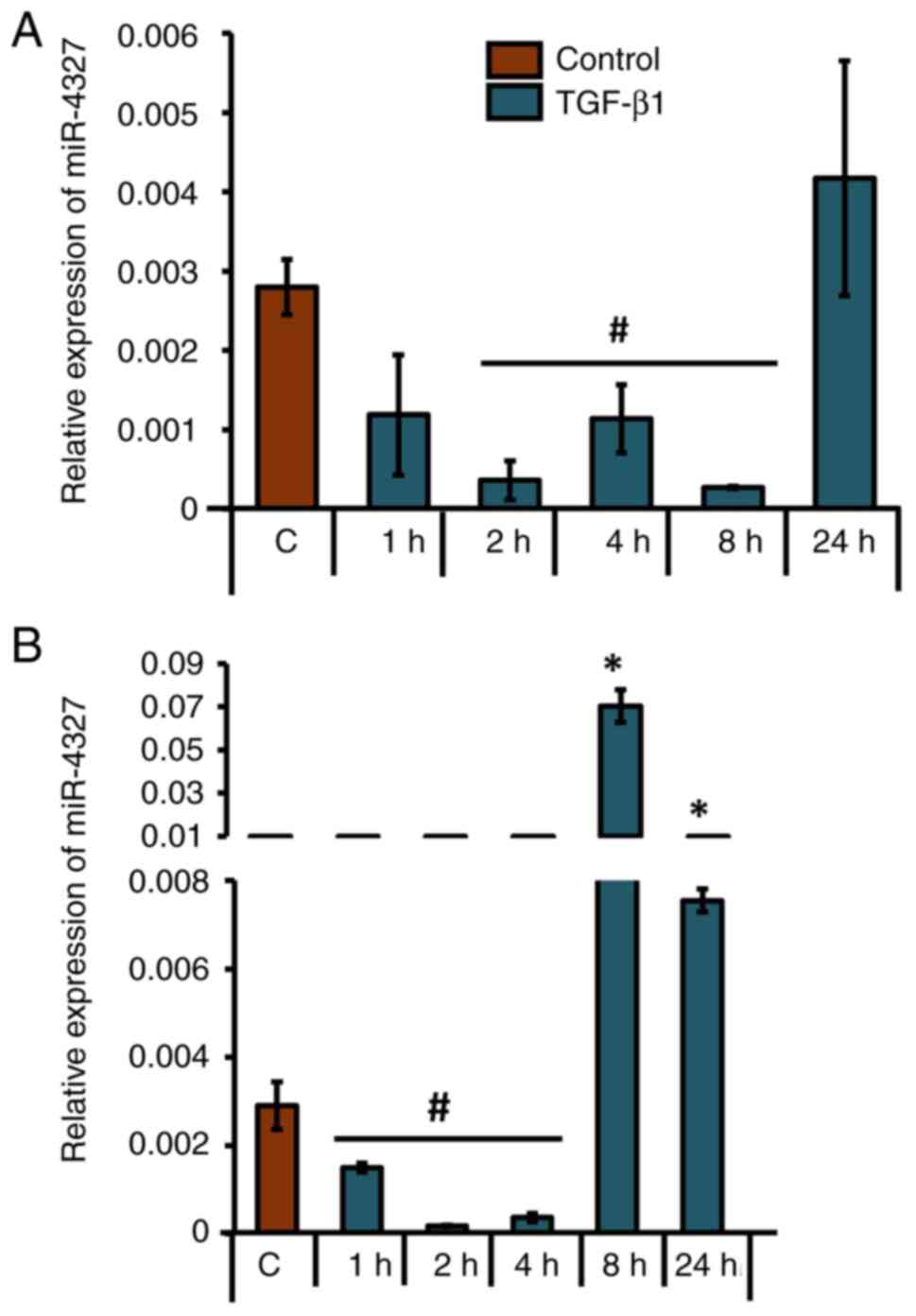
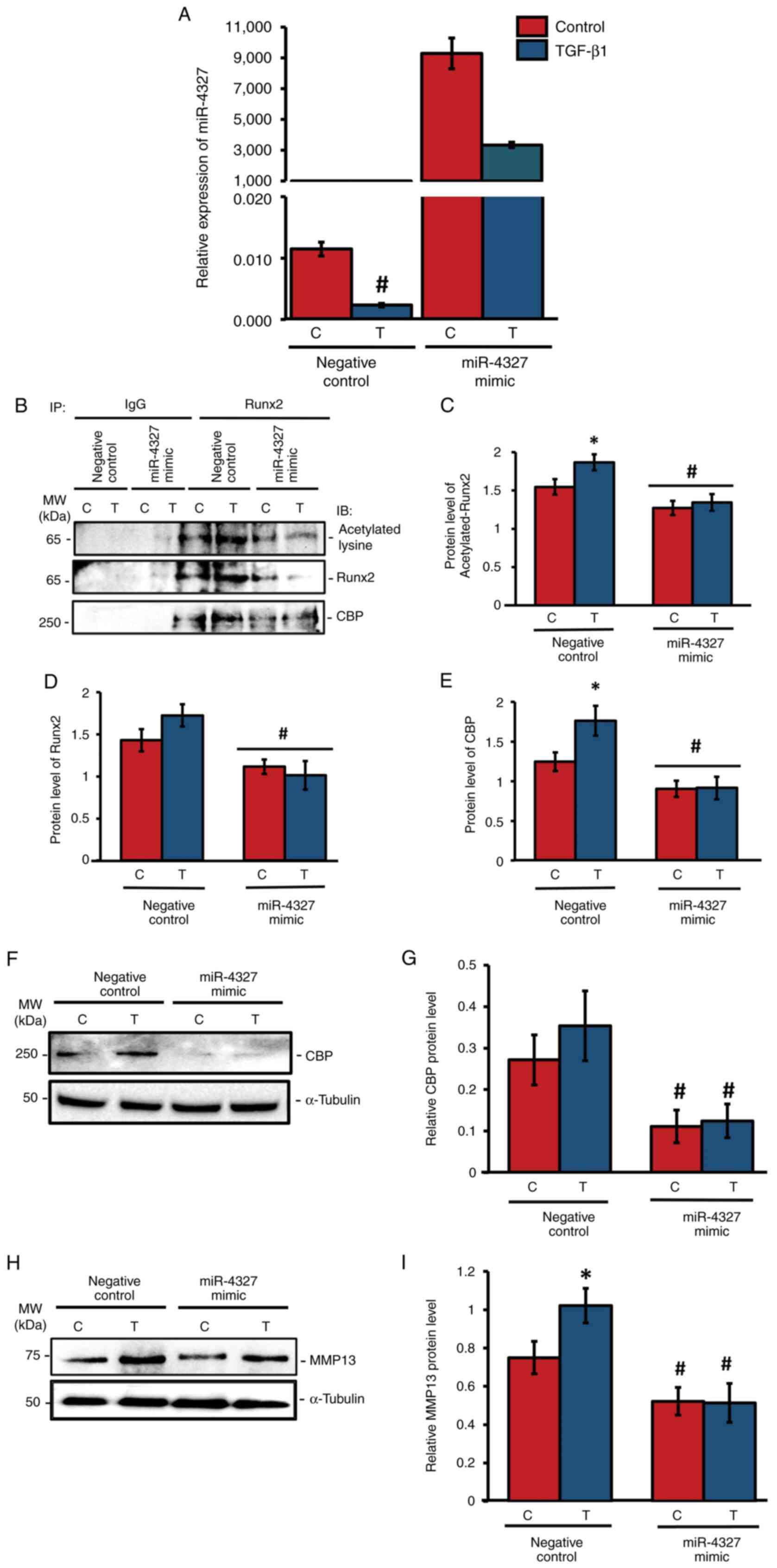
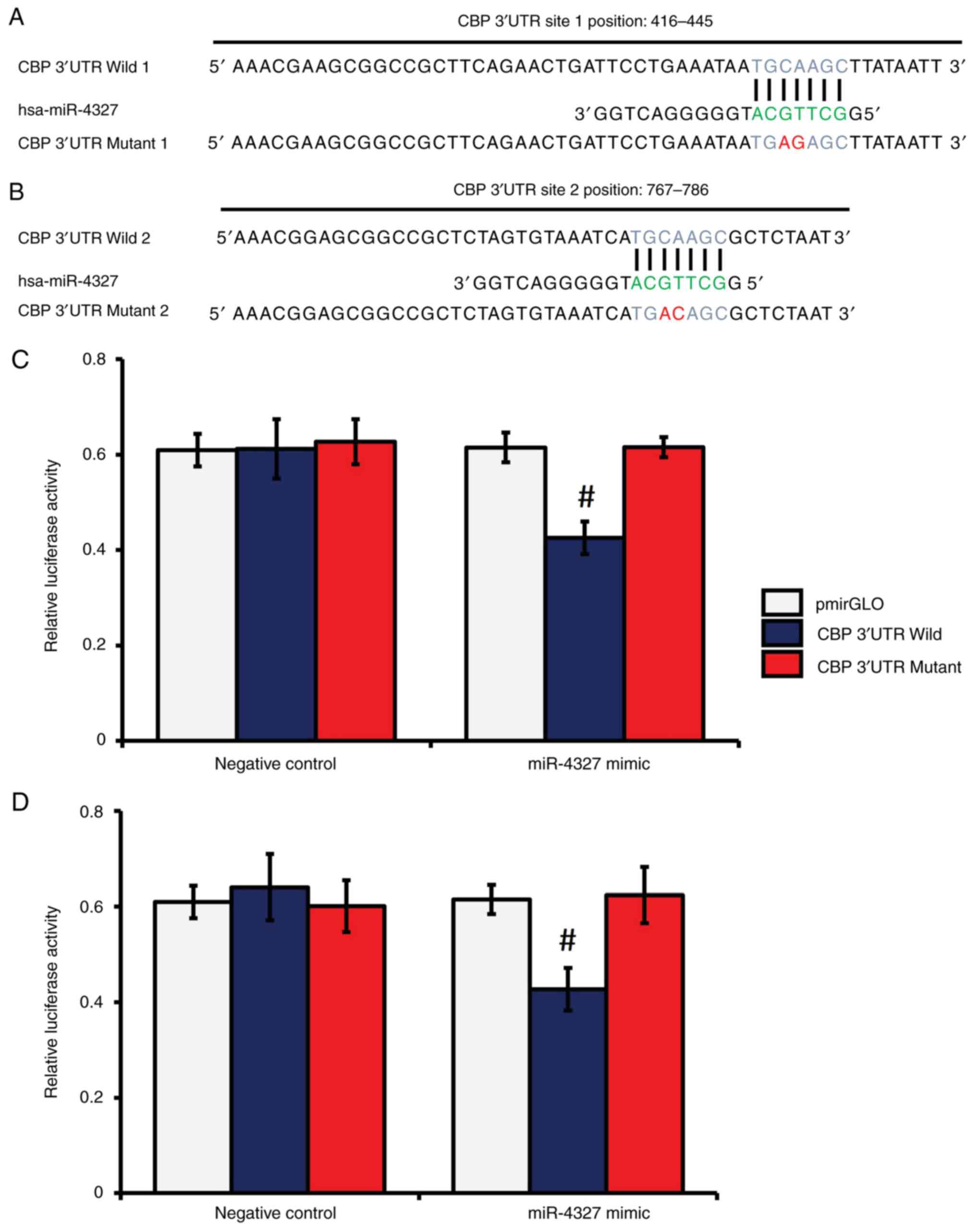
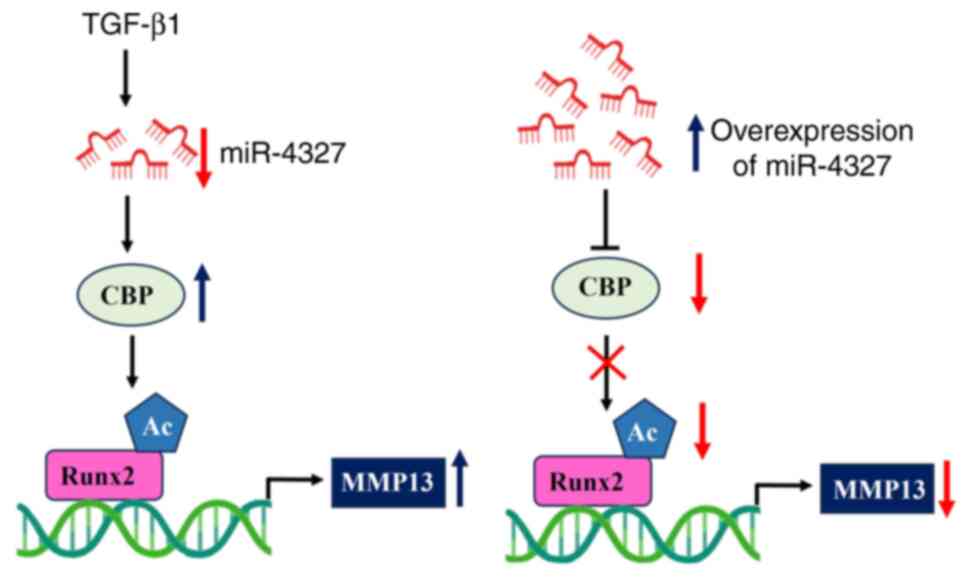
|
1
|
Oton-Gonzalez L, Mazziotta C, Iaquinta MR,
Mazzoni E, Nocini R, Trevisiol L, D'AgostiSno A, Tognon M, Rotondo
JC and Martini F: Genetics and epigenetics of bone remodeling and
metabolic bone diseases. Int J Mol Sci. 23(1500)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang H, Zheng X, Zhang Y, Huang J, Zhou W,
Li X, Tian H, Wang B, Xing D, Fu W, et al: The endocrine role of
bone: Novel functions of bone-derived cytokines. Biochem Pharmacol.
