Introduction
Gastric cancer is the predominant gastrointestinal malignancy at present. In 2020, there were >1 million new cases of gastric cancer globally and ~769,000 deaths. As a result, it ranks 5th in incidence and 4th in mortalities related to malignant tumors (1). Due to the invasiveness of endoscopic screening and the lack of ideal biomarkers, the early diagnosis rate of gastric cancer is <10% and the proportion of patients at the intermediate and late stage is higher (2). The 5-year survival rate is ~90% for early gastric cancer, while that of intermediate and advanced gastric cancer is only ~30% and the overall therapeutic effect is not ideal (3). Therefore, early detection and diagnosis are key to improving the survival rate and reducing the fatality rate of gastric cancer. However, early gastric cancer and precancerous lesions are mostly asymptomatic or cause nonspecific symptoms (4,5). Gastric cancer is often mistaken for benign diseases, such as gastritis and dyspepsia, in the early stage of the disease, and it fails to get the attention of patients so they easily miss the best treatment period. Most patients have developed advanced gastric cancer when diagnosed, which requires chemotherapy, radiotherapy, surgery and other treatments, increasing the physical and psychological burden of patients and endangering the safety of patients in serious cases (6,7). Currently, gastric cancer diagnosis primarily relies on gastroscopy and pathological diagnosis (8). However, gastroscopy and pathological biopsy are invasive examinations, which are difficult for patients to accept and have a high cost, and they also have certain requirements regarding the operation ability and experience of doctors, so the opportunity for early diagnosis is usually missed (9).
Gastric high-grade intraepithelial neoplasia (HGIN) is a precancerous lesion. Over 80% of gastric HGIN progresses to adenocarcinoma (10). If the lesions can be detected and treated in time at the gastric HGIN stage, the therapeutic effect on patients is significantly improved. Appropriate biomarkers can improve the rates of early diagnosis of gastric cancer and help achieve its prompt treatment (11). Although studies on biomarkers for gastric cancer have been gradually carried out at home and abroad, only a small number of biomarkers can be used for auxiliary diagnosis of clinical gastric cancer so far (12,13), such as carcinoembryonic antigen, carbohydrate antigen 19-9 (CA19-9), CA724 and α-fetoprotein, but the detection rate and accuracy are low, which are not sufficient to replace conventional pathological diagnosis. The gastric HGIN-related diagnostic sensitivity of of 77.8% and specificity of 81.8%, are still relatively low (14,15).
Blood is an ideal sample for detecting tumor biomarkers because some tumor-related specific proteins are released into the blood, and blood sampling is simple, rapid, and minimally invasive, so samples are relatively easy to obtain. Abnormal protein expression plays a principal function in the occurrence as well as the development of gastric cancer (16,17). Proteomic studies have been reported on gastric cancer, and certain differentially expressed proteins (DEPs), such as fibrinogen α-chain, apolipoprotein A-II, vitronectin and clusterin isoform 1, have been screened (18,19). Another study from Singapore, comparing plasma proteomics of normal individuals and patients with early and advanced gastric cancer found that the level of component 9 (C9) in patients with gastric cancer was significantly higher than that in normal subjects, and the subsequent blind test study on 119 plasma samples showed that the sensitivity of C9 could be as high as 90% at a specificity of 74% (20). However, due to different etiologies and different stages of the disease, the results of various research groups were different (21). In addition, proteomic data on gastric HGIN are still scarce.
Label-free quantification (LFQ) is one of the most popular high-throughput screening techniques in quantitative proteomics. The approach may detect >1,000 proteins in a serum sample with a single needle, after which it employs tandem mass spectrometric analysis to compare the relative protein content in up to 10 distinct samples concurrently (22). In addition, this technology has the advantages of high throughput, high sensitivity, low-abundance protein detection, accurate quantification, good repeatability, reliable identification results and detection of a wide range of proteins, and is widely used in the analysis and exploration of DEPs (23). In the present study, LFQ together with liquid chromatography with tandem mass spectrometry (LC-MS/MS) was employed to compare the peripheral blood's protein expression of patients with gastric HGIN and healthy controls, and the protein expression profile in serum was analyzed biologically, providing a basis for finding biomarkers for gastric HGIN.
Patients and methods
Patients and plasma samples
The present study was approved by the Ethics Committee of the Affiliated Hospital of Putian University (Putian, China; approval no. 202006) (24). All patients enrolled in this study were from the Affiliated Hospital of Putian University (Putian, China), with the enrollment period from January 2022 to March 2022. Each patient who participated signed a written informed consent form before the study began. All patients were divided into a gastric HGIN group and healthy control group, meeting the following inclusion and exclusion criteria: The gastric HGIN group was pathologically confirmed as HGIN, and endoscopic ultrasonography and computed tomography did not confirm metastasis, and there was no previous history of gastric malignancy. The healthy control group underwent gastroscopy to exclude gastric HGIN. The following conditions were excluded in both groups: Associations with other malignant tumors, serious systemic diseases and serious primary diseases. A total of 10 serum samples were collected and all personnel was from the Affiliated Hospital of Putian University. Male individuals with a higher incidence of gastric cancer were selected as the case group, and age-matched healthy individuals were chosen from the same period as the control group. There were 5 cases in the gastric HGIN group and 5 cases in the healthy control group. Among them, 5 patients in the gastric HGIN group underwent endoscopic submucosal dissection (ESD) at the Affiliated Hospital of Putian University, and were diagnosed with HGIN by postoperative pathology. The protocol was performed according to standard procedures. Centrifugation of blood samples was conducted at 1,300 x g for 10 min at 4˚C and immediately stored at -80˚C.
Protein extraction and trypsin digestion
The centrifugation process was crucial in the isolation of cellular debris contained in the serum sample (12,000 x g at 4˚C for 10 min). After centrifugation, the supernatant was transferred into another centrifuge tube. The Pierce™ Top 14 Abundant Protein Depletion Spin Columns Kit (Thermo Fisher Scientific, Inc.) was utilized to separate the top 14 high-abundance proteins. Afterward, the concentration of protein was ascertained utilizing a bicinchoninic acid kit (cat. no. P0011; Beyotime Institute of Biotechnology) as per the manufacturer's specifications.
The same amount of each sample protein was taken for enzymatic hydrolysis. An appropriate amount of standard protein (cat. no. 26620; Thermo Fisher Scientific, Inc.) was added and the volume was adjusted to be consistent with the lysate. A total of 5 mM dithiothreitol was utilized for the reduction of the protein solution to facilitate its digestion for 30 min at 56˚C. Subsequently, it underwent alkylation with 11 mM iodoacetamide for 15 min at room temperature, which was performed in the dark. The alkylated samples were transferred to ultrafiltration tubes, centrifuged at 12,000 x g for 20 min at room temperature, replaced with 8 M urea for 3 times and then replaced with displacement buffer for 3 times. Finally, trypsin (Promega Corporation) was introduced at 1:50 trypsin-to-protein mass ratios for digestion overnight at 37˚C. The peptides were subjected to centrifugation at 12,000 x g for 10 min at room temperature and then recovered once with ultrapure water, and the two peptide solutions were combined.
