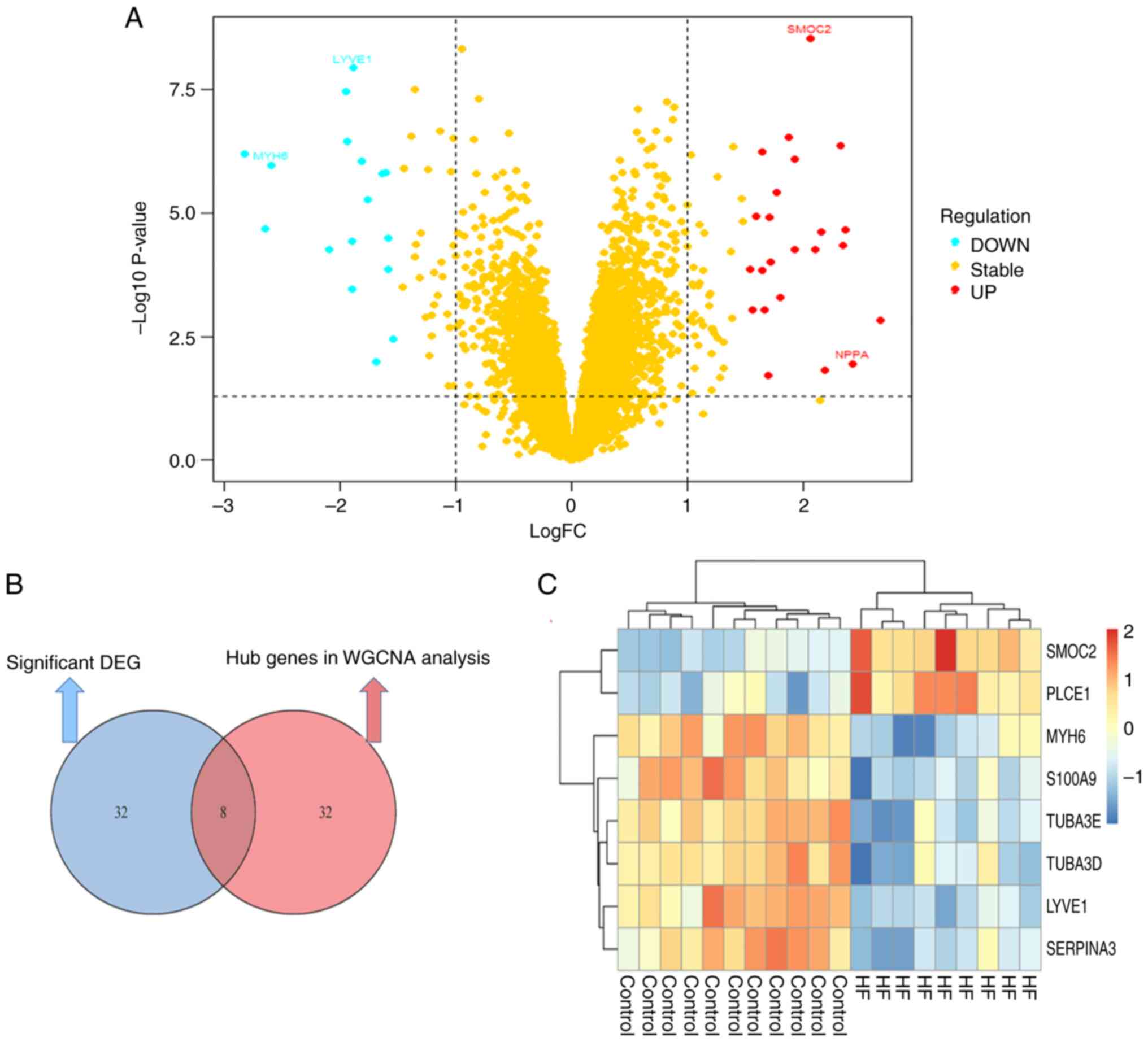
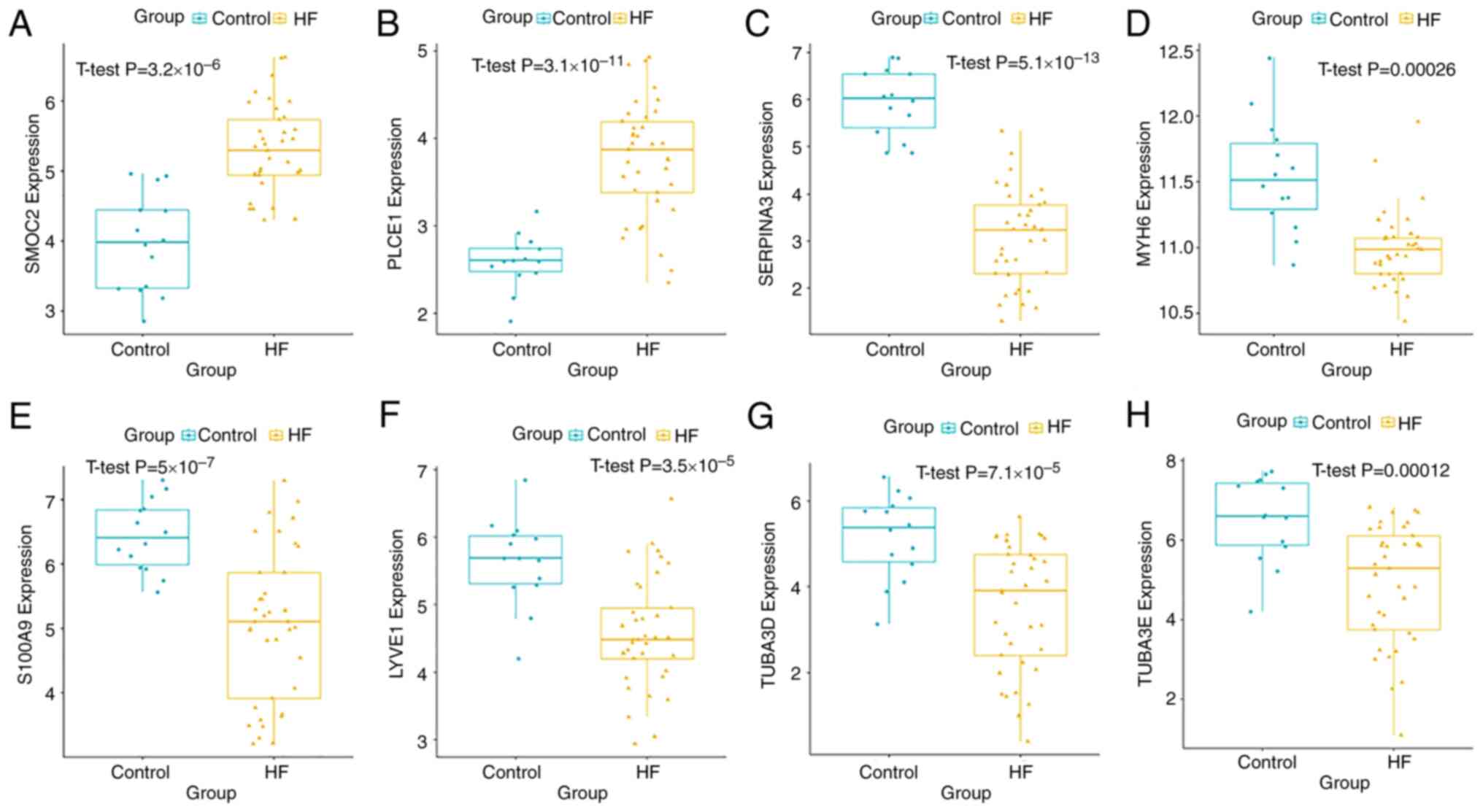
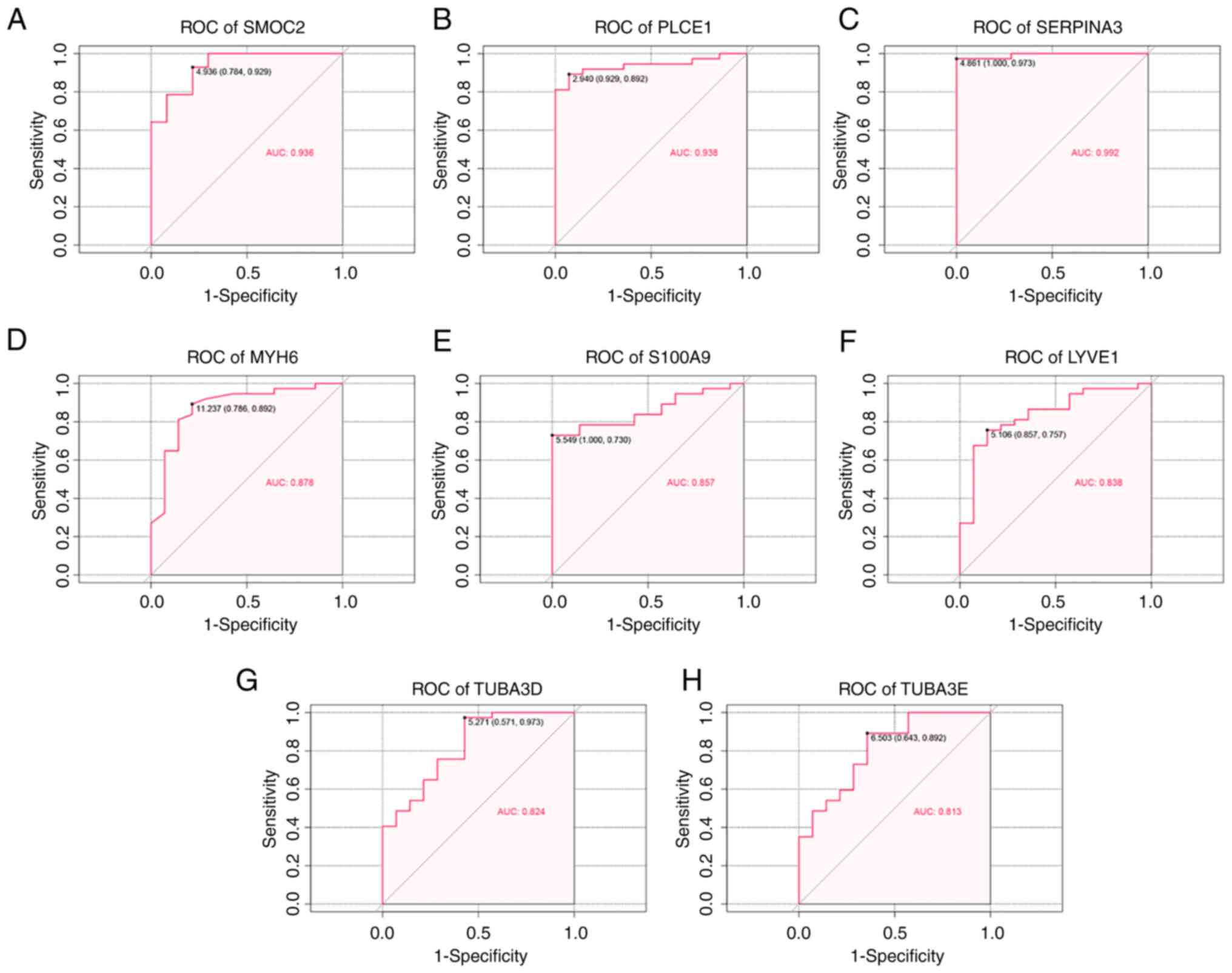
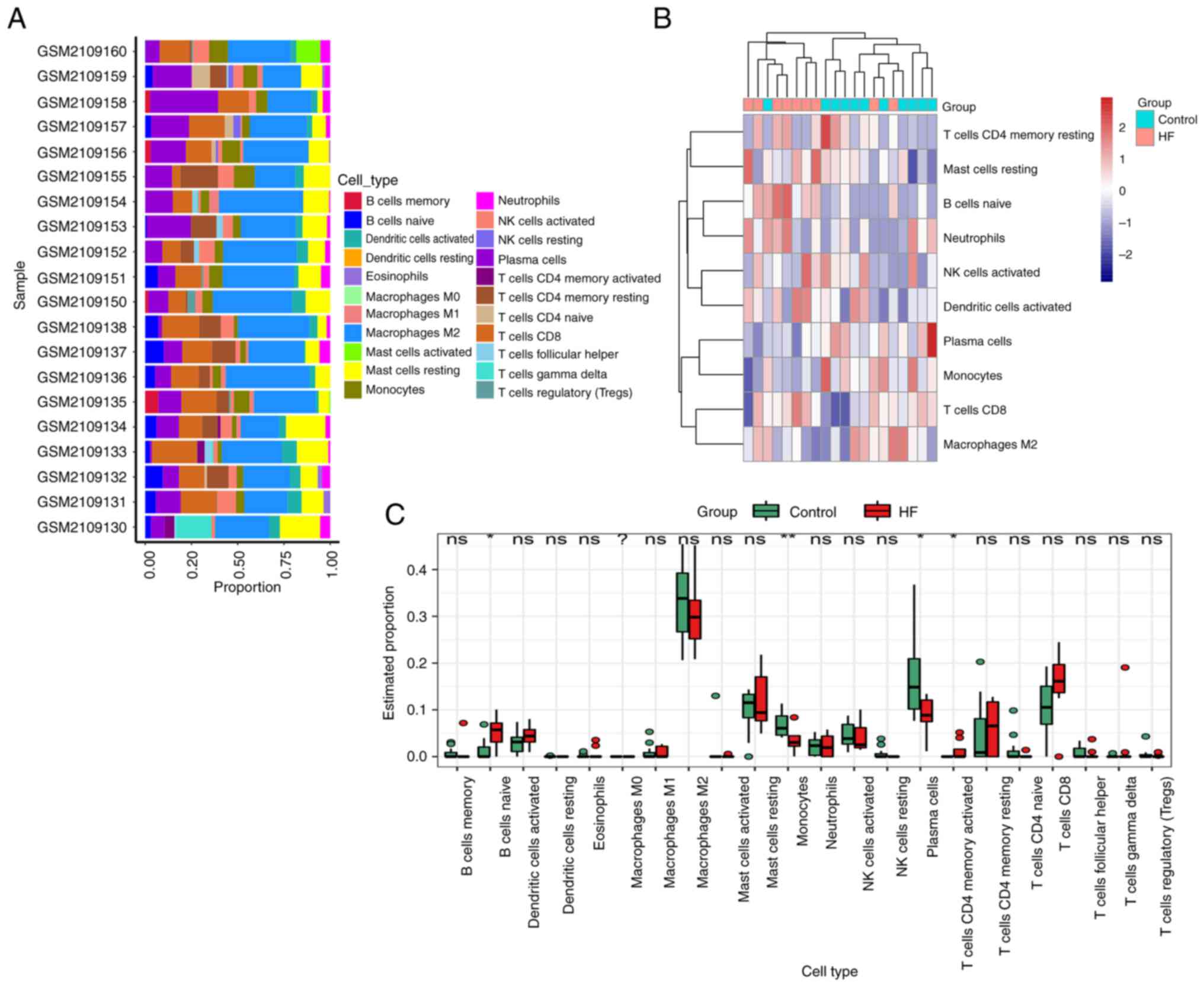
|
1
|
Savarese G and Lund LH: Global public
health burden of heart failure. Card Fail Rev. 3:7–11.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lumbers RT, Shah S, Lin H, Czuba T, Henry
A, Swerdlow DI, Mälarstig A, Andersson C, Verweij N, Holmes MV, et
al: The genomics of heart failure: Design and rationale of the
HERMES consortium. ESC Heart Fail. 8:5531–5541. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ziaeian B and Fonarow GC: Epidemiology and
aetiology of heart failure. Nat Rev Cardiol. 13:368–378.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Smith JG: Molecular epidemiology of heart
failure: Translational challenges and opportunities. JACC Basic
Transl Sci. 2:757–769. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Huang J, Yin H, Zhang M, Ni Q and Xuan J:
Understanding the economic burden of heart failure in China: Impact