183(114308)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Arias CF, Herrero MA, Echeverri LF, Oleaga
GE and López JM: Bone remodeling: A tissue-level process emerging
from cell-level molecular algorithms. PLoS One.
13(e0204171)2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bolamperti S, Villa I and Rubinacci A:
Bone remodeling: An operational process ensuring survival and bone
mechanical competence. Bone Res. 10(48)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ornitz DM and Marie PJ: Fibroblast growth
factor signaling in skeletal development and disease. Genes Dev.
29:1463–1486. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Bordukalo-Nikšić T, Kufner V and Vukičević
S: The role of BMPs in the regulation of osteoclasts resorption and
bone remodeling: From experimental models to clinical applications.
Front Immunol. 13(869422)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Crane JL, Xian L and Cao X: Role of TGF-β
signaling in coupling bone remodeling. Methods Mol Biol.
1344:287–300. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Trivedi T, Pagnotti GM, Guise TA and
Mohammad KS: The role of TGF-β in bone metastases. Biomolecules.
11(1643)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhao L and Hantash BM: TGF-β1 regulates
differentiation of bone marrow mesenchymal stem cells. Vitam Horm.
87:127–141. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li J, Ge L, Zhao Y, Zhai Y, Rao N, Yuan X,
Yang J, Li J and Yu S: TGF-β2 and TGF-β1 differentially regulate
the odontogenic and osteogenic differentiation of mesenchymal stem
cells. Arch Oral Biol. 135(105357)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yang W, Li HY, Wu YF, Mi RJ, Liu WZ, Shen
X, Lu YX, Jiang YH, Ma MJ and Shen HY: ac4C acetylation of RUNX2
catalyzed by NAT10 spurs osteogenesis of BMSCs and prevents
ovariectomy-induced bone loss. Mol Ther Nucleic Acids. 26:135–147.
