|
1
|
Sommer F, Anderson JM, Bharti R, Raes J
and Rosenstiel P: The resilience of the intestinal microbiota
influences health and disease. Nat Rev Microbiol. 15:630–638.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tumor and Microecology Committee of
Chinese Anti-Cancer Association. Chinese expert consensus on the
correlation between intestinal microecology and hematopoietic stem
cell transplantation. J Inte Oncol. 48:129–135. 2021.
|
|
3
|
Khoruts A, Staley C and Sadowsky MJ:
Faecal microbiota transplantation for Clostridioides difficile:
Mechanisms and pharmacology. Nat Rev Gastroenterol Hepatol.
18:67–80. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang J, Liang J, He M, Xie Q, Wu Q, Shen
G, Zhu B, Yu J, Yu L, Tan X, et al: Chinese expert consensus on
intestinal microecology and management of digestive tract
complications related to tumor treatment (version 2022). J Cancer
Res Ther. 18:1835–1844. 2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chahwan B, Kwan S, Isik A, van Hemert S,
Burke C and Roberts L: Gut feelings: A randomised, triple-blind,
placebo-controlled trial of probiotics for depressive symptoms. J
Affect Disord. 253:317–326. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Iida N, Dzutsev A, Stewart CA, Smith L,
Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S,
et al: Commensal bacteria control cancer response to therapy by
modulating the tumor microenvironment. Science. 342:967–970.
2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Microecology Branch of Chinese Preventive
Medicine Association. Expert consensus on clinical application of
microecological regulators in China (2020 edition). Chin J
Microecol. 32:953–965. 2020.
|
|
8
|
Johnson JS, Spakowicz DJ, Hong BY,
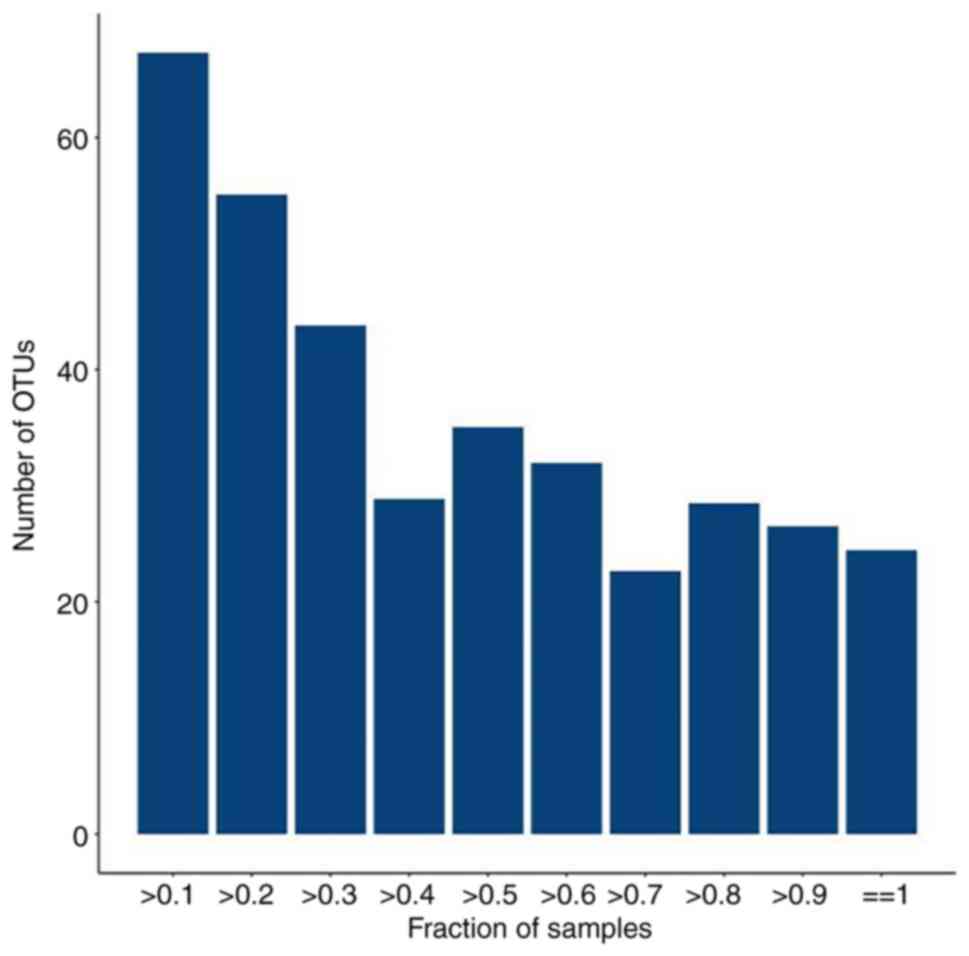
Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta
HO, Gerstein M, et al: Evaluation of 16S rRNA gene sequencing for
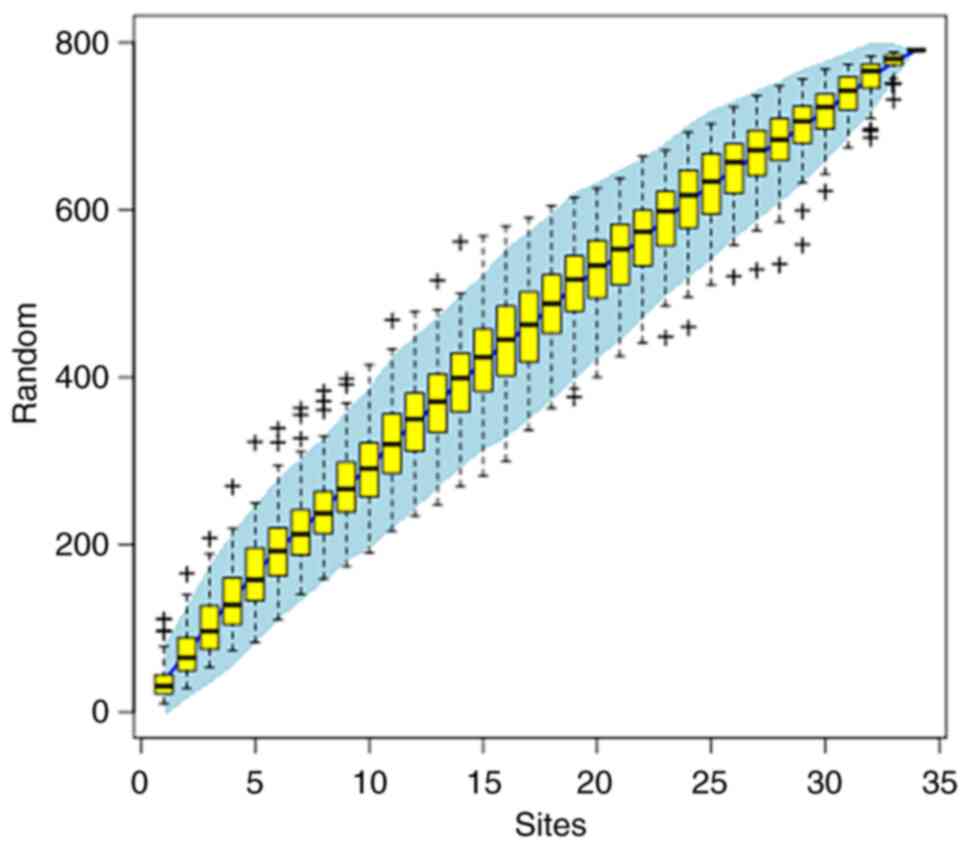
species and strain-level microbiome analysis. Nat Commun.
10(5029)2019.PubMed/NCBI View Article : Google Scholar
|
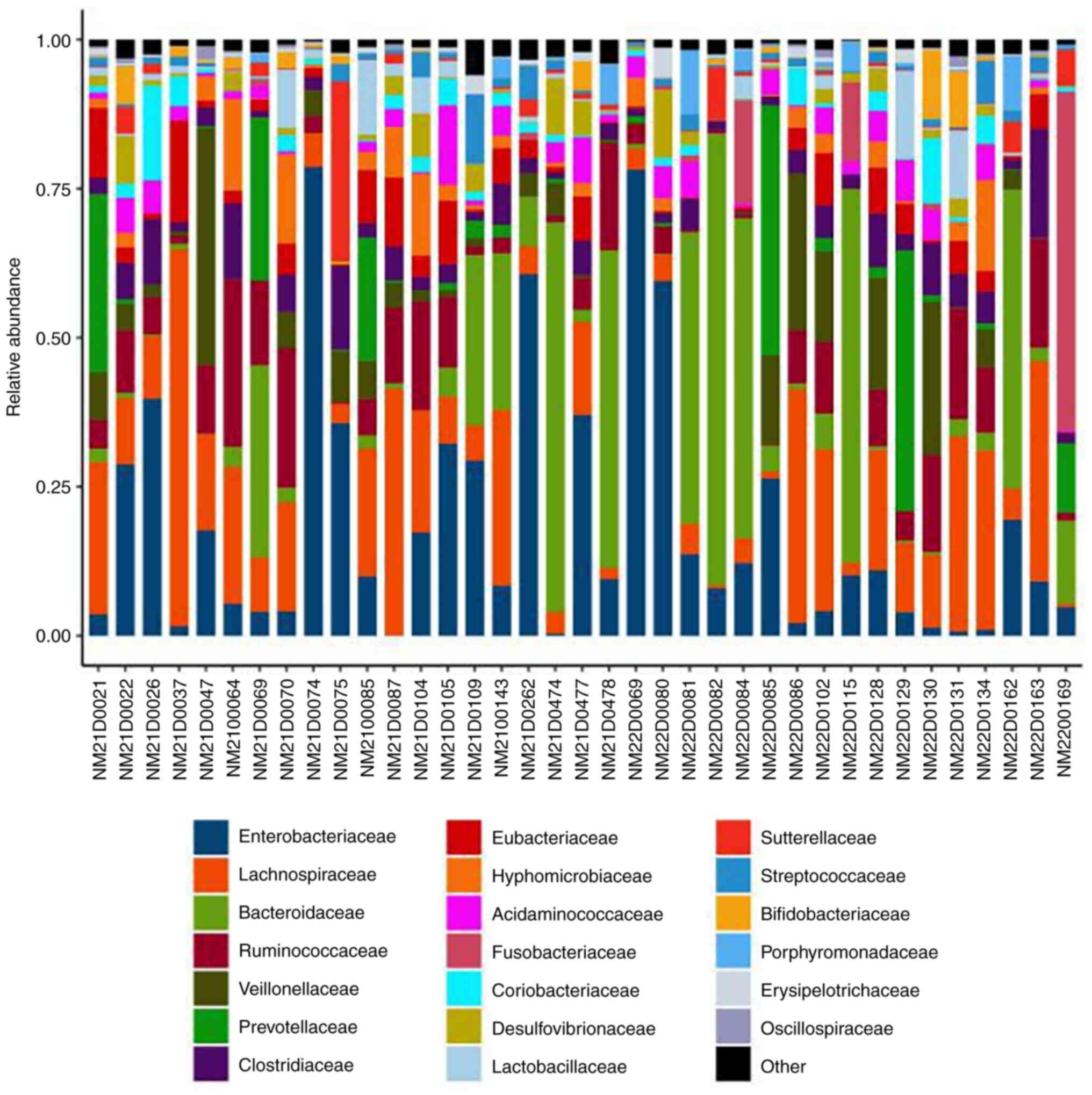
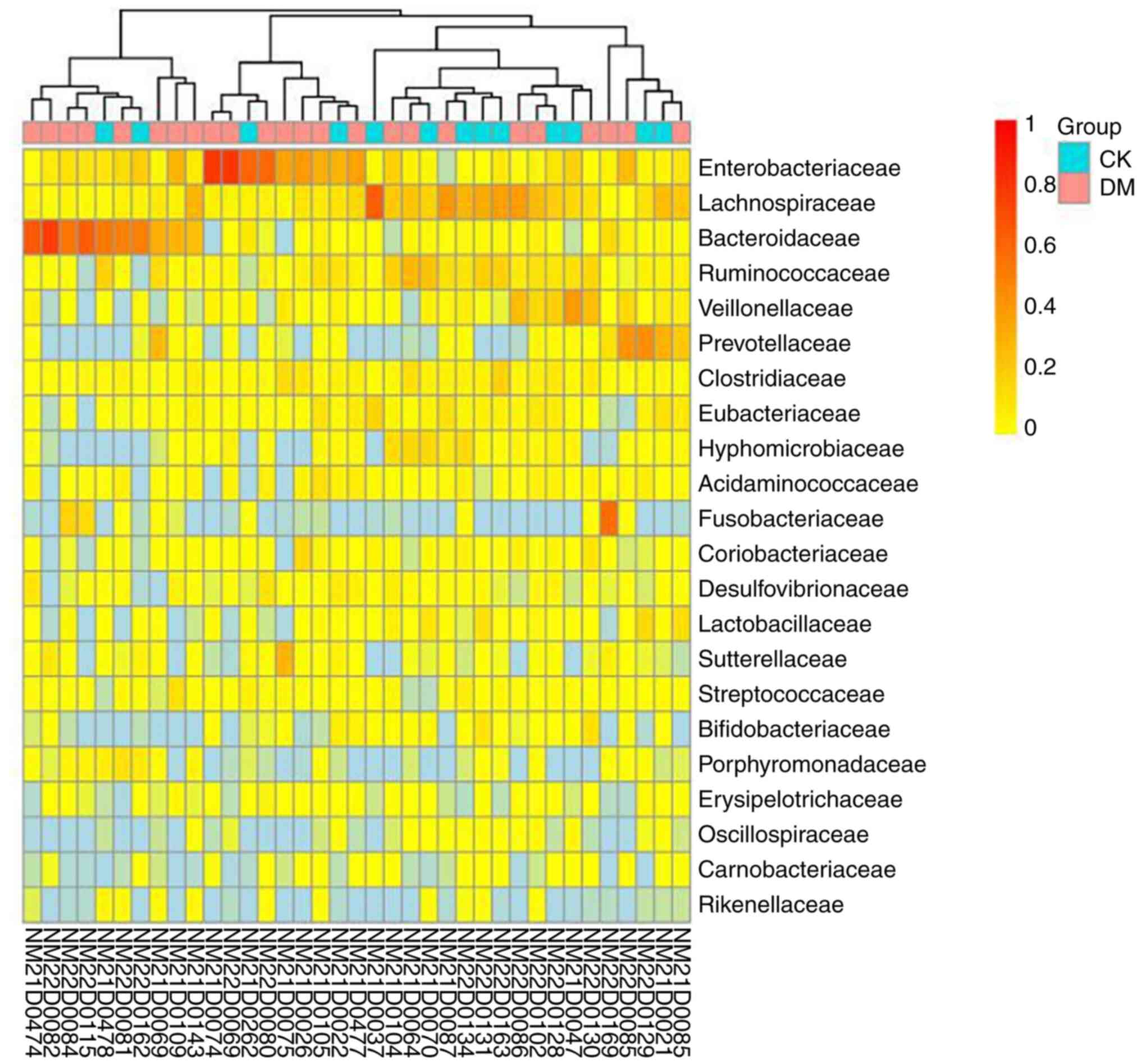
|
9
|
Zhang W, Fan X, Shi H, Li J, Zhang M, Zhao
J and Su X: Comprehensive Assessment of 16S rRNA gene amplicon
sequencing for microbiome profiling across multiple habitats.
Microbiol Spectr. 11(e0056323)2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Rutsch A, Kantsjö JB and Ronchi F: The
Gut-Brain axis: How microbiota and host inflammasome influence
