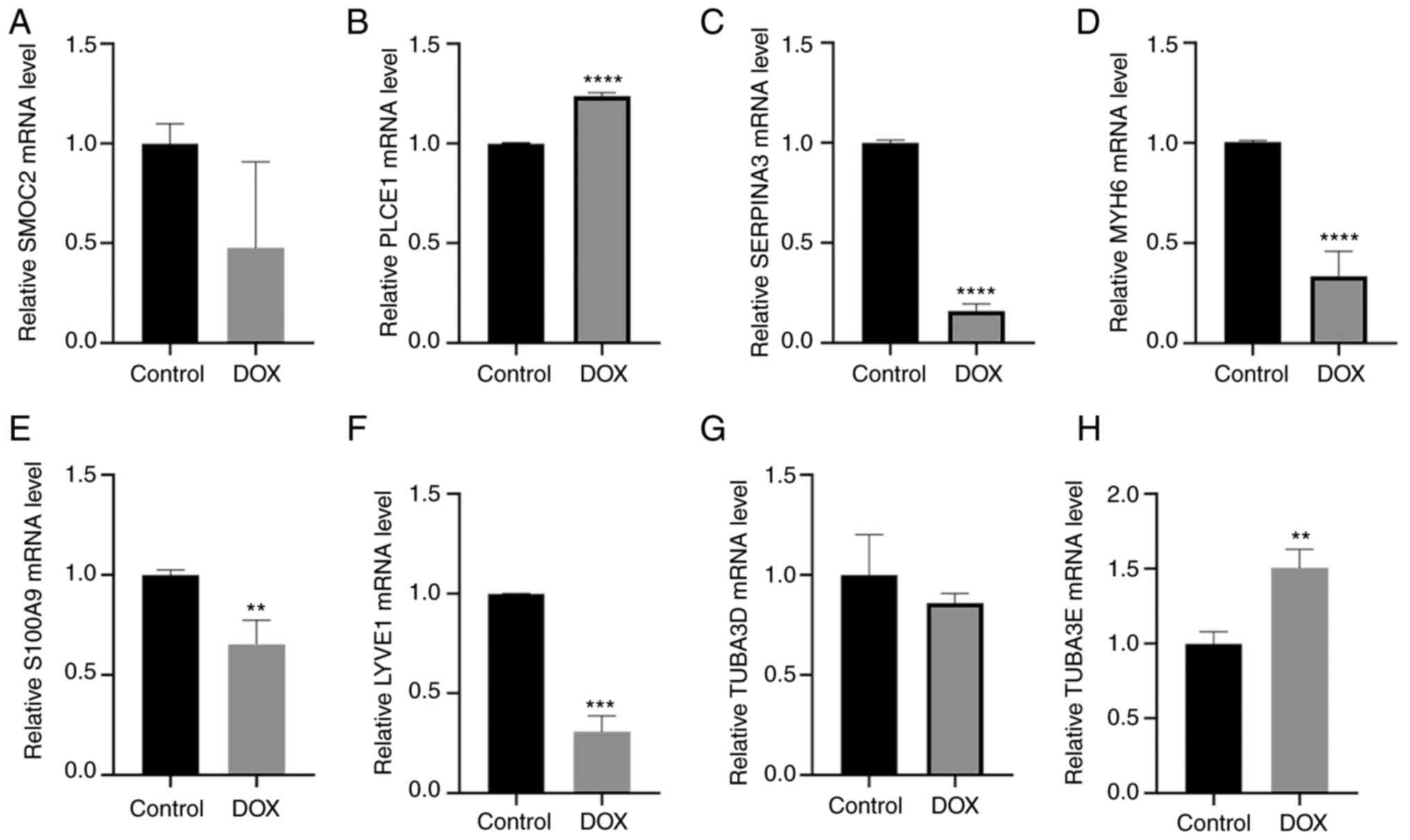 | Figure 11Relative mRNA expression levels of
key genes between control and DOX-induced cardiac injury cells.
mRNA expression levels for (A) SMOC2, (B) PLCE1, (C) SERPINA3, (D)
MYH6, (E) S100A9, (F) LYVE1, (G) TUBA3D, and (H) TUBA3E.