LC-MS/MS analysis
The tryptic peptides were dissolved in solvent A (0.1% formic acid, 2% acetonitrile/in water) and directly loaded onto a home-made reversed-phase analytical column (25 cm length, 100 µm inner diameter) (cat. no. ZTC18M096; MilliporeSigma). The liquid phase gradient was set as follows: 0-68 min, 4-20% solvent B (0.1% formic acid in 90% acetonitrile); 68-82 min, 20-32% B; 82-86 min, 32-80% B; 86-90 min, 80% B, all at a constant flow rate of 500 nl/min on an EASY-nLC 1200 ultra-performance LC system (Thermo Fisher Scientific, Inc.).
The separated peptides were analyzed in Exploris 480™ (Thermo Fisher Scientific, Inc.) with a nano-electrospray ion source. The electrospray voltage applied was 2.3 kV and the compensation voltages were -45 and -70 V. The full MS scan resolution was set to 60,000 for a scan range of 400-1,200 m/z. Up to 25 of the most abundant precursors were then selected for further MS/MS analyses with 30 sec dynamic exclusion. The higher-energy collisional dissociation fragmentation was performed at a normalized collision energy of 27%. The fragments were detected in the Orbitrap at a resolution of 30,000. The fixed first mass was set at 110 m/z. The automatic gain control target was set at 75%, with an intensity threshold of 1E4 ions/sec and the MS2 maximum injection time was set at 100 msec. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with database identifier PXD036991 (https://www.ebi.ac.uk/pride/profile/reviewer_pxd036991).
Database search
The resulting MS/MS data were processed using Proteome Discoverer search engine (v2.4.0.305) (https://www.thermofisher.cn/cn/zh/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/proteome-discoverer-software.html?erpType=Global_E1). Tandem mass spectra were searched against the Homo sapiens database (78,120 entries) concatenated with a reverse decoy database. Trypsin (Full) was specified as the cleavage enzyme, allowing up to 2 missing cleavages. The mass tolerance for precursor ions was set at 10 ppm in the first search and the mass tolerance for fragment ions was set at 0.02 Da. Carbamidomethyl on Cys was specified as a fixed modification, and oxidation (methionine; M), acetylation on protein N-term, Met-loss (M) and Met-loss+acetyl (M) were specified as variable modifications. The quantitative method was set as LFQ. The false discovery rate was adjusted to <1% and the minimum score for modified peptides was set to >40. The minimum peptide length was set at 6. All of the other parameters in Proteome Discoverer were set to default values.
Bioinformatics analysis
Gene Ontology (GO) annotations were acquired from the eggNOG database (v.5.0) (http://eggnog5.embl.de/#/app/home). The eggNOG-mapper software (v.2.0) (http://eggnog-mapper.embl.de/) analyzed these GO categories: Cellular component, molecular function and biological process. The Kyoto Encyclopedia of Genes and Genomes (KEGG) online service tool KAAS (v.2.1) (https://www.genome.jp/tools/kaas/) annotated the protein descriptions and then mapped using the KEGG mapper (v.5.0) (https://www.kegg.jp/kegg/mapper/) to their respective pathways. WoLFPSort (v.0.2) (https://www.genscript.com/wolf-psort.html) was used to investigate the subcellular localization. P<0.05 was deemed statistically significant for both GO terms and KEGG pathways. Gene count thresholds for the GO terms and KEGG pathways were established at ≥20 and ≥4, respectively.
Statistical analysis
Clinical parameter data were expressed as the mean ± standard deviation. Continuous variables were compared using Student's t-test and the Mann-Whitney U-test depending on whether the data were normally distributed. The protein data along with the intensity values were log-transformed with base 2 and median normalization was carried out to eliminate random errors and sample bias. DEPs were identified by a two-tailed Student's t-test. The pathway enrichment analysis was performed by a two-tailed Fisher's exact test. P<0.05 (two-tailed) was considered to indicate a statistically significant difference. Data were analyzed and visualized with SPSS 21.0 (IBM, Corp.) or GraphPad Prism 7.0 (GraphPad Software, Inc.).
Results
Characteristics of patients with gastric HGIN and healthy controls
A total of 10 serum samples were collected in this study. In the gastric HGIN group, 5 cases were male, with an average age of 62.6±3.8 years. All cases were diagnosed with HGIN after ESD. The healthy control group consisted of 5 cases, all male, with an average age of 59.8±0.8 years. There was no rmarked difference in terms of age between the patients with gastric HGIN and the healthy controls (P>0.05, two-tailed Student's t-test) (Table I).
 |
Table I
Demographics and clinicopathological characteristics of patients and healthy controls.
|
Table I
Demographics and clinicopathological characteristics of patients and healthy controls.
| Characteristics |
Patients |
Healthy controls |
P-value |
| Sex, n (%) |
|
|
1.000 |
| Male |
5(100) |
5(100) |
|
| Female |
0 (0) |
0 (0) |
|
| Mean age ± SD, years |
62.6±3.8 |
59.8±0.8 |
0.168 |
| Pathology, n (%) |
|
|
0.004 |
| Gastric HGIN |
5(100) |
0 |
|
| Normal |
0 |
5(100) |
|
Identification of DEPs
LFQ in conjunction with LC-MS/MS enabled the discovery of 1,192 serum proteins. Upregulated and downregulated DEPs were defined as proteins with expression ratio thresholds of >1.5-fold increase or <0.67-fold decrease (P<0.05, two-tailed Student's t-test), respectively. In comparison with the control group, 18 proteins were upregulated and 12 proteins were downregulated in the gastric HGIN group (Table II; Fig. 1).
