Introduction
Atopic dermatitis (AD) is a common chronic
inflammatory skin disease that is characterized by eczema-like
lesions accompanied by intense itching. This condition affects
individuals of all age groups and ethnicities, with 20% children
and 10% adults suffering from this condition in high-income
countries. Although this condition is typically non-fatal, it does
place significant burden on the patient (1,2). The
pathogenesis of AD involves a multitude of factors, including skin
barrier disorders, microbial dysbiosis and immune dysregulation.
These factors interact in a complex multidirectional network that
can exacerbate atopic skin diseases, although targeted therapies
can also alleviate the condition (3). Combating skin barrier dysfunction has
been an important aspect of clinical management for this disease,
with topical emollients being the first-line treatment for AD
(4).
The skin provides a key physical barrier between the
body and the external environment. This barrier structure consists
of the cuticle and tight junctions, which prevents transepithelial
water loss and the entry of external antigens (5). Damage to skin barrier function leads
to increased sensitivity of the body to environmental allergens and
various stimuli, triggering an inflammatory cascade reaction,
leading to immune disorders and eventually the onset of AD.
However, the specific mechanism underlying this process remains
unclear.
Skin lesions as a result of AD have been observed to
exhibit the dysregulated expression of several genes associated
with keratinocyte activity and T cell infiltration, including
Th2-related genes (such as IL-4, IL-10 and IL-13) and Th22-related
genes (such as IL-22) (6,7). In addition, cytokines serve an
important role in mediating inflammatory responses and regulating
the immune response. Transcriptomic sequencing is becoming
increasingly popular in recent years for analyzing the mechanism
underlying the roles mediated by differentially expressed genes
(DEGs) in AD and improving understanding into this disease.
MicroRNAs (miR/miRNA) can participate in various
regulatory processes, such as virus defense, cell proliferation,
cell apoptosis and organ formation, in addition to serving an
important role in regulating the inflammatory response in a number
of diseases, including AD (8).
miR-155 serves an important role in the pathogenesis
of AD. miR-155 is expressed by cutaneous T cells, dendritic cells
and mast cells. Previously studies have shown that miR-155 is
overexpressed in patients with AD, such that it is the most
significantly upregulated microRNA in terms of expressions
(9,10). It can enhance T cell proliferation
by inhibiting cytotoxic T lymphocyte associated antigen-4 (CTLA-4)
(11). In AD mouse models, miR-155
has been shown to target cAMP-dependent protein kinase inhibitor α
and regulate tight junction protein expression, which in turn
affect epithelial barrier function (10). In the skin model of AD, IL-32 has
been reported to promote the expression of Janus kinase 1 and
upregulate miR-155 expression, leading to the occurrence of AD
inflammation (9). Although
evidence on the role of miR-155 in the pathogenesis of AD has been
accumulating, the mechanism of miR-155 underlying the development
of AD remains unclear and requires further research.
To investigate the role of miR-155 in the
pathogenesis of AD, a human immortalized keratinocyte cell line
HaCaT was used as an in vitro model to screen for DEGs in
the epidermal immune microenvironment. In addition, immune cell
infiltration and prediction miR-155 target genes were assessed
using bioinformatic techniques. The aim of the present study was to
investigate the mechanism of miR-155 in immune infiltration in AD
and provide novel gene targets for diagnosis and treatment.
Materials and methods
Data acquisition and processing
The gene expression profile datasets were retrieved
from the GEO database (https://www.ncbi.nlm.nih.gov/gds/) according to the
following conditions: i) For AD; ii) the biological type was human;
and iii) the sample type was skin tissue. Finally,
GSE121212(12) and
GSE157194(13) were selected as
the subject of research and analysis. GSE121212 included skin
biopsy specimens from lesion and non-lesion sites from 21 patients.
By contrast, GSE157194 included lesion skin samples from 57
patients and non-lesion skin samples from 54 of these patients. The
gene expression matrices of GSE121212 and GSE157194 were downloaded
from the GEO database.
Screening for DEGs
DEGs were screened using the R-package (R version
4.2.3; http://www.R-project.org/) ‘limma-voom’
(version 3.40.6) (14) and the
screening criteria were as follows: log fold change >2,
P<0.05. Fold change is the fold change of the DEGs. The DEGs
after screening were visualized in the form of a heat map and a
volcano map.
Screening for differentially
co-expressed genes in the two datasets
The DEGs found in GSE121212 and GSE157194 were cut
from the VENNY 2.1.0 website (https://bioinfogp.cnb.csic.es/tools/venny/) to obtain
the differentially co-expressed genes.
Gene Ontology (GO) functional
enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analysis of differentially co-expressed
genes
The R software package ‘Cluster Profiler’ (version
4.8.3, https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
was used to perform GO function enrichment and KEGG pathway
enrichment analysis of differentially co-expressed genes, before
the enrichment results of differentially co-expressed genes were
obtained. P<0.05 was used as the evaluation standard and visual
analysis was performed. The GO analysis included the following
three aspects: Biological process (BP), molecular function (MF) and
cellular localization (CC).
Prediction of target genes of
miR-155-3p
The downstream target genes of miR-155-3p were
predicted using the miRNet 2.0 website (https://www.mirnet.ca/miRNet/home.xhtml). VENNY 2.1.0
site was used to screen for the intersection of the significantly
different co-expressed genes and the downstream target genes of
miR-155-3p, using which the genes designated for further studies
would be screened.
Construction of the protein-protein
interaction (PPI) network
As proteins rarely function alone, there was a need
to study the interactions among proteins. To identify potentially
important protein interactions, significantly differentially
co-expressing genes were screened using the adjusted log fold
change >2, P<0.05. The selected significantly differentially
coexpressing genes were imported into the STRING 12.0 database
(https://cn.string-db.org/) to construct
a PPI network.
Searching the gene expression data in
HaCaT cells
The Human Protein Atlas website (https://www.proteinatlas.org/) provides the RNA
expression data (normalized transcript per million values of cell
lines) of different genes in different cell lines; therefore, The
Human Protein Atlas was used to identify the expression levels of
target genes in HaCaT cells.
Cell culture and inflammatory cell
model
HaCaT cells (cat. no. CL-0090; Procell Life Science
& Technology Co., Ltd.) were cultured in minimal essential
medium (Procell Life Science & Technology Co., Ltd.)
supplemented with 10% FBS (Procell Life Science & Technology
Co., Ltd.) at 37˚C in 5% CO2. The cells were treated
with or without TNF-α and IFN-γ (5 ng/ml; PeproTech, Inc.) for 6 h.
At this time, the cells were termed the ‘TI’ group.
Transfection with the miR-155 mimics
or inhibitor
HaCaT cells were first seeded into six-well plates
at a density of 1x107 cells/ml. At 80% confluence, the
cells were treated with 100 pmol either miR-155 inhibitor
(5'-AACCCCUAUCACGAUUAGCAUUAA-3') or the inhibitor control
(5'-GUCCCUCACAUCAUAAGCUAAUAA-3'), or with miR-155 mimics (sense,
5'-UUAAUGCUAAUCGUGAUAGGGGUU-3' and antisense,
5'-CCCCUAUCACGAUUAGCAUUAAUU-3') or with the mimics control (sense,
5'-UUCUCCGAACGUGUCACGUTT-3' and antisense,
5'-ACGUGACACGUUCGGAGAATT-3') (Shanghai GenePharma Co., Ltd.) using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocols. The RNA-lipid complexes
were first added to the HaCaT cells before the medium was changed
after 6 h, and the cells continued to be transfected at 37˚C for 42
h.
Reverse transcription-quantitative PCR
(RT-qPCR)
Different groups of HaCaT cells were lysed with 1 ml
TRIzol® reagent (Thermo Fisher Scientific, Inc.) for 5
min. Then, 200 µl chloroform was added to the lysed samples for 3
min at room temperature. Following centrifugation at 13,400 x g for
15 min at 4˚C, the supernatant was collected and mixed with 500 µl
isopropanol. The mixture was then kept at 4˚C for 10 min. The
sample was removed and centrifuged at 13,400 x g for 15 min at 4˚C.
The supernatant was discarded and the pellet was washed twice with
500 µl 75% ethanol (centrifuged at 13,400 x g for 5 min at 4˚C).
Total RNA was obtained by adding 20 µl RNase-free ddH2O.
cDNA was synthesized with ReverTra Ace® qPCR RT Kit
(Toyobo Life Science), according to the manufacturer's protocol.
