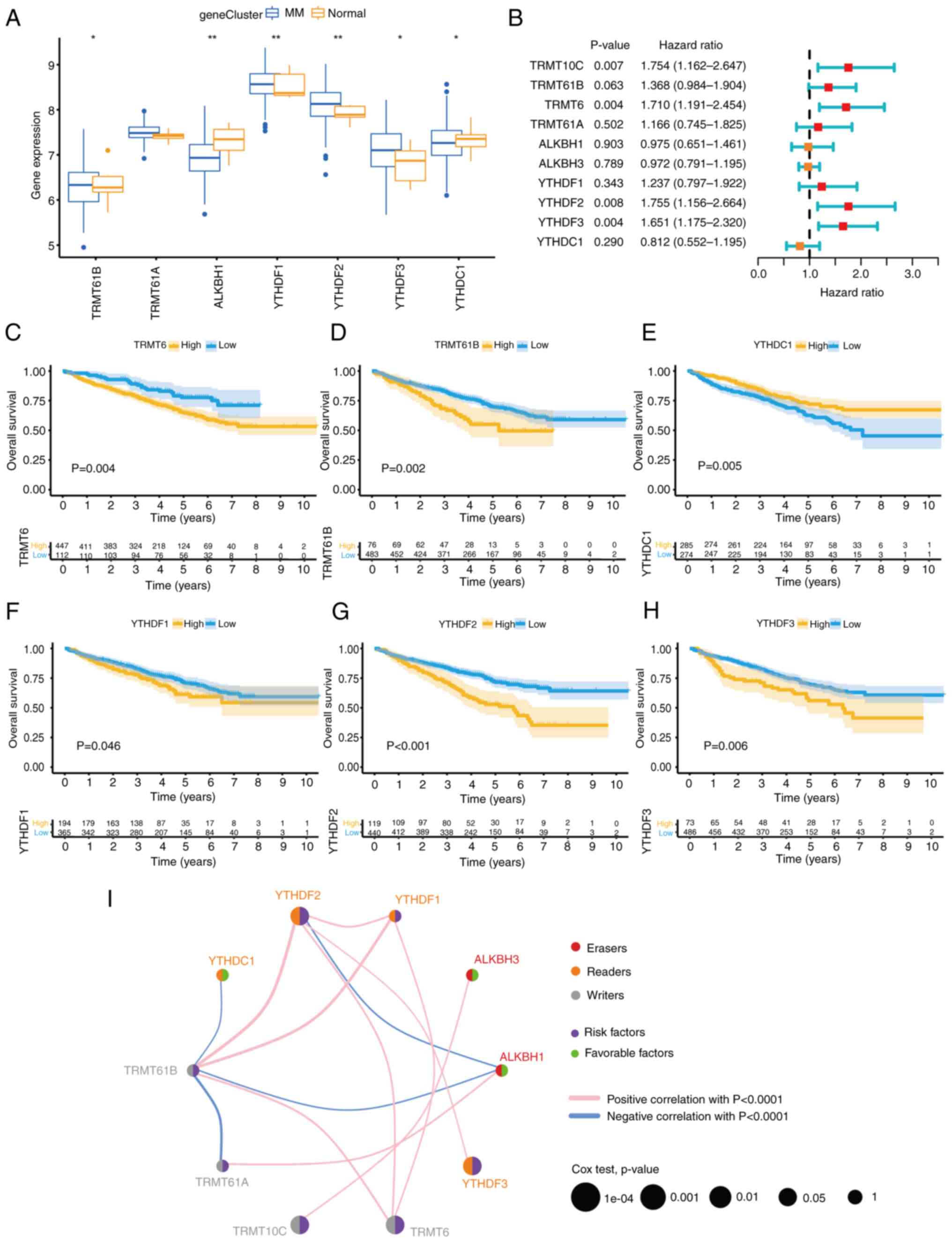
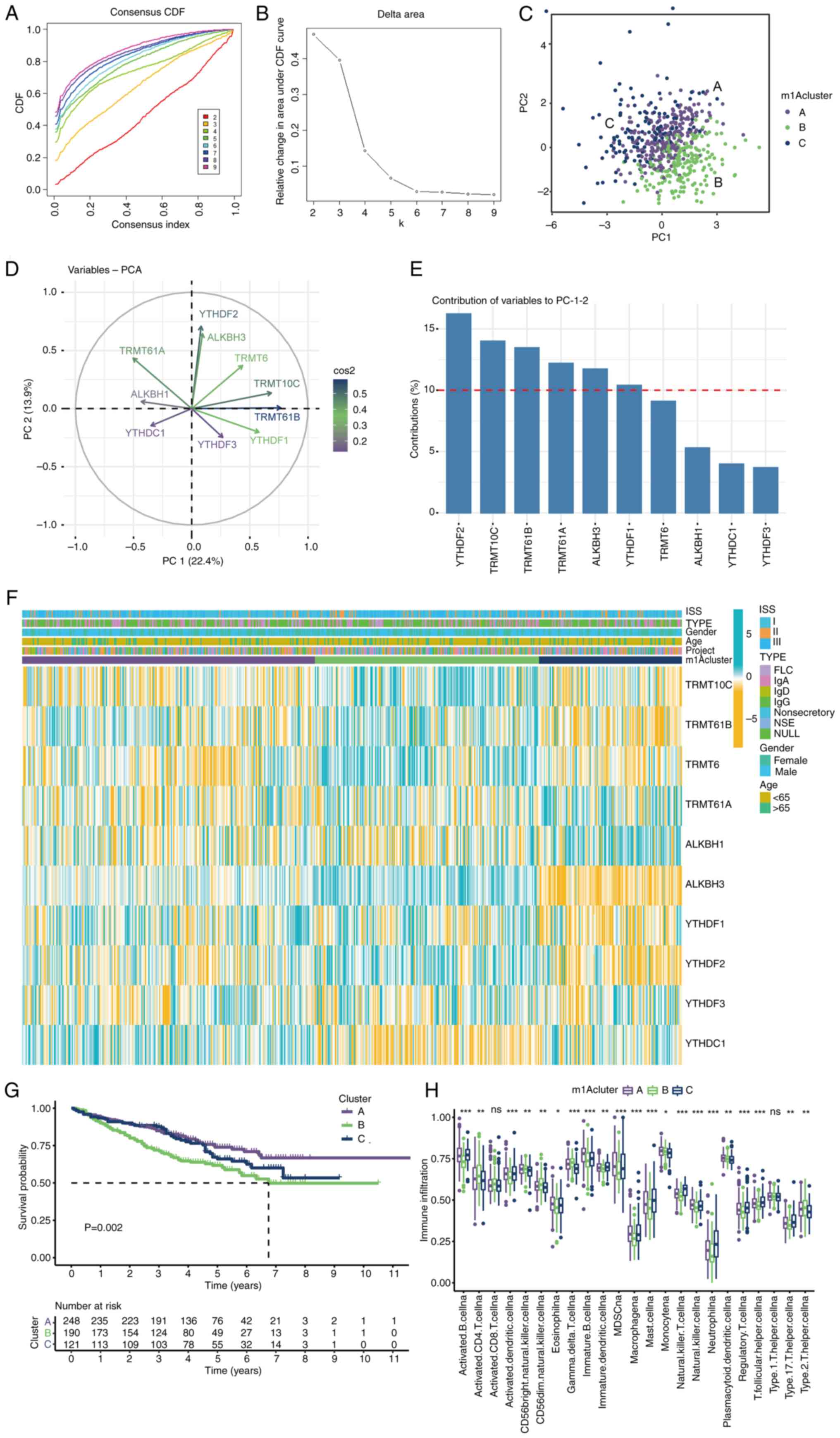
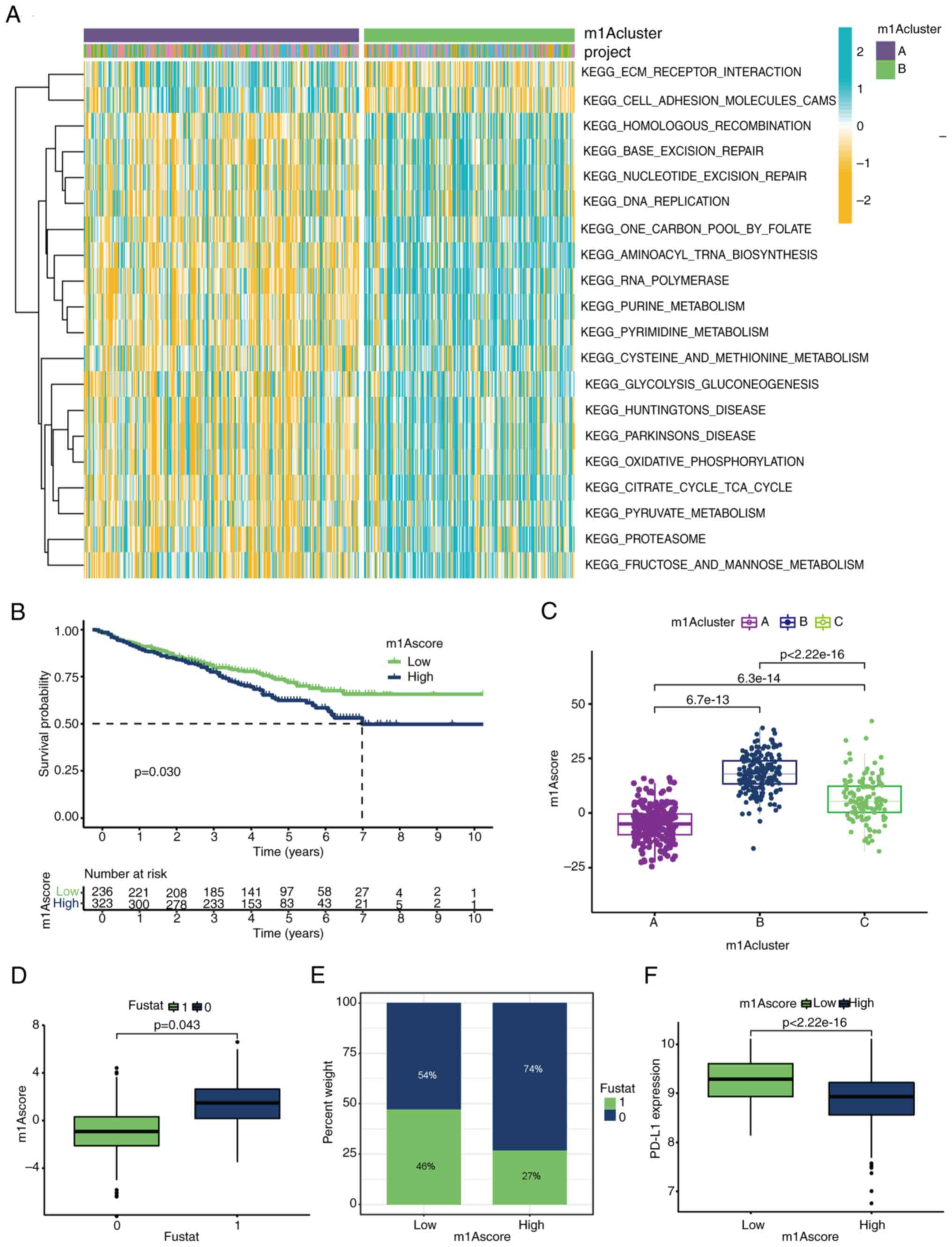
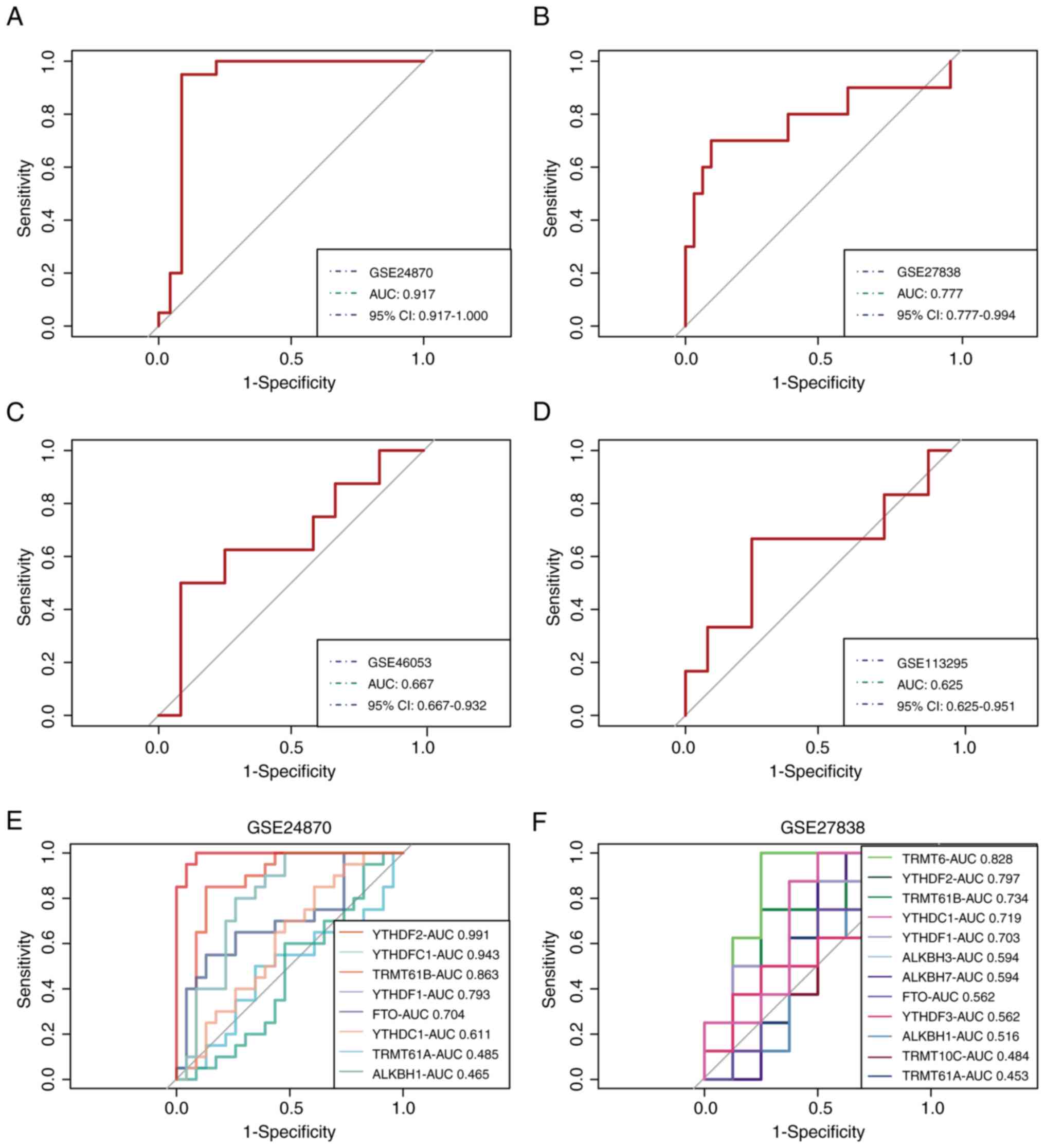
|
1
|
Zhan F, Huang Y, Colla S, Stewart JP,
Hanamura I, Gupta S, Epstein J, Yaccoby S, Sawyer J, Burington B,
et al: The molecular classification of multiple myeloma. Blood.
108:2020–2028. 2006.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Allegra A, Casciaro M, Barone P, Musolino
C and Gangemi S: Epigenetic crosstalk between malignant plasma
cells and the tumour microenvironment in multiple myeloma. Cancers
(Basel). 14(2597)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
He L, Yu C, Qin S, Zheng E, Liu X, Liu Y,
Yu S, Liu Y, Dou X, Shang Z, et al: The proteasome component PSMD14
drives myelomagenesis through a histone deubiquitinase activity.