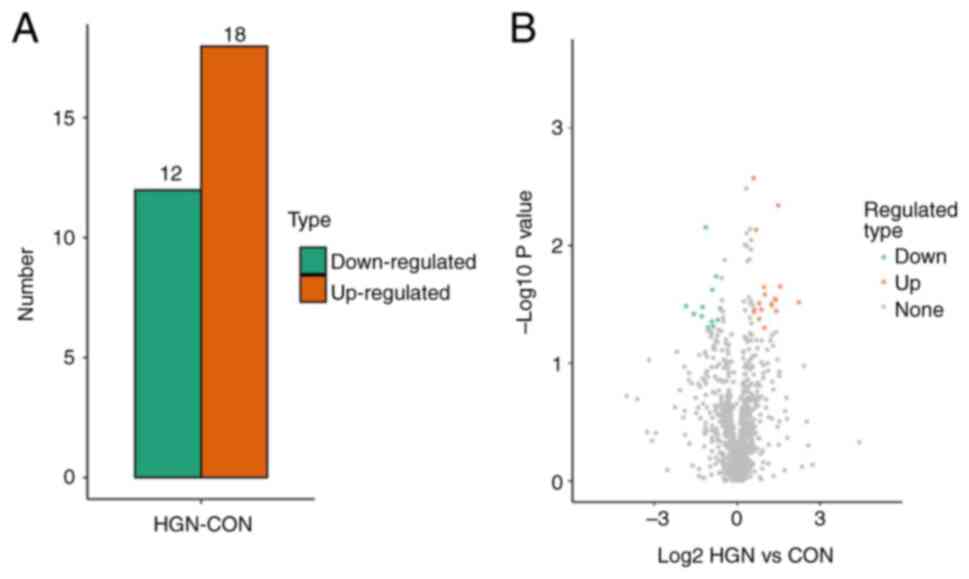 |
Figure 1
Quantification of DEPs. (A) The quantity of DEPs utilizing a threshold of 1.5-fold. (B) A volcanic scatter plot showing a threshold of 1.5. DEP, differentially expressed protein; HIGN, high-grade neoplasia; CON, control.
|
 |
Table II
Differentially expressed proteins between the gastric HGIN group and the control group.
|
Table II
Differentially expressed proteins between the gastric HGIN group and the control group.
| A, Upregulated proteins |
| Protein accession no. |
Protein description |
Gene name |
HGIN/CON ratio |
HGIN/CON P-value |
| F6SYF8 |
Dickkopf-related protein 3 |
DKK3 |
4.698 |
0.030 |
| P55259 |
Pancreatic secretory granulemembrane major glycoprotein GP2 |
GP2 |
2.938 |
0.022 |
| A0A0G2JMX5 |
Leukocyte immunoglobulin-like receptor subfamily B member 5 |
LILRB5 |
2.802 |
0.005 |
| O76076 |
CCN family member 5 |
CCN5 |
2.667 |
0.036 |
| A0A0B4J2D9 |
Immunoglobulin κ variable 1D-13 |
IGKV1D-13 |
2.639 |
0.029 |
| Q9NQ38 |
Serine protease inhibitor Kazal-type 5 |
SPINK5 |
2.585 |
0.029 |
| E7ENL6 |
Collagen α-3(VI) chain |
COL6A3 |
2.383 |
0.032 |
| H0YCU9 |
Transgelin (Fragment) |
TAGLN |
2.381 |
0.031 |
| Q92484 |
Acid sphingomyelinase-like phosphodiesterase 3a |
SMPDL3A |
2.009 |
0.026 |
| Q12864 |
Cadherin-17 |
CDH17 |
1.995 |
0.050 |
| A0A1W2PRS4 |
Junctional adhesion molecule-like |
JAML |
1.964 |
0.023 |
| Q9BRV8 |
Suppressor of IKBKE 1 |
SIKE1 |
1.836 |
0.035 |
| A0A0A0MSN4 |
Angiotensin-converting enzyme |
ACE |
1.748 |
0.031 |
| P17931 |
Galectin-3 |
LGALS3 |
1.738 |
0.042 |
| Q15828 |
Cystatin-M |
CST6 |
1.611 |
0.007 |
| P16070 |
CD44 antigen |
CD44 |
1.538 |
0.035 |
| P61769 |
β-2-microglobulin |
B2M |
1.53 |
0.037 |
| Q9NPY3 |
Complement component C1q receptor |
CD93 |
1.516 |
0.003 |
| B, Downregulated proteins |
| Protein accession no. |
Protein description |
Gene name |
HGIN/CON ratio |
HGIN/CON P-value |
| P30101 |
Protein disulfide-isomerase A3 |
PDIA3 |
0.659 |
0.034 |
| P28070 |
Proteasome subunit β type-4 |
PSMB4 |
0.618 |
0.043 |
| P26447 |
Protein S100-A4 |
S100A4 |
0.591 |
0.018 |
| A0A0J9YVY3 |
Immunoglobulin heavy variable 7-4-1 |
IGHV7-4-1 |
0.551 |
0.049 |
| Q12882 |
Dihydropyrimidine dehydrogenase [NADP(+)] |
DPYD |
0.539 |
0.024 |
| B5MDQ0 |
DNA excision repair protein ERCC-6-like |
ERCC6L |
0.531 |
0.044 |
| O95897 |
Noelin-2 |
OLFM2 |
0.484 |
0.050 |
| O14793 |
Growth/differentiation factor 8 |
MSTN |
0.456 |
0.007 |
| P04406 |
Glyceraldehyde-3-phosphate dehydrogenase |
GAPDH |
0.421 |
0.033 |
| Q9HBB8 |
Cadherin-related family member 5 |
CDHR5 |
0.415 |
0.040 |
| Q8N1N4 |
Keratin, type II cytoskeletal 78 |
KRT78 |
0.337 |
0.038 |
| P01705 |
Immunoglobulin λ variable 2-23 |
IGLV2-23 |
0.279 |
0.033 |
GO analysis
GO analysis evaluated the biological process, cellular component and molecular function terms enriched by the DEPs. Concerning the biological process analysis, the leading five modulated processes were ‘cellular’, ‘biological regulation’, ‘multicellular organismal’, ‘developmental’ and ‘reaction to stimulus processes’. Regarding the cellular component analysis, the most prevalent proteins were primarily linked to the ‘cell’, ‘intracellular’ and ‘protein-containing complex’. On the other hand, the molecular function analysis indicated that the prevalent molecular functions were ‘molecular function modulator’, ‘binding’ and ‘catalytic activity’ (Fig. 2A).