**P<0.01, ***P<0.001,
****P<0.0001 vs. control. Control, untreated cells;
DOX, doxorubicin; LYVE1, lymphatic vessel endothelial hyaluronic
acid receptor 1; MYH6, myosin heavy chain 6; PLCE1, phospholipase C
ε1; S100A9, S100 calcium binding protein A9; SERPINA3, serpin
family A member 3; SMOC2, cysteine-related modular calcium-binding
protein 2; TUBA3, tubulin α3. |
Discussion
DCM is one of the leading causes of HF worldwide and
is the most common indication for heart transplantation; it is
characterized by an enlarged heart with impaired contractility
(34). Individuals with the lowest
ejection fractions, often accompanied by circulatory collapse,
arrhythmias and thromboembolic events, have the worst prognosis
(35). Despite the emergence of
novel therapies, such as cardiac contractility modulation therapy
and stem cell therapy, DCM-induced HF still has a high morbidity
and mortality rate (36-40).
Therefore, elucidating the underlying mechanism of DCM-induced HF
may aid in the diagnosis and treatment of patients. The present
study aimed to investigate the gene co-expression networks in
DCM-induced HF. Using the WGCNA method, the blue module was
selected as the key module corresponding to DCM-induced HF, and
pathway enrichment analysis revealed that the AGE/RAGE signaling
pathway in diabetic complications, the p53 and MAPK signaling
pathways, adrenergic signaling in cardiomyocytes, and the JAK-STAT
and cGMP-PKG signaling pathways were associated with DCM-induced
HF.
Previous studies have shown that the AGE/RAGE
signaling pathway in diabetic complications can trigger fibroblast
activation in the heart, leading to fibroblast-mediated matrix
remodeling in HF (41,42). The present study further emphasizes
the crucial roles of the AGE/RAGE signaling pathway in diabetic
complications in the pathogenesis of DCM-induced HF. Previous
studies have indicated that aberrant JAK/STAT signaling may promote
progression from hypertrophy to HF (43-45).
In addition, the cGMP/PKG signaling pathway, adrenergic signaling
in cardiomyocytes, and the p53 signaling pathway are also
reportedly involved in the pathogenesis of HF (46-52).
The enrichment findings of the present study are in line with the
aforementioned conclusions, further strengthening the reliability
of the present results.
Using WGCNA and DEGs analysis, eight key genes were
identified in the blue module: SMOC2, SERPINA3, MYH6, S100A9,
TUBA3E, TUBA3D, LYVE1 and PLCE1. SMOC2, and PLCE1 were upregulated
in tissues from patients with DCM-induced HF compared with healthy
controls, whereas the expression of SERPINA3, MYH6, S100A9, TUBA3E,
TUBA3D and LYVE1 was lower. Furthermore, the expression of these
key genes was validated in the GSE116250 dataset. The expression
levels of SERPINA3, MYH6, S100A9, LYVE1 and PLCE1 were also
validated in vitro. The present study found that these genes
may perform a crucial role in the pathophysiology of DCM-induced
HF.
The present study not only identified the MYH6 gene
already involved in DCM and HF, but also provided new candidate
genes (SERPINA3, SMOC2, S100A9, LYVE1 and PLCE1) for further
experimental investigation. MYH6, encoding the α heavy chain
subunit of cardiac myosin, is expressed primarily in human cardiac
atria and performs crucial roles in cardiac muscle contraction
(53). Mutations in the MYH6 gene
are associated with dilated as well as hypertrophic phenotypes of
cardiomyopathy (54-57).
The subsequent structural changes of the myocardium induced by
alterations in MYH6 expression may eventually lead to cardiac
enlargement and dysfunction (58).
SERPINA3 is a member of the serpin superfamily of protease
inhibitors involved in a wide range of biological processes
(59). Previous studies have found
that SERPINA3 is downregulated in DCM and HF, and plasma SERPINA3
levels are associated with poor survival in patients with HF
(60-63).
It is mainly involved in regulating the inflammatory response and
oxidative stress (64,65); however, the function of SERPINA3 in
HF is unknown. We hypothesized that SERPINA3 may participate in the
progression of HF by regulating inflammatory activity. SMOC2, a
member of the SPARC family of matricellular proteins, modulates
cell-matrix interactions (66). It
has been previously reported that SMOC2 expression is higher in
right ventricular failure tissue (67). In addition, high expression of
SMOC2 is associated with cardiac fibrosis in chronic Chagas disease
cardiomyopathy (68). A number of
studies have reported that SMOC2 promotes tissue fibrosis by
regulating fibroblast-to-myofibroblast transformation (66,69,70).