Mol Cell. 83:4000–4016.e6. 2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Muylaert C, Van Hemelrijck LA, Maes A, De
Veirman K, Menu E, Vanderkerken K and De Bruyne E: Aberrant DNA
methylation in multiple myeloma: A major obstacle or an
opportunity? Front Oncol. 12(979569)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liu R, Shen Y, Hu J, Wang X, Wu D, Zhai M,
Bai J and He A: Comprehensive Analysis of m6A RNA methylation
regulators in the prognosis and immune microenvironment of multiple
myeloma. Front Oncol. 11(731957)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jin Z, MacPherson K, Liu Z and Vu LP: RNA
modifications in hematological malignancies. Int J Hematol.
117:807–820. 2023.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhao BS, Roundtree IA and He C:
Post-transcriptional gene regulation by mRNA modifications. Nat Rev
Mol Cell Biol. 18:31–42. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Dominissini D, Nachtergaele S,
Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni
A, Salmon-Divon M, Clark WC, et al: The dynamic
N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature.
530:441–446. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yang Y, Hsu PJ, Chen YS and Yang YG:
Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers
and functions in RNA metabolism. Cell Res. 28:616–624.
2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Liu Y, Zhang S, Gao X, Ru Y, Gu X and Hu
X: Research progress of N1-methyladenosine RNA modification in
cancer. Cell Commun Signal. 22(79)2024.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zou Z, Sepich-Poore C, Zhou X, Wei J and
He C: The mechanism underlying redundant functions of the YTHDF
proteins. Genome Biol. 24(17)2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J,
Lu Z, Zheng Z, Dai Q and Wang H: Transfer RNA demethylase ALKBH3
promotes cancer progression via induction of tRNA-derived small
RNAs. Nucleic Acids Res. 47:2533–2545. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu R, Miao J, Jia Y, Kong G, Hong F, Li
F, Zhai M, Zhang R, Liu J, Xu X, et al: N6-methyladenosine reader
YTHDF2 promotes multiple myeloma cell proliferation through
EGR1/p21cip1/waf1/CDK2-Cyclin E1 axis-mediated cell
cycle transition. Oncogene. 42:1607–1619. 2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Che F, Ye X, Wang Y, Wang X, Ma S, Tan Y,
Mao Y and Luo Z: METTL3 facilitates multiple myeloma tumorigenesis
by enhancing YY1 stability and pri-microRNA-27 maturation in
m6A-dependent manner. Cell Biol Toxicol. 39:2033–2050.
2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Su Z, Monshaugen I, Wilson B, Wang F,
Klungland A, Ougland R and Dutta A: TRMT6/61A-dependent base
methylation of tRNA-derived fragments regulates gene-silencing
activity and the unfolded protein response in bladder cancer. Nat
Commun. 13(2165)2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yang Z, Cai Z, Yang C, Luo Z and Bao X:
ALKBH5 regulates STAT3 activity to affect the proliferation and
tumorigenicity of osteosarcoma via an m6A-YTHDF2-dependent manner.
EBioMedicine. 80(104019)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xu A, Zhang J, Zuo L, Yan H, Chen L, Zhao
F, Fan F, Xu J, Zhang B, Zhang Y, et al: FTO promotes multiple
myeloma progression by posttranscriptional activation of HSF1 in an
m6A-YTHDF2-dependent manner. Mol Ther. 30:1104–1118.
2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Coira IF, Rincón R and Cuendet M: The
Multiple Myeloma Landscape: Epigenetics and Non-Coding RNAs.