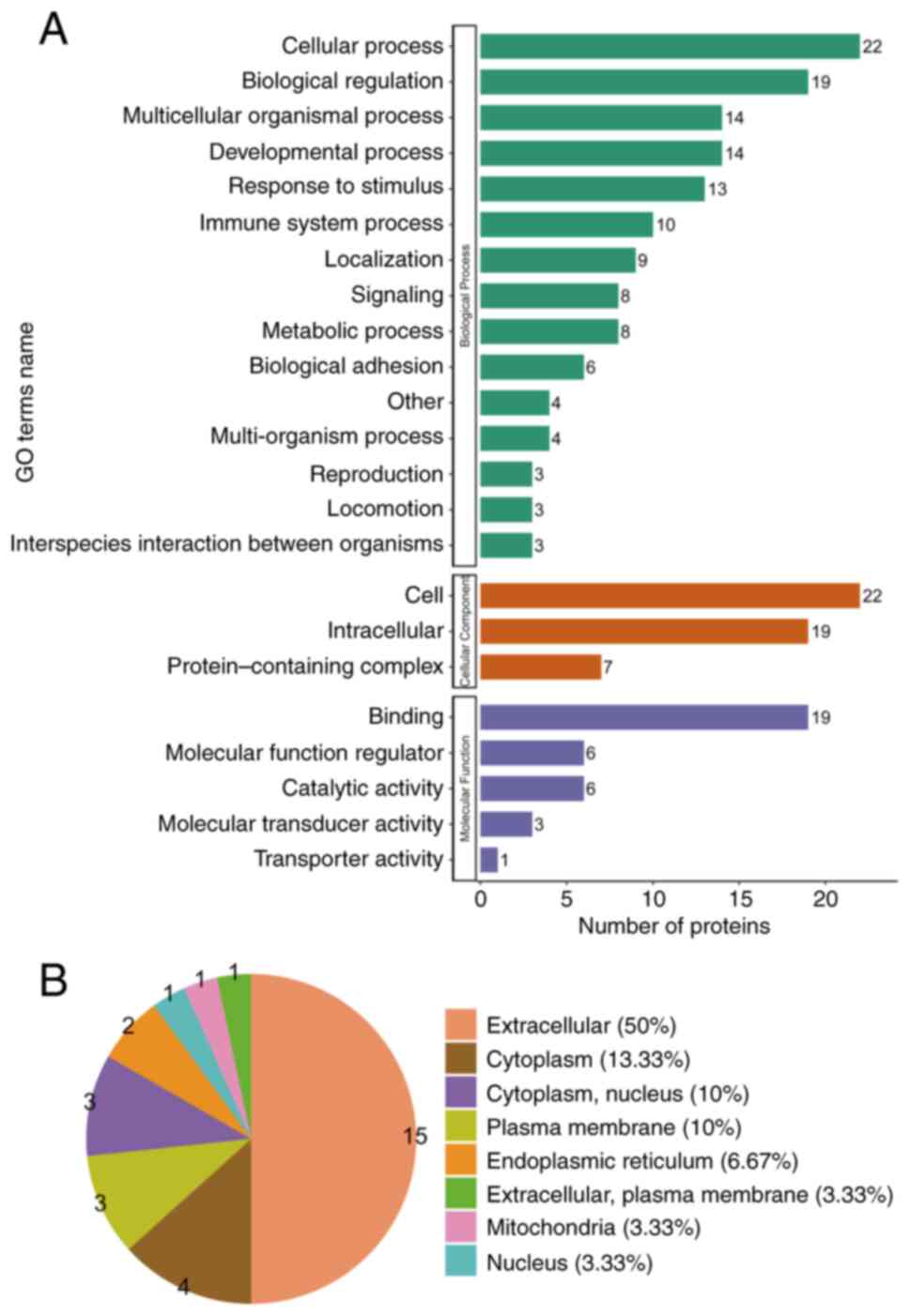 |
Figure 2
GO annotation analysis revealing the possible biomarkers principally engaged in the gastric HGIN group. (A) GO terms’ names, namely cellular component, molecular function and biological process in HGIN-linked proteins. (B) Subcellular structural localization evaluation in HGIN-linked proteins. GO, gene ontology; HGIN, high-grade intraepithelial neoplasia.
|
The major subcategories (>40%) for the gastric HGIN proteins were extracellular (50%), cytoplasm (13.33%), cytoplasm/nucleus (10%) and plasma membrane (10%) (Fig. 2B) as per the subcellular structural localization evaluations.
To determine whether the DEPs were remarkably enriched in certain functional terms, GO enrichment analysis was employed to analyze the DEPs. GO enrichment analysis identifies significant enrichment of DEPs in specific functional terms through statistical methods (P<0.05), whereas GO categorization describes the distribution profile of DEPs across GO hierarchy without emphasizing statistical significance. The core distinction resides in the application of statistical testing to assess functional specificity enrichment. In the category biological process, the most significantly enriched items were ‘mononuclear cell migration’, ‘positive modulation of cell killing’ and ‘hemopoiesis modulation’ (Fig. 3A). In cellular component, the most significantly enriched items were ‘brush border membrane’, ‘recycling endosome membrane’ and ‘microvillus’ (Fig. 3B). In the category molecular function, the most significantly enriched items were ‘transforming growth factor beta receptor binding’ and ‘phosphoric diester hydrolase activity’ (Fig. 3C).
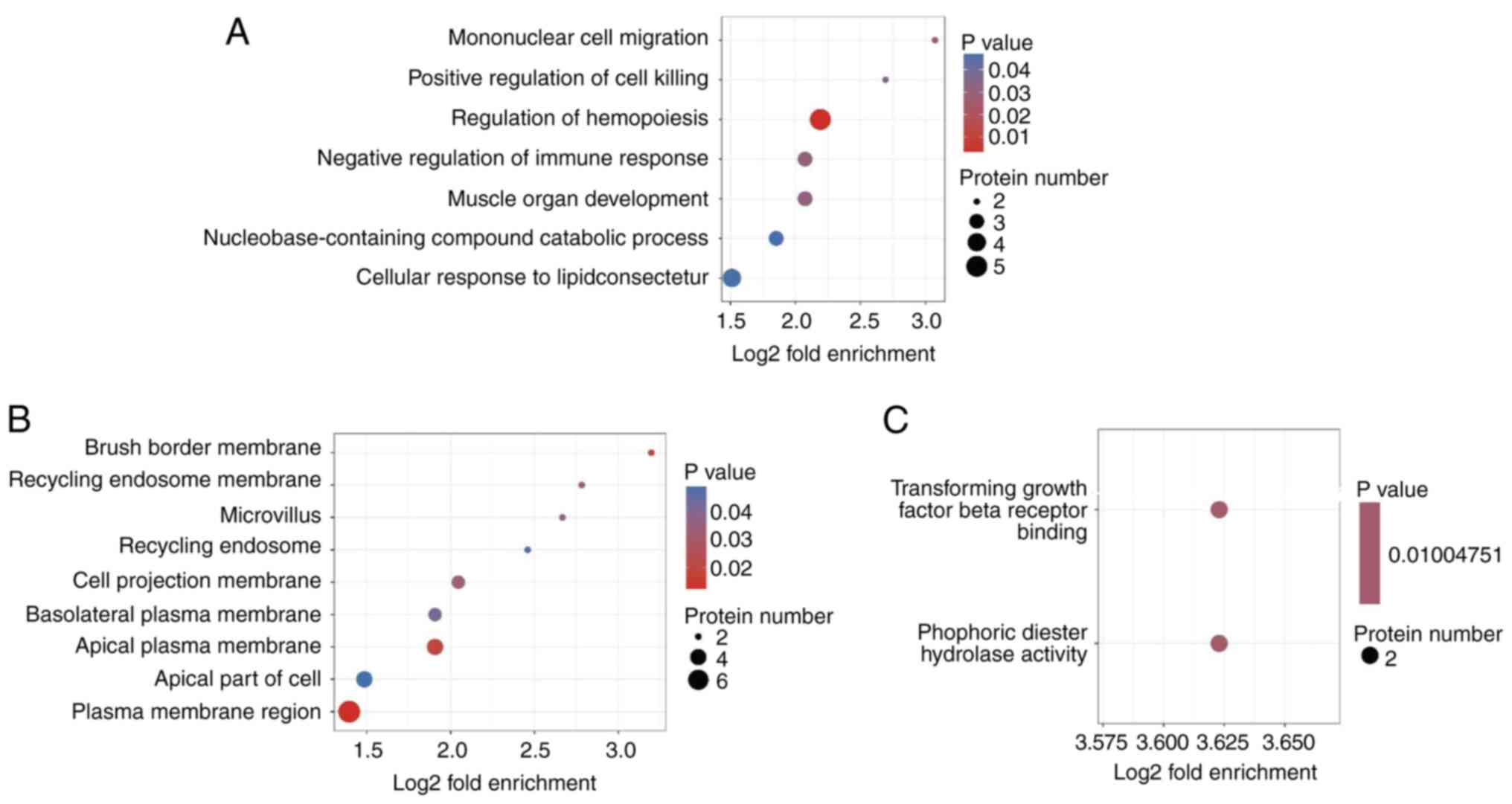 |
Figure 3
GO enrichment analyses. (A) GO enrichment analyses for biological process. (B) GO enrichment analyses for cellular component. (C) GO enrichment analyses for molecular function. GO, gene ontology.