2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gomathi K, Rohini M, Vairamani M and
Selvamurugan N: Identification and characterization of
TGF-β1-responsive Runx2 acetylation sites for matrix
Metalloproteinase-13 expression in osteoblastic cells. Biochimie.
201:1–6. 2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Krishnan RH, Sadu L, Akshaya RL, Gomathi
K, Saranya I, Das UR, Satishkumar S and Selvamurugan N:
Circ_CUX1/miR-130b-5p/p300 axis for parathyroid hormone-stimulation
of Runx2 activity in rat osteoblasts: A combined bioinformatic and
experimental approach. Int J Biol Macromol. 225:1152–1163.
2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Arumugam B, Vishal M, Shreya S, Malavika
D, Rajpriya V, He Z, Partridge NC and Selvamurugan N: Parathyroid
hormone-stimulation of Runx2 during osteoblast differentiation via
the regulation of lnc-SUPT3H-1:16 (RUNX2-AS1:32) and miR-6797-5p.
Biochimie. 158:43–52. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gomathi K, Rohini M, Partridge NC and
Selvamurugan N: Regulation of transforming growth
factor-β1-stimulation of Runx2 acetylation for matrix
metalloproteinase 13 expression in osteoblastic cells. Biol Chem.
403:305–315. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ma C, Gao J, Liang J, Dai W, Wang Z, Xia
M, Chen T, Huang S, Na J, Xu L, et al: HDAC6 inactivates Runx2
promoter to block osteogenesis of bone marrow stromal cells in
age-related bone loss of mice. Stem Cell Res Ther.
12(484)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Arumugam B, Vairamani M, Partridge NC and
Selvamurugan N: Characterization of Runx2 phosphorylation sites
required for TGF-β1-mediated stimulation of matrix
metalloproteinase-13 expression in osteoblastic cells. J Cell
Physiol. 233:1082–1094. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yi SJ, Lee H, Lee J, Lee K, Kim J, Kim Y,
Park JI and Kim K: Bone remodeling: Histone modifications as fate
determinants of bone cell differentiation. Int J Mol Sci.
20(3147)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sánchez-Molina S, Oliva JL, García-Vargas
S, Valls E, Rojas JM and Martínez-Balbás MA: The histone
acetyltransferases CBP/p300 are degraded in NIH 3T3 cells by
activation of Ras signalling pathway. Biochem J. 398:215–224.
2006.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Takahashi S, Tsuda M, Takahashi Y and
Asahara H: Transcriptional co-activator CBP/p300 regulates
chondrocyte-specific gene expression via association with Sox9.
Arthritis Res Ther. 5 (Suppl 3)(S78)2003.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang L, Zhu K, Xu J, Chen X, Sheng C,
Zhang D, Yang Y, Sun L, Zhao H, Wang X, et al: Acetyltransferases
CBP/p300 control transcriptional switch of β-catenin and stat1
promoting osteoblast differentiation. J Bone Miner Res.
38:1885–1899. 2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Martire S, Nguyen J, Sundaresan A and
Banaszynski LA: Differential contribution of p300 and CBP to
regulatory element acetylation in mESCs. BMC Mol Cell Biol.
21(55)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gou P and Zhang W: Protein lysine
acetyltransferase CBP/p300: A promising target for small molecules
in cancer treatment. Biomed Pharmacother.
171(116130)2024.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Luchian I, Goriuc A, Sandu D and Covasa M:
The role of matrix metalloproteinases (MMP-8, MMP-9, MMP-13) in
periodontal and peri-implant pathological processes. Int J Mol Sci.
23(1806)2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Arai Y, Choi B, Kim BJ, Park S, Park H,
Moon JJ and Lee SH: Cryptic ligand on collagen matrix unveiled by
MMP13 accelerates bone tissue regeneration via MMP13/Integrin
α3/RUNX2 feedback loop. Acta Biomater. 125:219–230. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Arai Y and Lee SH: MMP13-overexpressing
mesenchymal stem cells enhance bone tissue formation in the
presence of collagen hydrogel. Tissue Eng Regen Med. 20:461–471.