Cancers (Basel). 14(2348)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Agnelli L, Mosca L, Fabris S, Lionetti M,
Andronache A, Kwee I, Todoerti K, Verdelli D, Battaglia C, Bertoni
F, et al: A SNP microarray and FISH-based procedure to detect
allelic imbalances in multiple myeloma: An integrated genomics
approach reveals a wide gene dosage effect. Genes Chromosomes
Cancer. 48:603–614. 2009.PubMed/NCBI View Article : Google Scholar
|
|
20
|
López-Corral L, Corchete LA, Sarasquete
ME, Mateos MV, García-Sanz R, Fermiñán E, Lahuerta JJ, Bladé J,
Oriol A, Teruel AI, et al: Transcriptome analysis reveals molecular
profiles associated with evolving steps of monoclonal gammopathies.
Haematologica. 99:1365–1372. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Popovici V, Chen W, Gallas BG, Hatzis C,
Shi W, Samuelson FW, Nikolsky Y, Tsyganova M, Ishkin A, Nikolskaya
T, et al: Effect of training-sample size and classification
difficulty on the accuracy of genomic predictors. Breast Cancer
Res. 12(R5)2010.PubMed/NCBI View
Article : Google Scholar
|
|
22
|
Li J, Xie L, Xie Y and Wang F: Bregmannian
consensus clustering for cancer subtypes analysis. Comput Methods
Programs Biomed. 189(105337)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14(7)2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sotiriou C, Wirapati P, Loi S, Harris A,
Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, et al:
Gene expression profiling in breast cancer: Understanding the
molecular basis of histologic grade to improve prognosis. J Natl
Cancer Inst. 98:262–272. 2006.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang B, Wu Q, Li B, Wang D, Wang L and
Zhou YL: m6A regulator-mediated methylation modification patterns
and tumor microenvironment infiltration characterization in gastric
cancer. Mol Cancer. 19(53)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bruns I, Cadeddu RP, Brueckmann I, Fröbel
J, Geyh S, Büst S, Fischer JC, Roels F, Wilk CM, Schildberg FA, et
al: Multiple myeloma-related deregulation of bone marrow-derived
CD34(+) hematopoietic stem and progenitor cells. Blood.
120:2620–2630. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Garg TK, Szmania SM, Khan JA, Hoering A,
Malbrough PA, Moreno-Bost A, Greenway AD, Lingo JD, Li X, Yaccoby
S, et al: Highly activated and expanded natural killer cells for
multiple myeloma immunotherapy. Haematologica. 97:1348–1356.
2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Garcia-Gomez A, De Las Rivas J, Ocio EM,
Díaz-Rodríguez E, Montero JC, Martín M, Blanco JF, Sanchez-Guijo
FM, Pandiella A, San Miguel JF and Garayoa M: Transcriptomic
profile induced in bone marrow mesenchymal stromal cells after
interaction with multiple myeloma cells: Implications in myeloma
progression and myeloma bone disease. Oncotarget. 5:8284–8305.
2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Liu H, He J, Koh SP, Zhong Y, Liu Z, Wang
Z, Zhang Y, Li Z, Tam BT, Lin P, et al: Reprogrammed marrow
adipocytes contribute to myeloma-induced bone disease. Sci Transl
Med. 11(eaau9087)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Liu B, Li X, Liu F, Li F, Wei S, Liu J and
Lv Y: Expression and Significance of TRIM 28 in Squamous Carcinoma
of Esophagus. Pathol Oncol Res. 25:1645–1652. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Frapin M, Guignard S, Meistermann D, Grit
I, Moullé VS, Paillé V, Parnet P and Amarger V: Maternal protein
restriction in rats alters the expression of genes involved in
mitochondrial metabolism and epitranscriptomics in fetal
hypothalamus. Nutrients. 12(1464)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Costa F, Vescovini R, Marchica V, Storti
P, Notarfranchi L, Dalla Palma B, Toscani D, Burroughs-Garcia J,
Catarozzo MT, Sammarelli G and Giuliani N: PD-L1/PD-1 pattern of
expression within the bone marrow immune microenvironment in
smoldering myeloma and active multiple myeloma patients. Front
Immunol. 11(613007)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Huang T, Liu Z, Zheng Y, Feng T, Gao Q and
Zeng W: YTHDF2 promotes spermagonial adhesion through modulating
MMPs decay via m(6)A/mRNA pathway. Cell Death Dis.