brain physiology and pathology. Front Immunol.
11(604179)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ansaldo E, Farley TK and Belkaid Y:
Control of immunity by the microbiota. Annu Rev Immunol.
39:449–479. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Viaud S, Saccheri F, Mignot G, Yamazaki T,
Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ,
et al: The intestinal microbiota modulates the anticancer immune
effects of cyclophosphamide. Science. 342:971–976. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Routy B, Le Chatelier E, Derosa L, Duong
CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C,
Roberti MP, et al: Gut microbiome influences efficacy of PD-1-based
immunotherapy against epithelial tumors. Science. 359:91–97.
2028.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Gopalakrishnan V, Spencer CN, Nezi L,
Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman
K, Wei SC, et al: Gut microbiome modulates response to anti-PD-1
immunotherapy in melanoma patients. Science. 359:97–103.
2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tanoue T, Morita S, Plichta DR, Skelly AN,
Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, et
al: A defined commensal consortium elicits CD8 T cells and
anti-cancer immunity. Nature. 565:600–605. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Fluckiger A, Daillère R, Sassi M, Sixt BS,
Liu P, Loos F, Richard C, Rabu C, Alou MT, Goubet AG, et al:
Cross-reactivity between tumor MHC class I-restricted antigens and
an enterococcal bacteriophage. Science. 369:936–942.
2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sivan A, Corrales L, Hubert N, Williams
JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B,
Alegre ML, et al: Commensal Bifidobacterium promotes antitumor
immunity and facilitates anti-PD-L1 efficacy. Science.
350:1084–1089. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang T, Li Q, Cheng L, Buch H and Zhang
F: Akkermansia muciniphila is a promising probiotic. Microb
Biotechnol. 12:1109–1125. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Donald K and Finlay BB: Early-life
interactions between the microbiota and immune system: Impact on
immune system development and atopic disease. Nat Rev Immunol.
23:735–748. 2023.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Rinninella E, Raoul P, Cintoni M,
Franceschi F, Miggiano GAD, Gasbarrini A and Mele MC: What is the
Healthy gut microbiota composition? A changing ecosystem across
age, environment, diet, and diseases. Microorganisms.