As cardiac fibrosis is important in HF progression (71), it is reasonable to suggest that
high expression of SMOC2 may contribute to cardiac fibrosis in
DCM-induced HF. S100A9, a Ca2+-binding protein, has
anti-inflammatory and immunoregulatory actions, it serves key
immune response roles in inflammatory disorders, including
cardiovascular disease (72-75).
A study showed that recombinant S100A8/A9 attenuates cardiac
hypertrophy and fibrosis by suppressing the calcineurin/NFAT
pathway (76). Marinkovic et
al (77) found that short-term
S100A9 blockade reduces cardiac inflammation, limits myocardial
damage, and significantly improves cardiac function and
hemodynamics following myocardial ischemia; however, long-term
S100A9 blockade negatively impacts cardiac recovery. Marinkovic
et al (78) further found
that short-term S100A9 blockade improves cardiac function following
myocardial infarction by inhibiting inflammation. This suggested
that the downregulation of S100A9 may be closely associated with
the occurrence of DCM-induced HF. LYVE-1, a docking receptor for
hyaluronic acid-coated leukocytes, regulates the activation of
lymphocytes and the entry of immune cells from tissues into
lymphatic vessels (79,80). A previous study in mice reported
that LYVE-1 deletion leads to increased chronic inflammation and
long-term deterioration of cardiac function (80). PLCE1, a member of the
phosphoinositide-specific PLC family, is essential for
intracellular signaling through the catalyzation of membrane
phospholipid hydrolysis (81).
Overexpression of PLCE1 has been reported to promote inflammation
in myocardial ischemia-reperfusion injury through the activation of
the NF-κB signaling pathway (82).
However, the associations between these key genes and the
mechanisms of DCM-induced HF have not been identified and are worth
exploring in the future.
The diagnostic value of these genes was also
explored in the present study. The eight key genes showed a robust
predictive value in DCM-induced HF, indicating their potential use
as biomarkers. In addition, the characteristics of immune cell
infiltrations in DCM-induced HF were also explored, which revealed
a significant enrichment of naive B cells as well as
CD4-memory-activated T cells in the DCM-induced HF samples.
Previous studies have shown that CD4+ T cells are
abnormally activated in patients with DCM and have a direct
pathogenic role in the development of HF (83-85).
The present study has some limitations, including
the failure to assess BNP, NT-proBNP, TnI and TnT using western
blotting for in vitro phenotype validation. Additionally,
the present study did not validate the bioinformatics results in
DCM-induced HF and normal human tissues in vivo. Although
eight key genes associated with DCM-induced HF were identified, the
specific mechanism of these genes was not demonstrated. In future,
in vivo experiments will be performed to explore the
specific effects of these genes in DCM-induced HF.
In conclusion, the current study identified the
functional pathways, infiltrating immune cells and key genes
associated with DCM-induced HF, which may aid in our understanding
of the pathology and molecular mechanism of DCM-induced HF.
Supplementary Material
The primers used in this study.
Hub genes in WGCNA analysis.
Significant DEGs in GSE79962.
Acknowledgements
Not applicable.
Funding
Funding: This study was funded by The Natural Science Foundation
of Shanghai (grant. no. 19JC1415703) and the Foundation of Jinshan
Hospital of Fudan University (grant. no. JYQN-LC-202007).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from GEO at (https://www.ncbi.nlm.nih.gov/geo).
Authors' contributions
LZ and HG confirm the authenticity of all the raw
data. LZ and HG were responsible for study conception, as well as
writing the manuscript and performing data analyses. FP performed
GO and KEGG analysis. JXL was responsible for the bioinformatic
data collection, figure preparation and experimental data analysis.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Savarese G and Lund LH: Global public
health burden of heart failure. Card Fail Rev. 3:7–11.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lumbers RT, Shah S, Lin H, Czuba T, Henry
A, Swerdlow DI, Mälarstig A, Andersson C, Verweij N, Holmes MV, et
al: The genomics of heart failure: Design and rationale of the
HERMES consortium. ESC Heart Fail. 8:5531–5541. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ziaeian B and Fonarow GC: Epidemiology and
aetiology of heart failure. Nat Rev Cardiol. 13:368–378.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Smith JG: Molecular epidemiology of heart
failure: Translational challenges and opportunities. JACC Basic
Transl Sci. 2:757–769. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Huang J, Yin H, Zhang M, Ni Q and Xuan J:
Understanding the economic burden of heart failure in China: Impact
on disease management and resource utilization. J Med Econ.