Gene expression levels were determined in the CFX Connect Real-Time
System (Bio-Rad Laboratories, Inc.) using the SYBR Green PCR Master
Mix (Thermo Fisher Scientific, Inc.). The miR-155-5p and U6
specific bulge loop miRNA RT-qPCR primer sets (one RT primer and
one pair of qPCR primers in each set) were developed by Sangon
Biotech Co., Ltd. GAPDH was used as an internal control for mRNA
normalization and U6 was used as an internal control for miRNA
normalization. The thermocycling conditions were as follows:
Initial denaturation at 95˚C for 30 sec; followed by 40 cycles of
denaturation at 95˚C for 5 sec, and annealing and elongation at
60˚C for 30 sec. The mRNA primer sequences used for RT-qPCR (Sangon
Biotech Co., Ltd.) were as follows Human Elafin forward,
5'-CACTGTCAAAGGCCGTGTTC-3' and reverse,
5'-GCGGTTAGGGGGATTCAACAG-3'; human FOS-like 1, AP-1 transcription
factor subunit (FRA1 or FOSL) forward, 5'-CTGACCTACCCTCAGTACAGC-3'
and reverse, 5'-AAGTCGGTCAGTTCCTTCCTC-3'; human C-X-C motif
chemokine ligand (CXCL)1 forward, 5'-GGGAATTCACCCCAAGAACATC-3' and
reverse, 5'-GGATGCAGGATTGAGGCAAGC-3'; human CXCL-8 forward,
5'-CACTGCGCCAACACAGAAAT-3' and reverse,
5'-GCCCTCTTCAAAAACTTCTCCAC-3' and human GAPDH forward,
5'-AATTCCATGGCACCGTCAAG-3' and reverse, 5'-AGCATCGCCCCACTTGATTT-3'.
Gene expression was normalized to that of GAPDH or U6 expression
and relative expression was calculated using the 2-ΔΔCq
method (15).
Measurement of cell proliferation
HaCaT cells were seeded into 96-well plates at a
density of 1.5x104 cells/well. Cell Count Kit-8 (CCK8;
Dojindo Laboratories, Inc.) reagent was added at 90% confluence of
the cells according to the manufacturer's protocols. After
incubation at 37˚C for 2 h, the absorbance value of each well was
measured at wavelength of 450 nm using a microplate reader.
Measurement of cytokine levels
The concentrations of IL-1β (cat. no. E-EL-H0149c),
IL-6 (cat. no. E-EL-H6156), IL-10 (cat. no. E-EL-H6154), IL-15
(cat. no. E-EL-H0222c), CXCL1 (cat. no. E-EL-H0045c) and CXCL8
(cat. no. E-EL-H6008) in cell culture supernatants were measured
using ELISA kits (Elabscience Biotechnology, Inc.) according to the
manufacturer's protocols.
Western blotting analysis
The cells were scraped following the addition of the
RIPA protein lysis solution (RIPA: phenylmethylsulfonyl fluoride,
100:1). Samples were then collected into microcentrifuge tubes and
lysed for 20 min. The protein concentrations of the samples were
determined with a BCA protein assay kit (Beyotime, Institute of
Biotechnology). Total protein extracts (25 µg) were separated by
SDS-PAGE on 10% gels and transferred to PVDF membranes. The
membranes were blocked with a Blocking Buffer (Beyotime Institute
of Biotechnology) for 1.5 h at room temperature and washed three
times for 15 min each with TBS -0.1% Tween at room temperature. The
membranes were then incubated overnight at 4˚C with antibodies
directed against Elafin (cat. no. ab184972; 1:1,000 dilution;
Abcam), FRA1 (cat. no. 5281; 1:1,000 dilution; Cell Signaling
Technology, Inc.) or β-tubulin (cat. no. 10094-1-AP; 1:200,000
dilution; ProteinTech, Inc.). After the membranes were washed, they
were probed with a HRP-linked goat anti-rabbit IgG antibody (cat.
no. A0208; 1:1,000 dilution; Beyotime Institute of Biotechnology)
or the HRP-linked goat anti-mouse IgG antibody (cat. no. A0216;
1:1,000 dilution; Beyotime Institute of Biotechnology) for 1 h at
room temperature. Protein bands were detected with the Immobilon
Western Chemiluminescent HRP Substrate (cat. no. P0018S, Beyotime
Institute of Biotechnology) and protein expression was quantified
with a gel analysis software. The density of each specific band was
measured using ImageJ software V1.53t (National Institutes of
Health).
Statistical analysis
Data are expressed as means ± standard deviations
(SD). Data with several groups were compared using one-way analysis
of variance followed by Tukey's test, whereas student's t-test was
used to compare two groups, using the GraphPad Prism 8 (GraphPad
Software, Inc.; Dotmatics) software. One-way ANOVA followed by
Tukey's post-hoc test was used to compare multiple treatment
groups. P<0.05 was considered to indicate a statistically
significant difference. All experiments were repeated three times.
Statistical significance was set at *P<0.05,
**P<0.01 and ***P<0.001.
Results
Screening of DEGs
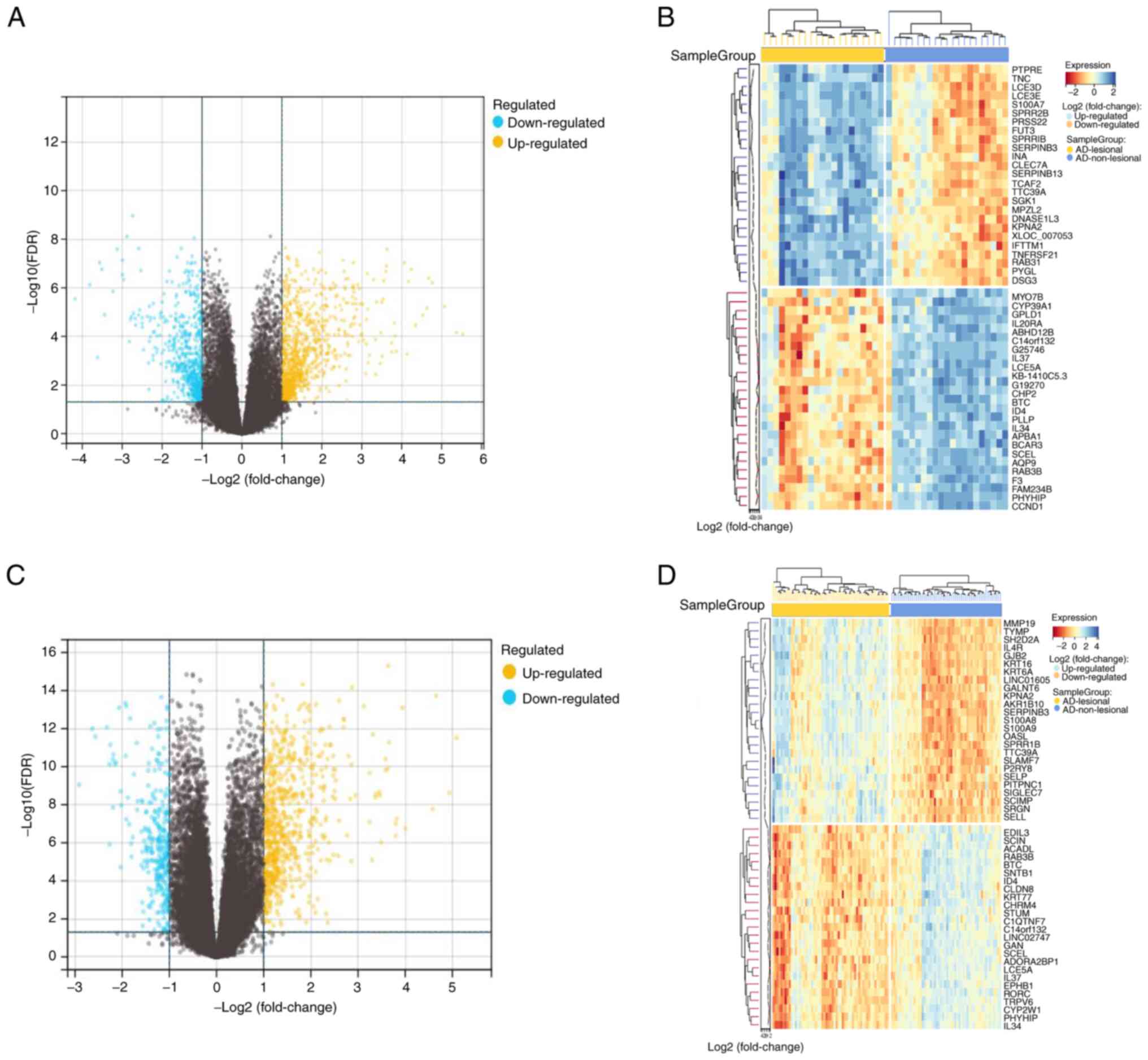
The high-throughput sequencing datasets of GSE121212
and GSE157194 was obtained from GEO and analyzed using the
R-packages limma (version 3.40.6) to target DEGs based on a
criteria of log-fold change >2 and P<0.05. A total of 1,547
DEGs were identified in the GSE121212 dataset, with 920 genes found
to be upregulated and 627 downregulated. A total of 1,031 DEGs were
identified in the GSE157194 dataset, with 731 genes being
upregulated and 300 downregulated. The results were visualized by
generating volcano and heat maps of the DEGs (Fig. 1).