|
KEGG pathway enrichment analysis
The proteins were integrated into the KEGG database and Fisher's exact test P-values were important to determine the proteins' enrichment levels in order to identify the pathways influenced by the DEPs. The KEGG pathway enrichment analysis manifested that the DEPs were predominantly enriched in ‘antigen processing and presentation’, ‘diabetic cardiomyopathy’, ‘Epstein-Barr virus infection’, ‘herpes simplex virus 1 infection’, ‘human immunodeficiency virus 1 infection’ and ‘human cytomegalovirus infection’ (Fig. 4).
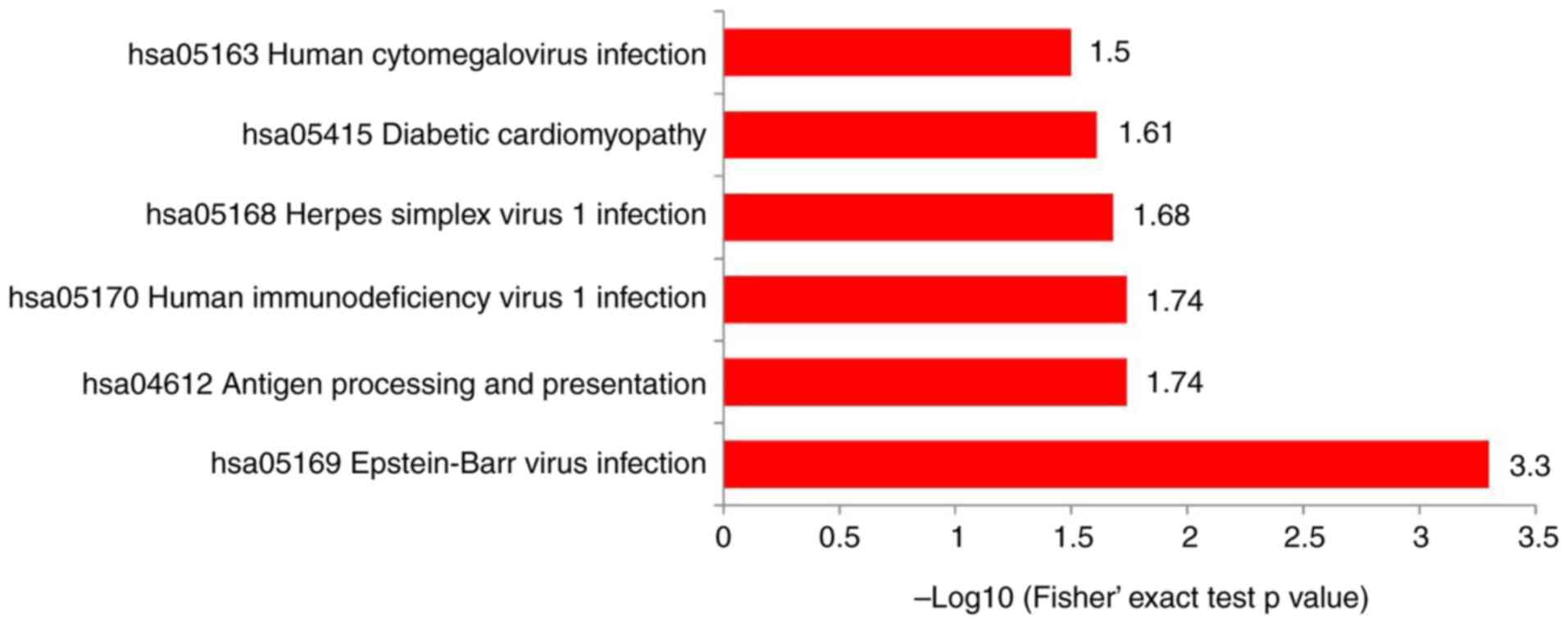 |
Figure 4
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis comparing differentially expressed proteins.
|
Discussion
In the present study, 30 DEPs were identified in the gastric HGIN group in comparison with the healthy control group, including 18 and 12 upregulated and downregulated proteins, correspondingly. Bioinformatics analysis showed that the functions of the DEPs mainly included ‘cellular process’, ‘biological regulation’, ‘multicellular organismal process’, ‘binding, molecular function regulator’ and ‘catalytic activity’. The major signaling pathways included ‘antigen processing and presentation’, ‘diabetic cardiomyopathy’, ‘Epstein-Barr virus infection’, ‘herpes simplex virus 1 infection’, ‘human immunodeficiency virus 1 infection’ and ‘human cytomegalovirus infection’. DEP functions, as well as the signaling pathways engaged, can avail an experimental foundation for determining the mechanism of occurrence and advancement of gastric HGIN. The proteomic characteristics and data of gastric HGIN analyzed in the study were different from those of previous gastric cancer proteomics and were heterogeneous (25,26). The discrepancy may be linked to the different stages of gastric cancer in each study.