20:549–553. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Klein S, Jiang S, Morey JR, Pai A, Mancini
DM, Lala A and Ferket BS: Estimated health care utilization and
expenditures in individuals with heart failure from the medical
expenditure panel survey. Circ Heart Fail.
14(e007763)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yingchoncharoen T, Wu TC, Choi DJ, Ong TK,
Liew HB and Cho MC: Economic burden of heart failure in asian
countries with different healthcare systems. Korean Circ J.
51:681–693. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gomes CPC, Schroen B, Kuster GM, Robinson
EL, Ford K, Squire IB, Heymans S, Martelli F, Emanueli C and Devaux
Y: EU-CardioRNA COST Action (CA17129). Regulatory RNAs in Heart
Failure. Circulation. 141:313–328. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Guo Q, Zhang Y, Zhang S, Jin J, Pang S, Wu
X, Zhang W, Bi X, Zhang Y, Zhang Q and Jiang F: Genome-wide
translational reprogramming of genes important for myocyte
functions in overload-induced heart failure. Biochim Biophys Acta
Mol Basis Dis. 1866(165649)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pepin ME, Drakos S, Ha CM,
Tristani-Firouzi M, Selzman CH, Fang JC, Wende AR and Wever-Pinzon
O: DNA methylation reprograms cardiac metabolic gene expression in
end-stage human heart failure. Am J Physiol Heart Circ Physiol.
317:H674–H84. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
van der Pol A, Hoes MF, de Boer RA and van
der Meer P: Cardiac foetal reprogramming: A tool to exploit novel
treatment targets for the failing heart. J Internal Med.
288:491–506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bondue A, Arbustini E, Bianco A,
Ciccarelli M, Dawson D, De Rosa M, Hamdani N, Hilfiker-Kleiner D,
Meder B, Leite-Moreira AF, et al: Complex roads from genotype to
phenotype in dilated cardiomyopathy: Scientific update from the
Working Group of Myocardial Function of the European Society of
Cardiology. Cardiovasc Res. 114:1287–1303. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cannata A, Fabris E, Merlo M, Artico J,
Gentile P, Pio Loco C, Ballaben A, Ramani F, Barbati G and Sinagra
G: Sex Differences in the Long-term prognosis of dilated
cardiomyopathy. Can J Cardiol. 36:37–44. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Merlo M, Cannata A, Gobbo M, Stolfo D,
Elliott PM and Sinagra G: Evolving concepts in dilated
cardiomyopathy. Eur J Heart Fail. 20:228–239. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Jefferies JL and Towbin JA: Dilated
cardiomyopathy. Lancet. 375:752–762. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Clarke R, Peden JF, Hopewell JC, Kyriakou
T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, et
al: Genetic variants associated with Lp(a) lipoprotein level and
coronary disease. N Engl J Med. 361:2518–2528. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kuehl U, Lassner D, Gast M, Stroux A,
Rohde M, Siegismund C, Wang X, Escher F, Gross M, Skurk C, et al:
Differential Cardiac MicroRNA expression predicts the clinical
course in human enterovirus cardiomyopathy. Circ Heart Fail.
8:605–618. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Roselli C, Chaffin MD, Weng LC,
Aeschbacher S, Ahlberg G, Albert CM, Almgren P, Alonso A, Anderson
CD, Aragam KG, et al: Multi-ethnic genome-wide association study
for atrial fibrillation. Nat Genet. 50:1225–1233. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tabibiazar R, Wagner RA, Liao A and
Quertermous T: Transcriptional profiling of the heart reveals
chamber-specific gene expression patterns. Circ Res. 93:1193–1201.
2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9(559)2008.PubMed/NCBI View Article : Google Scholar
|
|
21
|
To KY: Identification of differential gene
expression by high throughput analysis. Comb Chem High Throughput
Screen. 3:235–241. 2000.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Dang H, Ye Y, Zhao X and Zeng Y:
Identification of candidate genes in ischemic cardiomyopathy by
gene expression omnibus database. BMC Cardiovasc Disord.
20(320)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fan G and Wei J: Identification of
potential novel biomarkers and therapeutic targets involved in
human atrial fibrillation based on bioinformatics analysis.