7(14)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Parikh K, Antanaviciute A, Fawkner-Corbett
D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen
HH, Alham NK, et al: Colonic epithelial cell diversity in health
and inflammatory bowel disease. Nature. 567:49–55. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yu J, Yin Y, Yu Y, Cheng M, Zhang S, Jiang
S and Dong M: Effect of concomitant antibiotics use on patient
outcomes and adverse effects in patients treated with ICIs.
Immunopharmacol Immunotoxicol. 45:386–394. 2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu X, Tong X, Zou Y, Lin X, Zhao H, Tian
L, Jie Z, Wang Q, Zhang Z, Lu H, et al: Mendelian randomization
analyses support causal relationships between blood metabolites and
the gut microbiome. Nat Genet. 54:52–61. 2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zitvogel L, Ma Y, Raoult D, Kroemer G and
Gajewski TF: The microbiome in cancer immunotherapy: Diagnostic
tools and therapeutic strategies. Science. 359:1366–1370.
2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Matson V, Fessler J, Bao R, Chongsuwat T,
Zha Y, Alegre ML, Luke JJ and Gajewski TF: The commensal microbiome
is associated with anti-PD-1 efficacy in metastatic melanoma
patients. Science. 359:104–108. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Mager LF, Burkhard R, Pett N, Cooke NCA,
Brown K, Ramay H, Paik S, Stagg J, Groves RA, Gallo M, et al:
Microbiome-derived inosine modulates response to checkpoint
inhibitor immunotherapy. Science. 369:1481–1489. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kim J and Lee HK: Potential Role of the
gut microbiome in colorectal cancer progression. Front Immunol.
12(807648)2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang N and Fang JY: Fusobacterium
nucleatum, a key pathogenic factor and microbial biomarker for
colorectal cancer. Trends Microbiol. 31:159–172. 2023.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yang Y, Du L, Shi D, Kong C, Liu J, Liu G,
Li X and Ma Y: Dysbiosis of human gut microbiome in young-onset
colorectal cancer. Nat Commun. 12(6757)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kartal E, Schmidt TSB, Molina-Montes E,
Rodríguez-Perales S, Wirbel J, Maistrenko OM, Akanni WA, Alashkar
Alhamwe B, Alves RJ, Carrato A, et al: A faecal microbiota
signature with high specificity for pancreatic cancer. Gut.
71:1359–1372. 2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lee JWJ, Plichta D, Hogstrom L, Borren NZ,
Lau H, Gregory SM, Tan W, Khalili H, Clish C, Vlamakis H, et al:
Multi-omics reveal microbial determinants impacting responses to
biologic therapies in inflammatory bowel disease. Cell Host
Microbe. 29:1294–1304. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Young VB: The role of the microbiome in
human health and disease: An introduction for clinicians. BMJ.
356(j831)2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jian Y, Zhang D, Liu M, Wang Y and Xu ZX:
The impact of gut microbiota on radiation-induced enteritis. Front
Cell Infect Microbiol. 11(586392)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wang L, Wang X, Zhang G, Ma Y, Zhang Q, Li
Z, Ran J, Hou X, Geng Y, Yang Z, et al: The impact of pelvic
radiotherapy on the gut microbiome and its role in
radiation-induced diarrhoea: A systematic review. Radiat Oncol.
16(187)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Samarkos M, Mastrogianni E and
Kampouropoulou O: The role of gut microbiota in Clostridium
difficile infection. Eur J Intern Med. 50:28–32.
2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Shogbesan O, Poudel DR, Victor S, Jehangir
A, Fadahunsi O, Shogbesan G and Donato A: A systematic review of
the efficacy and safety of fecal microbiota transplant for
clostridium difficile infection in immunocompromised
patients. Can J Gastroenterol Hepatol. 2018(1394379)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Boulangé CL, Neves AL, Chilloux J,
Nicholson JK and Dumas ME: Impact of the gut microbiota on
inflammation, obesity, and metabolic disease. Genome Med.
8(42)2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li S, He M, Lei Y, Liu Y, Li X, Xiang X,
Wu Q and Wang Q: Oral microbiota and Tumor-A new perspective of
tumor pathogenesis. Microorganisms. 10(2206)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Diakite A, Dubourg G, Dione N, Afouda P,
Bellali S, Ngom II, Valles C, Tall ML, Lagier JC and Raoult D:
Optimization and standardization of the culturomics technique for
human microbiome exploration. Sci Rep. 10(9674)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Goren E, Wang C, He Z, Sheflin AM,
Chiniquy D, Prenni JE, Tringe S, Schachtman DP and Liu P: Feature
selection and causal analysis for microbiome studies in the
presence of confounding using standardization. BMC Bioinformatics.