on disease management and resource utilization. J Med Econ.
20:549–553. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Klein S, Jiang S, Morey JR, Pai A, Mancini
DM, Lala A and Ferket BS: Estimated health care utilization and
expenditures in individuals with heart failure from the medical
expenditure panel survey. Circ Heart Fail.
14(e007763)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yingchoncharoen T, Wu TC, Choi DJ, Ong TK,
Liew HB and Cho MC: Economic burden of heart failure in asian
countries with different healthcare systems. Korean Circ J.
51:681–693. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gomes CPC, Schroen B, Kuster GM, Robinson
EL, Ford K, Squire IB, Heymans S, Martelli F, Emanueli C and Devaux
Y: EU-CardioRNA COST Action (CA17129). Regulatory RNAs in Heart
Failure. Circulation. 141:313–328. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Guo Q, Zhang Y, Zhang S, Jin J, Pang S, Wu
X, Zhang W, Bi X, Zhang Y, Zhang Q and Jiang F: Genome-wide
translational reprogramming of genes important for myocyte
functions in overload-induced heart failure. Biochim Biophys Acta
Mol Basis Dis. 1866(165649)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pepin ME, Drakos S, Ha CM,
Tristani-Firouzi M, Selzman CH, Fang JC, Wende AR and Wever-Pinzon
O: DNA methylation reprograms cardiac metabolic gene expression in
end-stage human heart failure. Am J Physiol Heart Circ Physiol.
317:H674–H84. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
van der Pol A, Hoes MF, de Boer RA and van
der Meer P: Cardiac foetal reprogramming: A tool to exploit novel
treatment targets for the failing heart. J Internal Med.
288:491–506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bondue A, Arbustini E, Bianco A,
Ciccarelli M, Dawson D, De Rosa M, Hamdani N, Hilfiker-Kleiner D,
Meder B, Leite-Moreira AF, et al: Complex roads from genotype to
phenotype in dilated cardiomyopathy: Scientific update from the
Working Group of Myocardial Function of the European Society of
Cardiology. Cardiovasc Res. 114:1287–1303. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cannata A, Fabris E, Merlo M, Artico J,
Gentile P, Pio Loco C, Ballaben A, Ramani F, Barbati G and Sinagra
G: Sex Differences in the Long-term prognosis of dilated
cardiomyopathy. Can J Cardiol. 36:37–44. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Merlo M, Cannata A, Gobbo M, Stolfo D,
Elliott PM and Sinagra G: Evolving concepts in dilated
cardiomyopathy. Eur J Heart Fail. 20:228–239. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Jefferies JL and Towbin JA: Dilated
cardiomyopathy. Lancet. 375:752–762. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Clarke R, Peden JF, Hopewell JC, Kyriakou
T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, et
al: Genetic variants associated with Lp(a) lipoprotein level and
coronary disease. N Engl J Med. 361:2518–2528. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kuehl U, Lassner D, Gast M, Stroux A,
Rohde M, Siegismund C, Wang X, Escher F, Gross M, Skurk C, et al:
Differential Cardiac MicroRNA expression predicts the clinical
course in human enterovirus cardiomyopathy. Circ Heart Fail.