2023.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hu Q and Ecker M: Overview of MMP-13 as a
promising target for the treatment of osteoarthritis. Int J Mol
Sci. 22(1742)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang M, Sampson ER, Jin H, Li J, Ke QH, Im
HJ and Chen D: MMP13 is a critical target gene during the
progression of osteoarthritis. Arthritis Res Ther.
15(R5)2013.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Krishnan RH, Sadu L, Das UR, Satishkumar
S, Pranav Adithya S, Saranya I, Akshaya RL and Selvamurugan N: Role
of p300, a histone acetyltransferase enzyme, in osteoblast
differentiation. Differentiation. 124:43–51. 2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Saranya I, Akshaya RL, Gomathi K,
Mohanapriya R, He Z, Partridge NC and Selvamurugan N:
Circ_ST6GAL1-mediated competing endogenous RNA network regulates
TGF-β1-stimulated matrix Metalloproteinase-13 expression via Runx2
acetylation in osteoblasts. Noncoding RNA Res. 9:153–164.
2023.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lee M and Partridge NC: Parathyroid
hormone activation of matrix metalloproteinase-13 transcription
requires the histone acetyltransferase activity of p300 and PCAF
and p300-dependent acetylation of PCAF. J Biol Chem.
285:38014–38022. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kolipaka R, Magesh I, Bharathy MRA,
Karthik S, Saranya I and Selvamurugan N: A potential function for
MicroRNA-124 in normal and pathological bone conditions. Noncoding
RNA Res. 9:687–694. 2024.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li D, Yang C, Yin C, Zhao F, Chen Z, Tian
Y, Dang K, Jiang S, Zhang W, Zhang G and Qian A: LncRNA, important
player in bone development and disease. Endocr Metab Immune Disord
Drug Targets. 20:50–66. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ping J, Li L, Dong Y, Wu X, Huang X, Sun
B, Zeng B, Xu F and Liang W: The role of long non-coding RNAs and
circular RNAs in bone regeneration: Modulating miRNAs function. J
Tissue Eng Regen Med. 16:227–243. 2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Patil S, Dang K, Zhao X, Gao Y and Qian A:
Role of LncRNAs and CircRNAs in bone metabolism and osteoporosis.
Front Genet. 11(584118)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Puppo M, Valluru MK and Clézardin P:
MicroRNAs and their roles in breast cancer bone metastasis. Curr
Osteoporos Rep. 19:256–263. 2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhang Y, Cao X, Li P, Fan Y, Zhang L, Ma
X, Sun R, Liu Y and Li W: microRNA-935-modified bone marrow
mesenchymal stem cells-derived exosomes enhance osteoblast
proliferation and differentiation in osteoporotic rats. Life Sci.
272(119204)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Qiu M, Zhai S, Fu Q and Liu D: Bone marrow
mesenchymal stem cells-derived exosomal MicroRNA-150-3p promotes
osteoblast proliferation and differentiation in osteoporosis. Hum
Gene Ther. 32:717–729. 2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Long D, Lee R, Williams P, Chan CY, Ambros
V and Ding Y: Potent effect of target structure on microRNA
function. Nat Struct Mol Biol. 14:287–294. 2007.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Rohini M, Arumugam B, Vairamani M and
Selvamurugan N: Stimulation of ATF3 interaction with Smad4 via
TGF-β1 for matrix metalloproteinase 13 gene activation in human
breast cancer cells. Int J Biol Macromol. 134:954–961.
2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Malavika D, Shreya S, Raj Priya V, Rohini
M, He Z, Partridge NC and Selvamurugan N: miR-873-3p targets HDAC4
to stimulate matrix metalloproteinase-13 expression upon
parathyroid hormone exposure in rat osteoblasts. J Cell Physiol.
235:7996–8009. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Rohini M, Gokulnath M, Miranda PJ and
Selvamurugan N: miR-590-3p inhibits proliferation and promotes
apoptosis by targeting activating transcription factor 3 in human
breast cancer cells. Biochimie. 154:10–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Komori T: Whole aspect of Runx2 functions
in skeletal development. Int J Mol Sci. 23(5776)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Franceschi RT and Xiao G: Regulation of
the osteoblast-specific transcription factor, Runx2: Responsiveness
to multiple signal transduction pathways. J Cell Biochem.