Screening for differentially
co-expressed genes in the two datasets
The online bioinformatics analysis tool VENNY was
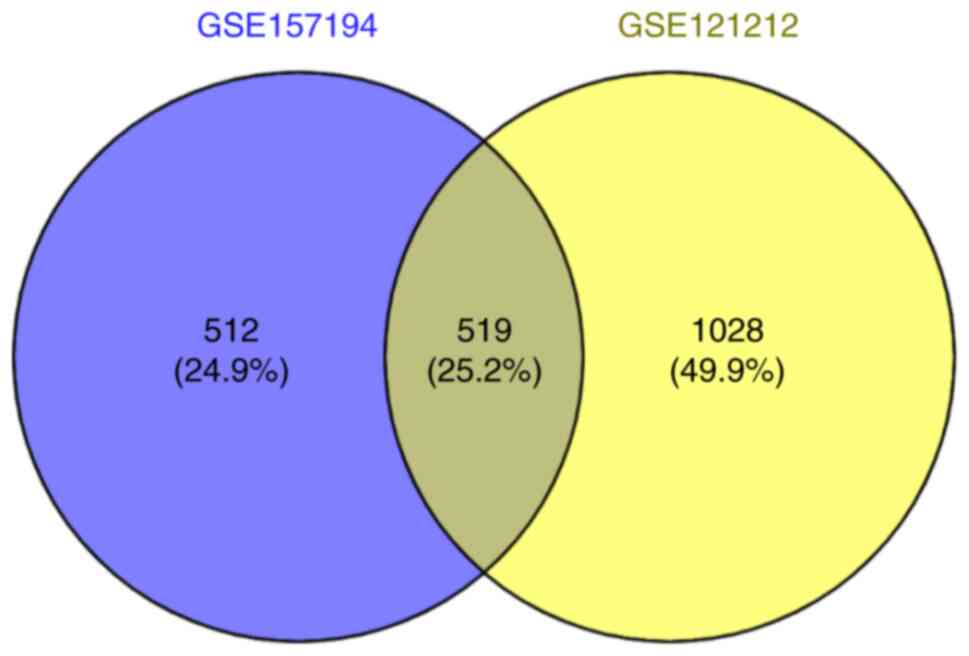
used to screen for the differentially co-expressed genes. A total
of 519 co-expressed differential genes were screened out (Fig. 2).
Analysis of functional GO and KEGG
pathway-enriched co-expressed differential genes
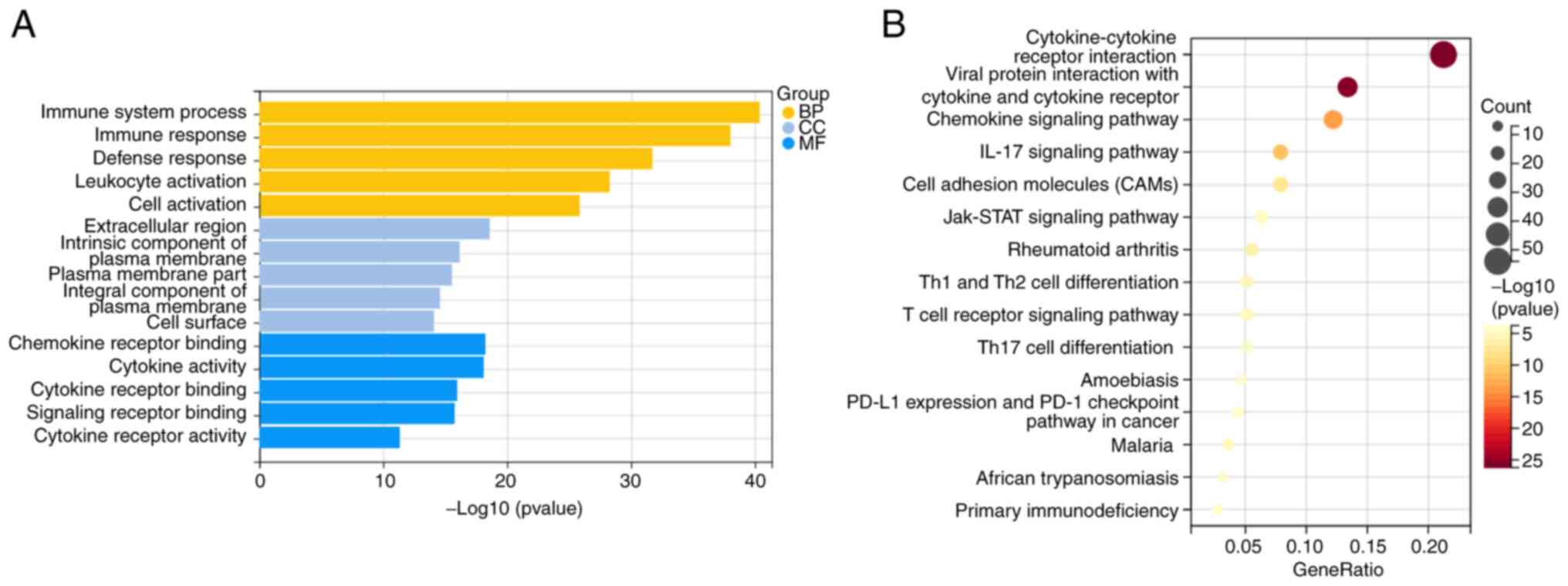
The 519 differentially co-expressed genes were then
underwent functional GO accumulation analysis and KEGG pathway
accumulation analysis. In total 1,023 functions were subjected to
GO analysis, with annotations categorized into BP, CC and MF. For
BP, the co-expressed differential genes were found significantly
enriched in the ‘process of the immune system’ and ‘immune
response.’ In terms of CC and MF, the co-expressed differential
genes were significantly enriched in the ‘extracellular region’ and
‘chemokine receptor binding’, respectively (Fig. 3A). In addition, critical signaling
pathways were identified using KEGG enrichment analysis. The
co-expressed differential genes were found to be significantly
enriched in the ‘cytokine-cytokine receptor interaction’ (Fig. 3B).
Prediction of target genes of
miR-155-3p
The miRNet website was used to predict the
downstream target genes of miR-155-3p. miRNet predicted 4,724
possible downstream targets. To assess the reliability of the
results, the GSE121212 and GSE157194 datasets were screened on a
smaller scale. The significant DEGs were identified using the
criteria log-fold change >2 and P<0.05. A total of 210
significant DEGs were identified in the GSE121212 dataset, with 158
genes being upregulated and 52 downregulated. By contrast, 122
significant DEGs were screened from the GSE157194 dataset, of which
106 genes were upregulated and 16 genes were downregulated.