8:605–618. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Roselli C, Chaffin MD, Weng LC,
Aeschbacher S, Ahlberg G, Albert CM, Almgren P, Alonso A, Anderson
CD, Aragam KG, et al: Multi-ethnic genome-wide association study
for atrial fibrillation. Nat Genet. 50:1225–1233. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tabibiazar R, Wagner RA, Liao A and
Quertermous T: Transcriptional profiling of the heart reveals
chamber-specific gene expression patterns. Circ Res. 93:1193–1201.
2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9(559)2008.PubMed/NCBI View Article : Google Scholar
|
|
21
|
To KY: Identification of differential gene
expression by high throughput analysis. Comb Chem High Throughput
Screen. 3:235–241. 2000.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Dang H, Ye Y, Zhao X and Zeng Y:
Identification of candidate genes in ischemic cardiomyopathy by
gene expression omnibus database. BMC Cardiovasc Disord.
20(320)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fan G and Wei J: Identification of
potential novel biomarkers and therapeutic targets involved in
human atrial fibrillation based on bioinformatics analysis.
Kardiologia Polska. 78:694–702. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yifan C, Jianfeng S and Jun P: Development
and validation of a random forest diagnostic model of acute
myocardial infarction based on Ferroptosis-related genes in
circulating endothelial cells. Front Cardiovasc Med.
8(663509)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Matkovich SJ, Al Khiami B, Efimov IR,
Evans S, Vader J, Jain A, Brownstein BH, Hotchkiss RS and Mann DL:
Widespread Down-regulation of cardiac mitochondrial and sarcomeric
genes in patients with sepsis. Crit Care Med. 45:407–414.
2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Schwientek P, Ellinghaus P, Steppan S,
D'Urso D, Seewald M, Kassner A, Cebulla R, Schulte-Eistrup S,
Morshuis M, Röfe D, et al: Global gene expression analysis in
nonfailing and failing myocardium pre- and postpulsatile and
nonpulsatile ventricular assist device support. Physiol Genomics.
42:397–405. 2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sweet ME, Cocciolo A, Slavov D, Jones KL,
Sweet JR, Graw SL, Reece TB, Ambardekar AV, Bristow MR, Mestroni L
and Taylor MRG: Transcriptome analysis of human heart failure
reveals dysregulated cell adhesion in dilated cardiomyopathy and
activated immune pathways in ischemic heart failure. BMC Genomics.
19(812)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chen B, Khodadoust MS, Liu CL, Newman AM
and Alizadeh AA: Profiling tumor infiltrating immune cells with
CIBERSORT. Methods Mol Biol. 1711:243–259. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Davidson MM, Nesti C, Palenzuela L, Walker
WF, Hernandez E, Protas L, Hirano M and Isaac ND: Novel cell lines
derived from adult human ventricular cardiomyocytes. J Mol Cell
Cardiol. 39:133–147. 2005.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Sachinidis A: Cardiotoxicity and heart
failure: Lessons from human-induced pluripotent stem cell-derived
cardiomyocytes and anticancer drugs. Cells. 9(1001)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhong Z, Tian Y, Luo X, Zou J, Wu L and
Tian J: Extracellular vesicles derived from human umbilical cord
mesenchymal stem cells protect against DOX-induced heart failure
through the miR-100-5p/NOX4 pathway. Front Bioeng Biotechnol.
9(703241)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Reichart D, Magnussen C, Zeller T and
Blankenberg S: Dilated cardiomyopathy: From epidemiologic to
genetic phenotypes: A translational review of current literature. J
Internal Med. 286:362–372. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Weintraub RG, Semsarian C and Macdonald P:
Dilated cardiomyopathy. Lancet. 390:400–414. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Diaz-Navarro R, Urrutia G, Cleland JG,
Poloni D, Villagran F, Acosta-Dighero R, Bangdiwala SI, Rada G and
Madrid E: Stem cell therapy for dilated cardiomyopathy. Cochrane
Database Syst Rev. 7(CD013433)2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Haas J, Frese KS, Peil B, Kloos W, Keller
A, Nietsch R, Feng Z, Müller S, Kayvanpour E, Vogel B, et al: Atlas
of the clinical genetics of human dilated cardiomyopathy. Eur Heart
J. 36:1123–1135a. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kadhi A, Mohammed F and Nemer G: The
genetic pathways underlying immunotherapy in dilated
cardiomyopathy. Front Cardiovasc Med. 8(613295)2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Merlo M, Pivetta A, Pinamonti B, Stolfo D,
Zecchin M, Barbati G, Di Lenarda A and Sinagra G: Long-term
prognostic impact of therapeutic strategies in patients with
idiopathic dilated cardiomyopathy: Changing mortality over the last
30 years. Eur J Heart Fail. 16:317–324. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Linde C, Grabowski M, Ponikowski P, Rao I,
Stagg A and Tschope C: Cardiac contractility modulation therapy
improves health status in patients with heart failure with
preserved ejection fraction: A pilot study (CCM-HFpEF). Eur J Heart
Fail. 24:2275–2284. 2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Liang B, Zhou Z, Yang Z, Liu J, Zhang L,
He J, Li H, Huang Y, Yang Q, Xian S and Wang L: AGEs-RAGE axis
mediates myocardial fibrosis via activation of cardiac fibroblasts
induced by autophagy in heart failure. Exp Physiol. 107:879–891.