88:446–454. 2003.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Choi YH, Kim YJ, Jeong HM, Jin YH, Yeo CY
and Lee KY: Akt enhances Runx2 protein stability by regulating
Smurf2 function during osteoblast differentiation. FEBS J.
281:3656–3666. 2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Kim HJ, Kim WJ and Ryoo HM:
Post-translational regulations of transcriptional activity of
RUNX2. Mol Cells. 43:160–167. 2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Selvamurugan N, Shimizu E, Lee M, Liu T,
Li H and Partridge NC: Identification and characterization of Runx2
phosphorylation sites involved in matrix metalloproteinase-13
promoter activation. FEBS Lett. 583:1141–1146. 2009.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wang X, Manner PA, Horner A, Shum L, Tuan
RS and Nuckolls GH: Regulation of MMP-13 expression by RUNX2 and
FGF2 in osteoarthritic cartilage. Osteoarthritis Cartilage.
12:963–973. 2004.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Takahashi A, de Andrés MC, Hashimoto K,
Itoi E, Otero M, Goldring MB and Oreffo ROC: DNA methylation of the
RUNX2 P1 promoter mediates MMP13 transcription in chondrocytes. Sci
Rep. 7(7771)2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Behonick DJ, Xing Z, Lieu S, Buckley JM,
Lotz JC, Marcucio RS, Werb Z, Miclau T and Colnot C: Role of matrix
metalloproteinase 13 in both endochondral and intramembranous
ossification during skeletal regeneration. PLoS One.
2(e1150)2007.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Chen Q, Yang B, Liu X, Zhang XD, Zhang L
and Liu T: Histone acetyltransferases CBP/p300 in tumorigenesis and
CBP/p300 inhibitors as promising novel anticancer agents.
Theranostics. 12:4935–4948. 2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
He ZX, Wei BF, Zhang X, Gong YP, Ma LY and
Zhao W: Current development of CBP/p300 inhibitors in the last
decade. Eur J Med Chem. 209(112861)2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Gomathi K, Akshaya N, Srinaath N, Rohini M
and Selvamurugan N: Histone acetyl transferases and their
epigenetic impact on bone remodeling. Int J Biol Macromol.
170:326–335. 2021.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Waddell AR, Huang H and Liao D: CBP/p300:
Critical co-activators for nuclear steroid hormone receptors and
emerging therapeutic targets in prostate and breast cancers.
Cancers (Basel). 13(2872)2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lakshmanan MD and Shaheer K: Endocrine
disrupting chemicals may deregulate DNA repair through estrogen
receptor mediated seizing of CBP/p300 acetylase. J Endocrinol
Invest. 43:1189–1196. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Szymczak-Pajor I, Drzewoski J, Świderska
E, Strycharz J, Gabryanczyk A, Kasznicki J, Bogdańska M and
Śliwińska A: Metformin induces apoptosis in human pancreatic cancer
(PC) cells accompanied by changes in the levels of histone
acetyltransferases [Particularly, p300/CBP-associated factor (PCAF)
protein levels]. Pharmaceuticals (Basel). 16(115)2023.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Gao Y, Patil S and Qian A: The role of
MicroRNAs in bone metabolism and disease. Int J Mol Sci.
21(6081)2020.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Bhushan R, Grünhagen J, Becker J, Robinson
PN, Ott CE and Knaus P: miR-181a promotes osteoblastic
differentiation through repression of TGF-β signaling molecules.