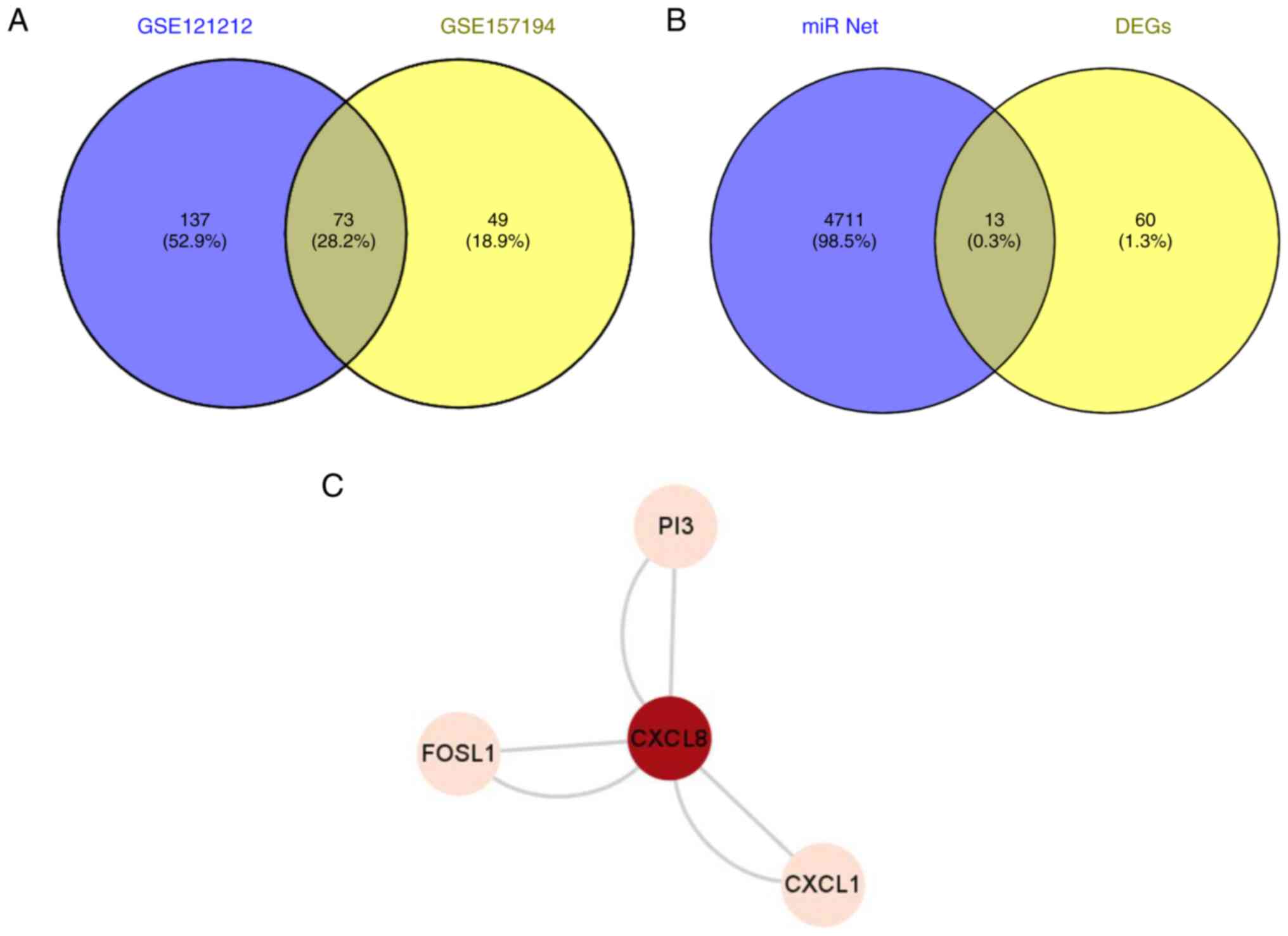
The significant DEGs of the two datasets were then
cut using the online bioinformatics analysis tool VENNY 2.1.0, from
which 73 significantly differentially co-expressed genes were
obtained (Fig. 4A). The
miR-net-predicted intersection of 73 significant DEGs and possible
miR-155-3p downstream target genes was then obtained from the VENNY
2.1.0 site, yielding 13 genes (Fig.
4B). These genes included CXCL8, keratin 6B, selectin E, PI3,
glutathione-Specific γ-glutamylcyclotransferase 1, transcobalamin
1, 2'-5'-oligoadenylate synthetase-like, CXCL1, C6orf223, insulin
growth factor-like family member 1, MMP1, aldo-keto reductase
family 1 member B10 and FOSL1.
Using The Human Protein Atlas website, the
transcript levels of these 13 proteins in HaCaT cells were searched
(Table I). The results showed that
among the 13 proteins, PI3, FOSL1, CXCL8 and CXCL1 were the four
proteins with the highest expression levels in HaCaT cells, where
possible interaction among these four proteins was predicted
(Fig. 4C).
 | Table IRNA expression data of genes in HaCaT
cells. |
Table I
RNA expression data of genes in HaCaT
cells.
| Gene | Normalized TPM
value |
|---|
| CXCL8 | 48.2 |
| CXCL1 | 38.6 |
| FOS-like 1, AP-1
transcription factor subunit | 60.6 |
| Peptidase Inhibitor
3 | 109.8 |
| Keratin 6B | 3.9 |
| Selectin E | 0 |
|
Glutathione-Specific
γ-glutamylcyclotransferase 1 | 6.0 |
| Transcobalamin
1 | 5.5 |
|
2'-5'-Oligoadenylate synthetase-like | 6.4 |
| C6orf223 | 0 |
| Insulin growth
factor-like family member 1 | 6.9 |
| MMP1 | 4.6 |
| Aldo-keto reductase
family 1 member B10 | 6.2 |
Through functional GO accumulation analysis and KEGG
pathway accumulation analysis, chemokines appeared to serve a
particularly important role in the skin immune microenvironment in
the setting of AD. Therefore, it was speculated that PI3, FOSL1,
CXCL8 and CXCL1 can serve an important role in the epidermal immune
microenvironment of patients with AD. The effects of miR-155 on
PI3, FOSL1, CXCL8 and CXCL1 were therefore next focused upon for
subsequent in vitro cell experiments.
miR-155 can inhibit the proliferation
of HaCaT cells and promote the secretion of pro-inflammatory
cytokines
To study the role of miR-155 in human keratinocytes
(HaCaT cells), miR-155 mimics, mimics NC, miR-155 inhibitor and
inhibitor NC we transfected with Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.) transient transfection. After
transfection of miR-155 mimics, mimics NC, miR-155 inhibitor and
inhibitor NC in HaCaT cells for 24 h, the molecular level of
miR-155 in HaCaT cells was detected by RT-qPCR. The transfection
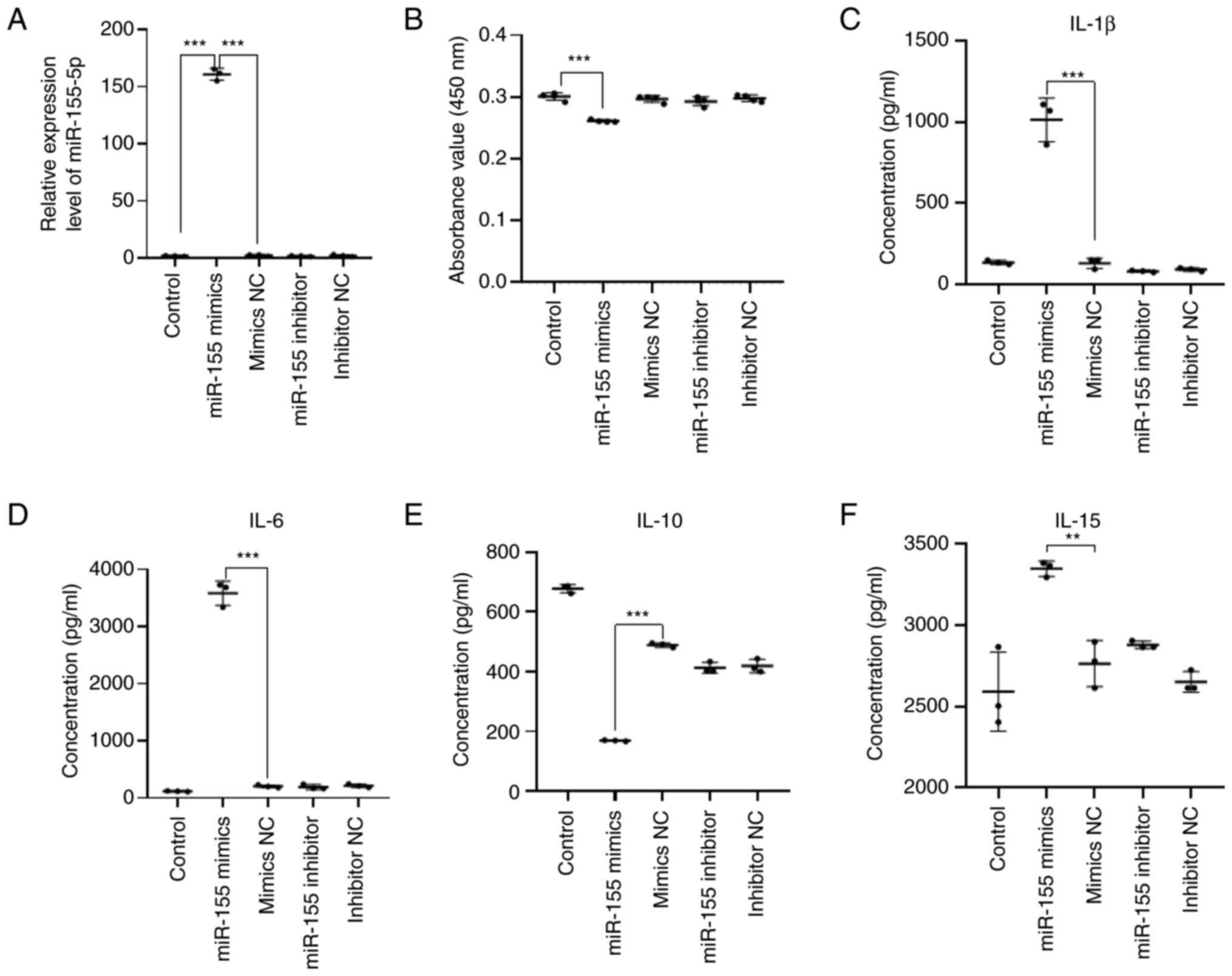
efficiency of miR-155 was detected. Compared with the mimics NC
group and the control group, the relative expression of miR-155 was
significantly increased in the miR-155 mimics group (P<0.001).