22(362)2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Laudadio I, Fulci V, Palone F, Stronati L,
Cucchiara S and Carissimi C: Quantitative assessment of shotgun
metagenomics and 16S rRNA amplicon sequencing in the study of human
gut microbiome. OMICS. 22:248–254. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kryukov K, Imanishi T and Nakagawa S:
Nanopore sequencing data analysis of 16S rRNA genes using the
GenomeSync-GSTK system. Methods Mol Biol. 2632:215–226.
2023.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhang T, Li H, Ma S, Cao J, Liao H, Huang
Q and Chen W: The newest oxford Nanopore R10.4.1 full-length 16S
rRNA sequencing enables the accurate resolution of species-level
microbial community profiling. Appl Environ Microbiol.
89(e0060523)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Deng X, Achari A, Federman S, Yu G,
Somasekar S, Bártolo I, Yagi S, Mbala-Kingebeni P, Kapetshi J,
Ahuka-Mundeke S, et al: Metagenomic sequencing with spiked primer
enrichment for viral diagnostics and genomic surveillance. Nat
Microbiol. 5:443–454. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Shi Y, Wang G, Lau HC and Yu J:
Metagenomic sequencing for Microbial DNA in human samples: Emerging
technological advances. Int J Mol Sci. 23(2181)2022.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Qin J, Li R, Raes J, Arumugam M, Burgdorf
KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al: A
human gut microbial gene catalogue established by metagenomic
sequencing. Nature. 464:59–65. 2010.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Johnson S, Lavergne V, Skinner AM,
Gonzales-Luna AJ, Garey KW, Kelly CP and Wilcox MH: Clinical
practice guideline by the infectious diseases society of America
(IDSA) and society for healthcare epidemiology of America (SHEA):
2021 Focused update guidelines on management of clostridioides
difficile infection in adults. Clin Infect Dis. 73:e1029–e1044.
2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Eltokhy MA, Saad BT, Eltayeb WN, El-Ansary
MR, Aboshanab KM and Ashour MSE: A metagenomic nanopore sequence
analysis combined with conventional screening and spectroscopic
methods for deciphering the antimicrobial metabolites produced by
alcaligenes faecalis soil isolate MZ921504. Antibiotics (Basel).
10(1382)2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Guan H, Pu Y, Liu C, Lou T, Tan S, Kong M,
Sun Z, Mei Z, Qi Q, Quan Z, et al: Comparison of fecal collection
methods on variation in gut metagenomics and untargeted
metabolomics. mSphere. 6(e0063621)2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Mordant A and Kleiner M: Evaluation of
sample preservation and storage methods for metaproteomics analysis
of intestinal microbiomes. Microbiol Spectr.
9(e0187721)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Haindl R, Totzauer L and Kulozik U:
Preservation by lyophilization of a human intestinal microbiota:
Influence of the cultivation pH on the drying outcome and
re-establishment ability. Microb Biotechnol. 15:886–900.
2022.PubMed/NCBI View Article : Google Scholar
|
|
52
|
China Preventive Medicine Association.
General Guidelines for Pathogen Screening based on High-throughput
Sequencing (T/CMPA 010-2020). J Pathogen Biology. 16:738–740.
2021.
|
|
53
|
Xiao W, Ren L, Chen Z, Fang LT, Zhao Y,
Lack J, Guan M, Zhu B, Jaeger E, Kerrigan L, et al: Toward best
practice in cancer mutation detection with whole-genome and
whole-exome sequencing. Nat Biotechnol. 39:1141–1150.
2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Poulsen CS, Ekstrøm CT, Aarestrup FM and
Pamp SJ: Library preparation and sequencing platform introduce bias
in metagenomic-based characterizations of microbiomes. Microbiol
Spectr. 10(e0009022)2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Kong N, Ng W, Thao K, Agulto R, Weis A,
Kim KS, Korlach J, Hickey L, Kelly L, Lappin S and Weimer BC:
Automation of PacBio SMRTbell NGS library preparation for bacterial
genome sequencing. Stand Genomic Sci. 12(27)2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Sanschagrin S and Yergeau E:
Next-generation sequencing of 16S ribosomal RNA gene amplicons. J
Vis Exp. 29(51709)2014.PubMed/NCBI View
Article : Google Scholar
|
|
57
|
Matsuo Y: Full-Length 16S rRNA gene
analysis using Long-Read nanopore sequencing for rapid
identification of bacteria from clinical specimens. Methods Mol
Biol. 2632:193–213. 2023.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Jin J, Yamamoto R and Shiroguchi K:
High-throughput identification and quantification of bacterial
cells in the microbiota based on 16S rRNA sequencing with
single-base accuracy using BarBIQ. Nat Protoc. 9:207–239.