11(37)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 61:507–519.
2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Dai X, Wang T, Gonzalez G and Wang Y:
Identification of YTH Domain-Containing Proteins as the Readers for
N1-Methyladenosine in RNA. Anal Chem. 90:6380–6384. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Seo KW and Kleiner RE: YTHDF2 Recognition
of N1-Methyladenosine (m1A)-Modified RNA Is Associated with
Transcript Destabilization. ACS Chem Biol. 15:132–139.
2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Paris J, Morgan M, Campos J, Spencer GJ,
Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA,
Sepulveda C, et al: Targeting the RNA m6A Reader YTHDF2 selectively
compromises cancer stem cells in acute myeloid leukemia. Cell Stem
Cell. 25:137–148. e6. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Hua Z, Wei R, Guo M, Lin Z, Yu X, Li X, Gu
C and Yang Y: YTHDF2 promotes multiple myeloma cell proliferation
via STAT5A/MAP2K2/p-ERK axis. Oncogene. 41:1482–1491.
2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Sha Y, Wu J, Paul B, Zhao Y, Mathews P, Li
Z, Norris J, Wang E, McDonnell DP and Kang Y: PPAR agonists
attenuate lenalidomide's anti-myeloma activity in vitro and in
vivo. Cancer Lett. 545(215832)2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Aouali N, Broukou A, Bosseler M, Keunen O,
Schlesser V, Janji B, Palissot V, Stordeur P and Berchem G:
Epigenetic activity of peroxisome proliferator-activated receptor
gamma agonists increases the anticancer effect of histone
deacetylase inhibitors on multiple myeloma cells. PLoS One.
10(e0130339)2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yu JT, Hu XW, Chen HY, Yang Q, Li HD, Dong
YH, Zhang Y, Wang JN, Jin J, Wu YG, et al: DNA methylation of FTO
promotes renal inflammation by enhancing m6A of PPAR-α in
alcohol-induced kidney injury. Pharmacol Res.
163(105286)2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Long JC and Caceres JF: The SR protein
family of splicing factors: Master regulators of gene expression.
Biochem J. 417:15–27. 2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Longman D, Johnstone IL and Cáceres JF:
Functional characterization of SR and SR-related genes in
caenorhabditis elegans. EMBO J. 19:1625–1637. 2000.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Shkreta L, Delannoy A, Salvetti A and
Chabot B: SRSF10: An atypical splicing regulator with critical
roles in stress response, organ development, and viral replication.
RNA. 27:1302–1317. 2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Jobbins AM, Haberman N, Artigas N, Amourda
C, Paterson HAB, Yu S, Blackford SJI, Montoya A, Dore M, Wang YF,
et al: Dysregulated RNA polyadenylation contributes to metabolic
impairment in non-alcoholic fatty liver disease. Nucleic Acids Res.
50:3379–3393. 2022.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Maimaiti A, Tuersunniyazi A, Meng X, Pei
Y, Ji W, Feng Z, Jiang L, Wang Z, Kasimu M, Wang Y and Shi X:
N6-methyladenosine RNA methylation regulator-related alternative
splicing gene signature as prognostic predictor and in immune
microenvironment characterization of patients with low-grade
glioma. Front Genet. 13(872186)2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lai S, Wang Y, Li T, Dong Y, Lin Y, Wang
L, Weng S, Zhang X and Lin C: N6-methyladenosine-mediated CELF2
regulates CD44 alternative splicing affecting tumorigenesis via
ERAD pathway in pancreatic cancer. Cell Biosci.
12(125)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Yuan J, Lv T, Yang J, Wu Z, Yan L, Yang J
and Shi Y: HDLBP-stabilized lncFAL inhibits ferroptosis
vulnerability by diminishing Trim69-dependent FSP1 degradation in
hepatocellular carcinoma. Redox Biol. 58(102546)2022.PubMed/NCBI View Article : Google Scholar
|