Kardiologia Polska. 78:694–702. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yifan C, Jianfeng S and Jun P: Development
and validation of a random forest diagnostic model of acute
myocardial infarction based on Ferroptosis-related genes in
circulating endothelial cells. Front Cardiovasc Med.
8(663509)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Matkovich SJ, Al Khiami B, Efimov IR,
Evans S, Vader J, Jain A, Brownstein BH, Hotchkiss RS and Mann DL:
Widespread Down-regulation of cardiac mitochondrial and sarcomeric
genes in patients with sepsis. Crit Care Med. 45:407–414.
2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Schwientek P, Ellinghaus P, Steppan S,
D'Urso D, Seewald M, Kassner A, Cebulla R, Schulte-Eistrup S,
Morshuis M, Röfe D, et al: Global gene expression analysis in
nonfailing and failing myocardium pre- and postpulsatile and
nonpulsatile ventricular assist device support. Physiol Genomics.
42:397–405. 2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sweet ME, Cocciolo A, Slavov D, Jones KL,
Sweet JR, Graw SL, Reece TB, Ambardekar AV, Bristow MR, Mestroni L
and Taylor MRG: Transcriptome analysis of human heart failure
reveals dysregulated cell adhesion in dilated cardiomyopathy and
activated immune pathways in ischemic heart failure. BMC Genomics.
19(812)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chen B, Khodadoust MS, Liu CL, Newman AM
and Alizadeh AA: Profiling tumor infiltrating immune cells with
CIBERSORT. Methods Mol Biol. 1711:243–259. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Davidson MM, Nesti C, Palenzuela L, Walker
WF, Hernandez E, Protas L, Hirano M and Isaac ND: Novel cell lines
derived from adult human ventricular cardiomyocytes. J Mol Cell
Cardiol. 39:133–147. 2005.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Sachinidis A: Cardiotoxicity and heart
failure: Lessons from human-induced pluripotent stem cell-derived
cardiomyocytes and anticancer drugs. Cells. 9(1001)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhong Z, Tian Y, Luo X, Zou J, Wu L and
Tian J: Extracellular vesicles derived from human umbilical cord
mesenchymal stem cells protect against DOX-induced heart failure
through the miR-100-5p/NOX4 pathway. Front Bioeng Biotechnol.
9(703241)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Reichart D, Magnussen C, Zeller T and
Blankenberg S: Dilated cardiomyopathy: From epidemiologic to
genetic phenotypes: A translational review of current literature. J
Internal Med. 286:362–372. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Weintraub RG, Semsarian C and Macdonald P:
Dilated cardiomyopathy. Lancet. 390:400–414. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Diaz-Navarro R, Urrutia G, Cleland JG,
Poloni D, Villagran F, Acosta-Dighero R, Bangdiwala SI, Rada G and
Madrid E: Stem cell therapy for dilated cardiomyopathy. Cochrane
Database Syst Rev. 7(CD013433)2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Haas J, Frese KS, Peil B, Kloos W, Keller
A, Nietsch R, Feng Z, Müller S, Kayvanpour E, Vogel B, et al: Atlas
of the clinical genetics of human dilated cardiomyopathy. Eur Heart
J. 36:1123–1135a. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kadhi A, Mohammed F and Nemer G: The
genetic pathways underlying immunotherapy in dilated
cardiomyopathy. Front Cardiovasc Med. 8(613295)2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Merlo M, Pivetta A, Pinamonti B, Stolfo D,
Zecchin M, Barbati G, Di Lenarda A and Sinagra G: Long-term
prognostic impact of therapeutic strategies in patients with
idiopathic dilated cardiomyopathy: Changing mortality over the last
30 years. Eur J Heart Fail. 16:317–324. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Linde C, Grabowski M, Ponikowski P, Rao I,
Stagg A and Tschope C: Cardiac contractility modulation therapy
improves health status in patients with heart failure with
preserved ejection fraction: A pilot study (CCM-HFpEF). Eur J Heart
Fail. 24:2275–2284. 2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Liang B, Zhou Z, Yang Z, Liu J, Zhang L,
He J, Li H, Huang Y, Yang Q, Xian S and Wang L: AGEs-RAGE axis
mediates myocardial fibrosis via activation of cardiac fibroblasts
induced by autophagy in heart failure. Exp Physiol. 107:879–891.