2024.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Ranjan R, Rani A, Metwally A, McGee HS and
Perkins DL: Analysis of the microbiome: Advantages of whole genome
shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun.
469:967–977. 2016.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Laboratory Medicine Branch of Chinese
Medical Association. Expert Consensus on the standardization of
clinical application of high-throughput metagenomic sequencing
technology to detect pathogenic microorganisms. Chin J Laboratory
Med. 43:1181–1195. 2020.
|
|
61
|
Nash AK, Auchtung TA, Wong MC, Smith DP,
Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, et
al: The gut mycobiome of the human microbiome project healthy
cohort. Microbiome. 5(153)2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Huang C, Callahan BJ, Wu MC, Holloway ST,
Brochu H, Lu W, Peng X and Tzeng JY: Phylogeny-guided microbiome
OTU-specific association test (POST). Microbiome.
10(86)2022.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Mokhtari EB and Ridenhour BJ: Filtering
ASVs/OTUs via mutual information-based microbiome network analysis.
BMC Bioinformatics. 23(380)2022.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Mao CX and Li J: Comparing species
assemblages via species accumulation curves. Biometrics.
65:1063–1067. 2009.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Sæther BE, Engen S and Grøtan V: Species
diversity and community similarity in fluctuating environments:
Parametric approaches using species abundance distributions. J Anim
Ecol. 82:721–738. 2013.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Shahi SK, Zarei K, Guseva NV and Mangalam
AK: Microbiota analysis using Two-step PCR and Next-generation 16S
rRNA gene sequencing. J Vis Exp: Oct 15, 2019 doi:
10.3791/59980.
|
|
67
|
Xun W, Li W, Xiong W, Ren Y, Liu Y, Miao
Y, Xu Z, Zhang N, Shen Q and Zhang R: Diversity-triggered
deterministic bacterial assembly constrains community functions.
Nat Commun. 10(3833)2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Wilmanski T, Rappaport N, Earls JC, Magis
AT, Manor O, Lovejoy J, Omenn GS, Hood L, Gibbons SM and Price ND:
Blood metabolome predicts gut microbiome α-diversity in humans. Nat
Biotechnol. 37:1217–1228. 2019.PubMed/NCBI View Article : Google Scholar
|
|
69
|
R Core Team (RELEASE YEAR-year version of
R used was released). R: A language and environment for statistical
computing. R Foundation for Statistical Computing, Vienna,
Austria.
|
|
70
|
Yu X, Jiang W, Kosik RO, Song Y, Luo Q,
Qiao T, Tong J, Liu S, Deng C, Qin S, et al: Gut microbiota changes
and its potential relations with thyroid carcinoma. J Adv Res.
35:61–70. 2021.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Yu X, Jiang W, Kosik RO, Song Y, Luo Q,
Qiao T, Tong J, Liu S, Deng C, Qin S, et al: Gut microbiota changes
and its potential relations with thyroid carcinoma. J Adv Res.
35:61–70. 2021.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Schlaberg R: Microbiome Diagnostics. Clin
Chem. 66:68–76. 2020.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Bharti R and Grimm DG: Current challenges
and best-practice protocols for microbiome analysis. Brief
Bioinform. 22:178–193. 2021.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Aly AM, Adel A, El-Gendy AO, Essam TM and
Aziz RK: Gut microbiome alterations in patients with stage 4
hepatitis C. Gut Pathog. 8(42)2016.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Gao Y and Wu M: Accounting for 16S rRNA
copy number prediction uncertainty and its implications in
bacterial diversity analyses. ISME Commun. 3(59)2023.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Karpiński TM, Ożarowski M and Stasiewicz
M: Carcinogenic microbiota and its role in colorectal cancer
development. Semin Cancer Biol. 86:420–430. 2022.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Lee M and Chang EB: Inflammatory bowel
diseases (IBD) and the Microbiome-Searching the crime scene for
clues. Gastroenterology. 160:524–537. 2021.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Aron-Wisnewsky J, Warmbrunn MV, Nieuwdorp
M and Clément K: Metabolism and metabolic disorders and the
microbiome: The intestinal microbiota associated with obesity,
lipid metabolism, and metabolic Health-Pathophysiology and
therapeutic strategies. Gastroenterology. 160:573–599.