Int J Biochem Cell Biol. 45:696–705. 2013.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Bai Y, Liu Y, Jin S, Su K, Zhang H and Ma
S: Expression of microRNA-27a in a rat model of osteonecrosis of
the femoral head and its association with TGF-β/Smad7 signalling in
osteoblasts. Int J Mol Med. 43:850–860. 2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Vimalraj S, Partridge NC and Selvamurugan
N: A positive role of microRNA-15b on regulation of osteoblast
differentiation. J Cell Physiol. 229:1236–1244. 2014.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Yin N, Zhu L, Ding L, Yuan J, Du L, Pan M,
Xue F and Xiao H: MiR-135-5p promotes osteoblast differentiation by
targeting HIF1AN in MC3T3-E1 cells. Cell Mol Biol Lett.
24(51)2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Gan L and Denecke B: Profiling
pre-MicroRNA and mature MicroRNA expressions using a single
microarray and avoiding separate sample preparation. Microarrays
(Basel). 2:24–33. 2013.PubMed/NCBI View Article : Google Scholar
|
|
64
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9(402)2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Akshaya RL, Rohini M, He Z, Partridge NC
and Selvamurugan N: MiR-4638-3p regulates transforming growth
factor-β1-induced activating transcription factor-3 and cell
proliferation, invasion, and apoptosis in human breast cancer
cells. Int J Biol Macromol. 222:1974–1982. 2022.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Lee EJ, Baek M, Gusev Y, Brackett DJ,
Nuovo GJ and Schmittgen TD: Systematic evaluation of microRNA
processing patterns in tissues, cell lines, and tumors. RNA.
14:35–42. 2008.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Varedi K SM, Ventura AC, Merajver SD and
Lin XN: Multisite phosphorylation provides an effective and
flexible mechanism for switch-like protein degradation. PLoS One.
5(e14029)2010.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Zhang YJ, Gendron TF, Xu YF, Ko LW, Yen SH
and Petrucelli L: Phosphorylation regulates proteasomal-mediated
degradation and solubility of TAR DNA binding protein-43 C-terminal
fragments. Mol Neurodegener. 5(33)2010.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Drazic A, Myklebust LM, Ree R and Arnesen
T: The world of protein acetylation. Biochim Biophys Acta.
1864:1372–1401. 2016.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Wang CY, Yang SF, Wang Z, Tan JM, Xing SM,
Chen DC, Xu SM and Yuan W: PCAF acetylates Runx2 and promotes
osteoblast differentiation. J Bone Miner Metab. 31:381–389.
2013.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Westendorf JJ: Transcriptional
co-repressors of Runx2. J Cell Biochem. 98:54–64. 2006.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Jensen ED, Schroeder TM, Bailey J,
Gopalakrishnan R and Westendorf JJ: Histone deacetylase 7
associates with Runx2 and represses its activity during osteoblast
maturation in a deacetylation-independent manner. J Bone Miner Res.
23:361–372. 2008.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Jin Y, Chen Z, Liu X and Zhou X:
Evaluating the microRNA targeting sites by luciferase reporter gene
assay. Methods Mol Biol. 936:117–127. 2013.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Ryoo HM, Kang HY, Lee SK, Lee KE and Kim
JW: RUNX2 mutations in cleidocranial dysplasia patients. Oral Dis.
16:55–60. 2010.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Lou Y, Javed A, Hussain S, Colby J,
Frederick D, Pratap J, Xie R, Gaur T, van Wijnen AJ, Jones SN, et
al: A Runx2 threshold for the cleidocranial dysplasia phenotype.
Hum Mol Genet. 18:556–568. 2009.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Xu W, Chen Q, Liu C, Chen J, Xiong F and
Wu B: A novel, complex RUNX2 gene mutation causes cleidocranial
dysplasia. BMC Med Genet. 18(13)2017.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Aiken A and Khokha R: Unraveling
metalloproteinase function in skeletal biology and disease using
genetically altered mice. Biochim Biophys Acta. 1803:121–132.
2010.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Burrage PS, Mix KS and Brinckerhoff CE:
Matrix metalloproteinases: Role in arthritis. Front Biosci.
11:529–543. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
79
|
Young DA, Barter MJ and Wilkinson DJ:
Recent advances in understanding the regulation of
metalloproteinases. F1000Res. 8(F1000 Faculty
Rev-195)2019.PubMed/NCBI View Article : Google Scholar
|