2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Burr SD and Stewart JA Jr: Extracellular
matrix components isolated from diabetic mice alter cardiac
fibroblast function through the AGE/RAGE signaling cascade. Life
Sci. 250(117569)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Boengler K, Hilfiker-Kleiner D, Drexler H,
Heusch G and Schulz R: The myocardial JAK/STAT pathway: From
protection to failure. Pharmacol Ther. 120:172–185. 2008.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Okonko DO, Marley SB, Anker SD,
Poole-Wilson PA and Gordon MY: Erythropoietin resistance
contributes to anaemia in chronic heart failure and relates to
aberrant JAK-STAT signal transduction. Int J Cardiol. 164:359–364.
2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Terrell AM, Crisostomo PR, Wairiuko GM,
Wang M, Morrell ED and Meldrum DR: Jak/STAT/SOCS signaling circuits
and associated cytokine-mediated inflammation and hypertrophy in
the heart. Shock. 26:226–234. 2006.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chen SN, Lombardi R, Karmouch J, Tsai JY,
Czernuszewicz G, Taylor MRG, Mestroni L, Coarfa C, Gurha P and
Marian AJ: DNA damage Response/TP53 pathway is activated and
contributes to the pathogenesis of dilated cardiomyopathy
associated with LMNA (Lamin A/C) mutations. Circ Res. 124:856–873.
2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Das B, Young D, Vasanji A, Gupta S, Sarkar
S and Sen S: Influence of p53 in the transition of
myotrophin-induced cardiac hypertrophy to heart failure. Cardiovasc
Res. 87:524–534. 2010.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Fujita T and Ishikawa Y: Apoptosis in
Heart Failure-The role of the beta-adrenergic receptor-mediated
signaling pathway and p53-mediated signaling pathway in the
apoptosis of cardiomyocytes. Circ J. 75:1811–1818. 2011.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Irie T, Sips PY, Kai S, Kida K, Ikeda K,
Hirai S, Moazzami K, Jiramongkolchai P, Bloch DB, Doulias PT, et
al: S-Nitrosylation of Calcium-handling proteins in cardiac
adrenergic signaling and hypertrophy. Circ Res. 117:793–803.
2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Persoon S, Paulus M, Hirt S, Jungbauer C,
Dietl A, Luchner A, Schmid C, Maier LS and Birner C: Cardiac
unloading by LVAD support differentially influences components of
the cGMP-PKG signaling pathway in ischemic and dilated
cardiomyopathy. Heart Vessels. 33:948–957. 2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Pleger ST, Boucher M, Most P and Koch WJ:
Targeting myocardial beta-adrenergic receptor signaling and calcium
cycling for heart failure gene therapy. J Card Fail. 13:401–414.
2007.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Port JD and Bristow MR: Altered
beta-adrenergic receptor gene regulation and signaling in chronic
heart failure. J Mol Cell Cardiol. 33:887–905. 2001.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Razmara E and Garshasbi M: Whole-exome
sequencing identifies R1279X of MYH6 gene to be associated with
congenital heart disease. BMC Cardiovasc Disord.
18(137)2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Carniel E, Taylor MR, Sinagra G, Di
Lenarda A, Ku L, Fain PR, Boucek MM, Cavanaugh J, Miocic S, Slavov
D, et al: Alpha-myosin heavy chain: A sarcomeric gene associated
with dilated and hypertrophic phenotypes of cardiomyopathy.
Circulation. 112:54–59. 2005.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Hao E, Zhang G, Mu L, Ma N and Wang T:
Establishment of a human MYH6 compound heterozygous knockout hESC
line to model cardiomyopathy and congenital heart defects by
CRISPR/Cas9 system. Stem Cell Res. 50(102128)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Hershberger RE, Norton N, Morales A, Li D,
Siegfried JD and Gonzalez-Quintana J: Coding sequence rare variants
identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312
patients with familial or idiopathic dilated cardiomyopathy. Circ
Cardiovasc Genet. 3:155–161. 2010.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Merlo M, Sinagra G, Carniel E, Slavov D,
Zhu X, Barbati G, Spezzacatene A, Ramani F, Salcedo E, Di Lenarda
A, et al: Poor prognosis of rare sarcomeric gene variants in
patients with dilated cardiomyopathy. Clin Transl Sci. 6:424–428.
2013.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Chen JH, Wang LL, Tao L, Qi B, Wang Y, Guo
YJ and Miao L: Identification of MYH6 as the potential gene for
human ischaemic cardiomyopathy. J Cell Mol Med. 25:10736–10746.