2021.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Youssef NH, Couger MB, McCully AL, Criado
AE and Elshahed MS: Assessing the global phylum level diversity
within the bacterial domain: A review. J Adv Res. 6:269–282.
2015.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Zou X, Wang L, Xiao L, Wang S and Zhang L:
Gut microbes in cerebrovascular diseases: Gut flora imbalance,
potential impact mechanisms and promising treatment strategies.
Front Immunol. 13(975921)2022.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Adak A and Khan MR: An insight into gut
microbiota and its functionalities. Cell Mol Life Sci. 76:473–493.
2019.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Dash NR, Khoder G, Nada AM and Al Bataineh
MT: Exploring the impact of Helicobacter pylori on gut microbiome
composition. PLoS One. 14(e0218274)2019.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Lun H, Yang W, Zhao S, Jiang M, Xu M, Liu
F and Wang Y: Altered gut microbiota and microbial biomarkers
associated with chronic kidney disease. Microbiologyopen.
8(e00678)2019.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H,
Xie H, Chen X, Shao L, Zhang R, et al: Gut microbiome analysis as a
tool towards targeted non-invasive biomarkers for early
hepatocellular carcinoma. Gut. 68:1014–1023. 2019.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Wang Q, Lei Y, Wang J, Xu X, Wang L, Zhou
H and Guo Z: Tumor and Microecology Committee of China Anti-Cancer
Association. Expert consensus on the relevance of intestinal
microecology and hematopoietic stem cell transplantation. Clin
Transplant. 38(e15186)2023.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Li S, Wang T, Ren Y, Liu Z, Gao J and Guo
Z: Prognostic impact of oral microbiome on survival of
malignancies: A systematic review and meta-analysis. Syst Rev.
13(41)2024.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Lu J, Zhang L, Zhai Q, Zhao J, Zhang H,
Lee YK, Lu W, Li M and Chen W: Chinese gut microbiota and its
associations with staple food type, ethnicity, and urbanization.
NPJ Biofilms Microbiomes. 7(71)2021.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Yang Q, Liang Q, Balakrishnan B,
Belobrajdic DP, Feng QJ and Zhang W: Role of dietary nutrients in
the modulation of gut microbiota: A narrative review. Nutrients.
12(381)2020.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Corbin KD, Carnero EA, Dirks B, Igudesman
D, Yi F, Marcus A, Davis TL, Pratley RE, Rittmann BE,
Krajmalnik-Brown R and Smith SR: Host-diet-gut microbiome
interactions influence human energy balance: A randomized clinical
trial. Nat Commun. 14(3161)2023.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Jacobs JP, Lagishetty V, Hauer MC, Labus
JS, Dong TS, Toma R, Vuyisich M, Naliboff BD, Lackner JM, Gupta A,
et al: Multi-omics profiles of the intestinal microbiome in
irritable bowel syndrome and its bowel habit subtypes. Microbiome.
11(5)2023.PubMed/NCBI View Article : Google Scholar
|
|
91
|
He J, Chu Y, Li J, Meng Q, Liu Y, Jin J,
Wang Y, Wang J, Huang B, Shi L, et al: Intestinal
butyrate-metabolizing species contribute to autoantibody production
and bone erosion in rheumatoid arthritis. Sci Adv.
8(eabm1511)2022.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Rhoads DD, Sintchenko V, Rauch CA and
Pantanowitz L: Clinical microbiology informatics. Clin Microbiol
Rev. 27:1025–1047. 2014.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Patel R: New developments in clinical
bacteriology laboratories. Mayo Clin Proc. 91:1448–1459.
2016.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Rozas M, Brillet F, Callewaert C and
Paetzold B: MinION™ nanopore sequencing of skin microbiome 16S and
16S-23S rRNA gene amplicons. Front Cell Infect Microbiol.
11(806476)2022.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Hoffmann DE, von Rosenvinge EC, Roghmann
MC, Palumbo FB, McDonald D and Ravel J: The DTC microbiome testing
industry needs more regulation. Science. 383:1176–1179.
2024.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Kalokairinou L, Borry P and Howard HC:
‘It's much more grey than black and white’: Clinical geneticists’
views on the oversight of consumer genomics in Europe. Per Med.
17:129–140. 2020.PubMed/NCBI View Article : Google Scholar
|