Collagen alpha-3 (VI) chain (COL6A3) is among the collagen type VI, which is the predominant structural extracellular matrix protein (27). It is primarily expressed in stromal cancer-associated fibroblasts (CAFs) (28). CAFs are a central determinant in the progression of malignancies, as affirmed by a previous study (29). COL6A3 expression was remarkably varied in the normal mucosa in contrast with cancerous tissues, with the greatest and lowest expression levels in primary colorectal cancer and adenoma, respectively. COL6A3 expression was positively directly proportional to the stage of prostate, breast and colorectal cancer (30). Certain studies have reported that the level of expression of COL6A3 in gastric cancer tissue is higher than in normal tissue (31,32). Therefore, COL6A3 could be used as a tumor diagnostic marker. COL6A3 functioned in tumorigenesis as well as in the progression of cholangiocarcinoma via the E2F1/LMCD1-AS1/miR-345-5p/COL6A3 axis (33). In a study on colon cancer, stromal COL6A3 could enhance tumor growth by regulating Hippo and Wnt signaling (34). COL6A3 knockout lowered cell proliferation and invasion, and enhanced cell apoptosis in cancer cell lines (28), further confirming the relationship between COL6A3 and tumor progression. In this study, COL6A3 expression was upregulated, indicating that it may be engaged in the development of gastric HGIN, but the specific mechanism of action remains to be further studied.
Cadherin-17 (CDH17), also referred to as human peptide transporter-1 or liver-intestine cadherin, encompasses seven homologous repeated domains. As one of the cadherin superfamily members, it functions in intercellular conjunction (35). CDH17 can regulate the Ca2+-dependent homophilic cell-cell adhesion, suggesting that it may be related to the occurrence of tumors (36). In general, CDH17 is expressed not only on the enterocytes’ basolateral surfaces but also on goblet cells in the small and large intestines selectively and is rarely discovered in the stomach or liver of a healthy adult (37,38). Previous documentation affirms that the elevated expression level of CDH17, contrary to the lowered levels of various classical cadherins, is linked to cholangiocarcinoma as well as gastric, liver, colorectal and pancreatic cancers, highlighting that CDH17 may have a central function in tumor progression (39). In many of these malignancies, CDH17 expression is linked to tumor stage or unfavorable survival of patients (40). Therefore, CDH17 can be utilized as a sensitive tumor marker for the identification of metastasis of unclear primary origin as well as their subsequent assignation to the gastrointestinal tract (41). Multiple studies on gastric cancer have affirmed that overexpression of CDH17 was positively linked to the histological stage and tumor invasion of gastric cancer, demonstrating that CDH17 expression can be utilized as a valuable indicator for anticipating the progression of gastric cancer (42). High expression of CDH17 can promote cell adhesion and proliferation and participates in the formation and development of gastric cancer mainly via three signaling pathways in the cells, i.e. the Wnt/β-catenin pathway, the NF-κB signaling pathway and the Ras/Raf/MEK/ERK MAPK signaling pathway (43). In the present study, the expression of CDH17 was upregulated in the gastric HGIN group, suggesting that CDH17 influences the occurrence and development of the disease. However, since we are studying precancerous lesions of gastric cancer, which is at a different disease stage from previous studies, whether it has the same mechanism of action needs to be proved by further studies.
The crucial element of the extracellular matrix (ECM), hyaluronic acid (HA) is essentially linked to a complex transmembrane adhesion glycoprotein known as cluster of differentiation 44 (CD44) (44). Numerous studies have affirmed that CD44 is not only important in the various physiological processes, for instance, diverse immune functions, organ development and hematopoiesis (45,46), but also in pathological processes, particularly tumors (47). Increasing evidence infers that CD44 is remarkably overexpressed in various cancer types such as lymphoma, breast, colon, endometrial, prostate, ovarian, gallbladder and oral squamous cell carcinoma, which is linked to aggressive biological behavior (48,49). CD44 expression was also upregulated in previous studies on gastric precancerous lesions and gastric cancer (50). CD44 interacts with different ECM components, cytokines and proteins that exist in the tumor microenvironment. The interaction between ligands, including osteopontin and HA, in addition to matrix metalloproteinases, with the CD44 receptor, can trigger numerous cellular signaling pathways, thus enhancing tumor advancement and aggressiveness (51). In the present study, CD44 expression was upregulated in the gastric HGIN group. Although the role of CD44 in gastric HGIN remains elusive, it may be involved in the formation of HGIN.
The protein disulfide isomerase A3 (PDIA3) was identified as the most remarkable downregulated protein based on the various ratios in protein expression and the related P-values. PDIA3, also referred to as endoplasmic reticulum resident protein 57, is among the principal members of the PDI gene family. It has been discovered as one of the primary enzymatic chaperones that are essential in the reconstruction of misfolded proteins inside the endoplasmic reticulum (52). It has been ascertained that in some malignancies, the expression of PDIA3 is negatively linked to the degree of differentiation. In uterocervical cancer, PDIA3 expression was downregulated in contrast with normal tissue, as well as cervical intraepithelial neoplasia (53). Another study showed that low expression of PDIA3 was significantly linked to poor cause-specific survival in patients with papillary thyroid carcinoma (54). In cancer immunity, PDIA3 creates a complex with major histocompatibility complexes (MHC) class I to enhance antigen processing, thus triggering an immune response against cancerous cells (55). On the contrary, when MHC class I expression is inadequate, cancerous cells can evade the cytotoxic impacts of immune cells that can be seen in gastric cancer (56). In the present study, downregulation of PDIA3 was detected, which indicated the possibility of inactivation of gastric HGIN as a tumor suppressor during the development of HGIN, but its potential impact requires further study.
The present study has certain limitations. First of all, the sample size included in this study is small and the screened DEPs have not been further verified. In the future, it is necessary to further expand the sample and use enzyme-linked immunosorbent assay and other experimental methods to verify the DEPs. Secondly, due to the small sample size, in order to take into account the reliability of the experimental data, only males with a relatively higher incidence of gastric cancer were selected as experimental subjects in this study (57), and it is required to expand the sample to further include females in the study.
To conclude, utilizing LFQ coupled with LC-MS/MS, it was found that certain proteins, such as COL6A3, CDH17, CD44 and PDIA3, could be candidate protein biomarkers for gastric HGIN. However, additional studies are indispensable to ascertain the possible functions of these candidate proteins in the development of gastric HGIN. The present data provide additional biological information regarding the formation of gastric HGIN and provide clues for further research on the pathogenesis of early gastric cancer.
Acknowledgements
Not applicable.
Funding
Funding: This research was funded by the Science and Technology Project Plan of Putian city (grant no. 2020S3F001).
Availability of data and materials
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with database identifier PXD036991 (https://www.ebi.ac.uk/pride/profile/reviewer_pxd036991).