2021.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Chelbi ST, Wilson ML, Veillard AC, Ingles
SA, Zhang J, Mondon F, Gascoin-Lachambre G, Doridot L, Mignot TM,
Rebourcet R, et al: Genetic and epigenetic mechanisms collaborate
to control SERPINA3 expression and its association with placental
diseases. Hum Mol Genet. 21:1968–1978. 2012.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Asakura M and Kitakaze M: Global gene
expression profiling in the failing myocardium. Circ J.
73:1568–1576. 2009.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Delrue L, Vanderheyden M, Beles M,
Paolisso P, Di Gioia G, Dierckx R, Verstreken S, Goethals M,
Heggermont W and Bartunek J: Circulating SERPINA3 improves
prognostic stratification in patients with a de novo or worsened
heart failure. ESC Heart Fail. 8:4780–4790. 2021.PubMed/NCBI View Article : Google Scholar
|
|
62
|
di Salvo TG, Yang KC, Brittain E, Absi T,
Maltais S and Hemnes A: Right ventricular myocardial biomarkers in
human heart failure. J Card Fail. 21:398–411. 2015.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Jiang Z, Guo N and Hong K: A three-tiered
integrative analysis of transcriptional data reveals the shared
pathways related to heart failure from different aetiologies. J
Cell Mol Med. 24:9085–9096. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Lok SI, van Mil A, Bovenschen N, van der
Weide P, van Kuik J, van Wichen D, Peeters T, Siera E, Winkens B,
Sluijter JP, et al: Post-transcriptional regulation of
α-1-antichymotrypsin by microRNA-137 in chronic heart failure and
mechanical support. Circ Heart Fail. 6:853–861. 2013.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Sanchez-Navarro A, Gonzalez-Soria I,
Caldino-Bohn R and Bobadilla NA: An integrative view of serpins in
health and disease: The contribution of SerpinA3. Am J Physiol Cell
Physiol. 320:C106–C108. 2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Gerarduzzi C, Kumar RK, Trivedi P, Ajay
AK, Iyer A, Boswell S, Hutchinson JN, Waikar SS and Vaidya VS:
Silencing SMOC2 ameliorates kidney fibrosis by inhibiting
fibroblast to myofibroblast transformation. JCI Insight.
2(e90299)2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Williams JL, Cavus O, Loccoh EC, Adelman
S, Daugherty JC, Smith SA, Canan B, Janssen PML, Koenig S, Kline
CF, et al: Defining the molecular signatures of human right heart
failure. Life Sci. 196:118–126. 2018.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Laugier L, Frade AF, Ferreira FM, Baron
MA, Teixeira PC, Cabantous S, Ferreira LRP, Louis L, Rigaud VOC,
Gaiotto FA, et al: Whole-Genome Cardiac DNA methylation fingerprint
and gene expression analysis provide new insights in the
pathogenesis of chronic chagas disease cardiomyopathy. Clin Infect
Dis. 65:1103–1111. 2017.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Luo L, Wang CC, Song XP, Wang HM, Zhou H,
Sun Y, Wang XK, Hou S and Pei FY: Suppression of SMOC2 reduces
bleomycin (BLM)-induced pulmonary fibrosis by inhibition of
TGF-β1/SMADs pathway. Biomed Pharmacother. 105:841–847.
2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Schmidt IM, Colona MR, Kestenbaum BR,
Alexopoulos LG, Palsson R, Srivastava A, Liu J, Stillman IE, Rennke
HG, Vaidya VS, et al: Cadherin-11, Sparc-related modular calcium
binding protein-2, and Pigment epithelium-derived factor are
promising non-invasive biomarkers of kidney fibrosis. Kidney Int.
100:672–683. 2021.PubMed/NCBI View Article : Google Scholar
|
|
71
|
McLellan MA, Skelly DA, Dona MSI, Squiers
GT, Farrugia GE, Gaynor TL, Cohen CD, Pandey R, Diep H, Vinh A, et
al: High-resolution transcriptomic profiling of the heart during
chronic stress reveals cellular drivers of cardiac fibrosis and
hypertrophy. Circulation. 142:1448–1463. 2020.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Agra RM, Fernandez-Trasancos A, Sierra J,
Gonzalez-Juanatey JR and Eiras S: Differential association of
S100A9, an inflammatory marker, and p53, a cell cycle marker,
expression with epicardial adipocyte size in patients with
cardiovascular disease. Inflammation. 37:1504–1512. 2014.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Marinkovic G, Koenis DS, de Camp L,
Jablonowski R, Graber N, de Waard V, de Vries CJ, Goncalves I,
Nilsson J, Jovinge S and Schiopu A: S100A9 links inflammation and
repair in myocardial infarction. Circ Res. 127:664–676.