2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Burr SD and Stewart JA Jr: Extracellular
matrix components isolated from diabetic mice alter cardiac
fibroblast function through the AGE/RAGE signaling cascade. Life
Sci. 250(117569)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Boengler K, Hilfiker-Kleiner D, Drexler H,
Heusch G and Schulz R: The myocardial JAK/STAT pathway: From
protection to failure. Pharmacol Ther. 120:172–185. 2008.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Okonko DO, Marley SB, Anker SD,
Poole-Wilson PA and Gordon MY: Erythropoietin resistance
contributes to anaemia in chronic heart failure and relates to
aberrant JAK-STAT signal transduction. Int J Cardiol. 164:359–364.
2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Terrell AM, Crisostomo PR, Wairiuko GM,
Wang M, Morrell ED and Meldrum DR: Jak/STAT/SOCS signaling circuits
and associated cytokine-mediated inflammation and hypertrophy in
the heart. Shock. 26:226–234. 2006.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chen SN, Lombardi R, Karmouch J, Tsai JY,
Czernuszewicz G, Taylor MRG, Mestroni L, Coarfa C, Gurha P and
Marian AJ: DNA damage Response/TP53 pathway is activated and
contributes to the pathogenesis of dilated cardiomyopathy
associated with LMNA (Lamin A/C) mutations. Circ Res. 124:856–873.
2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Das B, Young D, Vasanji A, Gupta S, Sarkar
S and Sen S: Influence of p53 in the transition of
myotrophin-induced cardiac hypertrophy to heart failure. Cardiovasc
Res. 87:524–534. 2010.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Fujita T and Ishikawa Y: Apoptosis in
Heart Failure-The role of the beta-adrenergic receptor-mediated
signaling pathway and p53-mediated signaling pathway in the
apoptosis of cardiomyocytes. Circ J. 75:1811–1818. 2011.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Irie T, Sips PY, Kai S, Kida K, Ikeda K,
Hirai S, Moazzami K, Jiramongkolchai P, Bloch DB, Doulias PT, et
al: S-Nitrosylation of Calcium-handling proteins in cardiac
adrenergic signaling and hypertrophy. Circ Res. 117:793–803.
2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Persoon S, Paulus M, Hirt S, Jungbauer C,
Dietl A, Luchner A, Schmid C, Maier LS and Birner C: Cardiac
unloading by LVAD support differentially influences components of
the cGMP-PKG signaling pathway in ischemic and dilated
cardiomyopathy. Heart Vessels. 33:948–957. 2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Pleger ST, Boucher M, Most P and Koch WJ:
Targeting myocardial beta-adrenergic receptor signaling and calcium
cycling for heart failure gene therapy. J Card Fail. 13:401–414.
2007.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Port JD and Bristow MR: Altered
beta-adrenergic receptor gene regulation and signaling in chronic
heart failure. J Mol Cell Cardiol. 33:887–905. 2001.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Razmara E and Garshasbi M: Whole-exome
sequencing identifies R1279X of MYH6 gene to be associated with
congenital heart disease. BMC Cardiovasc Disord.
18(137)2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Carniel E, Taylor MR, Sinagra G, Di
Lenarda A, Ku L, Fain PR, Boucek MM, Cavanaugh J, Miocic S, Slavov
D, et al: Alpha-myosin heavy chain: A sarcomeric gene associated
with dilated and hypertrophic phenotypes of cardiomyopathy.