Authors' contributions
NL, CH and JG designed the study. LL and XH collected data and patients. NL, LL and XH performed the research. NL and CH analyzed the data. NL, CH and JG wrote the manuscript. All authors contributed to editorial changes to the manuscript. NL and LL confirm the authenticity of the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the Affiliated Hospital of Putian University (approval no. 202006). Informed consent was obtained from all subjects involved in the study. Patients were fully aware of the purpose, methods, potential therapeutic benefits and possible adverse reactions of the study, and willingly agreed to participate and fully cooperate with the doctors.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wu D, Zhang P, Ma J, Xu J, Yang L, Xu W, Que H, Chen M and Xu H: Serum biomarker panels for the diagnosis of gastric cancer. Cancer Med. 8:1576–1583. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Liu K, Song S, Fu T, Liu Y, Zhang H, Yan M, He Z, Zhang W, Su H, Li Z, et al: Spatiotemporal trends in the incidence of gastrointestinal neoplasms in Wuwei City of Northwestern China from 1995 to 2016: A hospital-based retrospective observational study. Front Oncol. 11(712857)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Shetty A, Balaraju G, Shetty S and Pai CG: Diagnostic utility of alarm features in predicting malignancy in patients with dyspeptic symptoms. Indian J Gastroenterol. 40:183–188. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Repetto O, Maiero S, Magris R, Miolo G, Cozzi MR, Steffan A, Canzonieri V, Cannizzaro R and De Re V: Quantitative proteomic approach targeted to fibrinogen β Chain in tissue gastric carcinoma. Int J Mol Sci. 19(759)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Machlowska J, Baj J, Sitarz M, Maciejewski R and Sitarz R: Gastric cancer: Epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 21(4012)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liu C, Wang Y, Li L, He D, Chi J, Li Q, Wu Y, Zhao Y, Zhang S, Wang L, et al: Engineered extracellular vesicles and their mimetics for cancer immunotherapy. J Control Release. 349:679–698. 2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu Z, Su W, Ao J, Wang M, Jiang Q, He J, Gao H, Lei S, Nie J, Yan X, et al: Instant diagnosis of gastroscopic biopsy via deep-learned single-shot femtosecond stimulated Raman histology. Nat Commun. 13(4050)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Delgado-Guillena PG, Morales-Alvarado VJ, Elosua-González A, Murcia Pomares O, Pérez-Aisa A, Córdova H, Alcedo J, Calvet X and Fernández-Esparrach G: Gastroenterologists' attitudes on the detection and management of gastric premalignant conditions: Results of a nationwide survey in Spain. Eur J Cancer Prev. 30:431–436. 2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ray-Offor E and Obiorah CC: Helicobacter pylori and precancerous lesions of the stomach in a Nigerian Metropolis: A cohort study. Niger J Clin Pract. 21:375–379. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ji L, Liu Z, Zhou B, Cai Y, An F, Wang L, Lv Z, Xia M, Yang J, Yuan J, et al: Community-based pilot study of a screening program for gastric cancer in a Chinese population. Cancer Prev Res (Phila). 13:73–82. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Feng F, Tian Y, Xu G, Liu Z, Liu S, Zheng G, Guo M, Lian X, Fan D and Zhang H: Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer. 17(737)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Toh J, Hoppe MM, Thakur T, Yang H, Tan KT, Pang B, Ho S, Roy R, Ho KY, Yeoh KG, et al: Profiling of gastric cancer cell-surface markers to achieve tumour-normal discrimination. BMJ Open Gastroenterol. 7(e000452)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhao J, Zhou R, Zhang Q, Shu P, Li H, Wang X, Shen Z, Liu F, Chen W, Qin J and Sun Y: Establishment of risk evaluation model of peritoneal metastasis in gastric cancer and its predictive value. Zhonghua Wei Chang Wai Ke Za Zhi. 20:47–52. 2017.PubMed/NCBI(In Chinese).
|
|
15
|
Wang Z, Mo TM, Tian L and Chen JQ: Gastrin-17 combined with CEA, CA12-5 and CA19-9 improves the sensitivity for the diagnosis of gastric cancer. Int J Gen Med. 14:8087–8095. 2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Shi XJ, Wei Y and Ji B: Systems biology of gastric cancer: Perspectives on the omics-based diagnosis and treatment. Front Mol Biosci. 7(203)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Niclauss N, Gütgemann I, Dohmen J, Kalff JC and Lingohr P: Novel biomarkers of gastric adenocarcinoma: Current research and future perspectives. Cancers (Basel). 13(5660)2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Shi F, Wu H, Qu K, Sun Q, Li F, Shi C, Li Y, Xiong X, Qin Q, Yu T, et al: Identification of serum proteins AHSG, FGA and APOA-I as diagnostic biomarkers for gastric cancer. Clin Proteomics. 15(18)2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yoo MW, Park J, Han HS, Yun YM, Kang JW, Choi DY, Lee JW, Jung JH, Lee KY and Kim KP: Discovery of gastric cancer specific biomarkers by the application of serum proteomics. Proteomics. 17(1600332)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ahn HS, Sohn TS, Kim MJ, Cho BK, Kim SM, Kim ST, Yi EC and Lee C: SEPROGADIC-serum protein-based gastric cancer prediction model for prognosis and selection of proper adjuvant therapy. Sci Rep. 8(16892)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Rostami-Nejad M, Rezaei-Tavirani M, Mansouri V, Akbari Z and Abdi S: Impact of proteomics investigations on gastric cancer treatment and diagnosis. Gastroenterol Hepatol Bed Bench. 12 (Suppl 1):S1–S7. 2019.PubMed/NCBI
|
|
22
|
Panner Selvam MK, Finelli R, Agarwal A and Henkel R: Proteomics and metabolomics-current and future perspectives in clinical andrology. Andrologia. 53(e13711)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu D, Zhou L, Huang L, Zuo Z, Ho V, Jin L, Lu Y, Chen X, Zhao J, Qian D, et al: Microfluidic integrated capacitive biosensor for C-reactive protein label-free and real-time detection. Analyst. 146:5380–5388. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu Y, Sang J, Zhou J and Fang Y: Comparative analysis of differences between preoperative endoscopic biopsy and postoperative pathological examination for diagnosis of gastric intraepithelial neoplasia. J Int Med Res. 49(300060521994929)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Fernández-Coto DL, Gil J, Hernández A, Herrera-Goepfert R, Castro-Romero I, Hernández-Márquez E, Arenas-Linares AS, Calderon-Sosa VT, Sanchez-Aleman MÁ, Mendez-Tenorio A, et al: Quantitative proteomics reveals proteins involved in the progression from non-cancerous lesions to gastric cancer. J Proteomics. 186:15–27. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Huang A, Zhang M, Li T and Qin X: Serum proteomic analysis by tandem mass Tags (TMT) based quantitative proteomics in gastric cancer patients. Clin Lab. 64:855–866. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Svoronos C, Tsoulfas G, Souvatzi M and Chatzitheoklitos E: Prognostic value of COL6A3 in pancreatic adenocarcinoma. Ann Hepatobiliary Pancreat Surg. 24:52–56. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liu W, Li L, Ye H, Tao H and He H: Role of COL6A3 in colorectal cancer. Oncol Rep. 39:2527–2536. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Dai L, Li M, Zhang WL, Tang YJ, Tang YL and Liang XH: Fibroblasts in cancer dormancy: Foe or friend? Cancer Cell Int. 21(184)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang J and Pan W: The biological role of the collagen alpha-3 (VI) Chain and its cleaved C5 domain fragment endotrophin in cancer. Onco Targets Ther. 13:5779–5793. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lu Y, Kong X, Zhong W, Hu M and Li C: Diagnostic, therapeutic, and prognostic value of the thrombospondin family in gastric cancer. Front Mol Biosci. 8(647095)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bessa C, Matos P, Jordan P and Gonçalves V: Alternative splicing: Expanding the landscape of cancer biomarkers and therapeutics. Int J Mol Sci. 21(9032)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yu J, Zhang B, Zhang H, Qi Y and Wang Y, Wang W and Wang Y and Wang Y: E2F1-induced upregulation of long non-coding RNA LMCD1-AS1 facilitates cholangiocarcinoma cell progression by regulating miR-345-5p/COL6A3 pathway. Biochem Biophys Res Commun. 512:150–155. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Huang Y, Li G, Wang K, Mu Z, Xie Q, Qu H, Lv H and Hu B: Collagen type VI alpha 3 Chain promotes epithelial-mesenchymal transition in bladder cancer cells via transforming growth factor β (TGF-β)/Smad pathway. Med Sci Monit. 24:5346–5354. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lee CW, Lin SE, Tsai HI, Su PJ, Hsieh CH, Kuo YC, Sung CM, Lin CY, Tsai CN and Yu MC: Cadherin 17 is related to recurrence and poor prognosis of cytokeratin 19-positive hepatocellular carcinoma. Oncol Lett. 15:559–567. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Casal JI and Bartolomé RA: Beyond N-cadherin, relevance of cadherins 5, 6 and 17 in cancer progression and metastasis. Int J Mol Sci. 20(3373)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Valverde A, Povedano E, Ruiz-Valdepeñas Montiel V, Yáñez-Sedeño P, Garranzo-Asensio M, Rodríguez N, Domínguez G, Barderas R, Campuzano S and Pingarrón JM: Determination of cadherin-17 in tumor tissues of different metastatic grade using a single incubation-step amperometric immunosensor. Anal Chem. 90:11161–11167. 2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kang MH, Jeong GS, Smoot DT, Ashktorab H, Hwang CM, Kim BS, Kim HS and Park YY: Verteporfin inhibits gastric cancer cell growth by suppressing adhesion molecule FAT1. Oncotarget. 8:98887–98897. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liu X, Huang Y, Yuan H, Qi X, Manjunath Y, Avella D, Kaifi JT, Miao Y, Li M, Jiang K and Li G: Disruption of oncogenic liver-intestine cadherin (CDH17) drives apoptotic pancreatic cancer death. Cancer Lett. 454:204–214. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kmeid M, Lukose G, Hodge K, Cho D, Kim KA and Lee H: Aberrant expression of SATB2, CDX2, CDH17 and CK20 in hepatocellular carcinoma: A pathological, clinical and outcome study. Histopathology. 79:768–778. 2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Altree-Tacha D, Tyrrell J and Haas T: CDH17 is a more sensitive marker for gastric adenocarcinoma than CK20 and CDX2. Arch Pathol Lab Med. 141:144–150. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Jiang XJ, Lin J, Cai QH, Zhao JF and Zhang HJ: CDH17 alters MMP-2 expression via canonical NF-κB signalling in human gastric cancer. Gene. 682:92–100. 2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Long ZW, Zhou ML, Fu JW, Chu XQ and Wang YN: Association between cadherin-17 expression and pathological characteristics of gastric cancer: A meta-analysis. World J Gastroenterol. 21:3694–3705. 2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Thorne RF, Wang Y, Zhang Y, Jing X, Zhang XD, de Bock CE and Oliveira CS: Evaluating nuclear translocation of surface receptors: Recommendations arising from analysis of CD44. Histochem Cell Biol. 153:77–87. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chen C, Zhao S, Karnad A and Freeman JW: The biology and role of CD44 in cancer progression: Therapeutic implications. J Hematol Oncol. 11(64)2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Xu H, Niu M, Yuan X, Wu K and Liu A: CD44 as a tumor biomarker and therapeutic target. Exp Hematol Oncol. 9(36)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Senbanjo LT and Chellaiah MA: CD44: A multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front Cell Dev Biol. 5(18)2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Nimmakayala RK, Batra SK and Ponnusamy MP: Unraveling the journey of cancer stem cells from origin to metastasis. Biochim Biophys Acta Rev Cancer. 1871:50–63. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
He Y, Xue C, Yu Y, Chen J, Chen X, Ren F, Ren Z, Cui G and Sun R: CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Cancer Manag Res. 10:3857–3865. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Tongtawee T, Wattanawongdon W, Simawaranon T, Kaewpitoon S, Kaengpenkae S, Jintabanditwong N, Tangjanyatham P, Ratchapol W, Kangwantas K, Dechsukhum C, et al: Expression of cancer stem cell marker CD44 and its polymorphisms in patients with chronic gastritis, precancerous gastric lesion, and gastric cancer: A cross-sectional multicenter study in Thailand. Biomed Res Int. 2017(438482)2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wang L, Zuo X, Xie K and Wei D: The role of CD44 and cancer stem cells. Methods Mol Biol. 1692:31–42. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Hettinghouse A, Liu R and Liu CJ: Multifunctional molecule ERp57: From cancer to neurodegenerative diseases. Pharmacol Ther. 181:34–48. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Walsh RJ and Tan DSP: The role of immunotherapy in the treatment of advanced cervical cancer: Current status and future perspectives. J Clin Med. 10(4523)2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Kure S, Chiba T, Ebina A, Toda K, Jikuzono T, Motoda N, Mitani H, Sugitani I, Takeuchi K and Ohashi R: Correlation between low expression of protein disulfide isomerase A3 and lymph node metastasis in papillary thyroid carcinoma and poor prognosis: A clinicopathological study of 1,139 cases with long-term follow-up. Endocr J. 69:273–281. 2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Shimoda T, Wada R, Kure S, Ishino K, Kudo M, Ohashi R, Fujita I, Uchida E, Yoshida H and Naito Z: Expression of protein disulfide isomerase A3 and its clinicopathological association in gastric cancer. Oncol Rep. 41:2265–2272. 2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Song D, Liu H, Wu J, Gao X, Hao J and Fan D: Insights into the role of ERp57 in cancer. J Cancer. 12:2456–2464. 2021.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Morgan E, Arnold M, Camargo MC, Gini A, Kunzmann AT, Matsuda T, Meheus F, Verhoeven RHA, Vignat J, Laversanne M, et al: The current and future incidence and mortality of gastric cancer in 185 countries, 2020-40: A population-based modelling study. EClinicalMedicine. 47(101404)2022.PubMed/NCBI View Article : Google Scholar
|


