2020.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Pei XM, Tam BT, Sin TK, Wang FF, Yung BY,
Chan LW, Wong CS, Ying M, Lai CW and Siu PM: S100A8 and S100A9 are
associated with doxorubicin-induced Cardiotoxicity in the heart of
diabetic mice. Front Physiol. 7(334)2016.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Shah RD, Xue C, Zhang H, Tuteja S, Li M,
Reilly MP and Ferguson JF: Expression of Calgranulin Genes S100A8,
S100A9 and S100A12 Is modulated by n-3 PUFA during inflammation in
adipose tissue and mononuclear cells. PLoS One.
12(e0169614)2017.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Wei X, Wu B, Zhao J, Zeng Z, Xuan W, Cao
S, Huang X, Asakura M, Xu D, Bin J, et al: Myocardial hypertrophic
preconditioning attenuates cardiomyocyte hypertrophy and slows
progression to heart failure through upregulation of S100A8/A9.
Circulation. 131:1506–1517. 2015.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Marinković G, Koenis DS, de Camp L,
Jablonowski R, Graber N, de Waard V, de Vries CJ, Goncalves I,
Nilsson J, Jovinge S and Schiopu A: S100A9 links inflammation and
repair in myocardial infarction. Circ Res. 127:664–676.
2020.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Marinkovic G, Grauen Larsen H, Yndigegn T,
Szabo IA, Mares RG, de Camp L, Weiland M, Tomas L, Goncalves I,
Nilsson J, et al: Inhibition of pro-inflammatory myeloid cell
responses by short-term S100A9 blockade improves cardiac function
after myocardial infarction. Eur Heart J. 40:2713–2723.
2019.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Bizou M, Itier R, Majdoubi M, Abbadi D,
Pichery E, Dutaur M, Marsal D, Calise D, Garmy-Susini B,
Douin-Echinard V, et al: Cardiac macrophage subsets differentially
regulate lymphatic network remodeling during pressure overload. Sci
Rep. 11(16801)2021.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Vieira JM, Norman S, Villa Del Campo C,
Cahill TJ, Barnette DN, Gunadasa-Rohling M, Johnson LA, Greaves DR,
Carr CA, Jackson DG and Riley PR: The cardiac lymphatic system
stimulates resolution of inflammation following myocardial
infarction. J Clin Invest. 128:3402–3412. 2018.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Chen Y, Wang D, Peng H, Chen X, Han X, Yu
J, Wang W, Liang L, Liu Z, Zheng Y, et al: Epigenetically
upregulated oncoprotein PLCE1 drives esophageal carcinoma
angiogenesis and proliferation via activating the PI-PLCε-NF-κB
signaling pathway and VEGF-C/Bcl-2 expression. Mol Cancer.
18(1)2019.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Li W, Li Y, Chu Y, Wu W, Yu Q, Zhu X and
Wang Q: PLCE1 promotes myocardial ischemia-reperfusion injury in
H/R H9c2 cells and I/R rats by promoting inflammation. Biosci Rep.
39(BSR20181613)2019.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Youn JC, Jung MK, Yu HT, Kwon JS, Kwak JE,
Park SH, Kim IC, Park MS, Lee SK, Choi SW, et al: Increased
frequency of CD4+CD57+ senescent T cells in
patients with newly diagnosed acute heart failure: Exploring new
pathogenic mechanisms with clinical relevance. Sci Rep.
9(12887)2019.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Zeng Z, Wang K, Li Y, Xia N, Nie S, Lv B,
Zhang M, Tu X, Li Q, Tang T and Cheng X: Down-regulation of
microRNA-451a facilitates the activation and proliferation of
CD4+ T cells by targeting Myc in patients with dilated
cardiomyopathy. J Biol Chem. 292:6004–6013. 2017.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Rao M, Wang X, Guo G, Wang L, Chen S, Yin
P, Chen K, Chen L, Zhang Z, Chen X, et al: Resolving the
intertwining of inflammation and fibrosis in human heart failure at
single-cell level. Basic Res Cardiol. 116(55)2021.PubMed/NCBI View Article : Google Scholar
|