Circulation. 112:54–59. 2005.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Hao E, Zhang G, Mu L, Ma N and Wang T:
Establishment of a human MYH6 compound heterozygous knockout hESC
line to model cardiomyopathy and congenital heart defects by
CRISPR/Cas9 system. Stem Cell Res. 50(102128)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Hershberger RE, Norton N, Morales A, Li D,
Siegfried JD and Gonzalez-Quintana J: Coding sequence rare variants
identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312
patients with familial or idiopathic dilated cardiomyopathy. Circ
Cardiovasc Genet. 3:155–161. 2010.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Merlo M, Sinagra G, Carniel E, Slavov D,
Zhu X, Barbati G, Spezzacatene A, Ramani F, Salcedo E, Di Lenarda
A, et al: Poor prognosis of rare sarcomeric gene variants in
patients with dilated cardiomyopathy. Clin Transl Sci. 6:424–428.
2013.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Chen JH, Wang LL, Tao L, Qi B, Wang Y, Guo
YJ and Miao L: Identification of MYH6 as the potential gene for
human ischaemic cardiomyopathy. J Cell Mol Med. 25:10736–10746.
2021.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Chelbi ST, Wilson ML, Veillard AC, Ingles
SA, Zhang J, Mondon F, Gascoin-Lachambre G, Doridot L, Mignot TM,
Rebourcet R, et al: Genetic and epigenetic mechanisms collaborate
to control SERPINA3 expression and its association with placental
diseases. Hum Mol Genet. 21:1968–1978. 2012.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Asakura M and Kitakaze M: Global gene
expression profiling in the failing myocardium. Circ J.
73:1568–1576. 2009.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Delrue L, Vanderheyden M, Beles M,
Paolisso P, Di Gioia G, Dierckx R, Verstreken S, Goethals M,
Heggermont W and Bartunek J: Circulating SERPINA3 improves
prognostic stratification in patients with a de novo or worsened
heart failure. ESC Heart Fail. 8:4780–4790. 2021.PubMed/NCBI View Article : Google Scholar
|
|
62
|
di Salvo TG, Yang KC, Brittain E, Absi T,
Maltais S and Hemnes A: Right ventricular myocardial biomarkers in
human heart failure. J Card Fail. 21:398–411. 2015.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Jiang Z, Guo N and Hong K: A three-tiered
integrative analysis of transcriptional data reveals the shared
pathways related to heart failure from different aetiologies. J
Cell Mol Med. 24:9085–9096. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Lok SI, van Mil A, Bovenschen N, van der
Weide P, van Kuik J, van Wichen D, Peeters T, Siera E, Winkens B,
Sluijter JP, et al: Post-transcriptional regulation of
α-1-antichymotrypsin by microRNA-137 in chronic heart failure and
mechanical support. Circ Heart Fail. 6:853–861. 2013.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Sanchez-Navarro A, Gonzalez-Soria I,
Caldino-Bohn R and Bobadilla NA: An integrative view of serpins in
health and disease: The contribution of SerpinA3. Am J Physiol Cell
Physiol. 320:C106–C108. 2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Gerarduzzi C, Kumar RK, Trivedi P, Ajay
AK, Iyer A, Boswell S, Hutchinson JN, Waikar SS and Vaidya VS:
Silencing SMOC2 ameliorates kidney fibrosis by inhibiting
fibroblast to myofibroblast transformation. JCI Insight.
2(e90299)2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Williams JL, Cavus O, Loccoh EC, Adelman
S, Daugherty JC, Smith SA, Canan B, Janssen PML, Koenig S, Kline
CF, et al: Defining the molecular signatures of human right heart
failure. Life Sci. 196:118–126. 2018.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Laugier L, Frade AF, Ferreira FM, Baron
MA, Teixeira PC, Cabantous S, Ferreira LRP, Louis L, Rigaud VOC,
Gaiotto FA, et al: Whole-Genome Cardiac DNA methylation fingerprint
and gene expression analysis provide new insights in the
pathogenesis of chronic chagas disease cardiomyopathy. Clin Infect
Dis. 65:1103–1111. 2017.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Luo L, Wang CC, Song XP, Wang HM, Zhou H,
Sun Y, Wang XK, Hou S and Pei FY: Suppression of SMOC2 reduces
bleomycin (BLM)-induced pulmonary fibrosis by inhibition of
TGF-β1/SMADs pathway. Biomed Pharmacother. 105:841–847.
2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Schmidt IM, Colona MR, Kestenbaum BR,
Alexopoulos LG, Palsson R, Srivastava A, Liu J, Stillman IE, Rennke
HG, Vaidya VS, et al: Cadherin-11, Sparc-related modular calcium
binding protein-2, and Pigment epithelium-derived factor are
promising non-invasive biomarkers of kidney fibrosis. Kidney Int.