There was no significant difference in the relative expression of
miR-155 between mimics NC group, inhibitor NC group and control
group. There was no significant difference in the relative
expression of miR-155 between the miR-155 inhibitor group and the
inhibitor NC group (Fig. 5A). As
miR inhibitor can competitively bind to the downstream target genes
of mature miR and weaken the silencing effect of miR. Generally,
miRs are not degraded, so the molecular level of miR-155 can be
detected by RT-qPCR. This result suggested that transfection of
miR-155 mimics in HaCaT cells can significantly increase the
expression level of miR-155, which can be used for subsequent
experimental studies.
HaCaT cells were seeded into 96-well plates at a
density of 1.5x104 cells/well. Cell Count Kit-8 (CCK8)
reagent was then added at 90% cell confluency. After 2 h
incubation, the absorbance value of each well was measured at a
wavelength of 450 nm using a microplate reader. Compared with the
NC group, proliferation of HaCaT cells transfected with miR-155
mimics was found to be significantly inhibited (P<0.001;
Fig. 5B). This result suggested
that miR-155 can inhibit the proliferation of HaCaT cells.
Cytokines serve an important role in the
pathogenesis of AD. According to the aforementioned GO and KEGG
analyses, cytokines likely serve an important role in the epidermal
immune microenvironment of patients with AD. The potential effects
of miR-155 on cytokine secretion by HaCaT cells were therefore next
examined. Compared with those in the mimics NC group, the levels of
IL-1β (P<0.001), IL-6 (P<0.001) and IL-15 (P<0.01) in the
supernatant of HaCaT cells were found to be significantly increased
in the miR-155 mimics group, whilst those of IL-10 were
significantly decreased (P<0.001; Fig. 5C-F). These results suggest that the
overexpression of miR-155 in HaCaT cells can induce the secretion
of proinflammatory cytokines IL-1β, IL-6 and IL-15, whilst
inhibiting the secretion of the anti-inflammatory factor IL-10.
Effects of miR-155 on the predicted
target genes
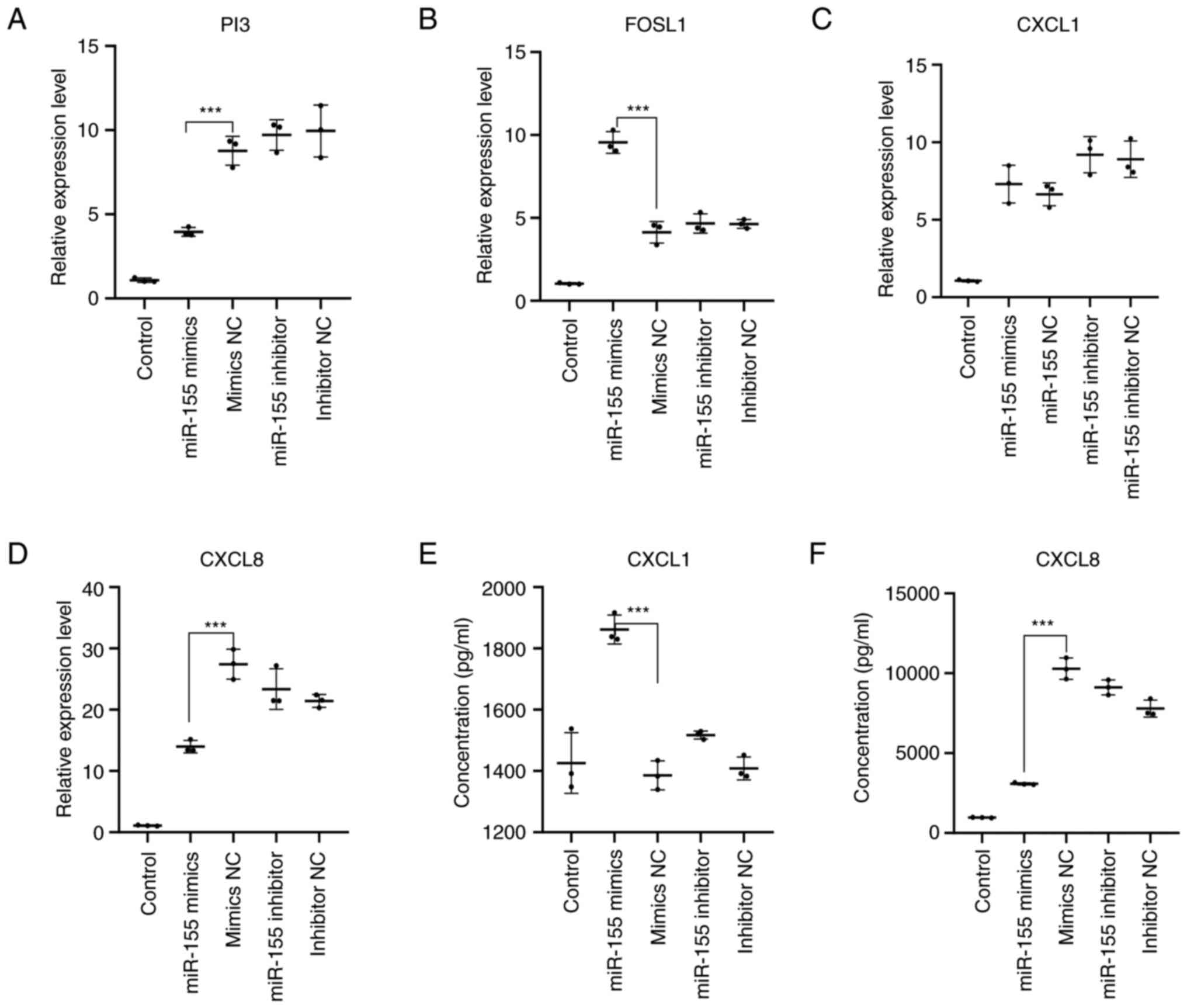
From the aforementioned experiments, high expression
of miR-155 can inhibit the proliferation of HaCaT cells while
promoting the secretion of pro-inflammatory cytokines. Through
bioinformatic analysis, several potential miR-155 target genes were
screened out, including PI3, FOSL1, CXCL1 and CXCL8. To determine
the effects of miR-155 on the expression of these potential target
genes and mechanism, changes in the expression of PI3, FOSL1, CXCL1
and CXCL8, in addition to the levels of CXCL1 and CXCL8 secretion
into the culture supernatant of HaCaT cells, were measured by
RT-qPCR and ELISA following miR-155 overexpression. RT-qPCR results
revealed that compared with those in the mimics NC group, the
expression levels of PI3 (P<0.001) and CXCL8 (P<0.001) were
significantly reduced in the miR-155 mimics group, whereas those of
FOSL1 (P<0.001) were increased. However, there was no
significant difference in the expression of CXCL1 in the miR-155
mimics group (Fig. 6A-D). ELISA
found that the level of CXCL1 secretion (P<0.001) was
significantly increased in the miR-155 mimics group compared with
that in the mimics NC group, whilst the secretion level of CXCL8
(P<0.001) was significantly decreased (Fig. 6E and F). These results suggested that miR-155
overexpression in HaCaT cells under physiological conditions
resulted in decreased PI3 expression, increased FOSL1 and CXCL1
secretion, but decreased CXCL8 expression and secretion.