100:672–683. 2021.PubMed/NCBI View Article : Google Scholar
|
|
71
|
McLellan MA, Skelly DA, Dona MSI, Squiers
GT, Farrugia GE, Gaynor TL, Cohen CD, Pandey R, Diep H, Vinh A, et
al: High-resolution transcriptomic profiling of the heart during
chronic stress reveals cellular drivers of cardiac fibrosis and
hypertrophy. Circulation. 142:1448–1463. 2020.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Agra RM, Fernandez-Trasancos A, Sierra J,
Gonzalez-Juanatey JR and Eiras S: Differential association of
S100A9, an inflammatory marker, and p53, a cell cycle marker,
expression with epicardial adipocyte size in patients with
cardiovascular disease. Inflammation. 37:1504–1512. 2014.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Marinkovic G, Koenis DS, de Camp L,
Jablonowski R, Graber N, de Waard V, de Vries CJ, Goncalves I,
Nilsson J, Jovinge S and Schiopu A: S100A9 links inflammation and
repair in myocardial infarction. Circ Res. 127:664–676.
2020.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Pei XM, Tam BT, Sin TK, Wang FF, Yung BY,
Chan LW, Wong CS, Ying M, Lai CW and Siu PM: S100A8 and S100A9 are
associated with doxorubicin-induced Cardiotoxicity in the heart of
diabetic mice. Front Physiol. 7(334)2016.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Shah RD, Xue C, Zhang H, Tuteja S, Li M,
Reilly MP and Ferguson JF: Expression of Calgranulin Genes S100A8,
S100A9 and S100A12 Is modulated by n-3 PUFA during inflammation in
adipose tissue and mononuclear cells. PLoS One.
12(e0169614)2017.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Wei X, Wu B, Zhao J, Zeng Z, Xuan W, Cao
S, Huang X, Asakura M, Xu D, Bin J, et al: Myocardial hypertrophic
preconditioning attenuates cardiomyocyte hypertrophy and slows
progression to heart failure through upregulation of S100A8/A9.
Circulation. 131:1506–1517. 2015.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Marinković G, Koenis DS, de Camp L,
Jablonowski R, Graber N, de Waard V, de Vries CJ, Goncalves I,
Nilsson J, Jovinge S and Schiopu A: S100A9 links inflammation and
repair in myocardial infarction. Circ Res. 127:664–676.
2020.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Marinkovic G, Grauen Larsen H, Yndigegn T,
Szabo IA, Mares RG, de Camp L, Weiland M, Tomas L, Goncalves I,
Nilsson J, et al: Inhibition of pro-inflammatory myeloid cell
responses by short-term S100A9 blockade improves cardiac function
after myocardial infarction. Eur Heart J. 40:2713–2723.
2019.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Bizou M, Itier R, Majdoubi M, Abbadi D,
Pichery E, Dutaur M, Marsal D, Calise D, Garmy-Susini B,
Douin-Echinard V, et al: Cardiac macrophage subsets differentially
regulate lymphatic network remodeling during pressure overload. Sci
Rep. 11(16801)2021.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Vieira JM, Norman S, Villa Del Campo C,
Cahill TJ, Barnette DN, Gunadasa-Rohling M, Johnson LA, Greaves DR,
Carr CA, Jackson DG and Riley PR: The cardiac lymphatic system
stimulates resolution of inflammation following myocardial
infarction. J Clin Invest. 128:3402–3412. 2018.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Chen Y, Wang D, Peng H, Chen X, Han X, Yu
J, Wang W, Liang L, Liu Z, Zheng Y, et al: Epigenetically
upregulated oncoprotein PLCE1 drives esophageal carcinoma
angiogenesis and proliferation via activating the PI-PLCε-NF-κB
signaling pathway and VEGF-C/Bcl-2 expression. Mol Cancer.
18(1)2019.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Li W, Li Y, Chu Y, Wu W, Yu Q, Zhu X and
Wang Q: PLCE1 promotes myocardial ischemia-reperfusion injury in
H/R H9c2 cells and I/R rats by promoting inflammation. Biosci Rep.
39(BSR20181613)2019.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Youn JC, Jung MK, Yu HT, Kwon JS, Kwak JE,
Park SH, Kim IC, Park MS, Lee SK, Choi SW, et al: Increased
frequency of CD4+CD57+ senescent T cells in
patients with newly diagnosed acute heart failure: Exploring new
pathogenic mechanisms with clinical relevance. Sci Rep.
9(12887)2019.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Zeng Z, Wang K, Li Y, Xia N, Nie S, Lv B,
Zhang M, Tu X, Li Q, Tang T and Cheng X: Down-regulation of
microRNA-451a facilitates the activation and proliferation of
CD4+ T cells by targeting Myc in patients with dilated
cardiomyopathy. J Biol Chem. 292:6004–6013. 2017.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Rao M, Wang X, Guo G, Wang L, Chen S, Yin
P, Chen K, Chen L, Zhang Z, Chen X, et al: Resolving the
intertwining of inflammation and fibrosis in human heart failure at
single-cell level. Basic Res Cardiol. 116(55)2021.PubMed/NCBI View Article : Google Scholar
|