Effects of TNF-α and IFN-γ treatment
on HaCaT cell proliferation and cytokine secretion
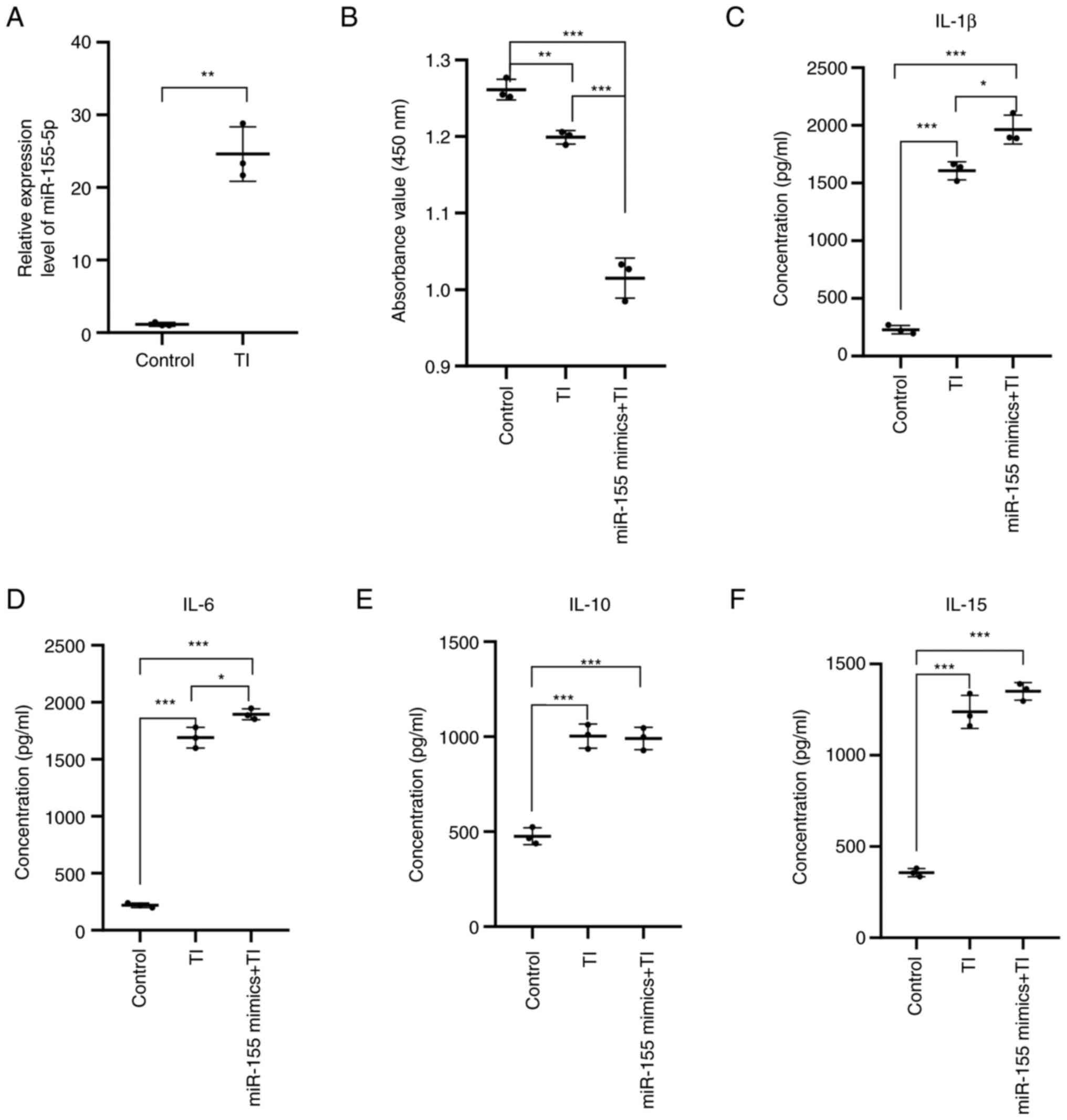
To examine the effect of TNF-α and IFN-γ on the
expression of miR-155 in HaCaT cells, 5 ng/ml TNF-α and IFN-γ were
added into the HaCaT cell culture medium for 6 h, before the medium
was changed and further incubation for 48 h. The expression level
of miR-155 in the cells was then detected by RT-qPCR. Compared with
that in the control group, the expression level of miR-155 was
significantly increased after TNF-α and IFN-γ stimulation
(P<0.01; Fig. 7A). These
results suggested that stimulation of HaCaT cells with TNF-α and
IFN-γ increased miR-155 expression levels in HaCaT cells.
The HaCaT cells treated with TNF-α and IFN-γ and
transfected were then divided into the following three groups:
Control group; TNF-α + IFN-γ group; and TNF-α + IFN-γ + miR-155
overexpression group.
Following the aforementioned treatments, the cells
were seeded into 96-well plates at a density of 1.5x104
cells/well, before proliferation of each group was detected by CCK8
assay. The results showed that cell proliferation was significantly
increased in the TNF-α + IFN-γ group (P<0.01) compared to the
control group, whereas that in the TNF-α + IFN-γ + miR-155
overexpression group (P<0.001) was significantly inhibited
compared with that in the control group. Compared with that in the
TNF-α + IFN-γ group, cell proliferation was significantly inhibited
in the TNF-α + IFN-γ + miR-155 overexpression group (P<0.001;
Fig. 7B). These results suggest
that miR-155 can inhibit the proliferation of HaCaT cells under the
TNF-α- and IFN-γ-induced inflammatory state.
The inflammatory cell model of HaCaT cells was
established by stimulating HaCaT cells with TNF-α and IFN-γ. The
supernatant of the cell culture medium in the three groups was then
collected before the level of IL-1β, IL-6, IL-10 and IL-15 were
detected using ELISA. Compared with that in the control group, the
secretion of IL-1β (P<0.001), IL-6 (P<0.001), IL-10
(P<0.001) and IL-15 (P<0.001) by HaCaT cells in the TNF-α +
IFN-γ and the TNF-α + IFN-γ + miR-155 overexpression groups were
significantly increased. Compared with that in the TNF-α + IFN-γ
induction group, the secretion of IL-1β (P<0.05) and IL-6
(P<0.05) was increased in the TNF-α + IFN-γ + miR-155
overexpressed group, whilst the secretion of IL-10 and IL-15 did
not change (Fig. 7C-F) These
results suggested that miR-155 can promote the secretion of the
proinflammatory cytokines IL-1β and IL-6 in HaCaT cells under the
induction of TNF-α and IFN-γ.
Effect of miR-155 on TNF-α- and IFN-γ-
induced target genes
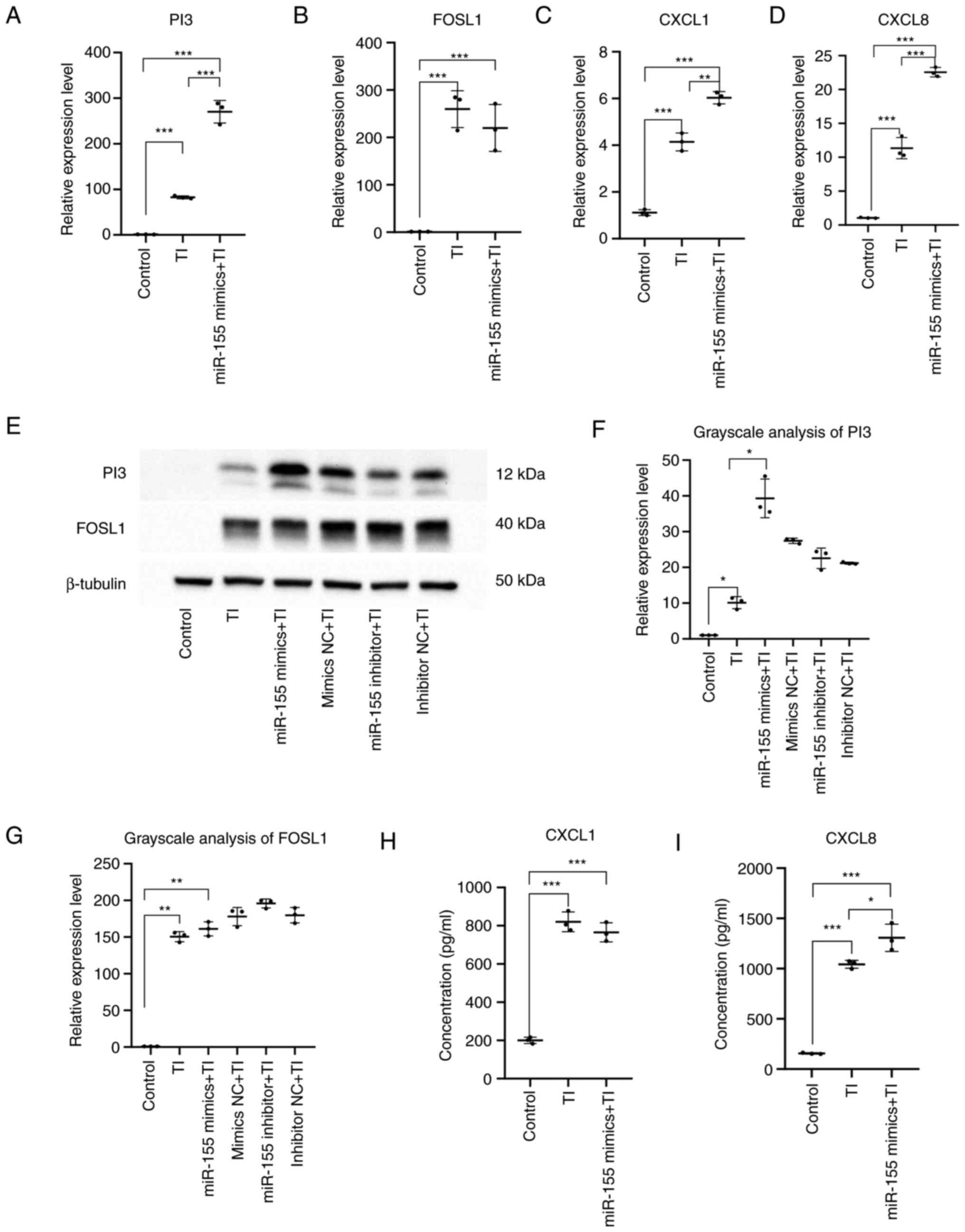
The inflammatory cell model of HaCaT cells was
constructed by stimulating HaCaT cells with TNF-α and IFN-γ. The
expression levels of PI3, FOSL1, CXCL1 and CXCL8 were then detected
by RT-qPCR, whereas western blotting was used to measure the
protein expression levels of PI3 and FOSL1. The levels of CXCL1 and
CXCL8 in the supernatant of HaCaT cells were detected by ELISA.
Compared with those in the control group, the mRNA
and protein expression levels of PI3 (P<0.05) and FOSL1
(P<0.01) whereas the mRNA expression levels and secretion of
CXCL1 (P<0.001) and CXCL8 (P<0.001) were significantly
increased in the TNF-α + IFN-γ group and the TNF-α + IFN-γ +
miR-155 overexpression group.
Compared to the TNF-α + IFN-γ group, the expression
of PI3 (P<0.001) and CXCL1 (P<0.01) were significantly
increased in the TNF-α + IFN-γ + miR-155 overexpression group. In
addition, there was a significant increase in the mRNA expression
level and secretion of CXCL8 (P<0.05) but no alterations could
be detected in the expression of FOSL1 mRNA (Fig. 8).
These results suggested that miR-155 overexpression
can increase PI3 and FOSL1 protein expression and CXCL1 and CXCL8
secretion, specially PI3 protein expression and CXCL8 secretion
under inflammatory conditions. This was mainly mediated by
significantly increasing the expression of proinflammatory factors
CXCL1 and CXCL8 and the anti-inflammatory factors PI3. It is
involved in the inflammatory response.
Discussion
AD is a chronic inflammatory skin disease that can
exert significant psychosocial effects on patients and their
families. AD can also increase the risk of asthma, allergic
rhinitis and food allergy. The pathogenesis of AD is complex and
involves the interaction among genetics, skin barrier and the
immune system. Destruction of the skin barrier function can lead to
skin barrier dysfunction, alter the molecular immune profile and
the function of immune cells in the epidermal microenvironment,
which then trigger skin inflammation in patients with AD. This in
turn results in immune system imbalance, which will further
aggravate skin barrier dysfunction.
Keratinocytes are an important component of the
skin. They act as a skin barrier and serve an immunomodulatory role
(16). Activated keratinocytes can
secrete a variety of cytokines and chemokines, such as IL-1, IL-6,
thymic stromal lymphopoietin and TNF-α., which serve an important
role in the regulation of the epidermal immune microenvironment and
skin barrier function (17). HaCaT
cells are widely used for in vitro studies of AD-associated
skin conditions.
A number of studies have shown that miR-155 is
involved in a variety of immune-related diseases by regulating
inflammatory responses (18,19).
In monocytes/macrophages, miR-155 has been reported to regulate
inflammatory responses that are activated by certain cytokines
(such as TNFα, IL-1β) or Toll-like receptor ligands in various cell
types (18,20). In addition, miR-155 can promote M1
macrophage polarization, leading to local inflammation in the heart
and even systemic inflammation in distant organs (21). miR-155 expression was found to be
significantly increased in skin lesions of patients with AD
(10). A previous study
demonstrated that miR-155 can enhance IL-1 production by targeting
SOCS17(18). It has also been
shown that miR-155 can coordinate with the NACHT domain-,
leucine-rich repeat- and PYD-containing protein 3 inflammasome to
drive IL-1β-mediated signaling, which further promotes IL-1β
release and miR-155 expression (20). The increase in peripheral blood
IL-6 levels in patients with AD is associated with the activation
of T cells (22). IL-1β can
mediate innate immune responses and skin inflammatory responses in
various skin diseases, including psoriasis, vitiligo, systemic
lupus erythematosus and AD (23,24).
The present study revealed that elevated expression levels of
miR-155 led to the release of the pro-inflammatory cytokines IL-1β,
IL-6 and IL-15 whilst inhibiting the secretion of the
anti-inflammatory cytokine IL-10. Although according to the present
study miR-155 can promote the immune response in skin lesions of
patients with AD, the underlying mechanism of action remain
unconfirmed.
TNF-α and IFN-γ are cytokines that have been
recognized to induce inflammation in HaCaT cells. To study the
immune microenvironment in AD lesions, an inflammatory model of
HaCaT cells was constructed. miR-155 has been shown to target and
bind annexin A2 to regulate microvascular integrity and endothelial
barrier function (25). The
present study showed that the expression level of miR-155 was
induced by TNF-α and IFN-γ in HaCaT cells and the expression level
of miR-155 was increased after TNF-α and IFN-γ stimulation in HaCaT
cells, whereas the high expression of miR-155 significantly
inhibited the proliferation of HaCaT cells.
In the present study, four potential target genes of
miR-155, PI3, FOSL1, CXCL1 and CXCL8, were identified by
bioinformatic analysis. Through GO and KEGG accumulation analysis,
it was found that chemokines and their signaling pathways probably
serve an important role in AD lesions. Furthermore, PI3, FOSL1 and
CXCL1 were each found to interact with CXCL8 following the
construction of the protein interaction network of the four genes.
The PI3 gene is a serine protease inhibitor that can inhibit
excessive damage to elastase released by neutrophils during the
inflammation process to resist the inflammatory response, antiviral
and immunomodulatory effects (26-30).
It has been previously reported that PI3 levels are increased in
the blood of children with AD (31). However, the mechanism of PI3 in AD
has not been studied. FOSL1 is a regulator of cell proliferation,
differentiation, inflammation, tumorigenesis and metastasis and is
involved in various proinflammatory responses (32-34).
Although FOSL1 can induce inflammation, the mechanism of FOSL1 in
AD remain unclear. CXCL1 and CXCL8 are important mediators of the
inflammatory response and are involved in proinflammatory cascades
mediating a number of inflammatory diseases (35-41).
However, the specific roles and regulatory mechanisms of CXCL1 and
CXCL8 in AD remain to be fully elucidated. Results from the present
study showed that when HaCaT cells were stimulated with TNF-α and
IFN-γ, the expression of miR-155, FOSL1, CXCL1 and CXCL8 was
increased whereas the expression of PI3 was also increased. miR-155
overexpression can induce inflammation. If inflammatory factors
persist in the environment, then the anti-inflammatory effect
caused by the increased anti-inflammatory proteins will weaken. If
miR-155 expression is increased, it can then induce immune cell
infiltration into the immune microenvironment, where they can
secrete a number of cytokines, such as IL-1, IL-6 and IL-15, whilst
reducing the secretion of IL-10 and other anti-inflammatory
cytokines, resulting in cytokine storm. Imbalance of inflammation
regulation mechanism and increases in the inflammatory response can
cause tissue and keratinocyte damage. Eventually, the skin's
protective function is destroyed, resulting in idiopathic
dermatitis.
Although the present study has found that miR-155
can regulate the production of PI3, FOSL1, CXCL1 and CXCL8 by HaCaT
cells when stimulated with TNF-α and IFN-γ, the specific regulatory
pathway downstream of miR-155 remain unclear. In addition, the
effects of miR-155 on the level of immune molecules, the number and
function of immune cells in the epidermal immune microenvironment
of patients with AD require further study.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by National Natural
Science Foundation of China (grant no. 31671092) and, Cooperative
Education between Industry and Education (Construction of New
Engineering, New Medical, New Agricultural and New Liberal Arts) of
the Department of Higher Education of the Ministry of Education
(grant no. 202102585001) and Hubei Education Department (grant no.
2020435). The research performed in this study followed the laws of
China and the authors' respective institutions.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WQ and KG conceived and designed the study. XW, LC
and CL conducted the data search. XW and XC performed the
statistical and experiments analysis. XW and WQ drafted the
manuscript. WQ and KG reviewed and edited the manuscript. XW and WQ
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Silverberg JI and Hanifin JM: Adult eczema
prevalence and associations with asthma and other health and
demographic factors: A US population-based study. J Allergy Clin
Immunol. 132:1132–1138. 2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hay RJ, Johns NE, Williams HC, Bolliger
IW, Dellavalle RP, Margolis DJ, Marks R, Naldi L, Weinstock MA,
Wulf SK, et al: The global burden of skin disease in 2010: An
analysis of the prevalence and impact of skin conditions. J Invest
Dermatol. 134:1527–1534. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Patrick GJ, Archer NK and Miller LS: Which
way do we go? Complex interactions in atopic dermatitis
pathogenesis. J Invest Dermatol. 141:274–284. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Langan SM, Irvine AD and Weidinger S:
Atopic dermatitis. Lancet. 396:345–360. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
De Benedetto A, Kubo A and Beck LA: Skin
barrier disruption: A requirement for allergen sensitization? J
Invest Dermatol. 132:949–963. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Bangert C, Rindler K, Krausgruber T, Alkon
N, Thaler FM, Kurz H, Ayub T, Demirtas D, Fortelny N,
Vorstandlechner V, et al: Persistence of mature dendritic cells,
T(H)2A and Tc2 cells characterize clinically resolved atopic
dermatitis under IL-4Rα blockade. Sci Immunol.
6(eabe2749)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wongvibulsin S, Sutaria N, Kannan S,
Alphonse MP, Belzberg M, Williams KA, Brown ID, Choi J, Roh YS,
Pritchard T, et al: Transcriptomic analysis of atopic dermatitis in
African Americans is characterized by Th2/Th17-centered cutaneous
immune activation. Sci Rep. 11(11175)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gross N, Kropp J and Khatib H: MicroRNA
signaling in embryo development. Biology (Basel).
6(34)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chang J, Zhou B, Wei Z and Luo Y: IL-32
promotes the occurrence of atopic dermatitis by activating the
JAK1/microRNA-155 axis. J Transl Med. 20(207)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang X, Chen Y, Yuan W, Yao L, Wang S, Jia
Z, Wu P, Li L, Wei P, Wang X and Hong M: MicroRNA-155-5p is a key
regulator of allergic inflammation, modulating the epithelial
barrier by targeting PKIα. Cell Death Dis. 10(884)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sonkoly E, Janson P, Majuri ML, Savinko T,
Fyhrquist N, Eidsmo L, Xu N, Meisgen F, Wei T, Bradley M, et al:
MiR-155 is overexpressed in patients with atopic dermatitis and
modulates T-cell proliferative responses by targeting cytotoxic T
lymphocyte-associated antigen 4. J Allergy Clin Immunol.
126:581–589. 2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tsoi LC, Rodriguez E, Degenhardt F,
Baurecht H, Wehkamp U, Volks N, Szymczak S, Swindell WR, Sarkar MK,
Raja K, et al: Atopic dermatitis is an IL-13-dominant disease with
greater molecular heterogeneity compared to psoriasis. J Invest
Dermatol. 139:1480–1489. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Möbus L, Rodriguez E, Harder I, Stölzl D,
Boraczynski N, Gerdes S, Kleinheinz A, Abraham S, Heratizadeh A,
Handrick C, et al: Atopic dermatitis displays stable and dynamic
skin transcriptome signatures. J Allergy Clin Immunol. 147:213–223.
2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Huang LY, Li ST, Lin SC, Kao CH, Hong CH,
Lee CH and Yang LT: Gasdermin a is required for epidermal
cornification during skin barrier regeneration and in an atopic
dermatitis-like model. J Invest Dermatol. 143:1735–1745.
2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Jiang Y, Tsoi LC, Billi AC, Ward NL, Harms
PW, Zeng C, Maverakis E, Kahlenberg JM and Gudjonsson JE:
Cytokinocytes: The diverse contribution of keratinocytes to immune
responses in skin. JCI Insight. 5(e142067)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Testa U, Pelosi E, Castelli G and Labbaye
C: miR-146 and miR-155: Two key modulators of immune response and
tumor development. Noncoding RNA. 3(22)2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jankauskas SS, Gambardella J, Sardu C,
Lombardi A and Santulli G: Functional role of miR-155 in the
pathogenesis of diabetes mellitus and its complications. Noncoding
RNA. 7(39)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Eissa MG and Artlett CM: The microRNA
miR-155 is essential in fibrosis. Noncoding RNA.
5(23)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hu C, Liao J, Huang R, Su Q and He L:
MicroRNA-155-5p in serum derived-exosomes promotes
ischaemia-reperfusion injury by reducing CypD ubiquitination by
NEDD4. ESC Heart Fail. 10:1144–1157. 2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Toshitani A, Ansel JC, Chan SC, Li SH and
Hanifin JM: Increased interleukin 6 production by T cells derived
from patients with atopic dermatitis. J Invest Dermatol.
100:299–304. 1993.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tang L and Zhou F: Inflammasomes in common
immune-related skin diseases. Front Immunol. 11(882)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bernard M, Carrasco C, Laoubi L, Guiraud
B, Rozières A, Goujon C, Duplan H, Bessou-Touya S, Nicolas JF,
Vocanson M and Galliano MF: IL-1β induces thymic stromal
lymphopoietin and an atopic dermatitis-like phenotype in
reconstructed healthy human epidermis. J Pathol. 242:234–245.
2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Barker KR, Lu Z, Kim H, Zheng Y, Chen J,
Conroy AL, Hawkes M, Cheng HS, Njock MS, Fish JE, et al: MiR-155
modifies inflammation, endothelial activation and blood-brain
barrier dysfunction in cerebral malaria. Mol Med. 23:24–33.
2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ollague JE and Nousari CH: Expression of
elafin in dermatitis herpetiformis. Am J Dermatopathol. 40:1–6.
2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Moreau T, Baranger K, Dade S,
Dallet-Choisy S, Guyot N and Zani ML: Multifaceted roles of human
elafin and secretory leukocyte proteinase inhibitor (SLPI), two
serine protease inhibitors of the chelonianin family. Biochimie.
90:284–295. 2008.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Teng G, Liu Z, Liu Y, Wu T, Dai Y, Wang H
and Wang W: Probiotic escherichia coli nissle 1917 expressing
elafin protects against inflammation and restores the gut
microbiota. Front Microbiol. 13(819336)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li K, Zhang F, Wei L, Han Z, Liu X, Pan Y,
Guo C and Han W: Recombinant human elafin ameliorates chronic
hyperoxia-induced lung injury by inhibiting nuclear factor-kappa B
signaling in neonatal mice. J Interferon Cytokine Res. 40:320–330.
2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Elgharib I, Khashaba SA, Elsaid HH and
Sharaf MM: Serum elafin as a potential inflammatory marker in
psoriasis. Int J Dermatol. 58:205–209. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Brunner PM, He H, Pavel AB, Czarnowicki T,
Lefferdink R, Erickson T, Canter T, Puar N, Rangel SM, Malik K, et
al: The blood proteomic signature of early-onset pediatric atopic
dermatitis shows systemic inflammation and is distinct from adult
long-standing disease. J Am Acad Dermatol. 81:510–519.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
He YY, Zhou HF, Chen L, Wang YT, Xie WL,
Xu ZZ, Xiong Y, Feng YQ, Liu GY, Li X, et al: The Fra-1: Novel role
in regulating extensive immune cell states and affecting
inflammatory diseases. Front Immunol. 13(954744)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mishra RK, Potteti HR, Tamatam CR,
Elangovan I and Reddy SP: c-Jun is required for nuclear
factor-kappaB-dependent, LPS-stimulated fos-related antigen-1
transcription in alveolar macrophages. Am J Respir Cell Mol Biol.
55:667–674. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Moon YM, Lee SY, Kwok SK, Lee SH, Kim D,
Kim WK, Her YM, Son HJ, Kim EK, Ryu JG, et al: The fos-related
antigen 1-JUNB/Activator protein 1 transcription complex, a
downstream target of signal transducer and activator of
transcription 3, induces t helper 17 differentiation and promotes
experimental autoimmune arthritis. Front Immunol.
8(1793)2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Korbecki J, Barczak K, Gutowska I, Chlubek
D and Baranowska-Bosiacka I: CXCL1: Gene, promoter, regulation of
expression, mRNA stability, regulation of activity in the
intercellular space. Int J Mol Sci. 23(792)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Korbecki J, Maruszewska A, Bosiacki M,
Chlubek D and Baranowska-Bosiacka I: The potential importance of
CXCL1 in the physiological state and in noncancer diseases of the
cardiovascular system, respiratory system and skin. Int J Mol Sci.
24(205)2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhu Y, Yang S, Zhao N, Liu C, Zhang F, Guo
Y and Liu H: CXCL8 chemokine in ulcerative colitis. Biomed
Pharmacother. 138(111427)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kamsteeg M, Jansen PA, van Vlijmen-Willems
IM, van Erp PE, Rodijk-Olthuis D, van der Valk PG, Feuth T, Zeeuwen
PL and Schalkwijk J: Molecular diagnostics of psoriasis, atopic
dermatitis, allergic contact dermatitis and irritant contact
dermatitis. Br J Dermatol. 162:568–578. 2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Hulshof L, Hack DP, Hasnoe Q, Dontje B,
Jakasa I, Riethmüller C, McLean WHI, van Aalderen WMC, Van't Land
B, Kezic S, et al: A minimally invasive tool to study immune
response and skin barrier in children with atopic dermatitis. Br J
Dermatol. 180:621–630. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T,
Chen Y, Han X and Wu K: The CXCL8-CXCR1/2 pathways in cancer.
Cytokine Growth Factor Rev. 31:61–71. 2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
DiStasi MR and Ley K: Opening the
flood-gates: How neutrophil-endothelial interactions regulate
permeability. Trends Immunol. 30:547–556. 2009.PubMed/NCBI View Article : Google Scholar
|